Abstract
We investigated the impact of low-level resistance to fluoroquinolones on the bactericidal activity of ciprofloxacin in a murine model of urinary tract infection. The susceptible Escherichia coli strain CFT073 (ciprofloxacin MIC [CIP MIC] of 0.008 μg/ml) was compared to its transconjugants harboring qnrA1 or qnrS1 and to an S83L gyrA mutant. The three derivatives showed similar low-level resistance to fluoroquinolones (CIP MICs, 0.25 to 0.5 μg/ml). Bactericidal activity measured in vitro after 1, 3, and 6 h of exposure to 0.5 μg/ml of ciprofloxacin was significantly lower for the derivative strains (P < 0.01). In the murine model of urinary tract infection (at least 45 mice inoculated per strain), mice were treated with a ciprofloxacin regimen of 2.5 mg/kg, given subcutaneously twice daily for 2 days. In mice infected with the susceptible strain, ciprofloxacin significantly decreased viable bacterial counts (log10 CFU/g of tissue) in the bladder (4.2 ± 0.5 versus 5.5 ± 1.3; P = 0.001) and in the kidney (3.6 ± 0.8 versus 5.0 ± 1.1; P = 0.003) compared with those of untreated mice. In contrast, no significant decrease in viable bacterial counts was observed with any of the three derivative strains. The area under the concentration-time curve from 0 to 24 h/MIC and the maximum concentration of drug in serum/MIC ratios measured in plasma were indeed equal to 827 and 147, respectively, for the parental strain, and only 12.4 to 24.8 and 2.2 to 4.4, respectively, for the derivative strains. In conclusion, low-level resistance to fluoroquinolones conferred by a qnr gene is associated with decreased bactericidal activity of ciprofloxacin, similar to that obtained with a gyrA mutation.
Urinary tract infection (UTI) due to Escherichia coli is the most common bacterial infection. Fluoroquinolones are commonly used for the treatment of UTI because isolated microorganisms are frequently resistant to aminopenicillins and trimethoprim-sulfamethoxazole (22), and fluoroquinolones are given orally. However, resistance to fluoroquinolones in E. coli has increased due to their large use (13, 23). Classical mechanisms of quinolone resistance are due to chromosomal mutations in the genes encoding their targets (quinolone resistance-determining regions of the type II topoisomerases) or in regulatory genes affecting permeability or efflux (15, 29). More recently, plasmid-mediated mechanisms were reported, such as those due to qnr genes (16, 25) encoding pentapeptide repeat proteins, aac(6′)-Ib-cr encoding a modified acetyltransferase (32), and qepA encoding an active efflux pump (27). The Qnr proteins protect DNA gyrase from quinolone inhibition.
Enterobacteriaceae with plasmid-mediated resistance to fluoroquinolones due to qnrA, qnrB, or qnrS have been described worldwide (31) and particularly among E. coli from UTI (5, 39). Acquisition of qnr genes increases fluoroquinolone MICs by between 8- and 64-fold; however, the final MICs remain below the susceptibility breakpoints, according to CLSI (1 μg/ml) (6) and to the European Committee on Antimicrobial Susceptibility Testing (0.5 μg/ml) (19). Although it has been suggested that the Qnr protein favors the selection of mutant resistance (25, 28), the therapeutic relevance of the acquisition of the qnr gene on the bactericidal activity of fluoroquinolones remains unclear. Therefore, the aim of our study was to evaluate the impact of low-level fluoroquinolone resistance conferred by qnr genes on ciprofloxacin bactericidal activity in vitro and in vivo. We measured ciprofloxacin bactericidal activity in vitro at the concentrations reached in humans and in vivo in a murine model of UTI. Results obtained with the susceptible strain of E. coli used for the experimental model were compared to those obtained with two of its transconjugants harboring a qnr gene, also taking into account the results obtained with its gyrA mutant that exhibits low-level fluoroquinolone resistance similar to that of the qnr transconjugants.
(This work was presented in part at the 48th Interscience Conference on Antimicrobial Agents and Chemotherapy, held in Washington, DC, in October 2008.)
MATERIALS AND METHODS
Bacterial strains, conjugation assays, and plasmid stability.
Experiments were performed with four isogenic strains derived from the quinolone-susceptible E. coli CFT073 strain used previously to set the murine model of pyelonephritis (20). A rifampin (rifampicin)-resistant mutant strain, E. coli CFT073-RR, was selected in vitro from E. coli CFT073 by plating 109 bacteria onto Mueller-Hinton (MH) agar containing a rifampin concentration of 100 μg/ml. E. coli CFT073-RR served as the quinolone-susceptible control strain. Two transconjugants were obtained after conjugation between the following E. coli CFT073-RR and clinical strains harboring qnr genes: E. coli Hm13 harboring qnrA1 and E. coli PS105 harboring qnrS1 (both resistant to amoxicillin [amoxicilline]). Plasmids of the clinical strains were negative for the presence of a second qnr gene, aac(6′)-Ib-cr or qepA. The transconjugants E. coli CFT073-RR Tc (pQnrA1) and E. coli CFT073 Tc (pQnrS1) were obtained after 40 min of mating in MH broth, as previously described (3). After incubation, transconjugants were selected by plating the conjugation mixture on MH agar supplemented with amoxicillin (100 μg/ml)-rifampin (100 μg/ml). The PCR experiments confirmed the presence of the transconjugants. In order to obtain a strain with low-level fluoroquinolone resistance from a different mechanism, we selected a strain with a single gyrA mutation from E. coli CFT073-RR by plating 109 bacteria onto MH agar containing nalidixic acid at a concentration of 40 μg/ml. PCR and DNA sequencing of the quinolone resistance-determining regions in the DNA gyrase and topoisomerase IV genes (4) confirmed a single gyrA S83L mutation in the clone, studied further as the E. coli CFT073-RR-gyrA mutant.
Plasmid stability in the transconjugants E. coli CFT073-RR Tc (pQnrA1) and E. coli CFT073 Tc (pQnrS1) was measured in vitro by daily subculturing for 4 weeks in antibiotic-free MH agar and by plating onto MH agar alone or containing 100 μg/ml of amoxicillin. After 4 weeks, the mean plasmid losses were 0.8% for E. coli CFT073-RR Tc (pQnrA1) and 0.3% for E. coli CFT073-RR Tc (pQnrS1).
Fluoroquinolone activities in vitro.
MICs were determined by the agar dilution method in accordance with CLSI guidelines (6). Time-kill curve kinetics were performed for each strain in 10 ml MH broth, with a ciprofloxacin concentration of 0.5 μg/ml and with a ciprofloxacin concentration equal to four times the MIC of the strain tested. Antimicrobial agent-free broth was evaluated in parallel as a control. Cultures were incubated at 37°C. Viable counts were determined by serial dilution after 0, 1, 3, 6, and 24 h of incubation and by plating 100 μl of the control, test cultures, or the dilution at the indicated times onto MH agar plates. Colony counts were determined after 24 h of incubation.
Selection of resistant mutants was performed at four times the MIC of ciprofloxacin against each strain, as described previously (4). Briefly, strains were grown at 37°C overnight in antibiotic-free MH broth and centrifuged, and the pellet was suspended in 5 ml of sterile broth, giving an inoculum of >1010 CFU/ml. Agar plates containing ciprofloxacin were inoculated with 100 μl of cell suspension and incubated at 37°C. The proportion of resistant mutants was calculated by dividing the number of CFU growing on MH agar plates with ciprofloxacin by the number of CFU growing on antibiotic-free MH agar.
The mutant prevention concentration (MPC) of ciprofloxacin was determined as previously described (1). Briefly, each strain was grown at 37°C overnight in antibiotic-free MH broth. Cultures (100 ml) were centrifuged at 4,000 × g for 15 min, and the pellet was suspended in 5 ml of sterile broth, giving an inoculum of >1010 CFU/ml. Agar plates containing ciprofloxacin concentrations ranging from 1× MIC to 16× MIC against each strain were inoculated with 100 μl of cell suspension and incubated at 37°C. The MPC was recorded as the lowest concentration of ciprofloxacin completely inhibiting bacterial growth after incubation at 37°C for 72 h.
All the in vitro experiments described above were repeated at least five times. Geometric means were used to express the results for MICs and MPCs, and the means ± standard deviations were calculated for CFU counts and rates of spontaneous mutant.
Mouse model of UTI.
The ascending unobstructed mouse model of UTI was used as previously described (20). Animal experiments were performed in accordance with prevailing regulations regarding the care and use of laboratory animals by the European Commission (10). The experimental protocol was approved by the Departmental Direction of Veterinary Services in Paris, France. Eight-week-old immunocompetent female CBA mice (weight, 20 to 23 g) were used. Inocula of different strains were obtained by overnight incubation in brain heart infusion broth, washing of the cells by centrifugation at 15,000 × g for 15 min in saline, and resuspension in saline to a final inoculum of 5 × 1010 CFU/ml. Pyelonephritis was induced during general anesthesia (with an intraperitoneal administration of 0.3 ml of 0.66% pentobarbital solution) by injecting 50 μl (i.e., 108 CFU) into the bladder through a urethral catheter.
In vivo growth experiments and plasmid stability.
Mice were inoculated with each of the four different strains. The growth rate of individual strains was studied in groups of at least 10 infected mice, in order to evaluate their potentials for performing stable pyelonephritis and to evaluate in vivo plasmid stability for the two transconjugants. Mice were sacrificed at 5 and 10 days after inoculation, and bladders and kidneys were aseptically taken out and homogenized in 1 ml of saline solution. One hundred microliters of the solution or its dilution was spread onto MH agar plates and incubated for 24 h. The number of CFU were counted and expressed as the number of CFU/g of tissue. Plasmid loss on day 5 and 10 was assessed for strains harboring plasmid pQnrA1 or pQnrS1 by comparing the number of CFU growing on MH plates alone or containing amoxicillin (100 μg/ml).
Antimicrobial treatment.
In order to determine the bactericidal activity of ciprofloxacin, at least 45 mice per strain were inoculated with strain CFT073-RR, CFT073-RR Tc (pQnrA1), CFT073-RR Tc (pQnrS1), or CFT073-RR-gyrA. Two days after inoculation, at least 15 mice in each group were treated with 2.5 mg/kg of ciprofloxacin injected subcutaneously twice a day for 2 days (four injections). This regimen was chosen because it provided peak levels of plasma that were in the range of those achieved in humans after an oral administration of 500 mg of ciprofloxacin (2) and because it provided peak concentration/MIC and area under the concentration-time curve from 0 to 24 h (AUC0-24)/MIC ratios against the susceptible parental strain that were above those required to achieve efficacy in humans (11, 30).
Mice were sacrificed 18 h after the last dose of ciprofloxacin. Bladders and kidneys were aseptically taken out and homogenized in 1 ml of saline solution. One hundred microliters of the solution or its dilution was spread onto MH agar plates and incubated for 24 h. The number of CFU were counted and expressed as the number of CFU/g of tissue. Kidney and bladder homogenates (0.1 ml of each) were also spread onto MH agar plates containing ciprofloxacin at concentrations of 1× MIC to 4× MIC and incubated at 37°C for 72 h in order to detect spontaneous, resistant mutants. Control mice were killed for organism quantification just before the start of ciprofloxacin therapy (15 to 19 mice, start-of-therapy controls) and 18 h after the end of therapy (15 to 17 mice, end-of-therapy controls).
Ciprofloxacin pharmacokinetics.
Single-dose pharmacokinetic studies were performed after an injection of 2.5 mg/kg of ciprofloxacin subcutaneously in infected mice. Blood samples of 200 μl were obtained by intracardiac puncturing from three to six anesthetized mice at 15, 30, 45, 60, 120, 240, 360, and 480 min after ciprofloxacin injection. Blood was centrifuged, and plasma samples were treated with a methanolic solution containing ofloxacin as an internal standard in order to precipitate proteins. Plasma ultrafiltrate was prepared from 200-μl plasma samples by centrifugation in a Centrifree micropartition unit (Amicon, Beverly, MA) at 2,000 × g for 30 min at 25°C. Ciprofloxacin concentrations were determined by liquid chromatography, with fluorimetric detection after deproteinization, as described previously (36). The method was linear over the concentration range from 0.1 to 40 μg/ml. Intra- and interday coefficients of variation obtained were less than 10%. The limit of quantitation used was 0.05 μg/ml. The peak concentration (maximum concentration of drug in serum [Cmax]) and the AUC0-24 were calculated using the WinNonlin pharmacokinetic program (Scientific Consulting).
Statistical analysis.
Results were expressed as means ± standard deviations for continuous variables. Continuous variables were compared by nonparametric testing (Mann-Whitney U test). A P value of less than 0.05 was considered significant. Analysis was performed using SAS statistical software (version 8.2; Cary, NC).
RESULTS
MICs, MPCs, and in vitro selection of ciprofloxacin-resistant mutants.
Ciprofloxacin MIC and MPC values against the four study strains are presented in Table 1. Acquisition of the plasmids pQnrA1 or pQnrS1 increased the ciprofloxacin MIC by 32 to 64 times and the MPC by 20 times. However, the strains harboring these plasmids remained susceptible to ciprofloxacin, according to currently accepted clinical breakpoints by CLSI or the European Committee on Antimicrobial Susceptibility Testing, and the MPC/MIC ratios were similar for the four strains (equaling 8). Results for the gyrA mutant were similar to those for the qnr transconjugants.
TABLE 1.
Ciprofloxacin MICs, MPCs, and proportion of resistant mutants against the four E. coli strains used in the study
| E. coli strain (plasmid) | MIC (μg/ml) | MPC (μg/ml) | MPC/MIC ratio (μg/ml) | PRMa |
|---|---|---|---|---|
| CFT703-RR | 0.0075 | 0.06 | 8 | 0.8 × 10−8 ± 0.4 × 10−8b |
| CFT073-RR Tc (pQnrA1) | 0.5 | 4 | 8 | 0.6 × 10−8 ± 0.7 × 10−8b |
| CFT073-RR Tc (pQnrS1) | 0.25 | 2 | 8 | 1.1 × 10−8 ± 1.5 × 10−8b |
| CT073-RR-gyrA | 0.25 | 2 | 8 | 0.3 × 10−8 ± 0.4 × 10−8c |
PRM, proportion of resistant mutants (selection by ciprofloxacin at four times the MIC). Each value is the mean ± standard deviation from five independent experiments.
P was 0.4 compared with E. coli strain CFT073-RR.
P was 0.2 compared with E. coli strain CFT073-RR.
No significant differences were observed in the proportion of ciprofloxacin-resistant mutant obtained at four times the MIC of each strain between E. coli CFT073-RR and the three derivative strains with either the qnr genes or a gyrA mutation (P > 0.4) (Table 1).
Time-kill curves.
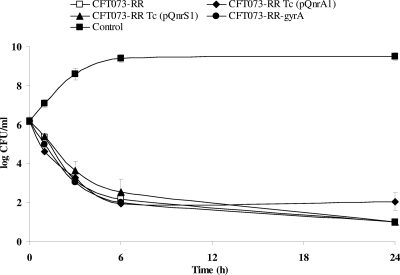
The viable counts of the four strains in time-kill curves are shown in Fig. 1. At a concentration of 0.5 μg/ml, the bactericidal activity of ciprofloxacin significantly decreased after 1, 3, and 6 h of antibiotic exposure against E. coli strains harboring plasmid pQnrA1 or pQnrS1 and against the E. coli CFT073-RR-gyrA mutant in comparison with the susceptible E. coli CFT073-RR strain (P < 0.01). At a concentration of four times the MIC of each strain, no significant difference in the bactericidal activity of ciprofloxacin was measured among any of the three strains with low-level resistance or the susceptible E. coli CFT073-RR strain (P of >0.2 after 1, 3, 6, and 24 h of antibiotic exposure for any of the three strains with low-level resistance compared with the parental strain) (Fig. 2).
FIG. 1.
Bactericidal activity of ciprofloxacin at a concentration of 0.5 μg/ml against the four different E. coli strains. P was <0.01 after 1, 3, and 6 h of incubation for any of the three strains with low-level of resistance compared with the susceptible E. coli CFT073-RR strain. P was <0.05 for E. coli CFT073-RR Tc (pQnrS1) and E. coli CFT073-RR-gyrA and 0.07 for E. coli CFT073-RR Tc (pQnrA1) compared with E. coli CFT073-RR after 24 h of incubation.
FIG. 2.
Bactericidal activity of ciprofloxacin at a concentration that was four times the MIC of each of the E. coli study strains. P was >0.2 at all times for the three strains with low-level resistance compared with the susceptible E. coli CFT073-RR strain.
Pharmacokinetics and pharmacodynamic parameters.
After a single subcutaneous injection of 2.5 mg/kg of ciprofloxacin, the mean Cmax value obtained was 1.1 ± 0.2 μg/ml at 15 min after the injection, and the mean AUC0-24 obtained was 6.2 ± 1.2 μg/h/ml. Corresponding Cmax/MIC and AUC0-24/MIC ratios for the different study strains are shown in Table 2. Because the level of serum protein binding for ciprofloxacin is near zero in mice (38), the free and total serum levels for ciprofloxacin were considered to be similar.
TABLE 2.
Pharmacokinetic-pharmacodynamic parameters of the ciprofloxacin dosing regimen used in experimental UTI against the four E. coli strains used in the study
| E. coli strain (plasmid) | Pharmacokinetic- pharmacodynamic ratiosb
|
|
|---|---|---|
| Cmax/MICa | AUC0-24/MICc | |
| CFT703-RR | 147 | 827 |
| CFT703-RR Tc (pQnrA1) | 2.2 | 12.4 |
| CFT703-RR Tc (pQnrS1) | 4.4 | 24.8 |
| CT073-RR-gyrA | 4.4 | 24.8 |
The mean Cmax for ciprofloxacin was 1.1 ± 0.2 μg/ml.
The level of serum protein binding for ciprofloxacin in mice was near 0% (38). Therefore, the levels of total and unbound drug binding were considered similar.
The AUC0-24 value used was 6.2 ± 1.2 μg/h/ml.
Mouse model of UTI.
Individual UTI experiment showed that the four derivative strains of E. coli CFT073 were all able to induce stable pyelonephritis in mice at least until day 10. On day 10, bacterial counts in kidneys and bladders (log10 CFU/g of tissue) were 5.0 ± 1.0 and 5.9 ± 1.9, respectively, for strain CFT073-RR; 3.0 ± 0.7 and 3.7 ± 0.7, respectively, for the strain harboring plasmid pQnrA1; 4.1 ± 0.5 and 5.9 ± 1.1, respectively, for the strain harboring plasmid pQnrS1; and 5.5 ± 1.0 and 5.4 ± 1.1, respectively, for the strain with a single gyrA mutation. The mean plasmid loss at day 10 was 4.5% for E. coli CFT073-RR harboring plasmid pQnrA1 and 4.2% for the strain harboring plasmid pQnrS1.
Therapeutic efficacy in experimental murine UTI.
In mice infected with E. coli strain CFT073-RR and treated with ciprofloxacin, a significant decrease of viable bacterial counts was observed in kidneys (Table 3) and in bladders (Table 4) compared with end-of-treatment control mice. In contrast, no statistically significant decrease in viable bacterial counts in kidneys (Table 3) and in bladders (Table 4) was observed when UTI was induced with any of the three resistant derivative E. coli strains between treated mice and end-of-treatment control mice (P > 0.2) (Tables 3 and 4). Of note, ciprofloxacin did not reduce the CFU counts in kidneys and bladders even against E. coli CFT073-RR harboring plasmid pQnrA1, which produced the lowest bacterial counts in start-of-treatment and end-of-treatment control mice. Therefore, treatment with ciprofloxacin did not influence, under our experimental conditions, the evolution of UTI due to strains harboring low-level resistance to fluoroquinolones due to qnr genes or a gyrA mutation.
TABLE 3.
Effect of ciprofloxacin on viable organisms in kidneys of mice infected with the four E. coli strains used in the study
| E. coli strain (plasmid) | Results (log CFU/g of kidney ± SD [no. of mice]) for mice treated with:
|
||
|---|---|---|---|
| Start-of-treatment control | End-of-treatment control | Ciprofloxacin | |
| CFT703-RR | 5.0 ± 0.5 (15) | 5.0 ± 1.1 (15) | 3.6 ± 0.8 (15)a |
| CFT703-RR Tc (pQnrA1) | 3.4 ± 0.9 (15) | 3.0 ± 0.8 (15) | 2.7 ± 0.6 (16)b |
| CFT703-RR Tc (pQnrS1) | 4.6 ± 1.0 (15) | 4.5 ± 1.1 (15) | 4.2 ± 0.7 (16)b |
| CFT703-RR-gyrA | 3.9 ± 1.4 (15) | 4.0 ± 1.3 (15) | 4.1 ± 1.4 (16)b |
P was 0.001 compared with the end-of-treatment control group.
P was >0.2 compared with the end-of-treatment control group.
TABLE 4.
Effect of ciprofloxacin on viable organisms in bladders of mice infected with the four E. coli strains used in the study
| E. coli strain (plasmid) | Results (log CFU/g of bladder ± SD [no. of mice]) for mice treated with:
|
||
|---|---|---|---|
| Start-of-treatment control | End-of-treatment control | Ciprofloxacin | |
| CFT703-RR | 6.2 ± 1.1 (19) | 5.5 ± 1.3 (17) | 4.2 ± 0.5 (15)a |
| CFT703-RR Tc (pQnrA1) | 5.1 ± 0.8 (16) | 4.7 ± 0.5 (15) | 4.7 ± 0.3 (20)b |
| CFT703-RR Tc (pQnrS1) | 6.6 ± 1.5 (15) | 5.0 ± 0.9 (15) | 5.0 ± 0.9 (16)b |
| CFT703-RR-gyrA | 5.4 ± 0.8 (15) | 5.4 ± 0.8 (15) | 5.4 ± 0.7 (16)b |
P was 0.003 compared with the end-of-treatment control group.
P was >0.6 compared with the end-of-treatment control group.
In vivo selection of resistant mutants.
No ciprofloxacin-resistant mutant was detected after antibiotic exposure when the experimental UTI was induced with any of the four E. coli strains.
DISCUSSION
The qnr gene is a novel plasmid-mediated gene used for fluoroquinolone resistance (25). The resistance level conferred is so low that qnr-positive strains remain susceptible to fluoroquinolones, according to international susceptibility breakpoints. Our in vitro and in vivo studies of isogenic strains of E. coli harboring qnrA or qnrS showed that the bactericidal activity of ciprofloxacin is markedly reduced against these strains.
Few studies have shown the therapeutic impact of low-level resistance to fluoroquinolones in Enterobacteriaceae either experimentally (12), in clinical reports (9, 26, 40), or in a retrospective clinical study (8). However, the qnr gene was not involved in any of these studies. The only report investigating the therapeutic impact of a qnr gene was performed in a murine model of pneumonia due to a strain of Klebsiella pneumoniae alone or expressing the qnrA1 gene (34). However, in contrast with our experiments, since the K. pneumoniae qnr-negative parental strain harbored several mechanisms of resistance to fluoroquinolones (has a gyrA mutation, deficient in porins, and has an active efflux pump), the derivative strain harboring the qnrA1 gene became resistant to fluoroquinolones in vitro, with a ciprofloxacin MIC of 4 μg/ml. Consequently, this led to an in vivo failure with ciprofloxacin.
In the present study, we showed that low-level resistance to fluoroquinolones due to a qnr gene is sufficient to significantly reduce the bactericidal activity of ciprofloxacin in vitro and in vivo. Mice infected with qnr-positive transconjugants of E. coli did not show any significant reduction in bacterial count when they were treated with ciprofloxacin compared with the counts of mice infected with the susceptible parental strain. We also showed that the therapeutic impact was similar to that observed with a derivative harboring a single gyrA mutation. However, two comments should be made concerning the initial bacterial inoculum used in our in vivo experiments. First, our experimental conditions were associated with a relatively low initial inoculum, since the largest bacterial counts we observed were ∼105 CFU in kidneys and ∼107 CFU in bladders. Thus, the relevance of our results with a higher inoculum deserves further investigations. Second, the CFU counts obtained before therapy in kidneys and bladders tended to be lower for qnr-containing strains than those for the parenteral strain or the derivative harboring a single gyrA mutation. To investigate that, we have performed competition studies in the same model, between the susceptible parental strain and each of the resistant derivative strains, in a 1/1 ratio. Results confirmed that the qnr-containing strains lost most of the competitions, while there was no difference for the gyrA derivative (data not shown). These data suggest a decrease in fitness for the strains harboring qnr but not for the strains with a gyrA mutation.
Our therapeutic results were obtained with a ciprofloxacin regimen, producing Cmax and AUC0-24 values in plasma that are in the lower range of those observed in humans with a ciprofloxacin dosing regimen of 500 mg twice a day given orally (14). In contrast, the proportion of active unbound ciprofloxacin was higher in mice than in humans, since the level of serum protein binding of ciprofloxacin is near 0% (38) at this range of ciprofloxacin concentrations in mice and ranges from 30% to 40% in humans (14, 37). The efficacy of ciprofloxacin treatment in mice infected with the parental quinolone-susceptible E. coli strain (CFT073-RR) and the reduced activity in mice infected with E. coli qnr-positive transconjugants can be explained by pharmacodynamic-pharmacokinetic indexes. In vivo, the parameters that best predict fluoroquinolone efficacy are the AUC0-24/MIC ratio (11) and the Cmax/MIC ratio (30). Values higher than 30 to 125 and 10 to 12, respectively, have been shown to be predictive of clinical and microbiological efficacy in different foci of infection (11, 30). In our experiments, the AUC0-24/MIC ratio of ciprofloxacin was higher than 125 for the susceptible E. coli strain CFT073-RR, whereas this ratio was below 30 for the three derivative strains. Likewise the Cmax/MIC ratio was widely higher than 12 for E. coli CFT073-RR and was below 10 for the three derivative strains.
In vitro, we showed that, at a concentration of 0.5 μg/ml, which is a concentration under the susceptibility breakpoint value for ciprofloxacin against Enterobacteriaceae (6) and one that is achievable in human serum during therapy with ciprofloxacin (14), bactericidal activity of ciprofloxacin was significantly decreased during the first 6 hours of exposure against the derivative strains harboring plasmid pQnrA1 or pQnrS1 and against E. coli CFT073-RR-gyrA in comparison with the susceptible E. coli CFT073-RR parental strain. Jacoby (17) observed similar results with E. coli J53, E. coli J53 with a plasmid carrying a qnrA1 gene, and E. coli J53 with a single mutation in gyrA. In contrast, at a concentration of four times the MIC of each strain, we did not find any significant difference in the bactericidal activity of ciprofloxacin against the parental and derivative strains, as shown by Mammeri et al. (24). The ciprofloxacin MPC was at least 20 times higher for the qnr transconjugants and for the gyrA mutant than for the parental strain, as previously reported (4, 35). However, MPC/MIC ratios remained similar for the four strains. This is in agreement with a similar proportion of resistant mutants for the four strains when exposed to a concentration of ciprofloxacin equal to four times the MIC. Acquisition of qnr genes has been reported to favor the selection of gyrA and parC mutations during a treatment with a fluoroquinolone and to select strains with higher levels of fluoroquinolone resistance (25, 28). In vivo, we have not detected ciprofloxacin-resistant mutants after antibiotic exposure. This result is probably due to the relatively low level of inoculum in our experimental model of UTI, which made the selection of resistant mutants unlikely.
The prevalence of microorganisms harboring the qnr gene is high, from 2% to 32% in Enterobacteriaceae resistant to expanded-spectrum cephalosporins (2, 7, 18, 21, 31, 33). The genes encoding Qnr proteins are usually located in multidrug-resistant, transferable plasmids. The antimicrobial treatment for the Enterobacteriaceae harboring these plasmids is often restricted to carbapenems and fluoroquinolones. Detection of strains with only one mechanism of resistance to quinolones, such as a single gyrase mutation, may benefit from susceptibility testing with quinolone nalidixic acid. In contrast, in strains harboring only qnr genes, the increase in MIC is lower for nalidixic acid than for fluoroquinolones, and fluoroquinolone MICs remain below the susceptibility breakpoints (6, 19).
In conclusion, our results suggest that low-level fluoroquinolone resistance conferred by qnr genes or a gyrA mutation is associated with a reduced bactericidal activity of ciprofloxacin in vivo. Therefore, detection of a mechanism of fluoroquinolone resistance resulting in a fluoroquinolone MIC below but close to breakpoint may become necessary for the treatment of severe infections due to Enterobacteriaceae.
Acknowledgments
We thank Louis Garry from INSERM E0339, Faculté de Médecine Xavier Bichat, Université Paris Diderot, for technical assistance with the experimental model.
Nicolas Allou was given a grant from Assistance Publique-Hôpitaux de Paris.
We have no conflict of interest to declare.
Footnotes
Published ahead of print on 27 July 2009.
REFERENCES
- 1.Blondeau, J. M., X. Zhao, G. Hansen, and K. Drlica. 2001. Mutant prevention concentrations of fluoroquinolones for clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 45:433-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brunner, M., H. Stab, J.-G. Möller, C. Schrolnberger, B. Erovic, U. Hollenstein, M. Zeitlinger, H. G. Eichler, and M. Müeller. 2002. Target site concentrations of ciprofloxacin after single intravenous and oral doses. Antimicrob. Agents Chemother. 46:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cambau, E., C. Lascols, W. Sougakoff, C. Bebear, R. Bonnet, J. D. Cavallo, L. Gutmann, M. C. Ploy, V. Jarlier, C. J. Soussy, and J. Robert. 2006. Occurrence of qnrA-positive clinical isolates in French teaching hospitals during 2002-2005. Clin. Microbiol. Infect. 12:1013-1020. [DOI] [PubMed] [Google Scholar]
- 4.Cesaro, A., R. R. Bettoni, C. Lascols, A. Merens, C. J. Soussy, and E. Cambau. 2008. Low selection of topoisomerase mutants from strains of Escherichia coli harbouring plasmid-borne qnr genes. J. Antimicrob. Chemother. 61:1007-1015. [DOI] [PubMed] [Google Scholar]
- 5.Chenia, H. Y., B. Pillay, and D. Pillay. 2006. Analysis of the mechanisms of fluoroquinolone resistance in urinary tract pathogens. J. Antimicrob. Chemother. 58:1274-1278. [DOI] [PubMed] [Google Scholar]
- 6.CLSI. 2005. Performance standards for antimicrobial susceptibility testing; 15th informational supplement. Document M100-S15. Clinical and Laboratory Standards Institute, Wayne, PA.
- 7.Corkill, J. E., J. J. Anson, and C. A. Hart. 2005. High prevalence of the plasmid-mediated quinolone resistance determinant qnrA in multidrug-resistant Enterobacteriaceae from blood cultures in Liverpool, UK. J. Antimicrob. Chemother. 56:1115-1117. [DOI] [PubMed] [Google Scholar]
- 8.Crump, J. A., K. Kretsinger, K. Gay, R. M. Hoekstra, D. J. Vugia, S. Hurd, S. D. Segler, M. Megginson, L. J. Luedeman, B. Shiferaw, S. S. Hanna, K. W. Joyce, E. D. Mintz, F. J. Angulo, et al. 2008. Clinical response and outcome of infection with Salmonella enterica serotype Typhi with decreased susceptibility to fluoroquinolones: a United States foodnet multicenter retrospective cohort study. Antimicrob. Agents Chemother. 52:1278-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Endimiani, A., F. Luzzaro, M. Perilli, G. Lombardi, A. Coli, A. Tamborini, G. Amicosante, and A. Toniolo. 2004. Bacteremia due to Klebsiella pneumoniae isolates producing the TEM-52 extended-spectrum beta-lactamase: treatment outcome of patients receiving imipenem or ciprofloxacin. Clin. Infect. Dis. 38:243-251. [DOI] [PubMed] [Google Scholar]
- 10.European Commission. 1986. Directive for the protection of vertebrate animals used for experiments and other scientific purposes (86/609/EEC). Off. J. Eur. Comm. L 358:1-29. [Google Scholar]
- 11.Forrest, A., D. E. Nix, C. H. Ballow, T. F. Goss, M. C. Birmingham, and J. J. Schentag. 1993. Pharmacodynamics of intravenous ciprofloxacin in seriously ill patients. Antimicrob. Agents Chemother. 37:1073-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuursted, K., and H. Schumacher. 2002. Significance of low-level resistance to ciprofloxacin in Klebsiella pneumoniae and the effect of increased dosage of ciprofloxacin in vivo using the rat granuloma pouch model. J. Antimicrob. Chemother. 50:421-424. [DOI] [PubMed] [Google Scholar]
- 13.Goettsch, W., W. van Pelt, N. Nagelkerke, M. G. Hendrix, A. G. Buiting, P. L. Petit, L. J. Sabbe, A. J. van Griethuysen, and A. J. de Neeling. 2000. Increasing resistance to fluoroquinolones in Escherichia coli from urinary tract infections in the Netherlands. J. Antimicrob. Chemother. 46:223-228. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez, M. A., F. Uribe, S. D. Moisen, A. P. Fuster, A. Selen, P. G. Welling, and B. Painter. 1984. Multiple-dose pharmacokinetics and safety of ciprofloxacin in normal volunteers. Antimicrob. Agents Chemother. 26:741-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hooper, D. C. 2000. Mechanisms of action and resistance of older and newer fluoroquinolones. Clin. Infect. Dis. 31(Suppl. 2):S24-S28. [DOI] [PubMed] [Google Scholar]
- 16.Jacoby, G., V. Cattoir, D. Hooper, L. Martinez-Martinez, P. Nordmann, A. Pascual, L. Poirel, and M. Wang. 2008. qnr gene nomenclature. Antimicrob. Agents Chemother. 52:2297-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jacoby, G. A. 2005. Mechanisms of resistance to quinolones. Clin. Infect. Dis. 41(Suppl. 2):S120-S126. [DOI] [PubMed] [Google Scholar]
- 18.Jacoby, G. A., N. Chow, and K. B. Waites. 2003. Prevalence of plasmid-mediated quinolone resistance. Antimicrob. Agents Chemother. 47:559-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kahlmeter, G., D. F. Brown, F. W. Goldstein, A. P. MacGowan, J. W. Mouton, I. Odenholt, A. Rodloff, C. J. Soussy, M. Steinbakk, F. Soriano, and O. Stetsiouk. 2006. European Committee on Antimicrobial Susceptibility Testing (EUCAST) technical notes on antimicrobial susceptibility testing. Clin. Microbiol. Infect. 12:501-503. [DOI] [PubMed] [Google Scholar]
- 20.Labat, F., O. Pradillon, L. Garry, M. Peuchmaur, B. Fantin, and E. Denamur. 2005. Mutator phenotype confers advantage in Escherichia coli chronic urinary tract infection pathogenesis. FEMS Immunol. Med. Microbiol. 44:317-321. [DOI] [PubMed] [Google Scholar]
- 21.Lavilla, S., J. J. Gonzalez-Lopez, M. Sabate, A. Garcia-Fernandez, M. N. Larrosa, R. M. Bartolome, A. Carattoli, and G. Prats. 2008. Prevalence of qnr genes among extended-spectrum beta-lactamase-producing enterobacterial isolates in Barcelona, Spain. J. Antimicrob. Chemother. 61:291-295. [DOI] [PubMed] [Google Scholar]
- 22.Lobel, B., A. Valot, V. Cattoir, O. Lemenand, and O. Gaillot. 2008. Comparison of antimicrobial susceptibility of 1,217 Escherichia coli isolates from women with hospital and community-acquired urinary tract infections. Presse Med. 37:746-750. (In French.) [DOI] [PubMed] [Google Scholar]
- 23.Mahamat, A., J. P. Lavigne, P. Fabbro-Peray, J. M. Kinowski, J. P. Daures, and A. Sotto. 2005. Evolution of fluoroquinolone resistance among Escherichia coli urinary tract isolates from a French university hospital: application of the dynamic regression model. Clin. Microbiol. Infect. 11:301-306. [DOI] [PubMed] [Google Scholar]
- 24.Mammeri, H., L. Poirel, and P. Nordmann. 2005. Bactericidal activity of fluoroquinolones against plasmid-mediated QnrA-producing Escherichia coli. Clin. Microbiol. Infect. 11:1048-1049. [DOI] [PubMed] [Google Scholar]
- 25.Martinez-Martinez, L., A. Pascual, and G. A. Jacoby. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797-799. [DOI] [PubMed] [Google Scholar]
- 26.McCarron, B., and W. C. Love. 1997. Acalculous nontyphoidal salmonellal cholecystitis requiring surgical intervention despite ciprofloxacin therapy: report of three cases. Clin. Infect. Dis. 24:707-709. [DOI] [PubMed] [Google Scholar]
- 27.Perichon, B., P. Courvalin, and M. Galimand. 2007. Transferable resistance to aminoglycosides by methylation of G1405 in 16S rRNA and to hydrophilic fluoroquinolones by QepA-mediated efflux in Escherichia coli. Antimicrob. Agents Chemother. 51:2464-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poirel, L., J. D. Pitout, L. Calvo, J. M. Rodriguez-Martinez, D. Church, and P. Nordmann. 2006. In vivo selection of fluoroquinolone-resistant Escherichia coli isolates expressing plasmid-mediated quinolone resistance and expanded-spectrum beta-lactamase. Antimicrob. Agents Chemother. 50:1525-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poole, K. 2000. Efflux-mediated resistance to fluoroquinolones in gram-negative bacteria. Antimicrob. Agents Chemother. 44:2233-2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Preston, S. L., G. L. Drusano, A. L. Berman, C. L. Fowler, A. T. Chow, B. Dornseif, V. Reichl, J. Natarajan, and M. Corrado. 1998. Pharmacodynamics of levofloxacin: a new paradigm for early clinical trials. JAMA 279:125-129. [DOI] [PubMed] [Google Scholar]
- 31.Robicsek, A., G. A. Jacoby, and D. C. Hooper. 2006. The worldwide emergence of plasmid-mediated quinolone resistance. Lancet Infect. Dis. 6:629-640. [DOI] [PubMed] [Google Scholar]
- 32.Robicsek, A., J. Strahilevitz, G. A. Jacoby, M. Macielag, D. Abbanat, C. H. Park, K. Bush, and D. C. Hooper. 2006. Fluoroquinolone-modifying enzyme: a new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 12:83-88. [DOI] [PubMed] [Google Scholar]
- 33.Robicsek, A., J. Strahilevitz, D. F. Sahm, G. A. Jacoby, and D. C. Hooper. 2006. qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob. Agents Chemother. 50:2872-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodriguez-Martinez, J. M., C. Pichardo, I. Garcia, M. E. Pachon-Ibanez, F. Docobo-Perez, A. Pascual, J. Pachon, and L. Martinez-Martinez. 2008. Activity of ciprofloxacin and levofloxacin in experimental pneumonia caused by Klebsiella pneumoniae deficient in porins, expressing active efflux and producing QnrA1. Clin. Microbiol. Infect. 14:691-697. [DOI] [PubMed] [Google Scholar]
- 35.Rodriguez-Martinez, J. M., C. Velasco, I. Garcia, M. E. Cano, L. Martinez-Martinez, and A. Pascual. 2007. Mutant prevention concentrations of fluoroquinolones for Enterobacteriaceae expressing the plasmid-carried quinolone resistance determinant qnrA1. Antimicrob. Agents Chemother. 51:2236-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubinstein, E., L. St Julien, J. Ramon, S. Dautrey, R. Farinotti, J. F. Huneau, and C. Carbon. 1994. The intestinal elimination of ciprofloxacin in the rat. J. Infect. Dis. 169:218-221. [DOI] [PubMed] [Google Scholar]
- 37.Schentag, J. J. 2000. Clinical pharmacology of the fluoroquinolones: studies in human dynamic/kinetic models. Clin. Infect. Dis. 31(Suppl. 2):S40-S44. [DOI] [PubMed] [Google Scholar]
- 38.Shandil, R. K., R. Jayaram, P. Kaur, S. Gaonkar, B. L. Suresh, B. N. Mahesh, R. Jayashree, V. Nandi, S. Bharath, and V. Balasubramanian. 2007. Moxifloxacin, ofloxacin, sparfloxacin, and ciprofloxacin against Mycobacterium tuberculosis: evaluation of in vitro and pharmacodynamic indices that best predict in vivo efficacy. Antimicrob. Agents Chemother. 51:576-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shin, J. H., H. J. Jung, J. Y. Lee, H. R. Kim, J. N. Lee, and C. L. Chang. 2008. High rates of plasmid-mediated quinolone resistance QnrB variants among ciprofloxacin-resistant Escherichia coli and Klebsiella pneumoniae from urinary tract infections in Korea. Microb. Drug Resist. 14:221-226. [DOI] [PubMed] [Google Scholar]
- 40.Vasallo, F. J., P. Martin-Rabadan, L. Alcala, J. M. Garcia-Lechuz, M. Rodriguez-Creixems, and E. Bouza. 1998. Failure of ciprofloxacin therapy for invasive nontyphoidal salmonellosis. Clin. Infect. Dis. 26:535-536. [DOI] [PubMed] [Google Scholar]