Abstract
We determined the complete nucleotide sequences of three plasmids that encode CTX-M extended-spectrum β-lactamases (ESBLs) in pulsed-field gel electrophoresis-defined United Kingdom variants (strains A, C, and D) of the internationally prevalent Escherichia coli O25:H4-ST131 clone. Plasmid pEK499 (strain A; 117,536 bp) was a fusion of type FII and FIA replicons and harbored the following 10 antibiotic resistance genes conferring resistance to eight antibiotic classes: blaCTX-M-15, blaOXA-1, blaTEM-1, aac6′-Ib-cr, mph(A), catB4, tet(A), and the integron-borne dfrA7, aadA5, and sulI genes. pEK516 (strain D; 64,471 bp) belonged to incompatibility group IncFII and carried seven antibiotic resistance genes: blaCTX-M-15, blaOXA-1, blaTEM-1, aac6′-Ib-cr, catB4, and tet(A), all as in pEK499. It also carried aac3-IIa, conferring gentamicin resistance, and was highly related to pC15-1a, a plasmid encoding the CTX-M-15 enzyme in Canada. By contrast, pEK204 (strain C; 93,732 bp) belonged to incompatibility group IncI1 and carried only two resistance genes, blaCTX-M-3 and blaTEM-1. It probably arose by the transposition of Tn3 and ISEcp1-blaCTX-M-3 elements into a pCOLIb-P9-like plasmid. We conclude that (i) United Kingdom variants of the successful E. coli ST131 clone have acquired different plasmids encoding CTX-M ESBLs on separate occasions, (ii) the blaCTX-M-3 and blaCTX-M-15 genes on pEK204 and pEK499/pEK516 represent separate escape events, and (iii) IncFII plasmids harboring blaCTX-M-15 have played a crucial role in the global spread of CTX-M-15 ESBLs in E. coli.
Escherichia coli strains producing CTX-M extended-spectrum β-lactamases (ESBLs) have emerged as major global pathogens, primarily associated with urinary tract infections, sometimes with contingent bacteremia (3, 25). Internationally disseminated clones have been recognized among these ESBL-producing E. coli strains through the application of multilocus sequence typing. These include E. coli lineages of sequence types ST131 and ST405, which are of particular public health concern (6, 27).
ST131 was first defined in 2008, though it is now known to have been in circulation at least from 2003, and has been reported widely across Europe, North America, and the Far East, often with the CTX-M-15 ESBL encoded by related IncFII plasmids that also encode OXA-1 and TEM-1 β-lactamases and the aminoglycoside/fluoroquinolone acetyltransferase AAC(6′)-Ib-cr (6, 16, 27). It belongs to phylogroup B2, is uropathogenic, and includes many variants with various complements of virulence determinants and divergent pulsed-field gel electrophoresis (PFGE) profiles, albeit generally related at ≥65% banding pattern similarity (15, 21, 23, 27, 27).
Allowing for its PFGE diversity, ST131 E. coli may be more widespread than is currently realized and may be prone to acquire prevalent resistance plasmids that are circulating in a particular area (35). Consistent with this view, ST131 variants in Japan have plasmids that encode group 2 or group 9 CTX-M enzymes, which are the dominant Asian types, rather than CTX-M-15 (31), whereas some United Kingdom members have the blaCTX-M-3 enzyme with different flanking sequences from the common blaCTX-M-15 gene. The prevalence of antibiotic-susceptible members of this clone is largely unknown, although one recent study identified carriage in 7% of healthy subjects in the Paris area (23).
In the United Kingdom, E. coli isolates with the CTX-M-15 ESBL have become prevalent since 2003. They include five PFGE-defined strains, A to E, which all belong to ST131 (21, 34), along with PFGE-diverse isolates, some of which also belong to ST131. Strain A, which is the most prevalent variant, is locally dominant (e.g., in Hampshire, Shropshire, and parts of Lancashire); strains B, C, and E are nationally scattered, whereas strain D is local to one center (34). The CTX-M-3 enzyme is associated with strain C isolates from Belfast: producers are less multiresistant than representatives of strain C with the CTX-M-15 enzyme from elsewhere in the United Kingdom.
We report here the complete nucleotide sequences of the blaCTX-M-15-harboring plasmids pEK499 and pEK516, from representative isolates of United Kingdom strains A and D, respectively, and of the blaCTX-M-3-harboring plasmid pEK204, from a Belfast representative of strain C.
(Parts of this work were presented as posters at the 17th [17] and 18th [14] European Congress of Clinical Microbiology and Infectious Diseases.).
MATERIALS AND METHODS
Bacterial isolates.
E. coli isolates EO499, H041280204, and EO516 all belong to the O25:H4-ST131 lineage (27, 35) and were selected to represent United Kingdom PFGE-defined strains A, C, and D, respectively (15, 16, 34). Isolates EO499 and EO516 produce CTX-M-15 ESBL, whereas H041280204 produces its close relative, CTX-M-3; their ESBL-encoding plasmids were designated pEK499, pEK516, and pEK204, respectively.
Plasmid sequencing.
Plasmids were transferred by conjugation or electroporation into E. coli strain DH5α or J53 (Table 1) and their sequences were determined by a shotgun cloning method (MWG, Planegg-Martinsried, Germany). Briefly, randomly sheared plasmid fragments of 2 to 3 kb were cloned into the pGEM-T Easy vector and then transformed into E. coli DH10b. Inserts were sequenced by BigDye Terminator chemistry. Sequences were assembled using the Staden package. Combinatorial PCRs, directed PCRs, and walking reads were used to assemble the contigs and to fill in gaps.
TABLE 1.
MICs of E. coli recipient strains J53 and DH5α and derivatives containing plasmids pEK499, pEK516, and pEK204
| Antibiotic | MIC (μg/ml) for indicated strain
|
||||
|---|---|---|---|---|---|
| J53 | J53(pEK499) | J53(pEK204) | DH5α | DH5α(pEK516) | |
| Ampicillin | 8 | >64 | >64 | 16 | >64 |
| Co-amoxiclav | 8 | 32 | 16 | 8 | 16 |
| Aztreonam | ≤0.125 | >64 | 8 | 0.25 | 32 |
| Cefotaxime | ≤0.125 | 256 | 64 | 0.25 | 128 |
| Cefotaxime + clavulanate | ≤0.060 | 0.125 | 0.125 | ≤0.060 | ≤0.060 |
| Ceftazidime | 0.25 | 64 | 2 | 0.25 | 16 |
| Ceftazidime + clavulanate | 0.25 | 0.5 | 0.25 | 0.25 | 0.25 |
| Cefpirome | ≤0.125 | >64 | 16 | ≤0.125 | 64 |
| Cefpirome + clavulanate | ≤0.060 | ≤0.060 | ≤0.060 | ≤0.060 | ≤0.060 |
| Cefoxitin | 8 | 8 | 8 | 8 | 8 |
| Piperacillin | 4 | >64 | >64 | 4 | 64 |
| Piperacillin-tazobactam | 4 | 16 | 4 | 4 | 4 |
| Imipenem | 0.25 | 0.25 | 0.25 | 0.5 | 0.25 |
| Meropenem | ≤0.060 | ≤0.060 | ≤0.060 | ≤0.060 | ≤0.060 |
| Ertapenem | ≤0.125 | ≤0.125 | ≤0.125 | ≤0.125 | ≤0.125 |
| Ciprofloxacin | ≤0.125 | ≤0.125 | ≤0.125 | ≤0.125 | ≤0.125 |
| Amikacin | ≤0.5 | 4 | ≤0.5 | 1 | 2 |
| Gentamicin | 0.25 | 0.25 | 0.25 | 0.5 | 8 |
| Tobramycin | 0.25 | 8 | 0.5 | 0.5 | 8 |
| Streptomycin | 1 | 32 | 2 | 1 | 0.5 |
| Chloramphenicol | 2 | 2 | 2 | 2 | 2 |
| Minocycline | 0.25 | 2 | 0.25 | 0.25 | 4 |
| Tetracycline | 1 | 32 | 1 | 1 | 32 |
| Tigecycline | ≤0.250 | ≤0.250 | ≤0.250 | ≤0.250 | ≤0.250 |
| Sulfamethoxazole | 16 | >1024 | 16 | 2 | 2 |
| Trimethoprim | 0.06 | >32 | 0.06 | 0.06 | 0.03 |
| Colistin | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.5 |
pMLST.
The plasmid multilocus sequence typing (pMLST) scheme for IncI1 plasmids (9) was used for pEK204. The relevant fragments of the pilL (254 bp), sogS (254 bp), ardA (343 bp), and repI1 (104 bp) genes and an 812-bp tnbA-pndC region were compared with known allelic variants (http://pubmlst.org/perl/mlstdbnet/mlstdbnet.pl?file=incI1_profiles.xml&page=oneseq).
Bioinformatics.
Open reading frames were predicted and annotated using the Bacterial Annotation System (BASys; http://wishart.biology.ualberta.ca/basys/cgi/submit.pl) (33) and confirmed with DNAMAN 5.2.10 software (Lynnon BioSoft, Lynnon Corporation; http://www.lynnon.com). Each predicted protein was compared against an all-protein database using BlastP (http://blast.ncbi.nlm.nih.gov/Blast.cgi) with a minimum cutoff of 30% identity over 80% length coverage, checking at least two best hits among the COG, KEGG, and nonredundant protein databases. Gene sequences were further compared and aligned with GenBank data using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and CLUSTAL W (http://www.ebi.ac.uk/clustalw) and with reference plasmids by two sequence alignments using the Blastnt-Blast2 algorithm (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The IncI1 plasmid R64 (GenBank accession no. NC_005014.1) was used as a reference for annotating pEK204, whereas IncF plasmids pU302L (NC_006816), pIP1206 (AM886293), pRSB107 (AJ851089), and pC15-1a (NC_005327) were used to annotate pEK499 and pEK516. Data files were compiled using Sequin (http://www.ncbi.nlm.nih.gov/Sequin/). The gross structures of whole plasmids were compared with WebACT (http://www.webact.org/WebACT/home) (1), and plasmid maps were prepared using the CGView server (http://stothard.afns.ualberta.ca/cgview_server/) (11).
Nucleotide sequence accession numbers.
The GenBank accession numbers for the three plasmids are EU935738 (pEK516), EU935739 (pEK499), and EU935740 (pEK204).
RESULTS AND DISCUSSION
Analysis of pEK499 (strain A).
Plasmid pEK499 was a circular molecule of 117,536 bp, belonged to incompatibility group F, and represented a fusion of two replicons of types FII and FIA. It harbored 185 predicted genes (Table 2), including 10 conferring resistance to eight antibiotic classes; pEK499 also included various modules that have been identified on other IncF plasmids and which are composed of resistance genes and insertion sequences. With the exception of blaTEM-1, all the antibiotic resistance genes were clustered in a 25-kb region. They included blaCTX-M-15 and blaOXA-1 as well as genes conferring resistance to aminoglycosides and ciprofloxacin (aac6′-Ib-cr), macrolides [mph(A)], chloramphenicol (catB4), and tetracycline [tet(A)]. A 1.8-kb class I integron was present within this multiresistance region and carried dfrA7 and aadA5, encoding trimethoprim and streptomycin resistances, respectively, and sulI, encoding sulfonamide resistance.
TABLE 2.
Open reading frames identified in pEK499
| Gene name | Nucleotide positiona | Function encoded |
|---|---|---|
| ccdA | 408-626 | Plasmid maintenance protein, antitoxin component |
| ccdB | 628-933 | Plasmid maintenance protein, toxin component |
| resD | 934-1740 | Site-specific resolvase that cleaves at the rfsF site |
| repE | 2514-3269 | Replication initiation protein of the FIA replicon |
| sopA | 3848-5023 | Partitioning protein |
| sopB | 5023-5994 | Partitioning protein |
| yccB | 6837-7763 | Hypothetical protein |
| yhdJ | 8148-8831 | DNA methylase |
| BASYS001 | 9067-9501 | Hypothetical protein |
| BASYS002 | 9501-10328 | Hypothetical protein |
| BASYS003 | Compl. 10634-11134 | Hypothetical protein |
| klcA | 10745-11170 | Putative antirestriction protein |
| BASYS004 | 11217-11639 | Hypothetical protein |
| BASYS005 | 11636-11827 | Hypothetical protein |
| BASYS006 | 11691-12026 | Hypothetical protein |
| BASYS007 | Compl. 12179-12601 | Hypothetical protein |
| BASYS008 | 12862-13092 | Hypothetical protein |
| BASYS009 | 13144-14505 | Hypothetical protein |
| BASYS0010 | 14552-15115 | Hypothetical protein |
| BASYS0011 | 14564-15220 | Hypothetical protein |
| ltrA | 16631-18532 | Putative reverse transcriptase |
| ssb | 18727-19167 | Single-stranded DNA-binding protein |
| BASYS0012 | Compl. 19022-19498 | Hypothetical protein |
| ykfF | 19196-19456 | Hypothetical protein |
| parB | 19509-21473 | ParB-like partitioning protein |
| psiB | 21528-21962 | Plasmid SOS inhibition protein B |
| psiA | 21959-22678 | Plasmid SOS inhibition protein A |
| mok | 22900-23108 | Modulator of Hok protein, Mok |
| hok | 22958-23116 | Postsegregational killing protein Hok |
| BASYS0013 | 23882-24010 | Hypothetical protein |
| BASYS0014 | Compl. 23990-24262 | Hypothetical protein |
| yubP | 24329-25264 | Hypothetical protein |
| gene X (orf39) | Compl. 25561-26208 | X-polypeptide; transglycosylation |
| traM | 26494-26877 | Mating signal |
| traJ | 27011-27757 | Regulation |
| traY | 27851-28078 | oriT nicking |
| traA | 28112-28474 | F pilin subunit |
| traL | 28479-28790 | F pilin assembly |
| traE | 28812-29378 | F pilin assembly |
| traK | 29365-30093 | F pilin assembly |
| traB | 30093-31520 | F pilin assembly |
| traP | 31510-32094 | Conjugal transfer protein |
| trbD | 32081-32401 | Conjugal transfer protein |
| trbG | 32394-32645 | Conjugal transfer protein |
| traV | 32642-33157 | F pilus assembly |
| traR | 33292-33513 | Conjugal transfer protein |
| traC | 33673-36303 | F pilus assembly |
| tnpA | 36350-37054 | Transposase of IS26 |
| tnpA | Compl. 37298-38560 | Transposase of ISEcp1 |
| tnpA | Compl. 38742-39119 | Transposase of Tn3, truncated by the insertion of ISEcp1 |
| tnpR | 39088-39681 | Resolvase of transposon Tn3 |
| blaTEM-1 | 39948-40676 | β-Lactamase TEM precursor |
| insA1 | 40727-41002 | Insertion element IS1 protein InsA |
| insB | 41047-41424 | Insertion element IS1 protein InsB |
| yigB | 41469-41930 | Putative endonuclease precursor |
| hha | 41976-42185 | Hemolysin expression-modulating protein |
| yihA | 42223-42813 | Hypothetical protein |
| repA2 | 43053-43307 | Negative regulator of repA1 expression, FII replicon |
| repA3 | 43414-43599 | Regulator of repA1 expression, FII replicon |
| repA1 | 43600-44469 | Replication initiation protein RepA1 of FII replicon |
| repA4 | 44832-45218 | Regulator of repA1 expression, FII replicon |
| tir | 45408-46061 | Inner-membrane protein |
| pemI | 46154-46411 | Stable plasmid inheritance, antitoxin |
| pemK | 46380-46745 | Stable plasmid inheritance, toxin |
| tnpA | Compl. 46882-49779 | Transposase of Tn501 |
| tnpR | 49874-50479 | Resolvase of Tn501 |
| tnpA | Compl. 50476-51321 | Transposase of Tn1721 truncated |
| pecM | 51786-52670 | Hypothetical protein |
| tet(A) | Compl. 52702-53976 | Tetracycline resistance protein, class A |
| tet(R) | 53980-54657 | Tetracycline repressor protein |
| tnpA | Compl. 54973-56562 | Transposase of Tn1721 |
| tnpA | 56553-57257 | Transposase of IS26 |
| aac6′-Ib-cr | 57263-57955 | Aminoglycoside N(6′)-acetyltransferase |
| blaOXA-1 | 58041-58916 | β-Lactamase OXA-1 precursor |
| catB4 | 59054-59602 | Chloramphenicol acetyltransferase |
| tnpA | Compl. 59548-60252 | Transposase of IS26 |
| tnpA | Compl. 60299-62314 | Transposase of IS26 |
| BASYS0015 | 62574-62906 | Hypothetical protein |
| blaCTX-M-15 | Compl. 62953-63888 | Extended-spectrum β-lactamase CTX-M-15 |
| tnpA | 63964-64669 | Transposase of IS26 |
| tnpM | Compl. 65014-65628 | Transposon Tn21 modulator protein |
| BASYS0016 | 65116-65580 | Hypothetical protein |
| int1I | Compl. 65567-66580 | Class 1 integrase |
| dhfrVII | 66537-67211 | Dihydrofolate reductase type VII |
| aadA5 | 67342-68130 | Aminoglycoside-3′-adenyltransferase |
| sulI | 68590-69516 | Dihydropteroate synthase type I |
| chrA | 70003-71208 | Putative chromate ion transporter |
| BASYS000716 | 71219-71524 | Hypothetical protein |
| tnpA | 71676-72497 | Putative transposase for insertion sequence |
| tnpA | 71715-72515 | Transposase of IS6100 |
| mph(R) | Compl. 73008-73592 | Erythromycin resistance repressor protein |
| mrx | Compl. 73592-74830 | Erythromycin resistance regulator protein |
| mph(A) | Compl. 75732-74827 | Macrolide 2-phosphotransferase |
| tnpA | Compl. 75854-76558 | Transposase of IS26 |
| nqrC | 76838-77599 | Na(+)-translocating NADH-quinone reductase subunit C |
| ywbL | 77886-79832 | High-affinity Fe2+/Pb2+ permease |
| tpd | 79870-80400 | Hypothetical protein |
| BASYS00056 | 80504-81883 | Hypothetical protein |
| ybjZ | 81886-83169 | Hypothetical protein |
| lolE | 83129-84289 | ABC transporter permease protein |
| ybbA | 84294-84989 | ABC transporter ATP-binding protein |
| BASYS00017 | Compl. 84600-85265 | Hypothetical protein |
| resA | 84958-85461 | Thiol-disulfide oxidoreductase |
| BASYS00018 | 85486-85971 | Hypothetical protein |
| agp | 86093-86647 | Glucose-1-phosphatase precursor |
| tnpA | Compl. 86681-87385 | Transposase of IS26 |
| BASYS000046 | 87386-87718 | Putative transposase OrfA of IS629, truncated |
| BASYS000047 | 88157-88606 | Putative transposase OrfA of IS629, truncated |
| tnpA | 88688-88861 | Hypothetical protein TnpA |
| tnpA | 88881-89246 | Transposase of IS4 |
| ugpB | Compl. 89305-90669 | Putative ABC transporter permease protein |
| ugpC | Compl. 90608-91732 | Putative ABC transporter ATP-binding protein |
| icc | Compl. 91687-92511 | Phosphodiesterase |
| araQ | Compl. 92522-93409 | Putative ABC transporter permease protein |
| ugpA | Compl. 93399-94277 | Putative ABC transporter permease protein |
| yigB | Compl. 94408-94638 | Oxidoreductase |
| yigB | Compl. 94666-94848 | Oxidoreductase |
| yihH | 95249-96139 | Dihydrodipicolinate synthase |
| tdcF | 96164-96544 | Translation initiation inhibitor |
| kdgT | 96577-97542 | Putative 2-keto-3-deoxygluconate permease |
| yfaX | Compl. 97588-98346 | Hypothetical protein |
| BASYS00018 | 98945-99229 | Hypothetical protein |
| BASYS00019 | 99229-99504 | Hypothetical protein |
| BASYS00020 | 99610-99903 | Hypothetical protein |
| BASYS00021 | Compl. 100099-101268 | Hypothetical protein |
| insA1 | 102357-102632 | Insertion sequence IS1 protein InsA |
| insA1 | 102677-103054 | Insertion sequence IS1 protein InsB |
| BASYS00016 | Compl. 103308-104330 | Hypothetical protein |
| vagD | Compl. 105955-106371 | Virulence-associated protein VagD |
| vagC | Compl. 106368-106598 | Virulence-associated protein VagC |
| pcar | 106949-110773 | Hypothetical protein |
| vagD | Compl. 110818-111234 | Virulence-associated protein VagD |
| vagC | Compl. 111231-111461 | Virulence-associated protein VagC |
| BASYS0020 | 111726-112226 | Hypothetical protein |
| BASYS0021 | 112239-113012 | Hypothetical protein |
| tnpA | Compl. 113223-114836 | Transposase of IS66, ORF3 |
| tnpA | Compl. 114867-115217 | Transposase of IS66, ORF2 |
| tnpA | Compl. 115214-115639 | Transposase of IS66, ORF1 |
| BASYS0022 | 115731-116852 | Hypothetical protein |
Compl., gene is the reverse complement of the positions shown.
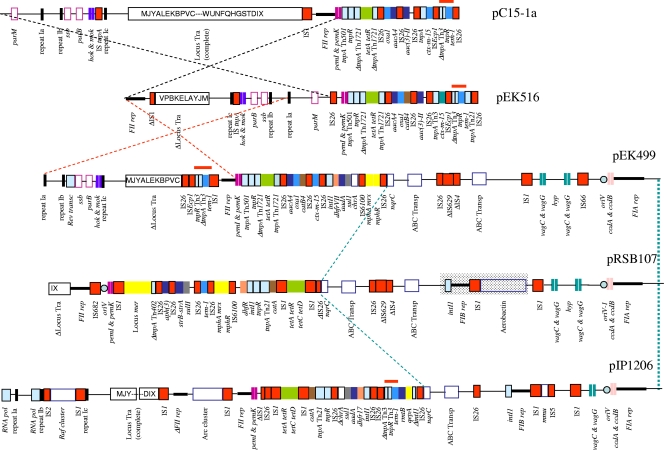
The structure of pEK499 was compared with the following two fully sequenced plasmids carrying the IncFII-FIA replicons: pRSB107 from a bacterium collected in a sewage treatment plant (32) and pIP1206 from an E. coli clinical isolate in a Belgian hospital (28). Neither of these encodes an ESBL (Fig. 1). pRSB107 encodes an aerobactin iron acquisition system and resistance to penicillins (blaTEM-1), aminoglycosides [aph(3) and strA-strB], sulfonamides (sulII), macrolides [mph(A)], chloramphenicol (catA), and tetracyclines [tet(A)]; it also carried the class 1 integron-borne trimethoprim resistance gene cassette dhfR (32). In common with pRSB107, pIP1206 also carried catA, tet(A), and blaTEM-1, but it also harbored (i) two class 1 integrons with the aadA4-dhfr17 and qepA gene cassettes, respectively (the latter gene confers resistance to hydrophilic fluoroquinolones), and (ii) rmtB, which encodes an 16S rRNA m7G methyltransferase that confers resistance to all aminoglycosides (28).
FIG. 1.
Major structural features of pEK499 (strain A) and pEK516 (strain D), encoding CTX-M-15 ESBLs, in comparison with IncFII-FIA plasmids pRSB107 (32) and pIP1206 (28) and the IncFII CTX-M-15-encoding plasmid pC15-1a (2). Resistance genes are indicated by colored boxes as follows: green, tetracycline resistance genes [tet(A), tet(R), tet(C), and tet(D)); brown, chloramphenicol resistance genes (catB4 and catA); azure, β-lactamase genes (blaOXA-1, blaTEM-1, and blaCTX-M-15); dark blue, aminoglycoside resistance genes [aacA4, aph(3), aadA1, strA-strB, and rmtB]; yellow, erythromycin, mercuric ion, and quinolone resistance genes [qepA, mer, mph(A), and mph(R)]; pale orange, trimethoprim resistance genes (dfhR, dhfRVII, and dhfr17); and gray, sulfonamide resistance genes (sul1). Transposon-related genes are indicated by black-squared boxes as follows: insertion sequences (IS) are colored in red, and other transposon-associated genes are colored in pale blue. Partitioning-associated genes (parM, ssb, parA, and parB) are indicated by pink-squared white boxes. Iron acquisition, type I DNA restriction, raffinose, arginine deaminase and ABC transporter clusters, and the homocysteine S-methyltransferase (mmu) and the NADH-ubiquinone oxidoreductase genes (nqrC) are indicated by blue-squared white boxes. The locus Tra is indicated by a squared white box with capital letters indicating the respective tra genes (i.e., V, traV; J, traJ; Y, traY; etc.). The antitoxin-toxin genes are indicated by violet (pemI-pemK), pink (ccdA-ccdB), blue (hok-mok), and green (vagC-vagD) double vertical lines, respectively. Repeats 1a, 1b, and 1c are indicated by black vertical lines. Replicons FIA, FIB, and FII are indicated by horizontal black lines. The origin or replication (oriV) is indicated by a circle. The red lines indicate the positions of the Tn3::blaTEM-1 transposons. Dashed lines represent the plasmid scaffold regions that are in common among plasmids; the black dashed lines indicate the inversion of the common region that occurred in plasmid pEK516 with respect to pC15-1a, the red dashed lines indicate the same inverted region with respect to pEK499; and the green dashed lines indicate the common region among the pEK499, pRSB107, and pIP1206 plasmids. The region of the FIB replicon of pRSB107 that is missing in the pEK499 plasmid is shadowed.
The pEK499 scaffold encoded no fewer than the following five systems to ensure stable plasmid inheritance and postsegregation killing: (i) the postsegregation killing protein Hok and its modulator Mok, located near the parB gene; (ii) the toxin-antitoxin system pemI-pemK, flanking the region of the replicon FII; (iii and iv) two copies of the vagC-vagD virulence-associated genes; and (v) one copy of the toxin-antitoxin system ccdA-ccdB, located in the region of the FIA replicon. This represents the largest number of addiction systems yet described on any IncF plasmid. For comparison, pRSB107 and pIP1206 carry four (two vagC-vagG, ccdA-ccdB, and pemI-pemK) and three (vagC-vagG, ccdA-ccdB, and pemI-pemK) systems, respectively (28, 32). These features seem likely to ensure that pEK499 is maintained in the absence of any antibiotic selective pressure. Plasmids pEK499 and pRSB107 also shared a region with two copies of the module that encodes permeases and ATP binding proteins of the ABC transporter family, and which is also partially present on pIP1206. The contribution of these transporters to virulence and plasmid maintenance has not yet been established.
pEK499 contained an incomplete transfer region composed of the genes traM to traC, but not traW to traX, and thus lacked functional conjugation machinery. Consistent with this observation, pEK499 was not transferable by conjugation in vitro; it was transformed prior to plasmid sequencing. pEK499 also lacked the aerobactin iron acquisition system, and the type I DNA restriction, raffinose and arginine deaminase clusters that have been described in the IncF plasmids pRSK107 and pIP1206, respectively (28, 32).
Analysis of pEK516 (strain D).
Plasmid pEK516 was a circular molecule of 64,471 bp, harboring 103 predicted genes (Table 3) and belonging to incompatibility group IncFII. It carried the following seven genes encoding antibiotic resistance clustered in a 22-kb region: blaCTX-M-15, blaOXA-1, blaTEM-1, aac6′-Ib-cr, aac3-IIa, catB4, and tet(A). An ISEcp1 element was located 48 bp upstream of blaCTX-M-15. pEK516 shared 75% of its DNA sequence with pEK499, albeit with considerable rearrangements (Fig. 1); notably, both plasmids carried the region containing the FII replicon and the hok-mok and parB genes. However, pEK516 was 53 kb (45%) smaller than pEK499 but carried the type I partitioning locus (parM and stbB), ensuring stable plasmid inheritance. Moreover, its Tn3::blaTEM-1 module flanked the FII replicon, whereas this module was located close to the deleted transfer region on pEK499 (Fig. 1, red line). The resistance region of pEK516 encoded an AAC(3) enzyme, absent from pEK499 and conferring resistance to gentamicin, but lacked the class 1 integron with genes conferring resistance to trimethoprim, streptomycin, and sulfonamides, also the macrolide resistance cluster. Consequently, it was similar to pC15-1a (2), from a widespread Canadian strain of E. coli with the CTX-M-15 ESBL and probably also belonging to the ST131 clone. In fact, three genetic events potentially accounted for all of the differences observed between pC15-1a and pEK516, namely, (i) the partial deletion of the tra region, (ii) the inversion of the region between the FII replicon and the parM gene, and (iii) the acquisition of catB4 close to blaOXA-1 within the resistance region of pEK516.
TABLE 3.
Open reading frames identified in pEK516
| Gene name | Nucleotide positiona | Function encoded |
|---|---|---|
| tnpA | Compl. 34-2931 | Transposase of Tn501 |
| tnpR | 3023-3630 | Resolvase of Tn21 |
| tnpA | 3627-4473 | Transposase of Tn1721 |
| pecM | 4938-5823 | Hypothetical protein |
| tet(A) | Compl. 5853-7128 | Tetracycline resistance protein, class A |
| tet(R) | 7132-7809 | Tetracycline resistance regulator protein |
| tnpA | Compl. 8124-9714 | Transposase of Tn1721 |
| tnpA | 9705-9980 | NH-2 terminal of the transposase of IS26, pseudogene |
| tnpA | 9980-10663 | COOH− terminal of the transposase of IS26, pseudogene |
| aac6′-Ib-cr | 10669-11361 | Aminoglycoside N(6′)- acetyltransferase |
| blaOXA-1 | 11447-12322 | β-Lactamase OXA-1 precursor |
| catB4 | 12460-13008 | Chloramphenicol acetyltransferase |
| tnpA | Compl. 12954-13658 | Transposase of IS26 |
| aacC3 | 13765-14625 | Aminoglycoside N(3′)-acetyltransferase III |
| BASYS0001 | 14638-15180 | Hypothetical protein |
| BASYS000006 | 15251-15583 | Hypothetical protein |
| insFI | 15580-16428 | Putative transposase of IS986/IS6110 |
| tnpA | Compl. 16374-17078 | Transposase of IS26 |
| tnpA | Compl. 17125-19140 | Transposase of Tn3, truncated |
| amb | 19400-19732 | Hypothetical protein |
| blaCTX-M-15 | Compl. 19779-20654 | Extended-spectrum β-lactamase CTX-M-15 |
| tnpA | Compl. 20910-22172 | Transposase ISEcp1 |
| tnpA | Compl. 22354-22731 | Transposase of Tn3 |
| tnpR | 22700-23293 | Resolvase of Tn3 |
| blaTEM-1 | 23476-24336 | β-Lactamase TEM precursor |
| tnpA | Compl. 2447-26954 | Transposase of Tn21 |
| tnpA | Compl. 27097-27801 | Transposase of IS26 |
| repA1 | Compl. 28926-29795 | Replication initiation protein RepA1 of the FII replicon |
| repA4 | Compl. 30088-30342 | Regulator of repA1 expression, FII replicon |
| yihA | Compl. 30582-31172 | Hypothetical protein |
| hha | Compl. 31210-31419 | Hemolysin expression-modulating protein |
| yigB | Compl. 31465-31938 | Putative endonuclease precursor |
| insB2 | Compl. 31971-32474 | Insertion element IS1 protein InsB |
| yfhA | Compl. 32697-33119 | Hypothetical protein |
| traR | Compl. 33112-33333 | Conjugal transfer protein |
| traV | Compl. 33468-33983 | F pilus assembly |
| trbD | Compl. 33980-34300 | Conjugal transfer protein |
| traP | Compl. 34287-34874 | Conjugal transfer protein |
| traB | Compl. 34843-36294 | F pilin assembly |
| traK | Compl. 36294-37022 | F pilin assembly |
| traE | Compl. 37009-37575 | F pilin assembly |
| traL | Compl. 37597-37908 | F pilin assembly |
| traA | Compl. 37923-38282 | F pilin subunit |
| traY | Compl. 38315-38542 | oriT nicking |
| traJ | Compl. 38679-39350 | Regulation |
| traM | Compl. 39544-40029 | Mating signal |
| gene X (orf39) | 40250-40852 | X− polypeptide; transglycosylation |
| yubP | Compl. 41149-41970 | Hypothetical protein |
| yeiA | Compl. 42081-42377 | Hypothetical protein |
| yehA | 42677-43054 | Hypothetical protein |
| tnpA | Compl. 43058-44461 | Transposase of ISPsy5 |
| ECs1338 | Compl. 44702-45052 | Hypothetical protein |
| ECs1337 | Compl. 45049-45474 | Hypothetical protein |
| hok | Compl. 45832-45990 | Postsegregational killing protein Hok |
| mok | Compl. 45836-46048 | Modulator of Hok protein, Mok |
| psiA | Compl. 46270-46989 | Plasmid SOS inhibition protein A |
| psiB | Compl. 46986-47423 | Plasmid SOS inhibition protein B |
| yefA | Compl. 47489-48064 | ParB-like nuclease |
| parB | Compl. 48091-49512 | ParB-like partitioning protein |
| BASYS00066 | 49537-49884 | Hypothetical protein |
| ydeA | Compl. 49573-49812 | Hypothetical protein |
| ssb | Compl. 49864-50430 | Single-strand binding protein |
| yddA | 50693-51154 | Hypothetical protein |
| ydcA | Compl. 51240-51803 | Hypothetical protein |
| ydbA | Compl. 51850-53211 | Hypothetical protein |
| SC | Compl. 54531-54722 | Hypothetical protein |
| ycjA | Compl. 54719-55141 | Hypothetical protein |
| yicB | Compl. 55188-55613 | Antirestriction protein KlcA |
| ychA | Compl. 56031-56858 | Hypothetical protein |
| ycgB | Compl. 56858-57292 | Hypothetical protein |
| P030 | Compl. 57306-57527 | Hypothetical protein |
| ycgA | Compl. 57528-58175 | Hypothetical protein |
| parM | 58728-59690 | Plasmid segregation protein |
| stbB | 59690-60043 | Stable plasmid inheritance protein |
| ycdB | Compl. 60166-60447 | Hypothetical protein |
| ycdA | 60476-60712 | Hypothetical protein |
| tnpA | 62389-63093 | Transposase of IS26 |
| tir | 63133-63684 | Inner-membrane protein |
| pemI | 63777-64034 | Stable plasmid inheritance, antitoxin |
| pemK | 63967-64368 | Stable plasmid inheritance, toxin |
Compl., gene is the reverse complement of the positions shown.
Both pEK516 and pC15-1a carried only two addiction systems, pemI-pemK and hok-mok, and resembled the IncFII portion of pEK499. pEK516 showed a greater deletion of the transfer region than pEK499, since the latter contained traC, which was missing in pEK516. Despite this, pEK516 was transferred by conjugation in vitro (16); so it seems likely that the tra deletion(s) occurred during subsequent storage and prior to plasmid sequencing.
In summary, pEK516 represented a highly related variant of the previously identified plasmid, pC15-1a (2). Plasmids of the FII group harboring blaCTX-M-15 have wide geographic scatter (4, 6) and have played a crucial role in the global spread of CTX-M-15 ESBLs in E. coli.
Analysis of pEK204 (strain C).
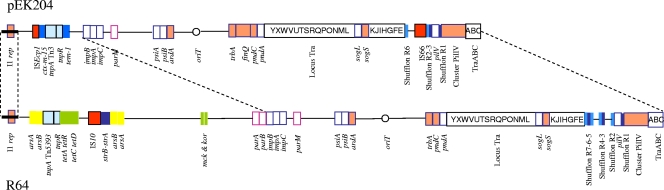
Plasmid pEK204 was a circular molecule of 93,732 bp, harboring 112 predicted genes (Table 4), and could be transferred by conjugation in vitro. It belonged to incompatibility group IncI1, which is characterized by the presence of a gene cluster encoding a thin, type IV pilus required for liquid matings (18) and the RepZ replicase gene. As such, it was distinct from two other fully sequenced plasmids encoding the CTX-M-3 enzyme, pCTX-M3, which is widespread in Poland (10, 26), and pK29 (5). pEK204 was assigned to a new IncI1 pMLST type (9), ST16 (I1, A5, S8, P6, and T10), with four unique alleles (pil6, sogS8, ardA5, and trbA-pndC10). However, its gross structure revealed strong similarity to IncI1 plasmid pCOLIb-P9 (GenBank AB021078), although the region that includes the colicin 1b gene (pCOLIb-P9 nucleotides 8310 to 20275) was absent, and to R64, which is the reference plasmid for the IncI1 group (GenBank accession no. AP005147; Fig. 2). In comparison with those of R64, pEK204 lacked the arsenic, tetracycline, and streptomycin resistance genes and the addiction systems mck-kor and parA-parB; this deleted region was substituted by its own resistance region. The colinearity between the pEK204 and R64 plasmid scaffolds was therefore well maintained in the transfer region, with the exception of the shufflons—a characteristic feature of IncI1 plasmids that may act as a biological switch (12, 19, 20)—which appeared to be rearranged in pEK204 with respect to R64 and also by the insertion of an IS66 element.
TABLE 4.
Open reading frames identified in pEK204
| Gene name | Nucleotide positiona | Function encoded |
|---|---|---|
| repY | 378-467 | Regulatory protein of RepZ |
| repZ | 455-1486 | Replication protein of the I1 replicon |
| yafA | Compl. 2556-3065 | Hypothetical protein |
| yafB | Compl. 3121-3681 | Hypothetical protein |
| yagA | 3966-5423 | Hypothetical protein |
| tnpA | Compl. 5711-5806 | Transposase of Tn3, truncated |
| tnpA | 5997-7259 | Transposase of ISEcp1 |
| blaCTX-M-3 | 7594-8469 | β-Lactamase CTX-M-3 precursor |
| tnpA | Compl. 8772-11930 | Transposase of Tn3, truncated |
| tnpR | 11899-12492 | Resolvase of Tn3 |
| blaTEM-1 | 12675-13535 | β-Lactamase TEM-1 precursor |
| impB | Compl. 15037-16308 | UV protection and mutation protein |
| impA | Compl. 16308-16745 | UV protection and mutation protein |
| impC | Compl. 16742-16990 | UV protection and mutation protein |
| yccA | 17098-17370 | Hypothetical protein |
| yccB | 17381-18310 | Hypothetical protein |
| ycdA | 18307-18628 | Hypothetical protein |
| parM | 19078-20058 | Plasmid segregation protein ParM |
| stbB | 20055-20483 | Stable plasmid inheritance protein |
| ycdB | 20874-21557 | Putative methylase |
| yceA | 21558-21779 | Hypothetical protein |
| yceB | 21793-22227 | Hypothetical protein |
| ycfA | 22272-23042 | Hypothetical protein |
| ycgB | 23455-23880 | Hypothetical protein |
| ycgC | 23927-24349 | Hypothetical protein |
| ychA | 24401-24703 | Hypothetical protein |
| ssb | 25322-25849 | Single-stranded DNA-binding protein |
| ykfF | 25880-26140 | Hypothetical protein |
| ycjA | 26193-28157 | Hypothetical protein |
| psiB | 28117-28644 | Plasmid SOS inhibition protein B |
| psiA | 28641-29360 | Plasmid SOS inhibition protein A |
| ygaA | 29357-29953 | Hypothetical protein |
| ygbA | 30025-30915 | Hypothetical protein |
| ardA | 30415-30915 | Antirestriction protein |
| ydfA | 31644-32078 | Hypothetical protein |
| ydfB | 32172-32438 | Hypothetical protein |
| BASYS00078 | 32615-33004 | Hypothetical protein |
| ygbB | 33001-33903 | Hypothetical protein |
| ygeA | Compl. 34202-34453 | Hypothetical protein |
| BASYS00079 | Compl. 34487-34798 | Hypothetical protein |
| ydiA | 35032-35880 | Hypothetical protein |
| yggA | Compl. 35966-36301 | Hypothetical protein |
| nikA | 36532-36867 | Relaxosome component protein |
| nikB | 36878-39577 | Relaxase |
| trbC | Compl. 39614-41905 | Hypothetical protein |
| trbB | Compl. 39614-41905 | Hypothetical protein |
| trbA | Compl. 42987-44195 | Hypothetical protein |
| finQ | Compl. 44502-45431 | Hypothetical protein |
| pndC | 45769-46053 | Postsegregation killing system, counter protein for PndA |
| pndA | 45908-46060 | Postsegregation killing protein |
| exc | Compl. 47893-48555 | Surface exclusion protein |
| traY | Compl. 48626-50863 | Integral membrane protein |
| traX | Compl. 50891-51475 | F pilin acetylation |
| traW | Compl. 51504-52706 | F pilus assembly |
| traV | Compl. 52673-53287 | F pilus assembly |
| traU | Compl. 53287-56331 | F pilus assembly |
| traT | Compl. 56421-57221 | Surface exclusion |
| traS | Compl. 57205-57393 | Surface exclusion |
| traR | Compl. 57457-57861 | Hypothetical protein |
| traQ | Compl. 58439-59143 | Conjugal transfer protein |
| traP | Compl. 58439-59143 | Conjugal transfer protein |
| traO | Compl. 59143-60432 | Hypothetical protein |
| traN | Compl. 60435-61418 | Aggregate stability |
| traM | Compl. 661429-62121 | Mating signal |
| traL | Compl. 62118-62465 | F pilus assembly |
| sogL | Compl. 62483-66286 | DNA primase |
| sogS | Compl. 62483-65020 | Regulative protein |
| nuc | Compl. 66340-66894 | EDTA-resistant nuclease |
| traK | Compl. 66906-67196 | F pilus assembly |
| traJ | Compl. 67193-68341 | ATP-binding protein |
| traI | Compl. 68338-69156 | DNA helicase |
| traH | Compl. 69153-69611 | F pilus assembly |
| traG | Compl. 70005-70589 | F pilus assembly |
| traF | Compl. 70649-71851 | F pilus assembly |
| traE | Compl. 71937-72761 | F pilus assembly |
| rci | Compl. 72912-74066 | Shufflon-specific DNA recombinase |
| tnpA | 75447-77060 | Transposase of IS66 |
| pilV | Compl. 77739-79163 | Type IV prepilin cluster |
| pilU | Compl. 79162-79819 | Type IV prepilin cluster, prepilin peptidase |
| pilT | Compl. 79804-80364 | Type IV prepilin cluster |
| pilS | Compl. 80374-80988 | Type IV prepilin cluster, prepilin |
| pilP | Compl. 80988-84132 | Type IV prepilin cluster |
| pilR | Compl. 81005-82103 | Type IV prepilin cluster, integral membrane protein |
| pilQ | Compl. 82115-83669 | TypeIV prepilin cluster, ATP-binding protein |
| pilP | Compl. 83680-84132 | Type IV prepilin cluster |
| pilO | Compl. 84179-85414 | Type IV prepilin cluster |
| pilN | Compl. 85407-87089 | Type IV prepilin cluster, secretin protein |
| pilM | Compl. 87103-87540 | Type IV prepilin cluster |
| pilL | Compl. 87540-88607 | Type IV prepilin cluster, lipoprotein |
| pilK | Compl. 88641-89234 | Type IV prepilin cluster |
| pilJ | Compl. 89231-89527 | Type IV prepilin cluster |
| pilI | Compl. 89694-89948 | Type IV prepilin cluster |
| BASYS00080 | Compl. 90108-90661 | Hypothetical protein |
| traC | Compl. 90834-91517 | F pilus assembly |
| traB | Compl. 91771-92304 | F pilus assembly |
| traA | Compl. 92309-92432 | F pilin subunit |
Compl., gene is the reverse complement of the positions shown.
FIG. 2.
Major structural features of pEK204 (strain C), encoding CTX-M-3 ESBL, in comparison with IncI1 plasmid R64. Resistance genes are indicated by colored boxes as follows: green, tetracycline resistance genes (tet(A), tet(R), tet(C), and tet(D)); azure, β-lactamase genes (blaTEM-1 and blaCTX-M-15); dark blue, aminoglycoside resistance genes (strA-strB); and yellow, arsenic resistance genes (arsA and arsB). Transposon-related genes are indicated by black-squared boxes as follows: insertion sequences (IS) are colored in red, and other transposon-associated genes are colored in pale blue. Partitioning-associated genes (parA, parB, and parM) are indicated by pink-squared white boxes. Blue-squared white boxes indicate characteristic IncI1 clusters (impABC, psiAB, pndAC, and pilV-IV). The Tra locus is indicated by a squared white box with capital letters indicating the respective tra genes (i.e., Y, traY; X, traX; W, traW; etc.). The antitoxin-toxin genes (mck-kor) are indicated by green double vertical lines. The shufflons are indicated by azure and dark blue vertical lines. The I1 replicon is indicated by a horizontal black line. The origin of transfer (oriT) is indicated by a circle. Dashed lines represent the plasmid scaffold regions that are common to both plasmids. Pale orange-colored boxes indicate the IncI1-pMLST target sites (repI1, ardA, trbA-pndC, SogS, and pilIV).
In contrast to multiresistance plasmids pEK499 and pEK516, pEK204 carried only two known resistance genes, blaCTX-M-3 and blaTEM-1 (Fig. 3). The blaCTX-M-3 gene, which encodes an ESBL that differs from CTX-M-15 by only a Asp240→Gly substitution, has previously been detected on plasmids belonging to different incompatibility groups and with broad host ranges (4), including IncL/M (pCTX-M-3; ca. 90 kb) in Poland (10, 26) and IncHI2 (pK29; ca. 270 kb) in Taiwan (5), as well as the IncA/C, IncFII, and IncN types (4). Large (>90-kb) IncI1 plasmids encoding the CTX-M-3 enzyme have been reported previously in diverse members of the Enterobacteriaceae in a university hospital in Taiwan (24). Moreover, IncI1 plasmids have been reported to encode myriad other β-lactamases besides the CTX-M-3 enzyme, including CTX-M-1, CTX-M-2, CTX-M-9, CTX-M-14, CTX-M-15, and CTX-M-24, as well as TEM-1 (as here), TEM-52, SHV-12, several CMY (acquired AmpC) enzymes, and the metallocarbapenemase VIM-1 (4, 5, 9).
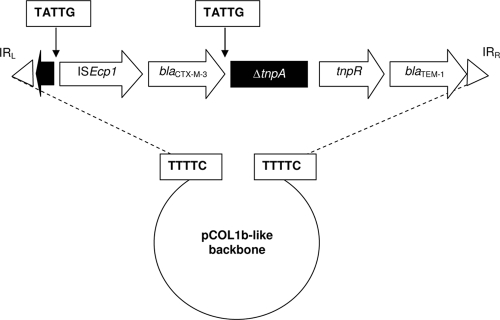
FIG. 3.
Detail of the resistance region of pEK204, which arose through the acquisition of Tn3 and ISEcp1-blaCTX-M-3 by a pCOL1b-P9-like plasmid. The 5-bp direct repeats consistent with transposition events are shown. The disrupted tnpA gene of the Tn3 element is shaded black.
The sequence data suggest that pEK204 arose by the transposition of Tn3 and ISEcp1-blaCTX-M-3 elements into a pCOLIb-P9-like plasmid. This Tn3 element inserted after the position equivalent to nucleotide 8269 of pCOLIb-P9 and was flanked by 5-bp direct repeats of TTTTC. Both of the Tn3 terminal inverted repeats, IRL and IRR, were intact, but the tnpA gene (encoding the transposase) was disrupted by ISEcp1-blaCTX-M-3 (Fig. 3). The ISEcp1 element was located 128 bp upstream of blaCTX-M-3, and the linking sequence was identical to that of pCTX-M-3 (10), though different from that (48 bp) between ISEcp1 and blaCTX-M-15 on pEK516 or the remnant of this IS element on pEK499. This underscores the point that, although they differ only by one nucleotide, the blaCTX-M-3 and blaCTX-M-15 genes in the United Kingdom plasmids represent separate escape events from Kluyvera spp.
The ISEcp1-blaCTX-M-3 element of pEK204 was flanked by 5-bp direct repeats of TATTG, consistent with ISEcp1-mediated transposition, which prefers AT-rich target sequences (29). Despite disruption, the Tn3 transposase (tnpA) gene on pEK204 was still predicted to encode a protein of 999 amino acids, comprising 970 (ca. 97%) “genuine” amino acids plus 29 “new” C-terminal amino acids resulting from the read-through of the in-frame ISEcp1-blaCTX-M-3 insertion. The genuine C-terminal residues of the Tn3 transposase were encoded by a ΔtnpA remnant located between IRL and ISEcp1 (Fig. 3). The disruption of transposons by other transposable elements is a feature common to many bacterial genomes and plasmids, including all of those reported here. The resistance region of pEK204 represents a simple example of this but raises the question of whether this plasmid evolved via one or two transposition events. If the predicted transposase of Tn3 on pEK204 remains functional, it would be expected to mediate the transposition of a new element, still defined by the Tn3 terminal inverted repeats IRL and IRR but encoding CTX-M-3 ESBL in addition to TEM-1 penicillinase (Fig. 3). The potential for the simultaneous transposition of two distinct β-lactamase genes from pEK204-like plasmids has public health importance and will be investigated further. It is relevant in context that Tn3 played a major role in the huge dissemination of TEM-1 β-lactamase in the 1960s and 1970s.
Concluding remarks.
We have determined the complete sequences of the CTX-M ESBL-encoding plasmids found in three United Kingdom variants (A, C, and D) of the pathogenic, multiresistant ST131 E. coli lineage. Strain A (pEK499), which is widespread and locally dominant in the United Kingdom, has been found in Austria (8) and, more recently, in Bolzano, northern Italy (R. Aschbacher, D. M. Livermore, and N. Woodford, unpublished data). Strain C is also widely found in the United Kingdom: some Belfast isolates of the strain have the CTX-M-3 enzyme encoded by pEK204, as here, but others, including many from the United Kingdom mainland, have CTX-M-15, possibly encoded by a different plasmid. In contrast to these widely disseminated strains, the third ST131 variant studied, strain D (pEK516), is prevalent only in Shropshire, a county on the English-Welsh border.
Clearly, differences exist among variants of the ST131 clone and these probably have significant impact on their relative success and prevalence. Success is likely to reflect a complex combination of characteristics, both intrinsic to the PFGE-defined variant or acquired on plasmids and mobile genetic elements, interacting with local pressures and opportunities relating to antibiotic usage and patient types. It is clear, however, that these strains and their multiresistance plasmids have been moving beyond the hospital environment in the United Kingdom for some years (34). A recent study identified ESBL-producing E. coli in the bowel flora of 40% of residents of long-term-care facilities in Belfast; almost half were colonized by strain A (30) and most of the remainder by diverse ST131 variants harboring pEK204-like plasmids (7). The multitude of addiction systems present on pEK499 will, in particular, ensure that it is maintained even in the absence of antibiotic selection.
Many patients with E. coli producing CTX-M ESBLs, most of them elderly, present with community-onset infections in increasing numbers, providing evidence of true community acquisition (25). This age distribution may change over time, leading to new problems. A study from Hong Kong found significant rates of ESBL production (particularly of the CTX-M-14 enzyme) among urinary isolates of E. coli from women of all ages; the prevalence was 7.3% among community-onset infections in the 18-to-35 age group (13). Another study, from Canada, of community-onset infections caused by ESBL-producing E. coli recorded a much broader age distribution among patients deemed to have community-acquired infections versus those considered to have health care-associated infections (22). Rising rates of E. coli with CTX-M ESBLs in the genitourinary tracts of sexually active women raise the alarming possibility that these enzymes might “escape” into sexually transmitted bacterial pathogens, specifically Neisseria gonorrhoeae. Oral and intramuscular oxyimino-cephalosporins, such as cefixime and ceftriaxone, are widely used as a first-line treatment for uncomplicated gonorrhea, and any evolution of ESBL-producing gonococci would be a catastrophic development.
In summary, United Kingdom E. coli strains A, C, and D belonging to the internationally disseminated O25:H4-ST131 clone (21, 27, 34) have acquired different plasmids encoding CTX-M ESBLs on separate occasions.
Acknowledgments
We thank AstraZeneca for supporting this work.
Footnotes
Published ahead of print on 17 August 2009.
REFERENCES
- 1.Abbott, J. C., D. M. Aanensen, K. Rutherford, S. Butcher, and B. G. Spratt. 2005. WebACT—an online companion for the Artemis Comparison Tool. Bioinformatics 21:3665-3666. [DOI] [PubMed] [Google Scholar]
- 2.Boyd, D. A., S. Tyler, S. Christianson, A. McGeer, M. P. Muller, B. M. Willey, E. Bryce, M. Gardam, P. Nordmann, and M. R. Mulvey. 2004. Complete nucleotide sequence of a 92-kilobase plasmid harboring the CTX-M-15 extended-spectrum β-lactamase involved in an outbreak in long-term-care facilities in Toronto, Canada. Antimicrob. Agents Chemother. 48:3758-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Canton, R., and T. M. Coque. 2006. The CTX-M β-lactamase pandemic. Curr. Opin. Microbiol. 9:466-475. [DOI] [PubMed] [Google Scholar]
- 4.Carattoli, A. 23 March 2009. Resistance plasmid families in Enterobacteriaceae. Antimicrob. Agents Chemother. doi: 10.1128/AAC.01707-08. [DOI] [PMC free article] [PubMed]
- 5.Chen, Y. T., T. L. Lauderdale, T. L. Liao, Y. R. Shiau, H. Y. Shu, K. M. Wu, J. J. Yan, I. J. Su, and S. F. Tsai. 2007. Sequencing and comparative genomic analysis of pK29, a 269-kilobase conjugative plasmid encoding CMY-8 and CTX-M-3 β-lactamases in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 51:3004-3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coque, T. M., A. Novais, A. Carattoli, L. Poirel, J. Pitout, L. Peixe, F. Baquero, R. Canton, and P. Nordmann. 2008. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum β-lactamase CTX-M-15. Emerg. Infect. Dis. 14:195-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dhanji, H., N. Woodford, R. Hope, A. Loughrey, P. Rooney, and D. M. Livermore. 2008. Diversity of Escherichia coli with CTX-M ESBLs in Long-Term Care Facilities (LTCFs) in Belfast, abstr. C2-3890. Abstr. 48th Annu. Intersci. Conf. Antimicrob. Agents Chemother. (ICAAC)-Infect. Dis. Soc. Am. (IDSA) 46th Annu. Meet. American Society for Microbiology and Infectious Diseases Society of America, Washington, DC.
- 8.Eisner, A., E. J. Fagan, G. Feierl, H. H. Kessler, E. Marth, D. M. Livermore, and N. Woodford. 2006. Emergence of Enterobacteriaceae isolates producing CTX-M extended-spectrum β-lactamase in Austria. Antimicrob. Agents Chemother. 50:785-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garcia-Fernandez, A., G. Chiaretto, A. Bertini, L. Villa, D. Fortini, A. Ricci, and A. Carattoli. 2008. Multilocus sequence typing of IncI1 plasmids carrying extended-spectrum β-lactamases in Escherichia coli and Salmonella of human and animal origin. J. Antimicrob. Chemother. 61:1229-1233. [DOI] [PubMed] [Google Scholar]
- 10.Golebiewski, M., I. Kern-Zdanowicz, M. Zienkiewicz, M. Adamczyk, J. Zylinska, A. Baraniak, M. Gniadkowski, J. Bardowski, and P. Ceglowski. 2007. Complete nucleotide sequence of the pCTX-M3 plasmid and its involvement in spread of the extended-spectrum β-lactamase gene blaCTX-M-3. Antimicrob. Agents Chemother. 51:3789-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grant, J. R., and P. Stothard. 2008. The CGView server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36:W181-W184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gyohda, A., N. Furuya, A. Ishiwa, S. Zhu, and T. Komano. 2004. Structure and function of the shufflon in plasmid R64. Adv. Biophys. 38:183-213. [PubMed] [Google Scholar]
- 13.Ho, P. L., W. W. N. Poon, S. L. Loke, M. S. T. Leung, K. H. Chow, R. C. W. Wong, K. S. Yip, E. L. Lai, and K. W. T. Tsang on behalf of the COMBAT Study Group. 2007. Community emergence of CTX-M type extended-spectrum β-lactamases among urinary Escherichia coli from women. J. Antimicrob. Chemother. 60:140-144. [DOI] [PubMed] [Google Scholar]
- 14.Karisik, E., M. J. Ellington, D. M. Livermore, and N. Woodford. 2008. Complete nucleotide sequences of plasmids pEK204 and pEK516, encoding CTX-M enzymes in two major UK Escherichia coli strains, abstr. P2003. Abstr. 18th Eur. Congr. Clin. Microbiol. Infect. Dis., Barcelona, Spain. http://www.blackwellpublishing.com/eccmid18/abstract.asp?id=69900.
- 15.Karisik, E., M. J. Ellington, D. M. Livermore, and N. Woodford. 2008. Virulence factors in Escherichia coli with CTX-M-15 and other extended-spectrum β-lactamases in the UK. J. Antimicrob. Chemother. 61:54-58. [DOI] [PubMed] [Google Scholar]
- 16.Karisik, E., M. J. Ellington, R. Pike, R. E. Warren, D. M. Livermore, and N. Woodford. 2006. Molecular characterization of plasmids encoding CTX-M-15 β-lactamases from Escherichia coli strains in the United Kingdom. J. Antimicrob. Chemother. 58:665-668. [DOI] [PubMed] [Google Scholar]
- 17.Karisik, E., A. Underwood, M. J. Ellington, D. M. Livermore, and N. Woodford. 2007. Complete nucleotide sequence of pEK499, a multi-drug resistance plasmid from the UK's most prevalent Escherichia coli strain with CTX-M-15 β-lactamase, abstr. 1733_362. Abstr. 17th Eur. Congr. Clin. Microbiol. Infect. Dis., Munich, Germany. http://www.blackwellpublishing.com/eccmid17/abstract.asp?id=56661.
- 18.Kim, S. R., and T. Komano. 1997. The plasmid R64 thin pilus identified as a type IV pilus. J. Bacteriol. 179:3594-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Komano, T., S. R. Kim, and T. Nisioka. 1987. Distribution of shufflon among IncI plasmids. J. Bacteriol. 169:5317-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Komano, T. 1999. SHUFFLONS: multiple inversion systems and integrons. Annu. Rev. Genet. 33:171-191. [DOI] [PubMed] [Google Scholar]
- 21.Lau, S. H., M. E. Kaufmann, D. M. Livermore, N. Woodford, G. A. Willshaw, T. Cheasty, K. Stamper, S. Reddy, J. Cheesbrough, F. J. Bolton, A. J. Fox, and M. Upton. 2008. UK epidemic Escherichia coli strains A-E, with CTX-M-15 β-lactamase, all belong to the international O25:H4-ST131 clone. J. Antimicrob. Chemother. 62:1241-1244. [DOI] [PubMed] [Google Scholar]
- 22.Laupland, K. B., D. L. Church, J. Vidakovich, M. Mucenski, and J. D. D. Pitout. 2008. Community-onset extended-spectrum β-lactamase (ESBL) producing Escherichia coli: importance of international travel. J. Infect. 57:441-448. [DOI] [PubMed] [Google Scholar]
- 23.Leflon-Guibout, V., J. Blanco, K. Amaqdouf, A. Mora, L. Guize, and M. H. Nicolas-Chanoine. 2008. Absence of CTX-M enzymes but high prevalence of clones, including clone ST131, among fecal Escherichia coli isolates from healthy subjects living in the area of Paris, France. J. Clin. Microbiol. 46:3900-3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu, S. Y., L. H. Su, Y. L. Yeh, C. Chu, J. C. Lai, and C. H. Chiu. 2007. Characterisation of plasmids encoding CTX-M-3 extended-spectrum β-lactamase from Enterobacteriaceae isolated at a university hospital in Taiwan. Int. J. Antimicrob. Agents 29:440-445. [DOI] [PubMed] [Google Scholar]
- 25.Livermore, D. M., R. Canton, M. Gniadkowski, P. Nordmann, G. M. Rossolini, G. Arlet, J. Ayala, T. M. Coque, I. Kern-Zdanowicz, F. Luzzaro, L. Poirel, and N. Woodford. 2007. CTX-M: changing the face of ESBLs in Europe. J. Antimicrob. Chemother. 59:165-174. [DOI] [PubMed] [Google Scholar]
- 26.Mierzejewska, J., A. Kulinska, and G. Jagura-Burdzy. 2007. Functional analysis of replication and stability regions of broad-host-range conjugative plasmid CTX-M3 from the IncL/M incompatibility group. Plasmid 57:95-107. [DOI] [PubMed] [Google Scholar]
- 27.Nicolas-Chanoine, M. H., J. Blanco, V. Leflon-Guibout, R. Demarty, M. P. Alonso, M. Caniça, Y.-J. Park, J.-P. Lavigne, J. Pitout, and J. R. Johnson. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61:273-281. [DOI] [PubMed] [Google Scholar]
- 28.Perichon, B., P. Bogaerts, T. Lambert, L. Frangeul, P. Courvalin, and M. Galimand. 2008. Sequence of conjugative plasmid pIP1206 mediating resistance to aminoglycosides by 16S rRNA methylation and to hydrophilic fluoroquinolones by efflux. Antimicrob. Agents Chemother. 52:2581-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poirel, L., M. F. Lartigue, J. W. Decousser, and P. Nordmann. 2005. ISEcp1B-mediated transposition of blaCTX-M in Escherichia coli. Antimicrob. Agents Chemother. 49:447-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rooney, P. J., M. C. O'Leary, A. C. Loughrey, M. McCalmont, B. Smyth, P. Donaghy, M. Badri, N. Woodford, E. Karisik, and D. M. Livermore. 2009. Nursing homes as a reservoir of multi-drug resistant Escherichia coli. J. Antimicrob. Chemother. 64:635-641. [DOI] [PubMed]
- 31.Suzuki, S., N. Shibata, K. Yamane, J. I. Wachino, K. Ito, and Y. Arakawa. 2009. Change in the prevalence of extended-spectrum-β-lactamase-producing Escherichia coli in Japan by clonal spread. J. Antimicrob. Chemother. 63:72-79. [DOI] [PubMed] [Google Scholar]
- 32.Szczepanowski, R., S. Braun, V. Riedel, S. Schneiker, I. Krahn, A. Puhler, and A. Schluter. 2005. The 120,592 bp IncF plasmid pRSB107 isolated from a sewage-treatment plant encodes nine different antibiotic-resistance determinants, two iron-acquisition systems and other putative virulence-associated functions. Microbiology 151:1095-1111. [DOI] [PubMed] [Google Scholar]
- 33.Van Domselaar, G. H., P. Stothard, S. Shrivastava, J. A. Cruz, A. Guo, X. Dong, P. Lu, D. Szafron, R. Greiner, and D. S. Wishart. 2005. BASys: a web server for automated bacterial genome annotation. Nucleic Acids Res. 33:W455-W459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woodford, N., M. E. Ward, M. E. Kaufmann, J. Turton, E. J. Fagan, D. James, A. P. Johnson, R. Pike, M. Warner, T. Cheasty, A. Pearson, S. Harry, J. B. Leach, A. Loughrey, J. A. Lowes, R. E. Warren, and D. M. Livermore. 2004. Community and hospital spread of Escherichia coli producing CTX-M extended-spectrum β-lactamases in the UK. J. Antimicrob. Chemother. 54:735-743. [DOI] [PubMed] [Google Scholar]
- 35.Woodford, N. 2008. Successful, multiresistant bacterial clones. J. Antimicrob. Chemother. 61:233-234. [DOI] [PubMed] [Google Scholar]