Abstract
Centrosome duplication must remain coordinated with cell cycle progression to ensure the formation of a strictly bipolar mitotic spindle, but the mechanisms that regulate this coordination are poorly understood. Previous work has shown that prolonged S-phase is permissive for centrosome duplication, but prolonging either G2 or M-phase cannot support duplication. To examine whether G1 is permissive for centrosome duplication, we release serum-starved G0 cells into mimosine, which delays the cell cycle in G1.We find that in mimosine, centrosome duplication does occur, albeit slowly compared with cells that progress into S-phase; centrosome duplication in mimosine-treated cells also proceeds in the absence of a rise in Cdk2 kinase activity normally associated with the G1/S transition.CHO cells arrested with mimosine can also assemble more than four centrioles (termed “centrosome amplification”), but the extent of centrosome amplification during prolonged G1 is decreased compared to cells that enter S-phase and activate the Cdk2-cyclin complex. Together, our results suggest a model, which predicts that entry into S-phase and the rise in Cdk2 activity associated with this transition are not absolutely required to initiate centrosome duplication, but rather, serve to entrain the centrosome reproduction cycle with cell cycle progression.
The single interphase centrosome duplicates exactly once in a cell cycle dependent fashion, yielding two daughter centrosomes, each of which contributes to the assembly of a mitotic spindle pole (reviewed in Stearns, 2001; Delattre and Gönczy, 2004). The two initial events in the centrosome duplication cycle are commonly thought to be the “disorientation” or “disengagement” of the parental centriole pair (termed the diplosome), which prepares the centrosome for duplication, followed by the assembly of short daughter centrioles—called pro-centrioles—at right angles to the pre-existing centrioles (Kuriyama and Borisy, 1981; Kochanski and Borisy, 1990). Centrosome duplication is coordinated with nuclear events during cell cycle progression (reviewed in Hinchcliffe and Sluder, 2001a). This is important because centrosomes play a dominant role in spindle pole organization; there must be two and only two centrosomes present as the cell enters mitosis, otherwise the potential exists to assemble a multipolar spindle, resulting in an increased frequency of aneuploidy and tumor cell progression (reviewed in Brinkley, 2001; Fisk et al., 2002; Sluder and Nordberg, 2004).
Understanding the cell cycle control of centrosome duplication has been difficult, primarily because there is no good marker to signify when duplication begins. By the time morphologically distinct centrioles have formed, and are recognizable in the electron microscope, the initial events of centrosome duplication may have already taken place (discussed in Hinchcliffe and Sluder, 1998). In order to examine the cell cycle regulation of centrosome duplication, several studies have relied on arresting cell cycle progression in a particular phase, and then assaying whether or not centrosomes are capable of duplicating one or more times (reviewed in Hinchcliffe and Sluder, 2001a). The results of this work have experimentally defined those stages that are capable of supporting centrosome duplication, and those that cannot. In cells that are driven into G0, the centrosome does not duplicate until serum-released (Tucker et al., 1979; Okuda et al., 2000). During S-phase arrest, centrosomes can clearly undergo repeated rounds of duplication in both zygotes and certain transformed somatic cells (Kuriyama et al., 1986; Sluder and Lewis, 1987; Raff and Glover, 1988; Balczon et al., 1995; Hinchcliffe et al., 1998). In cells arrested in G2 with topoisomerase inhibitors, the duplicated centrosome cannot undergo further rounds of duplication (Balczon et al., 1995). When mitosis is prolonged in a variety of cell types, centrosome duplication cannot proceed (Hinchcliffe et al., 1998; Vidwans et al., 1999), although the diplosome can undergo disengagement (Tsou and Stearns, 2006).
While the aforementioned studies have provided information about which cell cycle stages can or cannot support centrosome duplication, there are conflicting reports about whether or not centrosome duplication can occur during G1. This phase of the cell cycle is particularly important, because centrosome duplication is said to normally occur at the G1/S phase transition (Robbins et al., 1968; Rattner and Phillips, 1973; Kuriyama and Borisy, 1981; Vorobjev and Chentsov, 1982; Alvey, 1985). The implication is that as the cell cycle proceeds into S-phase, the signals that initiate DNA replication also drive the duplication of the centrosome (discussed in Sluder and Rieder, 1996). However, it remains a formal possibility that centrosome duplication could be initiated without the activation of these signals, and then continues in parallel as the cell cycle transitions into S-phase. Several studies have suggested that centrosome duplication cannot occur until after the G1/S phase transition (Robbins et al., 1968; Vorobjev and Chentsov, 1982; Marshall et al., 2001), while other have revealed that it can proceed prior to entry into S-phase (Rattner and Phillips, 1973; Winey and Byers, 1993; Fukasawa et al., 1996; Schutz et al., 1997; Hinchcliffe et al., 1998; Uzawa et al., 2004). However, many of these studies have examined when centrosome duplication occurs in cycling cells, rather than examining the effect of prolonging the cell cycle in G1, and assaying for centrosome duplication. Put another way, centrosome duplication may begin in G1, and by the time morphologically distinct centrioles are recognizable in the electron microscope, the cell cycle has proceeded into S-phase. The consequences of prolonging G1 on the centrosome duplication cycle have not been experimentally tested in mammalian somatic cells. To address this, we use both transformed and non-transformed cultured Chinese hamster cells to directly test whether or not centriole reproduction can occur when the cell cycle is prolonged in G1.
Materials and Methods
Unless otherwise noted all reagents were obtained from Sigma Chemical (St. Louis, MO).
Cell culture
Chinese hamster ovary (CHO-K1) cells were obtained from ATCC (Manassas, VA), and cultured in Ham’s F-12, with 10% FCS (Gibco, Grand Island, NY) and 1 mg/ml pen-strep. Low passage (<9) Chinese hamster embryonic fibroblasts (CHEF IIC9 cells) were obtained from ECACC (Wiltshire, UK), and cultured in 1:1 MEM/Ham’s F-12, with 10% FCS (Gibco) and 1 mg/ml pen/strep.
To drive cells into G0, cells were cultured in Ham’s F-12/0.05% FCS for 48 h. To re-induce proliferation, G0 cells were trypsinized and re-plated at lower density into media containing 10% FCS. To induce G1 arrest, mimosine was added to cells released from G0 at a final conc. of 600 µM. For S-phase arrest, cells were treated with either 10 µg/ml aphidicolin (Aph), or 2 mM hydroxyurea (HU). To wash out drugs, cells were transferred to fresh media, and given five changes over a 30-min period.
Centrin-GFP expressing cells
CHO cells expressing GFP-centrin 2 were generated by transfection with human GFP-centrin 2 plasmid (pJLS 148 in Dh5-α cells, described in Salisbury et al., 2002) using FuGene 6 (Roche, Indianapolis, IN) and selected with 2 mg/ml G 418. Resistant cells were sub-cloned in 24-well plates, and screened by fluorescence microscopy. Final colonies were plated as single cells onto a feeder layer of PtK2 cells, re-screened and frozen in LN2.
Fluorescence microscopy
Cells on coverslips were fixed in −20°C methanol, which preserved the centrin-GFP fluorescence. Fixed cells were labeled with a polyclonal anti-γ tubulin (Sigma) followed by 2° coupled to Alexa 594 (Molecular Probes, Eugene, OR). For centrin-2 immunoflorescence, cells were labeled with monoclonal anti-human centrin 2 (Lingle et al., 1998) followed by a 2° antibody coupled to Alexa 488.
Immunofluorescent images were collected as a Z-series on a DM RXA2 upright microscope, with a 63 × 0.32 NA apochromatic oil immersion objective (Leica, Bannockburn, IL), and an ORCA-ER CCD camera (Hamamatsu, E. Bridgewater, NJ). Images were captured using Simple-PCI software (Compix, Cranberry Township, PA), and presented as maximal projections.
Live-cell microscopy
For time-lapse imaging, CHO-A8 cells were plated onto bio-cleaned glass coverslips in Imaging Media (Ham’s F-12 w/o phenol red; PromoCell GmbH, Heidelberg, Germany), containing 12 mM Hepes, pH 7.2 10% fetal bovine serum and assembled onto aluminum support slides, as described (Hinchcliffe et al., 2001; Uetake and Sluder, 2004). Time-lapse images were captured using a Leica DM RXA2 microscope stand, equipped with fluorescence and differential interference optics, enclosed in a custom-made Plexiglas box maintained at 37°C. Live-cell fluorescence images were captured using a Yokagawa CSU-10 spinning disk confocal head, as modified by McBain Industries (San Diego, CA). Illumination of the fluorescent images was done with a Coherent 488 nm 200 mW “Sapphire” continuous wave optically pumped solid state laser (CW-OPSL); the laser was connected through a fiber optic cable into the excitation port of the spinning disk confocal head, and shuttered via a TTL pulse through a Ludl MAC5000 shutter controller. Confocal fluorescent images were taken through a Leica Plan Apo 63×/1.3 NA 37°C glycerol immersion objective; the detector on the confocal microscope was a Hamamatsu ORCA-AG Digital CCD camera, and images were captured using Simple PCI imaging software.
Electron microscopy
Fixing, embedding and serial sectioning were done as described (Rieder and Cassels, 1999). 85 nm sections were cut on a diamond using an RMC MTX ultramicrotome (Boeckeler Instruments, Inc. Tucson, AR). Sections were collected on grids and post-stained using saturated Uranyl acetate and Reynold’s lead citrate, and viewed on an Hitachi H600 run at 75 kV.
Immunoprecipitation and Cdk2 in vitro kinase assay
In vitro kinase assays of anti-Cdk2 immunoprecipitates were done as previously described. Equal aliquots of cell lysates (2.5 × 106 cells each) were incubated with mouse anti-Cdk2 (Santa Cruz Biotechnology, Santa Cruz, CA), recovered with protein-A sepharose beads (Pharmacia Biotechnology, Uppsala, Sweden), and washed twice in 2× kinase buffer (40 mM Tris–HCl, pH 7.5, 8 mM MgCl2, 1 mM dithiothreitol). Sample was then resuspended in 2× kinase buffer containing 30 µM ATP, 5 mCi [γ32p]ATP and 75 µg Histone H1 (Sigma) at 30°C for 30 min. Radiolabeled proteins were separated by SDS–PAGE, and the dried gel exposed to film. Parallel samples were separated by SDS–PAGE and identified by immunoblot as a loading control.
Cell cycle measurements
To determine the onset of S-phase cells were treated with 5-bromo-2′-deoxyuridine (BrdU) at a final concentration of 300 µg/ml for 1/2 h. Cells on coverslips were then briefly washed in PBS, fixed in −20°C methanol, and post-fixed for 4 h in 4 M HCl at RT, neutralized in PBS, and labeled with an anti-BrdU antibody/Alexa 488 2° antibody (Roche).
To determine DNA content, a minimum of 10,000 cells per treatment were stained with 5 µg/ml propidium iodine and analyzed by flow cytometry, using a Coulter Epics XL cytometer (Coulter, Miami, FL), as described (Roy et al., 2005). The gating of the instrument was kept constant for each sample.
Results
Analyzing centrosome duplication
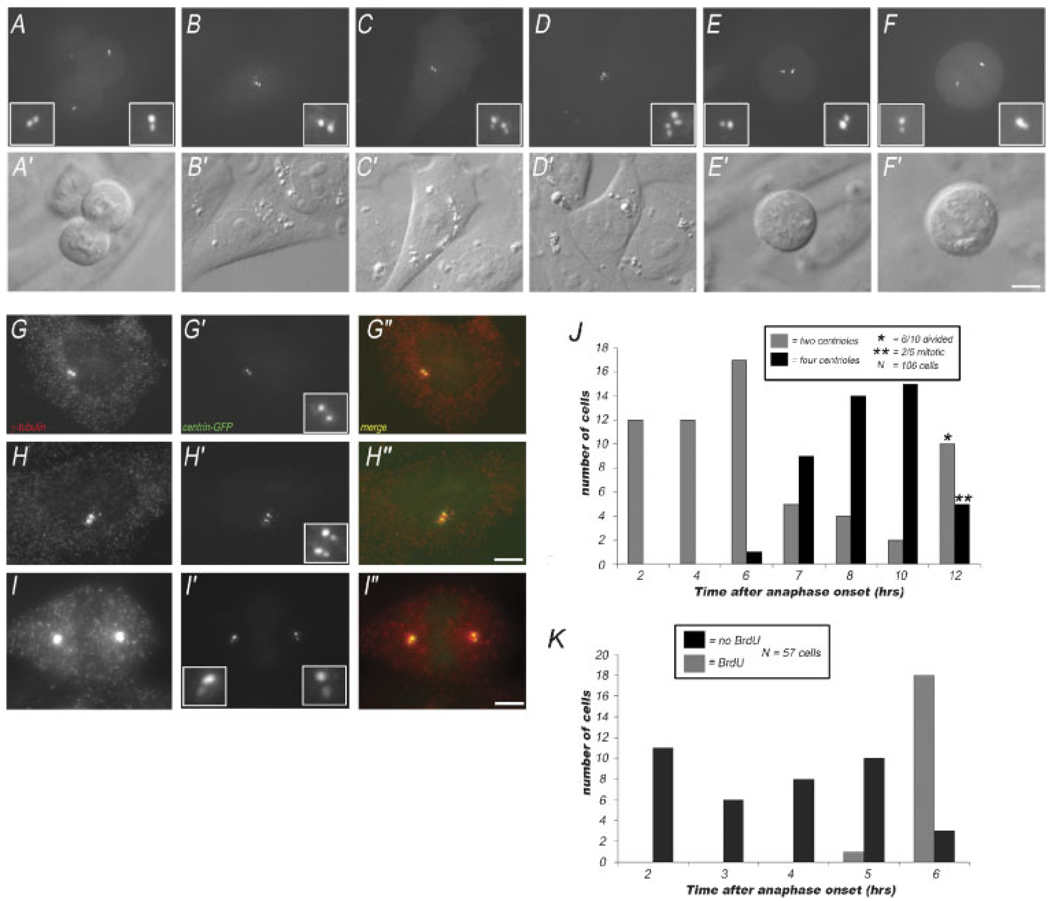
To analyze centrosome duplication in mammalian cells, we generated a line of CHO cells that constitutively expresses centrin 2-GFP (termed CHO-A8 cells). CHO-A8 cells exhibit similar growth characteristics to the parental CHO-K1 cell line, with an average growth cycle of 14 h (as judged by time-lapse video microscopy). We have used CHO cells for several reasons: (1) there is an extensive literature describing the morphological events of centrosome duplication in these cells (Kuriyama and Borisy, 1981), and (2) these cells are capable of supporting centrosome amplification during S-phase arrest, yet in a cycling population of CHO cells, there is little or no spontaneous centrosome amplification (Balczon et al., 1995). Centrin-GFP has been shown to accumulate in the lumen of the centrioles, and has been used previously as a marker for centriole number in a wide variety of mammalian somatic cell types (Piel et al., 2000; White et al., 2000; Salisbury et al., 2002; Uetake et al., 2007). We find that the centriole number in our cells can easily be counted using either live-cell imaging or fixed cell immunofluorescence imaging, as has been shown previously for other centrin-GFP expressing cell types. By live-cell imaging with a spinning disk confocal, we can identify the centriole number in both interphase and mitotic cells (Fig. 1). In anaphase cells, we find that there is a pair of centrin-GFP dots at each pole of the spindle (Fig. 1A). In early interphase cells, we find two centrin dots, often closely spaced, representing the un-duplicated centrosome (Fig. 1B). Cells were also observed in interphase having two pairs of centrin dots, representing the duplicated centrosome (Fig. 1C,D). When mitotic CHO-A8 cells were examined, we found that cells in prophase contain two pairs of closely spaced centrin dots (Fig. 1E), while those in prometaphase/metaphase had a pair of centrin-GFP dots coincident with each of the two spindle poles (Fig. 1F).
Fig. 1.
Centriole number in centrin-GFP expressing CHO cells. A–F: Maximal intensity projections of live CHO cells progressing from anaphase to interphase showing GFP-centrin 2 fluorescence (insets show centrioles). A: This anaphase cell has two centrin dots at each pole of the cell. B: Interphase cell, with two centrin dots, representing the centriole pair inherited after mitosis. C,D: Interphase cells, each with four centrin dots, representing the duplicated centrosome. The cell in (D) is in the process of condensing its chromosomes and breaking down its nuclear envelope. E: Prometaphase cell, with two pairs of centrin dots, representing the duplicated centrosomes, around which the spindle poles are assembling. F: Metaphase cell, with a pair of centrin dots at each spindle pole. A′–F′: Corresponding DIC images of cells in (A–F). G–I: Images of fixed CHO-A8 cells labeled with anti-γ tubulin. G: Interphase cell with un-duplicated centrosomes, (H) interphase cell with duplicated centrosomes, and (I) mitotic cell, with a centrosome at each spindle pole. Note that in either case, there are just two γ-tubulin foci. G′–I′: Centrin-GFP fluorescence in the same fixed cells, showing either a pair of centrioles (G), or two pairs of centrioles (H,I). G″–I″: Merged γ-tubulin/centrin-GFP. The centrin-GFP positive foci are co-incident with the γ-tubulin spots. Bar = 10 µm. J: Quantification of centriole number in cells followed from 2 to 12 h after anaphase onset. Cells in anaphase were identified, and followed by time-lapse video microscopy with DIC optics. After specified intervals, the centriole number in each cell was determined by collecting a Z-series through the cell using fluorescence microscopy. The graphs shows number of cells with two or four centrioles at each time point. N = 106 cells. Note, in some of the later time-points, the cells have entered into or completed the next round of mitosis. K: Time-course for the onset of S-phase, relative to the end of mitosis in cycling CHO cells. Fields of cells were followed by time-lapse video microscopy from anaphase onset, then treated with BrdU, and fixed. Graph shows number of BrdU positive cells, at each interval after anaphase onset. N = 57 cells. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
When the centrin-GFP fluorescence was examined in fixed cells, we observed a similar distribution. Interphase cells contained either a single pair of centrin dots, or two pairs of closely spaced dots, indicating un-duplicated, or duplicated centrosomes, respectively (Fig. 1G′,H′). In fixed mitotic cells, there were two pairs of centrin-GFP dots, one at each pole of the cell (Fig. 1I′). Importantly, the centrin dots in both interphase and mitosis co-localized with anti-γ tubulin staining, further confirming that the centrin-GFP dots in our CHO-A8 cells represent the position of the centrioles within the centrosome as a whole (Fig. 1G–I). Importantly, in interphase cells, there are two γ-tubulin foci, which can contain either two centrioles or four centrioles.
Next, we measured the timing of centrosome duplication in individual cells, relative to the onset of anaphase, which serves as a convenient cell cycle stage marker. Cells were imaged by time-lapse video microscopy using DIC. Two hours after anaphase onset, a Z-series through that cell was captured using fluorescence optics, to image centrin-GFP and determine the centriole number at that time-point (Fig. 1J). Because exposure to 488 nm light can have deleterious effects on cell cycle progression (Uetake et al., 2007), cells were not followed after centriole number was determined by fluorescent imaging. Instead, a new anaphase cell that had not been exposed to 488-nm light was identified in our imaging chamber, and a time-series for that cell was collected using DIC. This process was repeated for multiple time-points ranging from 0 to 12 h post-anaphase. By analyzing the centriole number for 106 cells, for which we had timing data relative to anaphase onset, we were able to determine that centrosome duplication begins between 6 and 8 h following mitotic exit (Fig. 1J). We note that several of the cells followed were either in mitosis or had completed division by the 12 h time-point. These cells were scored as having duplicated their centrosomes. Thus, at 12 h, 11 of the 15 cells had duplicated their centrosomes.
Next, the onset of S-phase in individual CHO-A8 cells was measured using time-lapse video microscopy and BrdU incorporation, which serves as a marker for entry into S-phase. Cells were filmed by DIC for intervals from 1½ to 5½ h. Any cells that underwent anaphase were used for analysis, because the interval from anaphase onset until fixation was known. Cells were processed for BrdU, and incorporation in individual cells was then compared to the video history of those cells. In this way, we were able to measure the timing from anaphase onset until to the onset of BrdU incorporation for a population of cells (N = 57 cells). We found that in cycling CHO-A8 cells, S-phase began between 5 and 6 h after anaphase onset (Fig. 1K). Thus, we conclude that centrosome duplication in CHO-A8 cells occurs around the time of the G1/S-phase transition, consistent with previous ultrastructural studies (Kuriyama and Borisy, 1981).
Centrosome duplication in G1-arrested CHO cells
We next examined the ability of CHO-A8 cells arrested prior to the onset of S phase to duplicate their centrioles. Unlike arresting the cell cycle in S-phase, which can easily be induced by treatment with pharmacological agents that inhibit DNA replication (and activate the replication checkpoint), there are few ways to reliably block the cell cycle in G1 (Alpan and Pardee, 1996; Krude, 1999). One method, which has been reported to prevent cell cycle progression into S-phase, is addition of the rare plant amino acid mimosine to cells released from a G0 arrest (Alpan and Pardee, 1996; Krude et al., 1997; Matsumoto et al., 1999). At high concentrations, mimosine reversibly arrests the cell cycle in late G1 by blocking the establishment of DNA replication foci, but is otherwise non-toxic to living cells, including CHO cells (Krude, 1999, 2000). The mode of mimosine arrest is not entirely clear; mimosine acts as a strong iron chelator, decreases cellular pools of dNTPs, and also up-regulates the Cdk inhibitory protein p21, even in the absence of p53 (Alpan and Pardee, 1996). Importantly, previous work used nuclear run-on replication assays to demonstrate that the cell cycle arrest induced by mimosine is distinctively different from the S-phase block induced by treatment with either aphidicolin or HU (Krude, 1999; Szüts and Krude, 2004). This work also showed that mimosine arrest in G1 is dependent upon the cells first being driven into G0, then released by addition of serum into culture medium containing mimosine. If mimosine is added to an asynchronous culture of cells, the cell cycle will arrest in both G1 and S-phase (Krude, 1999).
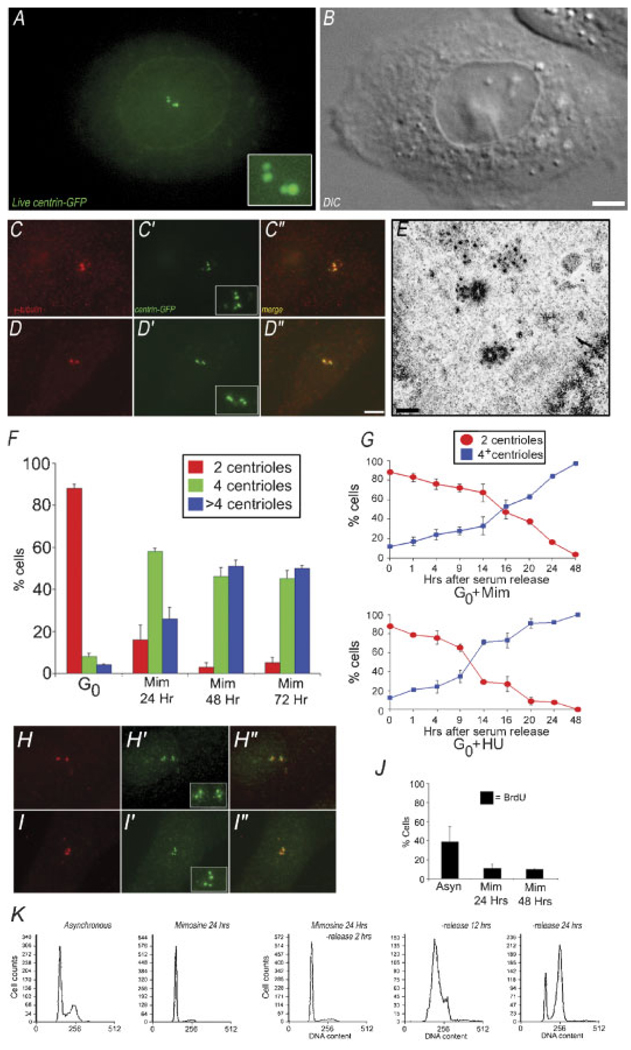
To induce a synchronized population of cells arrested in G1, we first drove CHO-A8 cells into G0 by serum-starvation for 48 h (Merrill, 1998). These were then released into serum-containing media with 600 mM mimosine. When CHO-A8 cells were examined by live-cell fluorescence microscopy after a 20-h treatment with mimosine, we found that many cells contained four centrin dots (Fig. 2A), suggesting that these cells had undergone centrosome duplication. To confirm this, we examined CHO-A8 cells treated with mimosine for 24, 48, or 72 h, and quantified the numbers of centrosomes in fixed samples (Fig. 2C,D). Figure 2C–C″,D–D″ shows two separate cells, fixed after 24 h in mimosine; each cell contains four centrin foci that co-localize with γ-tubulin. When we examined a population of CHO-A8 cells, we found that after 24 h in mimosine, 16% of cells contained two centrioles, 58% had four centrioles, and 26% had more than four centrioles (Fig. 2F). After 48 h in mimosine, the number of cells with two centrioles was only 3%, with 46% of the cells having four centrioles, and 51% having more than four. These numbers did not significantly change after 72 h in mimosine (Fig. 2F). Importantly, when a population of G0-arrested CHO-A8 cells was examined, we found that 88% of cells had two centrioles, 8% had four, and only 4% had more than four centrioles, indicating that there is not extensive centrosome amplification in these cells (Fig. 2F). Also, excluded from this analysis were any cells that were multinucleate, because these arise from cleavage failure, and by definition, have abnormal centrosome numbers.
Fig. 2.
G1 arrested CHO cells support centrosome duplication. A: Maximum intensity projection of centrin-GFP fluorescence in a live CHO-A8 cell, released from G0 in the presence of 600 mM mimosine for 20 h. There are two pairs of bright fluorescent dots, as well as centrin-GFP fluorescence in the nuclear region. B: Corresponding DIC image of cell in (A). C,D: Immunofluorescent images of CHO-A8 cells arrested in G1 for 24 h. Cells are labeled with anti-γ tubulin/centrin 2-GFP, showing two pairs of centrin-GFP fluorescent dots, co-incident with anti-γ tubulin foci. E: Transmission electron micrograph of a CHO-A8 cell released from G0 into mimosine for 20 h prior to fixation and embeddment. Each of the parental centrioles is assembling a short pro-centriole at right angles. F: Centriole number in a population of G0 cells, and G1 cells (24, 48, and 72 h after serum release into mimosine). Graph shows percentage of cells containing either 2, 4, or >4 centrioles per cell. Data are average of three experiments, 200 cells per condition. G: Centriole number in cells treated with mimosine or HU fixed at intervals following serum release from G0. Note that the number of cells with four centrioles begins to increase between 14 and 16 h for mimosine, and 9 h for HU. Average of three experiments, 200 cells per time point. H–H″/I–I″: Immunofluorescence images of two separate CHO-K1 cells, released from G0 into mimosine for 20 h, then fixed and labeled with anti-γ tubulin (H,I), and anti-centrin 2 (H′,I′).H″, I″: Merged images, showing co-localization of centrin pairs and γ-tubulin positive centrosomes. Each cell contains a pair of anti-centrin 2 positive foci coincident with the two γ-tubulin positive foci. J: BrdU incorporation in control versus cell cycle arrested cells. Graph shows % of cell positive for anti-BrdU fluorescence. Mimosine arrests cells prior to the onset of S-phase. K: Flow cytometry of control, and mimosine treated CHO-A8 cells. DNA content of mimosine-treated cells following drug wash-out as they progress into S-phase over 24 h. Bar in (B) = 10 µm, in (D) = 10 µm, and in (E) = 0.5 µm. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
To ensure that these centrin fluorescence foci represented true centrioles, we released cells from G0 into mimosine for 20 h, then fixed the cells, embedded and sectioned them, and analyzed centriole number by transmission electron microscopy. We examined eight separate cells and found that one contained two centrioles, five contained four centrioles, and two contained more than four centrioles. Figure 2E shows a mimosine-treated cell that contains two centrioles in the process of duplicating. In the electron micrograph, there are two parental centrioles, each with an associated pro-centriole, confirming that mimosine-treated cells can undergo centriole replication within 20 h of serum-release from G0.
Our results suggest that in CHO-A8 cells released from G0 into mimosine for 20 h, the centrioles can duplicate without coordinate entry into S-phase. To determine the timing of the centrosome duplication event, relative to cell cycle release into mimosine, we fixed mimosine-treated cells or HU-treated cells at intervals from 1 to 48 h following serum-release, and then quantified the number of centrioles per cell in the population at each time point (Fig. 2G). We found that over the first 14 h after release into mimosine, the number of cells that had undergone at least one round of centrosome duplication increased by only 19% (from an initial 14% at T = 0–33% at T = 14 h). However, from 14 to 24 h, that number increased from 33% to 78%. In contrast, centriole duplication in cells serum-released into HU began duplication around 9 h (Fig. 2G).
To rule out the possibility that this centrosome duplication was due to transfection with centrin-GFP, we also conducted G0 arrest and serum-release using the parental CHO-K1 strain. Centriole number was assayed by immunofluorescence microscopy using anti-centrin 2 antibody (Lingle et al., 1998). Figure 2H–H″,I–I″ shows two examples of non-transfected CHO-K1 cells after 24 h in mimosine. Each cell contains four centrioles (centrin dots) coincident with anti-γ tubulin. When we analyzed the number of centrin dots in CHO-K1 cells treated with mimosine for 24 h, we found results similar to those seen in CHO-A8 cells (22% contained two centrioles, 64% had four centrioles, and 14% had more than four). Thus, we conclude that CHO-K1 cells serum-released from G0 into mimosine can undergo centriole replication within 24 h.
To confirm that mimosine treatment results in a G1 arrest in our cells, we analyzed BrdU incorporation, a marker for entry into S-phase, in CHO-A8 cells released from G0 arrest into 600 µM mimosine. Figure 2J shows the extent of BrdU incorporation in G0-arrested cells released into mimosine. After 24 h, only 11% of cells incorporated BrdU, and there was no significant increase in BrdU incorporation at 48 h (10%).
We also examined the DNA content of 24 h mimosine-treated cells by flow cytometry, and compared it to cycling/asynchronous cells (Fig. 2K). The cycling population of cells shows a distribution of DNA content, with both 2 N and 4 N peaks. The mimosine-treated cells show only a 2 N peak. When CHO-A8 cells were released from mimosine arrest, they cycled into and through S-phase (Fig. 2K), consistent with previous findings that mimosine arrest is reversible (Krude, 1999).
Cdk2 kinase activity during mimosine arrest
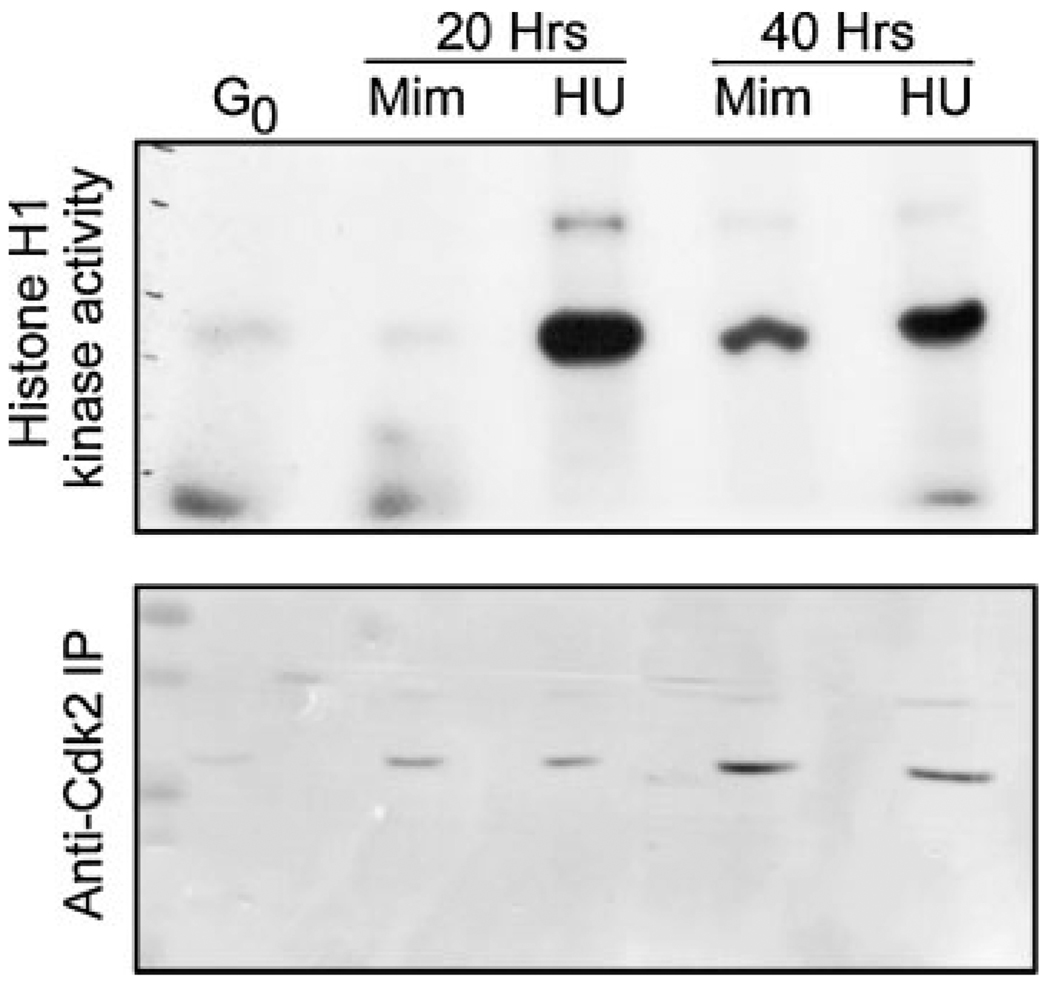
Previous work has shown that cyclin dependent kinase 2 associates with cyclin E and A, driving the G1/S transition and maintaining S-phase progression (Sherr and Roberts, 1999). Interestingly, the activities of both Cdk2-E and Cdk2-A have been implicated in the regulation of centrosome duplication, albeit by unknown mechanisms (reviewed in Hinchcliffe and Sluder, 2002). Importantly, it was shown that Cdk2 activity in CHO cells remains low during mimosine treatment (Matsumoto et al., 1999), consistent with previous studies (Alpan and Pardee, 1996). However, in this previous work, centrosome duplication was blocked by mimosine treatment. Because we see a difference in the ability of CHO cells treated with mimosine to undergo centrosome duplication, we measured the activity of immunoprecipitated Cdk2 kinase, using the same anti-Cdk2 antibody used in the previous study (Matsumoto et al., 1999). First, CHO cells were released from G0 arrest into either 600 mM mimosine or 2 mM HU, and cells were harvested for immunoprecipitation at 20 and 40 h following serum stimulation. Next, whole-cell lysates were prepared, and the kinase activities of anti-Cdk2 immunoprecipitates were measured, using Histone H1 as a substrate (Fig. 3). In cells released from G0 into mimosine for 20 h, Cdk2 kinase activity remained at or below basal levels (measured as the level of activity in G0 cells), consistent with previously published results (Matsumoto et al., 1999). In HU treated cells, the level of Cdk2 kinase activity rapidly rises following serum addition, and remains high at 40 h, also consistent with the previous work.
Fig. 3.
Cdk2 kinase activity in CHO cells serum-released into either mimosine or hydroxyurea (HU). Whole cell lysates were obtained from cells released into either mimosine (MIM) or HU. Cdk2 was immunoprecipated, and kinase activity was assay in vitro, using Histone H1 as a substrate. The phospho-H1 bands are shown for 20 and 40 h timepoints. G0 represents the kinase activity in cells immediately following addition of serum.
We note that after 40 h inmimosine, Cdk2-immunoprecipitated H1 kinase activity did rise above G0 levels, although the level was below the maximal reached in HU cells. However, at this time point, these cells still do not incorporate BrdU or replicate their DNA (not shown).
Centrosome amplification during G1 arrest
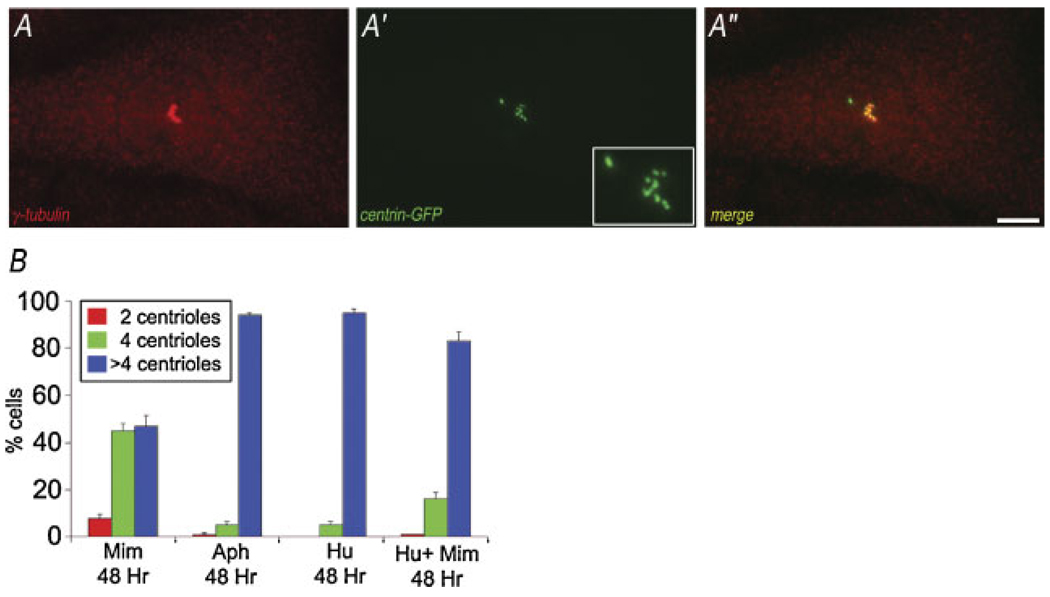
During prolonged G1-arrest, a significant percentage of mono-nucleated CHO cells assembled more than four centrioles (centrosome amplification: Fig. 2F), and the percentage of cells that show centrosome amplification increased with the length of cell cycle arrest in G1. Because the results of previous studies using transformed somatic cells, zygotes, and yeast suggested that centrosome amplification is linked with the cell cycle conditions of S-phase (Balczon et al., 1995; Hinchcliffe et al., 1998; Haase et al., 2001; Khodjakov et al., 2002; Wong and Stearns, 2003; LaTerra et al., 2005), we tested whether there is a quantitative difference between G1 and S-phase arrest in the ability to support centrosome amplification. To measure this, cells in G0 were released into mimosine alone (G1), HU alone (S-phase), or HU to first arrest the cell cycle in S-phase, followed by addition of mimosine (to determine if mimosine itself can indirectly inhibit centrosome duplication). After 48 h we counted the number of centrioles per cell assembled under each condition (Fig. 4B). In S-phase arrested cells, there is significant centrosome amplification: ~90% of cell released into either aphidicolin or HU show greater than four centrioles. This is in contrast to G1-arrested cells treated with mimosine, where only ~50% of cells had more than four centrioles. Importantly, the addition of mimosine to cells already arrested in S-phase did not significantly inhibit the ability of these cells to undergo centrosome amplification: >80% of HU/MIM-treated CHO cells contained more than four centrioles (Fig. 4B).
Fig. 4.
Centrosome amplification in G1 versus S-phase arrested CHO cells. A–A″: Maximal intensity projection of fluorescence of aCHO-A8 cell serum-released into 600 µM mimosine, labeled with anti-γ tubulin (A), and GFP-centrin 2 (A′). There is an increase in the number of γ-tubulin positive foci, reflecting the centrosome amplification in these cells. The merged image shows co-localization of the γ-tubulin and centrin, except for one bright region. B: Quantitation of centriole number in mimosine versus S-phase arrested CHO cells, treated for 48 h. Note the extent of centrosome amplification in mimosine-treated cells is decreased, compared to that seen for cells in S-phase by either Aph or HU. When mimosine is added to cells arrested S-phase, there does not appear to be a significant decrease in the ability of these cells to undergo centrosome amplification. Average of three experiments, 200 cells per condition/experiment. Bar in (A–A″) = 10 µm. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Centrosome duplication during G1 in non-transformed Chinese hamster cells
CHO cells have a mutation in their p53 gene and are transformed (Hu et al., 1999). Loss of p53 has been correlated with mis-regulation of the centrosome duplication cycle (Fukasawa et al., 1996). Thus, it is possible that the centriole duplication prior to S-phase that we observed in CHO cells could be in response to the loss of p53 activity, altering the dynamics of the centrosome duplication cycle. To test this possibility, we examined centriole reproduction in non-transformed Chinese hamster cells (Chinese Hamster Embryonic Fibroblasts—CHEF cells), which have wild-type p53 (Keenan et al., 2004). Centriole number was determined by methanol fixation and labeling with the anti-centrin 2 antibody (Lingle et al., 1998). Co-staining with anti-γ tubulin confirmed that the centrin-positive foci were centrioles within the centrosome. We identified interphase CHEF cells with either two anti-centrin dots (Fig. 5A–A″), or four anti-centrin dots (Fig. 5B–B″), confirming that anti-centrin 2 can be used to assay for centriole number in CHEF cells.
Fig. 5.
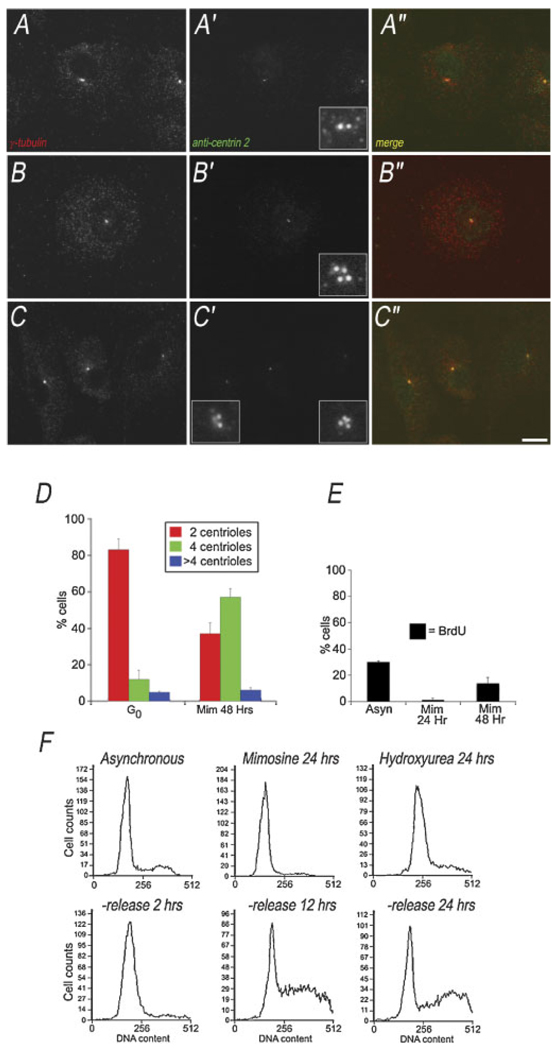
Centrosome duplication in G1 arrested CHEF cells. A,B: Maximum intensity projections of immunofluorescence showing control CHEF cells with either two or four centrioles. The anti-centrin 2 foci are co-incident with the two γ-tubulin foci. C–C″: Maximum intensity projection of immunofluorescence image stack, showing two CHEF cells serum-released into mimosine for 24 h, labeled with anti-γ tubulin and anti-centrin 2. One cell has two centrioles, the other has fourcentrioles. Insets show blow up of centriole region. D: Quantitation of centriole numberin G0, and G1 CHEF cells. Graph shows the percentage of cells with 2, 4 or >4 centrioles. Average of three experiments, N = 200 cells per condition. E: BrdU incorporation into CHEF cells treated with mimosine for 24 or 48 h, compared to cycling control cells. F: Flow cytometry measuring DNA content of CHEF cells in mimosine arrest, compared to asynchronous and S-phase arrested. Lower parts show DNA content in CHEF cells following wash-out of mimosine. Bar in (C) = 10 µm. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
Similar to the cell cycle analysis of CHO cells, we found that in CHEF cells, treatment with 600 µM mimosine—following the serum-release from G0—induces a prolonged G1 arrest. To confirm this, we used BrdU incorporation (Fig. 5E) and flow cytometry (Fig. 5F).
After 48 h of serum starvation, G0 CHEF cells have two centrioles in 83% of cases, 12% have four centrioles, and 5% have more than four centrioles (Fig. 5D), similar to serum-starved CHO cells. CHEF cells released from G0 into complete media containing 600 µM mimosine for 48 h have predominantly four centrioles (57% of cells), whereas some cells still contain only two centrioles (37%), and very few cells have more than four centrioles (6%). Thus, we find that centrioles can duplicate during G1 arrest in non-transformed Chinese hamster cells (Fig. 5C).
Discussion
Centrosome duplication without transition into S-phase
The coordination between centrosome duplication and progression of nuclear events during the cell cycle is absolutely essential, in order to ensure that the centrosome has reproduced prior to entry into mitosis. Understanding when during the cell cycle centrosome duplication is initiated is important in order to identify the cellular signals that control this process. It is important to note that the morphological events of centrosome reproduction (i.e., the assembly of daughter centrioles) may not represent the earliest stage in this process: pro-centriole formation may actually be a downstream manifestation of a reproductive event that occurred earlier in the cell cycle (discussed in Hinchcliffe and Sluder, 1998). To address when centrosome duplication occurs, many studies have relied on arresting or inhibiting cell cycle progression, in order to allow the reproductive event to occur, and then become manifest, without coordinate cell cycle progression (Gard et al., 1990; Sluder et al., 1990; Hinchcliffe et al., 1998; Vidwans et al., 1999; Khodjakov et al., 2002; Tsou and Stearns, 2006). Here we have extended these studies by examining whether or not centrosome duplication can proceed if the transition into S-phase is inhibited.
In the present study we find that centrosome duplication does occur during prolonged G1. Also, centriole replication occurs during G1 arrest in both transformed CHO cells, and non-transformed CHEF cells, indicating that this phenomenon is not a consequence of either cellular transformation, or the p53 mutation found in CHO cells. Centriole duplication during G1 arrest continues even though the kinase activity of Cdk2 remains at basal levels. These observations are consistent with previous work, which suggested that centrosome duplication does not absolutely require entry in S phase, or Cdk2 kinase activity in order to proceed (Hinchcliffe et al., 1998, 1999; Duensing et al., 2006).
Importantly, CHO cells arrested in G1 require a period of between 14 and 24 h following serum-release from G0 in order to replicate their centrioles. This is equivalent of 1–1.7 times the duration of a complete cell cycle in CHO cells. The slow kinetics of centrosome duplication during G1 arrest does not appear to be a direct consequence of the drug mimosine. If mimosine is added to cells already arrested in S-phase with HU, centrosome re-duplication continues, reaching levels similar to those seen for HU alone.
Centrosome duplication does not occur in cells that have exited the cell cycle (Kuriyama and Borisy, 1981). The slow duplication we observe during prolonged G1 suggests that following serum stimulation, centrosome duplication can be initiated during G1, and then slowly continues as the cell cycle progresses toward S-phase. This reveals that centrosome duplication can proceed without a true transition into S-phase, suggesting that G1 is permissive for duplication, if sufficiently prolonged.
Coordinating centrosome duplication with cell cycle progression
Our finding that centriole replication during prolonged G1 takes between 14 and 24 h suggests the idea that in the absence of a major “trigger”, the centrosome duplication cycle can proceed as an inherently slow process—following either exit from mitosis or serum stimulation from a G0 arrest. If centrosome duplication were to proceed via this mechanism, the cell would enter mitosis long before the duplication of the centrosome, resulting in abnormal division. How then does the cell ensure timely duplication of this organelle, coordinate with the progression of the division cycle? Based on our data, as well as that of previous work, we propose the following model. First, the centrosome inherited at the end of mitosis is competent to duplicate, having become functionally disengaged during the preceding mitosis (Tsou and Stearns, 2006). As the cell cycle proceeds into G1, this “competent” centrosome begins its duplication cycle. However, by the time the cell cycle enters S-phase, the centrosome may or may not have completed duplication, because during G1, the kinetics of duplication are slow. But, as the cell transitions through G1 into S-phase, the process of centrosome reproduction becomes entrained by key regulatory “activities” that are associated with the G1/S transition. These “activities” cause the centrosome duplication cycle to become accelerated, and the daughter centrosomes rapidly complete their duplication as the cell proceeds through S-phase. Thus, as the cell transitions from S-phase into G2, it has replicated its DNA and reproduced its centrosomes in preparation for mitosis. Our model predicts that one of the functions of the G1/S-transition is to entrain the slow, free-running centrosome duplication cycle, rather than initiate this cycle. Consistent with our model, we find that in normal cycling CHO cells, the morphological events of centrosome duplication (the assembly of daughter centrioles) are seen coordinate with the time that these cells are incorporating BrdU into newly synthesized DNA (Fig. 1). In the case of prolonged G1, we find that the centrosome will eventually duplicate, suggesting that the initiation of the process occurs during G1 (or even in the preceding M-phase; see Tsou and Stearns, 2006), and that during prolonged G1 there is sufficient time for the inherent centrosome cycle to run to completion.
Cdk2 kinase activity and centrosome duplication
Several studies have shown that centrosome duplication is regulated by the activity of cyclin dependant kinases (Hinchcliffe et al., 1999; Lacey et al., 1999; Matsumoto et al., 1999; Meraldi et al., 1999; Haase et al., 2001), but the mechanism of this regulation is poorly understood (reviewed in Winey, 1999; Hinchcliffe and Sluder, 2002). Here we show that centrosome duplication can occur even in the absence of a rise in Cdk2 kinase activity, which is consistent with previous work in mammalian cells (Duensing et al., 2006). Thus, it does not appear that a global rise in Cdk2 activity is required for the initiation of centriole replication by triggering centriole disorientation/disengagement, as previously thought (discussed in Tsou and Stearns, 2006). If Cdk2 activity is not required to initiate centrosome duplication, then the question remains, what is its role in regulating centrosome duplication? We imagine two possibilities, which are not mutually exclusive. First, we envision that Cdk2 activity could function as part of the entrainment process at the G1/S transition, which coordinates centrosome duplication with cell cycle progression. If the centrosome had failed to duplicate at the time of entry into S-phase, then the activation of Cdk2 at this time could drive the completion of duplication. If so, Cdk2 may be acting directly on the centrosome, by phosphorylating key centrosome subunits (see Okuda et al., 2000), or may be acting indirectly, by phosphorylating key down-stream regulatory molecules, which serve to entrain the duplication of the centrosome. Consistent with this notion, several candidate molecules have been identified, which have been shown to play a role in regulating centrosome duplication, and require Cdk2 in order to become activated. Both E2F (a family of transcription factors) and mouse Mps1p (a kinase) are required for centrosome duplication, and both require activation by Cdk2 (Meraldi et al., 1999; Fisk and Winey, 2001). Thus, one of functions of Cdk2 in regulating centrosome duplication appears to be in regulating down-stream effectors of this process (see Hinchcliffe and Sluder, 2001b).
A second possibility is that Cdk2 kinase functions to restore the ability of the centrosome to undergo duplication in the subsequent cell cycle. This idea is supported by work in both frog egg extracts and Cdk2−/− knock-out MEFs, in which loss of Cdk2 activity does not prevent one round of duplication, but does inhibit succeeding rounds (Hinchcliffe et al., 1999; Duensing et al., 2006). Both of these studies suggested that during centrosome re-duplication, Cdk2 is “licensing” the centrosome. However, whether this is a true licensing event, or simply reflects a requirement for Cdk2 activity to support repeated rounds of duplication is not known.
Our finding that centrosomes can undergo duplication in mimosine-treated CHO cells that have low Cdk2 kinase activity is in contrast to the findings of previous work, which showed that CHO cells treated with 400 µM mimosine have basal levels of Cdk2 activity, but cannot support centrosome duplication (Matsumoto et al., 1999). The obvious discrepancies between our two findings are easily explained. In the previous work, centrosome duplication was assayed for primarily by using anti-γ tubulin staining of CHO cells. In mimosine-treated cells, almost all exhibited two γ-tubulin spots, which was interpreted as un-duplicated centrosomes. Indeed, we also find predominantly two γ-tubulin spots in our mimosine-treated cells. However, when centrin distribution was examined—either by centrin-GFP, or by anti-centrin 2 antibody—we observed two pairs of centrioles, one pair lying within each γ-tubulin focus. Also, we find by electron microscopy that cells released into mimosine for 20 h contain centrioles that had disengaged and begun assembling pro-centrioles. Thus, the previous study was correct that the number of γ-tubulin foci did not increase in mimosine-treated cells, but this does not appear to represent the true number of centrioles within these cells. The previous work did find that the number of γ-tubulin foci increased during S-phase arrest, and this was inhibited by loss of Cdk2 activity, strongly supporting their overall findings.
Pathway for centriole assembly during G1 arrest: templated versus de novo formation
Recent work has shown that centrioles can arise de novo (Marshall et al., 2001; Khodjakov et al., 2002; LaTerra et al., 2005). It is interesting to speculate whether the centriole formation we observe in mimosine-treated cells arise via the templated pathway, or the de novo pathway. However, it appears that the pre-existing centriole pair functions to suppress the de novo pathway (Marshall et al., 2001). Indeed, if only one centriole remains in the cytoplasm, de novo formation is prevented (Khodjakov et al., 2002). In addition, de novo centriole formation will proceed only if the cell cycle has transitioned into S-phase (Khodjakov et al., 2002; LaTerra et al., 2005; Uetake et al., 2007). Here, we see centriole formation during G1, and this occurs in the presence of pre-existing centrioles, which should suppress the formation of de novo centrioles. Also, ultrastructural analysis revealed that the new centrioles assemble at right angles to the pre-existing parental centriole pair. Thus, it appears that the formation of daughter centrioles during mimosine treatment is occurs via the templated pathway.
Acknowledgments
We thank Jeff Salisbury for generously providing the centrin 2-GFP cDNA and anti-centrin monoclonal antibodies, Alexey Khodjakov for advice on generating stable GFP-expressing cells, Judy Navarez for help with flow cytometry, Polla Hergert, Sheila Adams and Bill Archer for assistance with serial section electron microscopy, and both Emily Tribble and Nick Collins for technical assistance. This work was supported by National Institutes of Health grants GM072754 to E. Hinchcliffe and GM60560 to K. Vaughan. EHH and KTV are Research Scholars of the American Cancer Society.
Contract grant sponsor: National Institute of General Medical Sciences;
Contract grant numbers: R01 GM072754, R01 GM60560.
Literature Cited
- Alpan RS, Pardee AB. p21WAF1/CIP1/SDI1 is elevated through a p53-independent pathway by mimosine. Cell Growth Differ. 1996;7:893–901. [PubMed] [Google Scholar]
- Alvey PL. An investigation of the centriole cycle using 3T3 and CHO cells. J Cell Sci. 1985;78:147–162. doi: 10.1242/jcs.78.1.147. [DOI] [PubMed] [Google Scholar]
- Balczon R, Bao L, Zimmer W, Brown K, Zinkowski RP, Brinkley BR. Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J Cell Biol. 1995;130:105–115. doi: 10.1083/jcb.130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkley BR. Managing the centrosome numbers game: From chaos to stability in cancer cell division. Trends Cell Biol. 2001;11:18–21. doi: 10.1016/s0962-8924(00)01872-9. [DOI] [PubMed] [Google Scholar]
- Delattre M, Gönczy P. The arithmetic of centrosome biogenesis. J Cell Sci. 2004;117:1619–1630. doi: 10.1242/jcs.01128. [DOI] [PubMed] [Google Scholar]
- Duensing A, Liu Y, Tseng M, Malumbres M, Barbacid M, Deunsing S. Cyclin-dependent kinase 2 is dispensable for normal centrosome duplication but required for oncogene-induced centrosome overduplication. Oncogene. 2006;25:2943–2949. doi: 10.1038/sj.onc.1209310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk HA, Winey M. The mouse Mps 1 p-like kinase regulates centrosome duplication. Cell. 2001;106:95–104. doi: 10.1016/s0092-8674(01)00411-1. [DOI] [PubMed] [Google Scholar]
- Fisk HA, Mattison CP, Winey M. Centrosomes and tumour suppressors. Curr Opin Cell Biol. 2002;14:700–705. doi: 10.1016/s0955-0674(02)00385-x. [DOI] [PubMed] [Google Scholar]
- Fukasawa K, Choi T, Kuriyama R, Rulong S, Vande Woude GF. Abnormal centrosome amplification in the absence of p53. Science. 1996;271:1744–1747. doi: 10.1126/science.271.5256.1744. [DOI] [PubMed] [Google Scholar]
- Gard DL, Hafezi S, Zhang T, Doxsey SJ. Centrosome duplication continues in cycloheximide-treated Xenopus blastulae in the absence of a detectable cell cycle. J Cell Biol. 1990;110:2033–2042. doi: 10.1083/jcb.110.6.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase SB, Winey M, Reed SI. Multi -step control of spindle pole body duplication by cyclin-dependent kinase. Nat Cell Biol. 2001;3:38–42. doi: 10.1038/35050543. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe EH, Sluder G. The apparent linkage between centriole replication and S phase of the cell cycle. Cell Biol Int. 1998;22:3–5. doi: 10.1006/cbir.1997.0249. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe EH, Sluder G. “It takes two to tango”: Understanding how centrosome duplication is regulated throughout the cell cycle. Genes Dev. 2001a;15:1167–1181. doi: 10.1101/gad.894001. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe EH, Sluder G. Centrosome duplication: Three kinases come up a winner. Curr Biol. 2001b;11:R698–R701. doi: 10.1016/s0960-9822(01)00412-2. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe EH, Sluder G. Two for two: Cdk2 and its role in centrosome doubling. Oncogene. 2002;21:6154–6160. doi: 10.1038/sj.onc.1205826. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe EH, Cassels GO, Rieder CL, Sluder G. The coordination of centrosome reproduction with nuclear events during the cell cycle in the sea urchin zygote. J Cell Biol. 1998;140:1417–1426. doi: 10.1083/jcb.140.6.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchcliffe EH, Li C, Thompson EA, Maller JL, Sluder G. Requirement of Cdk2 -Cyclin E ctivity for repeated centrosome reproduction in Xenopus egg extracts. Science. 1999;283:51–854. doi: 10.1126/science.283.5403.851. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe EH, Miller FJ, Cham M, Khodjakov A, Sluder G. Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science. 2001;291:1547–1550. doi: 10.1126/science.1056866. [DOI] [PubMed] [Google Scholar]
- Hu T, Miller CM, Ridder GM, Aardema MJ. Characterization of p53 in Chinese hamster cell lines CHO-K1, CHO-WBL, and CHL: Implications for genotoxicity testing. Mutat Res. 1999;426:51–62. doi: 10.1016/s0027-5107(99)00077-9. [DOI] [PubMed] [Google Scholar]
- Keenan SM, Lents N, Baldassare JJ. Expression of cyclin E renders cyclin D-Cdk4 dispensable for inactivation of the retinablastoma tumor suppressor protein, activation of E2F, and G1-S phase transition. J Biol Chem. 2004;279:5387–5396. doi: 10.1074/jbc.M310383200. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Rieder CL, Sluder G, Cassels G, Sibon O, Wang CL. De novo formation of centrosomes in vertebrate cells arrested during S phase. J Cell Biol. 2002;158:1171–1181. doi: 10.1083/jcb.200205102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochanski RS, Borisy GG. Mode of centriole duplication and distribution. J Cell Biol. 1990;110:1599–1605. doi: 10.1083/jcb.110.5.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krude T. Mimosine arrests proliferating human cells before onset of DNA replication in a dose-dependent manner. Exp Cell Res. 1999;247:148–159. doi: 10.1006/excr.1998.4342. [DOI] [PubMed] [Google Scholar]
- Krude T. Initiation of human DNA replication in vitro using nuclei from cells arrested at an initiation-competent state. J Biol Chem. 2000;275:13699–13707. doi: 10.1074/jbc.275.18.13699. [DOI] [PubMed] [Google Scholar]
- Krude T, Jackman M, Pines J, Laskey RA. Cyclin/Cdk-dependent initiation of DNA replication in a human cell-free system. Cell. 1997;88:109–119. doi: 10.1016/s0092-8674(00)81863-2. [DOI] [PubMed] [Google Scholar]
- Kuriyama R, Borisy GG. Centriole cycle in Chinese hamster ovary cells as determined by whole-mount electron microscopy. J Cell Biol. 1981;91:814–821. doi: 10.1083/jcb.91.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriyama R, Dagupta S, Borisy GG. Independence of centriole formation and initiation of DNA synthesis in Chinese hamster ovary cells. Cell Motil Cytoskeleton. 1986;6:355–362. doi: 10.1002/cm.970060402. [DOI] [PubMed] [Google Scholar]
- Lacey KR, Jackson PK, Stearns T. Cyclin-dependent kinase control of centrosome duplication. Proc Natl Acad Sci USA. 1999;96:2817–2822. doi: 10.1073/pnas.96.6.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaTerra S, English CN, Hergert P, McEwen BF, Sluder G, Khodjakov A. The de novo centriole assembly pathway in HeLa cells: Cell cycle progression and centriole assembly/maturation. J Cell Biol. 2005;168:713–722. doi: 10.1083/jcb.200411126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle WA, Lutz WH, Ingle JN, Maihle NJ, Salisbury JL. Centrosome hypertrophy in human breast tumors: Implications for genomic stability and cell polarity. Proc Natl Acad Sci USA. 1998;95:2950–2955. doi: 10.1073/pnas.95.6.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Vucica Y, Rosenbaum JL. Kinetics and regulation of de novo centriole assembly. Implications for the mechanism of centriole duplication. Curr Biol. 2001;11:308–317. doi: 10.1016/s0960-9822(01)00094-x. [DOI] [PubMed] [Google Scholar]
- Matsumoto Y, Hayashi K, Nishida E. Cyclin-dependent kinase 2 (Cdk2) is required for centrosome duplication in mammalian cells. Curr Biol. 1999;9:429–432. doi: 10.1016/s0960-9822(99)80191-2. [DOI] [PubMed] [Google Scholar]
- Meraldi P, Lukas J, Fry AM, Bartek J, Nigg EA. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-Cyclin A. Nat Cell Biol. 1999;1:88–93. doi: 10.1038/10054. [DOI] [PubMed] [Google Scholar]
- Merrill GF. Cell synchronization. Methods Cell Biol. 1998;57:229–249. [PubMed] [Google Scholar]
- Okuda M, Horn HF, Tarapore P, Tokuyama Y, Smulian AG, Chan PK, Knudsen ES, Hofmann IA, Snyder JD, Bove KE, Fukasawa K. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell. 2000;103:127–140. doi: 10.1016/s0092-8674(00)00093-3. [DOI] [PubMed] [Google Scholar]
- Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol. 2000;149:317–330. doi: 10.1083/jcb.149.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff JW, Glover DM. Nuclear and cyctoplasmic mitotic cycles continue in Drosophila embryos in which DNA synthesis is inhibited by aphidicolin. J Cell Biol. 1988;107:2009–2019. doi: 10.1083/jcb.107.6.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattner JB, Phillips SG. Independence of centriole formation and DNA synthesis. J Cell Biol. 1973;57:358–372. doi: 10.1083/jcb.57.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Cassels G. Correlative light and electron microscopy of mitotic cells in monolayer cultures. Methods Cell Biol. 1999;61:297–315. doi: 10.1016/s0091-679x(08)61987-1. [DOI] [PubMed] [Google Scholar]
- Robbins EL, Jentzsch G, Micali A. The centriole cycle in synchronized HeLa cells. J Cell Biol. 1968;36:329–339. doi: 10.1083/jcb.36.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SK, Packman K, Jeffrey R, Tenniswood M. Histone deacetylase inhibitors differentially stabilize acetylated p53 and induce cell cycle arrest or apoptosis in prostate cancer cells. Cell Death Differ. 2005;12:482–491. doi: 10.1038/sj.cdd.4401581. [DOI] [PubMed] [Google Scholar]
- Salisbury JL, Sunio KM, Busby R, Springett M. Centrin-2 is required for centriole duplication in mammalian cells. Curr Biol. 2002;12:1287–1292. doi: 10.1016/s0960-9822(02)01019-9. [DOI] [PubMed] [Google Scholar]
- Schutz AR, Giddings TH, Steiner E, Winey M. The yeast CDC37 gene interacts with MPS1 and is required for proper execution of spindle pole duplication. J Cell Biol. 1997;36:969–982. doi: 10.1083/jcb.136.5.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr CJ, Roberts JM. CDK inhibitors: Positive and negative regulators of G1-phase rogression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- Sluder G, Lewis K. Relationship between nuclear DNA synthesis and centrosome reproduction in sea urchin eggs. J Exp Zool. 1987;244:89–100. doi: 10.1002/jez.1402440111. [DOI] [PubMed] [Google Scholar]
- Sluder G, Nordberg JJ. The good, the bad and the ugly: The practical consequences of centrosome amplification. Curr Opin Cell Biol. 2004;16:49–54. doi: 10.1016/j.ceb.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Sluder G, Rieder CL. Controls for centrosome reproduction in animal cells: Issues and recent observations. Cell Motil Cytoskeleton. 1996;33:1–5. doi: 10.1002/(SICI)1097-0169(1996)33:1<1::AID-CM1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Sluder G, Miller FJ, Cole R, Rieder CL. Protein synthesis and the cell cycle: Centrosome reproduction in sea urchin eggs is not under translational control. J Cell Biol. 1990;110:2025–2032. doi: 10.1083/jcb.110.6.2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns T. Centrosome duplication. A centriolar pas de deux. Cell. 2001;105:417–420. doi: 10.1016/s0092-8674(01)00366-x. [DOI] [PubMed] [Google Scholar]
- Szüts D, Krude T. Cell cycle arrest at the initiation step of human chromosomal DNA replication causes DNA damage. J Cell Sci. 2004;117:4897–4908. doi: 10.1242/jcs.01374. [DOI] [PubMed] [Google Scholar]
- Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442:947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- Tucker RW, Pardee AB, Fujiwara K. Centriole ciliation is related to quiescence and DNA synthesis in 3T3 cells. Cell. 1979;17:527–535. doi: 10.1016/0092-8674(79)90261-7. [DOI] [PubMed] [Google Scholar]
- Uetake Y, Sluder G. Cell cycle progression after cleavage failure: Mammalian somatic cells do not possess a “tetraploidy checkpoint”. J Cell Biol. 2004;165:609–615. doi: 10.1083/jcb.200403014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y, Loncarek J, Nordberg JJ, English CN, LaTerra S, Khodjakov A, Sluder G. Cell cycle progression and de novo centriole assembly after centrosomal removal in untransformed human cells. J Cell Biol. 2007;176:173–182. doi: 10.1083/jcb.200607073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzawa S, Li F, Jin Y, McDonald KL, Braunfeld MB, Agard DA, Cande WZ. Spindle pole body duplication in fission yeast occurs at the G1/S boundary but maturation is blocked until exit from S by an event downstream of cdc10+ Mol Biol Cell. 2004;15:5219–5230. doi: 10.1091/mbc.E04-03-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidwans SJ, Wong ML, O’Farrell PH. Mitotic regulators govern progress through steps in the centrosome duplication cycle. J Cell Biol. 1999;147:1371–1378. doi: 10.1083/jcb.147.7.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev IA, Chentsov YS. Centrioles in the cell cycle I: Epithelial cells. J Cell Biol. 1982;93:938–949. doi: 10.1083/jcb.93.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RA, Pan Z, Salisbury JL. GFP-centrin as a marker for centriole dynamics in living cells. Microsc Res Tech. 2000;49:451–457. doi: 10.1002/(SICI)1097-0029(20000601)49:5<451::AID-JEMT7>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- Winey M. Cell cycle: Driving the centrosome cycle. Curr Biol. 1999;9:R449–R452. doi: 10.1016/s0960-9822(99)80279-6. [DOI] [PubMed] [Google Scholar]
- Winey M, Byers B. Assembly and functions of the spindle pole body in budding yeast. Trends Genet. 1993;9:300–304. doi: 10.1016/0168-9525(93)90247-f. [DOI] [PubMed] [Google Scholar]
- Wong C, Stearns T. Centrosome number is controlled by a centrosome-intrinsic block to reduplication. Nat Cell Biol. 2003;5:539–544. doi: 10.1038/ncb993. [DOI] [PubMed] [Google Scholar]