Abstract
In adult hippocampus, new neurons are continuously generated from neural stem cells (NSCs), but the molecular mechanisms regulating adult neurogenesis remain elusive. We found that Wnt signaling, together with the removal of Sox2, triggered the expression of NeuroD1 in mice. This transcriptional regulatory mechanism was dependent on a DNA element containing overlapping Sox2 and T-cell factor/lymphoid enhancer factor (TCF/LEF)-binding sites (Sox/LEF) in the promoter. Notably, Sox/LEF sites were also found in long interspersed nuclear element 1 (LINE-1) elements, consistent with their critical roles in the transition of NSCs to proliferating neuronal progenitors. Our results describe a previously unknown Wnt-mediated regulatory mechanism that simultaneously coordinates activation of NeuroD1 and LINE-1, which is important for adult neurogenesis and survival of neuronal progenitors. Moreover, the discovery that LINE-1 retro-elements embedded in the mammalian genome can function as bi-directional promoters suggests that Sox/LEF regulatory sites may represent a general mechanism, at least in part, for relaying environmental signals to other nearby loci to promote adult hippocampal neurogenesis.
In the neurogenic niche of the adult mammalian brain, self-renewing NSCs give rise to committed neuronal progenitors in the subgranular zone (SGZ) of the dentate gyrus1. Astrocytes are an essential cell population that defines the SGZ niche and astrocyte-derived factors have instructive effects to promote adult neurogenesis2,3. Recently, it has been shown that Wnt3 expression persists in the adult hippocampus and Wnt3 is released by astrocytes to regulate adult neurogenesis in vitro and in vivo4. In the canonical Wnt/β-catenin pathway, the TCF transcription factor transduces Wnt/β-catenin signals to activate downstream target genes4–9. However, the target genes of Wnt/β-catenin signaling that are responsible for promoting adult neurogenesis have not been identified. Moreover, the regulatory mechanism underlying Wnt-mediated neuronal differentiation has not yet been elucidated.
NeuroD1 is a proneural basic helix-loop-helix (bHLH) transcription factor that is essential for the development of the CNS, particularly for the generation of granule cells in the hippocampus and cerebellum10,11. Environmental signals regulate adult neurogenesis, at least in part, through the activation of NeuroD1 (refs. 12,13). Previously, we found that overexpression of NeuroD is sufficient to promote neuronal differentiation in adult hippocampal neural progenitors14, whereas deletion of NeuroD results in decreased survival and maturation of newborn neurons15. Thus, we hypothesized that astrocyte-derived Wnt signals may directly or indirectly regulate the transcription of NeuroD1 to control the transition of NSCs to committed neuronal progenitors.
The HMG-box transcription factor Sox2 is expressed in embryonic stem cells and most uncommitted cells in the developing CNS16–18. Sox2, which can be detected in cells of the mouse blastocyst, maintains precursor cells in a multipotent state19–21. During CNS development, Sox2 prevents neurogenesis22 and forced expression of Sox2 results in the loss of proneural cells23. Overexpression of Sox2 in neural progenitor cells derived from embryonic ventricular zone permitted the differentiation of progenitors into astroglia, but it inhibited neurogenesis24. Although these analyses indicate that Sox2 is a transcriptional repressor of neuronal target genes during development, the exact nature of Sox2 regulation during adult neurogenesis remains elusive.
Here, we found that the transcriptional activation of NeuroD1 is dependent on canonical Wnt/β-catenin activation and removal of Sox2 repression from the Neurod1 promoter in a sequence-specific manner. We discovered a previously unknown overlapping DNA-binding site corresponding to Sox2 and TCF/LEF (Sox/LEF) in the Neurod1 promoter. Using retrovirus gene delivery of Sox2–Cre-GFP (Cre-GFP fusion protein under the control of the Sox2 promoter, referred to here as Sox2CREGFP) into β-catenin conditional knockout (cKO) mice, we observed a significant loss (P < 0.001; data represent mean ± s.d., n = 6 per group) of NeuroD1-positive progenitors as well as a decrease in newborn granule neurons, with no effect on the stem/progenitor cell pool. These findings are extended to the regulation of LINE-1 retro-transposon expression through silencing and activation of Sox2 and Wnt/β-catenin, respectively, which is consistent with LINE-1 being critical during neuronal differentiation25. Together, these results suggest that Wnt-mediated activation of NeuroD1 and LINE-1 is coordinately regulated during adult neurogenesis, which may extend to other nearby genomic loci using the bi-directionality of Sox/LEF sites. These findings also suggest that crosstalk between Sox2 and Wnt/β-catenin signaling represents an important mechanism underlying neuronal differentiation.
RESULTS
The expression of Sox2 and NeuroD1 in adult neurogenesis
To characterize the expression of transcription factors in adult dentate gyrus, we performed immunohistochemical analysis in the Sox2–enhanced GFP (EGFP) transgenic mouse1,26. NeuroD1-positive cells were clearly detected in the SGZ region of the dentate gyrus and did not colocalize with Sox2-GFP or S100β, a marker of astrocytes (Fig. 1a). Moreover, Sox2-positive and NeuroD1-positive cells were mutually exclusive with mature neurons, suggesting that Sox2 and NeuroD1 counteract one another to regulate the early stages of adult neurogenesis (Fig. 1b).
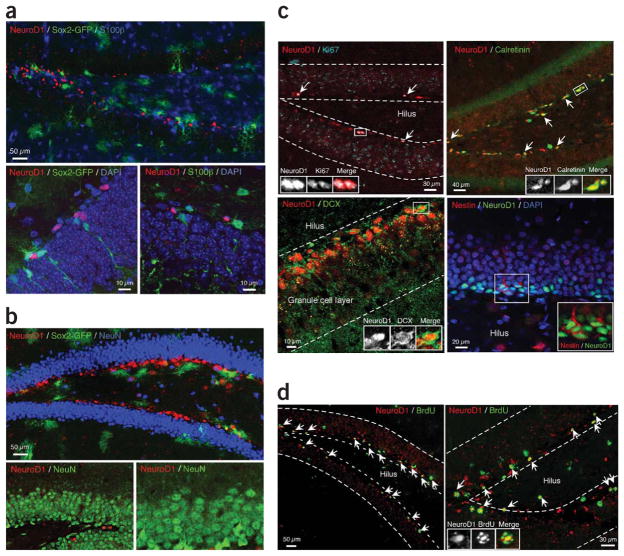
Figure 1.
Specific expression of the Neurod1 gene in early committed neurogenic cells in adult hippocampus. (a) Immunohistochemical analysis of neurogenic dentate gyrus area in adult hippocampus of the transgenic mouse that has a Sox2 promoter–driven EGFP reporter. Top, Sox2 is shown in green, and we stained for NeuroD1 (red) and S100β (blue). Bottom left, a higher-magnification image is shown with DAPI staining (blue) Bottom right, section stained for NeuroD1 (red), S100β (green) and DAPI (blue). (b) Distinct population of neuroblast cells expressing the Neurod1 gene and mature neurons in adult mouse hippocampus. Top, Sox2 is shown in green in adult hippocampus of a Sox2 promoter–driven EGFP reporter transgenic mouse; we also stained for NeuroD1 (red) and NeuN (blue). Bottom, NeuroD1-positive cells (red) and NeuN-positive cells (green) were exclusive to the inner layer of the dentate gyrus. (c) Immunocytochemical analysis of NeuroD1-positive cells in adult rat hippocampus. NeuroD1-positive cells (red) colocalized with Ki67 (cyan). White arrows indicate colocalizing cells; the region in the white square is magnified in a separate window. Bottom left, NeuroD1-positive cells (red) colocalized calretinin (green) and DCX (green). Bottom right, some NeuroD1-positive cells (green) colocalized with nestin (red). White arrows indicate populations of colocalized cells for both markers. (d) Proliferative status of NeuroD1-positive cells in adult hippocampus. BrdU (100 mg per kg of body weight) was injected for 1 week into Fisher 344 rats (7–8 weeks old). Cells that were double positive for NeuroD1 (red) and BrdU (green) are indicated by white arrows. The colocalizing cell (NeuroD1 and BrdU positive) in the white square is magnified on the right.
To further define the properties of NeuroD1-positive cells, we examined additional markers of stem/progenitor cells and immature granule neurons. NeuroD1-positive cells colocalized with nestin, calretinin and doublecortin (DCX; Fig. 1c). Sox2-GFP cells colocalized with the radial glial cell marker nestin, whereas only a few weakly stained GFP-positive cells colocalized with the weaker DCX-positive cells1. A majority of the NeuroD1-positive cells colocalized with DCX, whereas only a few cells were nestin positive, suggesting that NeuroD1-positive cells have recently transitioned from Sox2-positive NSCs to neuronal progenitors/immature neurons (Fig. 1c). In addition, NeuroD1-positive cells colocalized with Ki67 (Fig. 1c), indicating that they are among the proliferating population in adult dentate gyrus. To further examine their proliferative status, we treated Fischer 344 rats with BrdU and found that NeuroD1-positive cells colocalized with BrdU-positive cells in the SGZ (Fig. 1d). Taken together, these data indicate that NeuroD1 is transiently expressed in dividing progenitor cells and immature granule neurons in adult dentate gyrus and suggest that the expression of Sox2 and NeuroD1 must be coordinately regulated during adult neurogenesis.
Sox/LEF DNA recognition motif in the Neurod1 promoter
Next, we surveyed 3 kb of the regulatory region upstream of the Neurod1 and Neurod2 promoters and found binding sites for the TCF/LEF and Sox transcription factors (Fig. 2a). Notably, some Sox and TCF/LEF sequences were found to overlap with each other, forming a previously unknown binding motif, which we refer to as the Sox/LEF-binding site, that was conserved among humans, rats and mice (Fig. 2b).
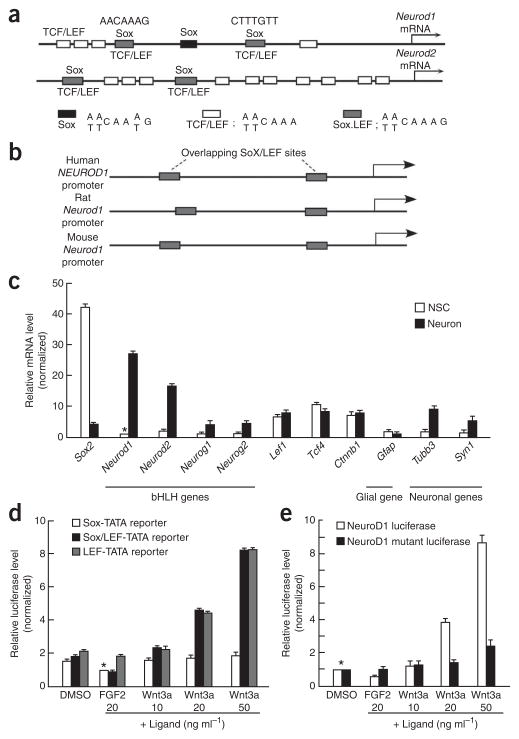
Figure 2.
Sox2/LEF DNA regulatory elements on the Neurod promoters. (a) Schematic representation of the binding sites of TCF/LEF and Sox transcription factors on the 3-kb promoters of the Neurod1 and Neurod2 genes. The sequences of the DNA regulatory elements recognized by Sox2 (black box) and TCF/LEF (white box) and the overlapping DNA regulatory consensus sequence (Sox/LEF; gray box) recognized by both Sox2 and TCF/LEF are shown (bottom). (b) Schematic representation of the Sox2/LEF-binding sites (gray boxes) in human, rat, and mouse Neurod1 promoters. (c) Comparison of expression levels between NSCs and differentiating neurons by quantitative real-time PCR (qRT-PCR) of genes related to Wnt signaling. The expression level of Gfap as a typical glial gene was assessed. Expression levels of β-tubulin III (Tubb3) and synapsin I (Syn1) were also analyzed as neuronal genes. Each mRNA value was normalized to that of Gapdh and then plotted as the fold increase of the sample of Neurod1 mRNA in NSCs (asterisk). (d) Simple reporter assay with the regulatory elements of Sox2, TCF/LEF and Sox/LEF transcription factors. The luciferase value was snormalized to a sample with a Sox-TATA reporter construct, with an FGF2 ligand (asterisk). (e) Activity of the Neurod1 promoter. A 1.5-kb Neurod1 promoter region (including a Sox/LEF site and a TCF/LEF site) was linked to the luciferase gene. The reporter construct with mutation at the Sox/LEF site on the Neurod1 promoter (NeuroD1 mutant luciferase) was also introduced to the adult NSCs. Luciferase value was normalized to sample with DMSO as a control ligand (asterisk).
Using an established adult rat hippocampal NSC line27, we compared the relative expression levels of Sox2, Neurod1 and other pro-neural genes. Adult NSCs expressed high levels of Sox2 in undifferentiated stages (Fig. 2c), but Sox-2 expression was reduced in neurons. In contrast, both NeuroD1 and NeuroD2 were significantly upregulated (P <0.001) in neurons (Fig. 2c). Notably, induction levels of NeuroD1 and NeuroD2 were higher than those of Neurog1 and Neurog2, which function as important bHLH proteins during neural development28–30. Gene activation by Wnt/β-catenin signaling requires stabilization of the β-catenin protein and nuclear association with TCF/LEF9. We found that the mRNA levels of β-catenin, TCF and LEF1 remained unchanged during neuronal differentiation, suggesting that Wnt and β-catenin are regulated at the post-transcriptional level (Fig. 2c).
To investigate the requirement of the Sox/LEF sequence, we prepared a set of reporter constructs (Sox-, Sox/LEF- and LEF-TATA; Online Methods). Incubation with fibroblast growth factor 2 (FGF2), which maintains cells in an undifferentiated state, reduced luciferase activity resulting from the Sox-TATA and Sox/LEF-TATA constructs, but not from the LEF-TATA construct, compared with the luciferase activity in cells treated with DMSO. This finding suggests that the Sox regulatory element has a negative role on transcription in the presence of FGF2, whereas the LEF regulatory element itself has no effect. When the Wnt3a ligand was introduced to cells, we observed a clear dose-dependent upregulation of luciferase in cells expressing the Sox/LEF-TATA and LEF-TATA constructs (Fig. 2d). The Sox regulatory element itself had almost no effect in the presence of Wnt3a, indicating that there is an apparent functional difference in the Sox and LEF regulatory elements with regard to ligand response.
Next, we introduced the Wnt3a ligand into NSCs and observed a dose-dependent upregulation of Neurod1 promoter activity (Fig. 2e). In contrast, when the mutant reporter construct (NeuroD1 mutant luciferase (Fig. 2e) contains a mutation in the Sox/LEF site to abolish Sox2 and TCF/LEF binding) was introduced into NSCs, we failed to observe either a reduction in luciferase activity in the undifferentiated state (FGF2 ligand) or an increase in luciferase activity by the addition of Wnt3a ligand (Fig. 2e). These data suggest that the Sox/LEF binding sequence in the Neurod1 promoter is important for discriminating between both TCF/LEF- and Sox2-mediated transcriptional regulation during adult neurogenesis.
Upregulation of NeuroD1 is dependent on the Sox/LEF site
To investigate the regulatory mechanism underlying Neurod1 gene transcription, we compared the expression of NeuroD1 with that of proteins in the Wnt signaling pathway. S100β-positive and glial fibrillary acidic protein (GFAP)-positive astrocytes were also positive for both Wnt3 and Wnt3a, consistent with published results4 (Supplementary Fig. 1). Following neuronal differentiation in vitro, NeuroD1 expression peaked at 1 d after neuronal induction and diminished by 4 d. In protein blotting experiments, we found that Sox2 was expressed in undifferentiated NSCs and downregulated on neuronal differentiation (Fig. 3a). In contrast, β-catenin was clearly stabilized and accumulated when NeuroD1 was highly expressed, although the level of β-catenin mRNA was unchanged (Fig. 2c). These results suggest that NeuroD1 expression is temporally regulated, consistent with NeuroD1 expression in vivo, and Wnt/β-catenin activation and de-silencing of Sox2 may be involved in promoting neuronal differentiation.
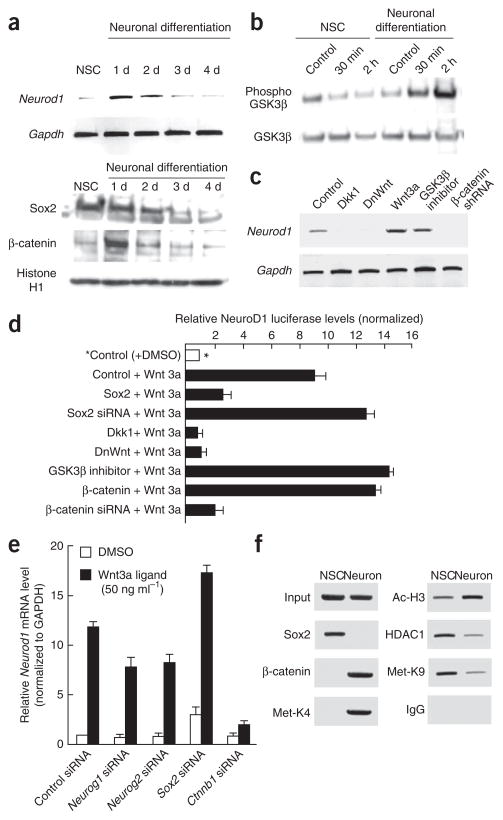
Figure 3.
Wnt signaling increases Neurod1 promoter activity during early neurogenesis. (a) Time course of Neurod1 mRNA expression during the early stages of neurogenesis in cultured adult NSCs. RT-PCR detection of Neurod1 and Gapdh is shown. Western blots of Sox2 and β-catenin during neurogenesis in cultured adult NSCs are shown in the lower panels. (b) Induction of a phosphorylated inactive form of GSK3β to stabilize β-catenin during Wnt signaling in early committed neurogenic cells. Western blots of both GSK3β and phosphorylated GSK3β were conducted using the same neuronal induction treatment. (c) The effect of Wnt signaling on the expression of Neurod1 mRNA. RT-PCR analysis using total RNA extracted from adult NSCs treated with Dkk1, DnWnt, Wnt3, TDZD8 or β-catenin shRNA. (d) The effect of Wnt signaling on the promoter activity of the Neurod1 gene. The luciferase value was normalized to that of cultured NSC sample with control vector and control ligand (DMSO, asterisk, white bar). (e) The effect of siRNAs targeting Neurog1, Neurog2, Sox2 and β-catenin on Wnt3a ligand–mediated induction on Neurod1 mRNA. qRT-PCR analysis for Neurod1 mRNA was plotted. The amount of mRNA present for each sample was normalized to that of Gapdh and then plotted as the fold increase over the control (control siRNA with DMSO). (f) ChIP analysis at the Neurod1 promoter in adult neurogenesis. PCR primers were designed to surround the Sox/LEF sequence on the rat Neurod1 promoter.
For Wnt-mediated transcriptional activation of target genes, the phosphorylated, inactive form of GSK3β is the canonical enhancer for the stabilization of β-catenin protein9. Thus, we assessed whether neuronal differentiation in adult NSCs influenced the phosphorylation of GSK3β. As hypothesized, GSK3β phosphorylation was triggered on neuronal differentiation, whereas total GSK3β levels remained unchanged (Fig. 3b).
To evaluate the effects of Wnt signaling on Neurod1 gene activity, we carried out gain-of-function studies using Wnt3a-expressing lentivirus4 and a pharmacological inhibitor of GSK3β (TDZD8). For loss-of-function studies, we used a full-length cDNA Sox2 construct, a secreted mutant Wnt (dominant-negative Wnt, DnWnt) construct4, a small hairpin RNA (shRNA) that was specific to β-catenin mRNA (Supplementary Fig. 2) and the Wnt antagonist Dickkopf1 (Dkk1). Neurod1 mRNA levels were strongly increased by both Wnt3a expression and the GSK3β inhibitor (Fig. 3c). In contrast, Dkk1, DnWnt and an shRNA to β-catenin reduced NeuroD1 expression. To further examine the effect of Wnt3a expression on NeuroD1 transcriptional activation, we introduced a NeuroD1 luciferase construct into adult NSCs. Sox2, Dkk1 and DnWnt expression all had negative effects on Wnt3a-mediated NeuroD1 transcriptional activation. In contrast, Neurod1 promoter activity was further enhanced by the constitutively active form of β-catenin and by TDZD8 (Fig. 3d).
To confirm that the transcriptional activation of NeuroD1 is dependent on the Wnt3a ligand, we introduced several sets of synthesized small interfering RNAs (siRNAs) into adult NSCs. Wnt3a ligand increased Neurod1 mRNA expression (~12-fold increase) in control siRNA-transfected cells (Fig. 3e). In contrast, β-catenin siRNA substantially reduced NeuroD1 activation on Wnt3a ligand treatment (~sixfold decrease). Notably, we did not observe substantial reduction of Wnt3a-mediated NeuroD1 activation by Neurog1 and Neurog2 siRNAs, indicating that Wnt3a effects on NeuroD1 may be direct. The amount of NeuroD1 mRNA increased in cells treated with Sox2 siRNA more than in cells treated with control siRNA, possibly as a result of residual endogenous Sox2 protein at the onset of neuronal induction (Fig. 3a), which may have a negative influence on NeuroD1 expression.
Finally, we performed chromatin immunoprecipitation (ChIP) analysis to assess protein association. We found that Sox2 and the histone deacetylase HDAC1 repressor protein were associated on the endogenous Neurod1 promoter, specifically at the Sox/LEF site, in undifferentiated NSCs, and that this association diminished when the neurons differentiated (Fig. 3f). Consistently, di-methylated histone H3 at Lys9 (K9), commonly associated with transcriptional repression31, was also present in NSCs. Moreover, β-catenin, acetylated histone H3 and methylated histone H3 at Lys4 (K4), all of which are associated with transcriptional activation, were observed on the Neurod1 promoter locus in neuronal cells, suggesting that Neurod1 gene transcription is mediated by an active process. These results suggest that the conversion of a Sox2 repressor complex to a β-catenin activator complex is associated with chromatin remodeling on neuronal induction.
Wnt3a-dependent activation of LINE-1 retrotransposon
Sox2 can suppress LINE-1 expression in adult NSCs25. The observation that Sox2 is downregulated at nearly the same time that β-catenin is upregulated raises the possibility that they may target the same Sox/LEF regulatory sequences in neuronal genes. Indeed, many Sox/LEF-binding sites were present throughout the entire LINE-1 sequence, including several sites in open reading frame 2 (ORF2; Fig. 4a). We next determined whether these sequences were functional using reporter assays in adult NSCs.
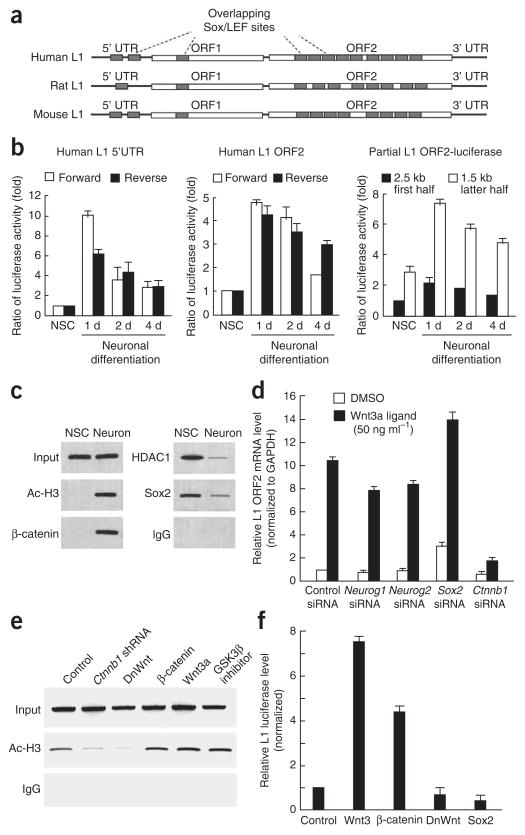
Figure 4.
Effect of Wnt signaling on the expression of NeuroD1 and LINE-1 during adult neurogenesis. (a) DNA regulatory elements recognized by Sox2 and TCF/LEF transcription in LINE-1. A schematic representation of the Sox/LEF DNA regulatory elements (gray boxes) in the human, rat, and mouse retrotransposon LINE-1 is shown. (b) Promoter activity of the 5′ UTR and ORF fragment of LINE-1 during adult neurogenesis. Luciferase constructs with 5′ UTR and LINE-1 ORF2 sequences linked to the luciferase gene, in both forward (white) and reverse (black) orientations, were introduced into adult NSCs by lentivirus infection (left and middle panels). Partial fragments of the LINE-1 ORF2, the first 2.5 kb (black columns) and the last 1.5 kb (white columns), were also linked to the luciferase gene in the reporter assay (right). (c) ChIP analysis of rat LINE-1. PCR primers were designed to surround the Sox/LEF DNA regulatory elements. (d) Wnt3a-mediated induced production of LINE-1 ORF2 mRNA. The induction level was measured by qRT-PCR with several synthesized siRNAs. The mRNA level was normalized to that of Gapdh and then plotted as the fold increase over the control (control siRNA with DMSO). (e) The effect of Wnt signaling on the chromatin remodeling of LINE-1. Activation of the chromatin state (acetylation of histone H3) in the LINE-1 sequences was assessed using several Wnt-related constructs. (f) The activation and repression of the LINE-1 promoter. The effect of Wnt on the activity of LINE-1–based promoters was examined using the LINE-1 luciferase construct. After 1 d in the culture, luciferase assays were performed on adult NSCs treated with each construct.
We previously reported that promoter activity in the human LINE-1 5′ untranslated region (UTR) is increased during neuronal differentiation compared with undifferentiated cells25. We confirmed this observation using a reporter construct containing the LINE-1 5′ UTR in both forward and reverse orientation upstream of the luciferase gene (Fig. 4b). Because ORF2 of LINE-1 contains several Sox/LEF-binding sites, we cloned the LINE-1 ORF2 portion and linked it to the luciferase gene to assess putative promoter activity. The ORF2 sequence demonstrated promoter activity in both forward and reverse orientation during neuronal differentiation. The activity was highest 1 d after neuronal induction and it gradually declined during neuronal differentiation. This transient upregulation immediately after neuronal induction was common to both LINE-1 5′ UTR– and ORF2-based reporter constructs (Fig. 4b), a finding that is also consistent with the expression dynamics of NeuroD1 (Fig. 3a).
LINE-1 sequences comprise a substantial part of the genome, and because the Sox/LEF sites are clustered in LINE-1 sequences, they are also prevalent in the genome. Using ChIP, we found that Sox2 and HDAC were associated in undifferentiated NSCs in which LINE-1 was silenced (Fig. 4c), similar to their association on the Neurod1 promoter (Fig. 3f). On the other hand, we observed that β-catenin and acetylated histone H3 associated with each other in neurons. To investigate Wnt3a ligand–mediated transcriptional activation of LINE-1 mRNAs, we compared the induction levels of LINE-1 in cells given control siRNA with the cells treated with several gene-specific siRNAs. Wnt3a ligand caused a tenfold increase in the amount of LINE-1 ORF2 mRNA in cells treated with control siRNA (Fig. 4d). When cells were treated with β-catenin siRNA, Wnt3a ligand–induced activation was almost abolished, whereas treatment with Neurog1 and Neurog2 siRNAs had almost no effect on Wnt3a-mediated stimulation. On introduction of Sox2 siRNA, endogenous LINE-1 was upregulated and Wnt3a-mediated stimulation of LINE-1 was increased (Fig. 4d).
Next, we examined the chromatin status (acetylated histone H3) in the LINE-1 element. β-catenin shRNA and DnWnt stopped the chromatin from switching from the silenced state to the activated state (Fig. 4e). The expression of endogenous LINE-1 mRNA was also downregulated by β-catenin shRNA (Supplementary Fig. 2), consistent with our ChIP data. The addition of Wnt3, β-catenin or TDZD8 increased acetylated histone H3 levels in the LINE-1 genomic region, indicating that Wnt signaling itself could induce the active chromatin state, directly or indirectly, in the LINE-1 locus (Fig. 4e).
We also examined the effect of Wnt3a on the activity of LINE-1–based promoters using the LINE-1 luciferase construct. β-catenin and Wnt3a enhanced LINE-1 luciferase activity significantly (P < 0.001) (Fig. 4f). In contrast, Sox2 and DnWnt did not promote LINE-1–promoter activity. These results suggest that Wnt signaling actively mediates the expression of LINE-1 in neurons and that the process is not merely a result of de-repression of LINE-1 transcription.
Because LINE-1 sequences are spread throughout the genome, we decided to investigate which genes might be under the influence of this molecular mechanism. Using computational analysis, we scanned downstream regions of human genes that were likely to influence transcription, looking for LINE-1 sequences that contained Sox2, LEF or Sox/LEF-binding sites. We identified 79, 84 and 25 LINE-1 elements within −6,000 and +1,000 base pairs of the start sites of the human, mouse and rat genes, respectively (Supplementary Tables 1–3). The LINE-1 elements from different species were not consistently near the same genes. Notably, we found genes encoding olfactory receptors in mouse containing Sox/LEF-binding sites upstream of LINE-1 elements. Furthermore, and of particular interest to us, we found several neuronally relevant genes that may be susceptible to the repressor/activator mechanism described above, including, for example, DCX and Neuregulin 4, or related to cell cycle (SCAPER (zinc finger protein 291) and mitogen-activated protein kinase 10; Supplementary Tables 1–3).
Role of β-catenin in neuronal differentiation of adult NSCs
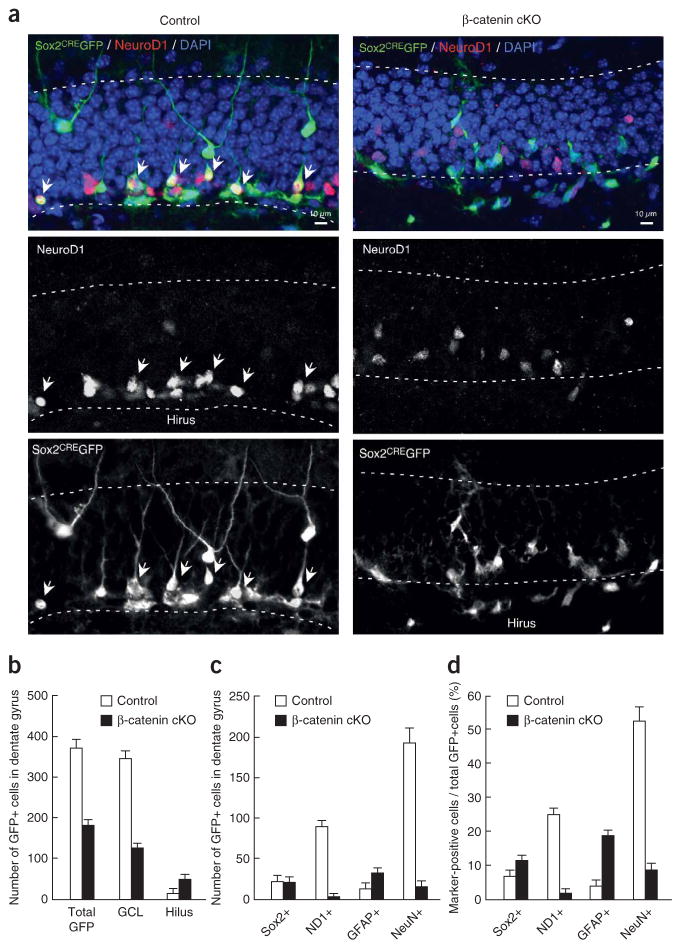
We further investigated the effect of Wnt/β-catenin signaling on cell fate choice in vivo. A retrovirus encoding Sox2 promoter–driven cre-gfp (Sox2CREGFP)1 was injected into the dentate gyrus of Ctnnb1loxP/loxP mice (β-catenin cKO; Supplementary Fig. 3)32. Because a retrovirus only transduces one of the two daughter cells of dividing cells, single-cell clones are generated by the conversion of Sox2-positive cells to differentiated lineages in β-catenin cKO mice.
To identify the composition of cell types in the Sox2 lineage, we carried out an immunohistochemical analysis (Figs. 5 and 6). The number of total GFP-positive cells in β-catenin cKO mice was decreased by about 50% compared with control mice (Fig. 5b). In control mice, many Sox2CREGFP-positive cells colocalized with NeuroD1-positive cells (Fig. 5a). In contrast, the proportion of ND1 and GFP double-positive cells in β-catenin cKO mice was decreased by 92% relative to that of control mice (Fig. 5c,d). In addition to NeuroD1, Sox2CREGFP-positive cells that colocalized with markers for newborn neurons, such as DCX (Supplementary Fig. 4) and TUJ1 (Supplementary Fig. 5), were significantly reduced (P <0.001) in the β-catenin cKO mice (Supplementary Fig. 6). In control mice, we readily observed Sox2CREGFP-positive cells that gave rise to mature neurons with extensive neurites (Fig. 5a). Quantification of Sox2CREGFP-positive cells in control mice revealed that ~50% of them became NeuN-positive mature granule neurons (Fig. 5d).
Figure 5.
Adult NSC cannot transition to immature and mature granule neurons in β-catenin cKO mice. Immunohistochemical analysis of the Sox2CREGFP cells. (a) Sox2CREGFP retrovirus was injected into the dentate gyrus of control mice (left) or β-catenin cKO mice (right). Immunohistochemical analysis of NeuroD1 (red), GFP (green) and DAPI (blue) in both groups is shown and GFP-positive cells colocalized with NeuroD1-positive cells in control mice (indicated by white arrows, left panels). In control mice, GFP-positive cells were present among the more differentiated neurons deeper in the granule cell layer (left). In β-catenin cKO mice, GFP-positive cells were observed more often near or in the hilus region, rather than among the more differentiated neurons deeper in the granule cell layer. (b) Numbers of GFP-positive cells in the dentate gyrus of control mice (white bars) and β-catenin cKO mice (black bars). (c) Numbers of marker and GFP double-positive cells in the dentage gyrus of control mice (white bars) and β-catenin cKO mice (black bars). (d) Percentages of marker-positive cells in the dentage gyrus of control mice (white bars) and β-catenin cKO mice (black bars).
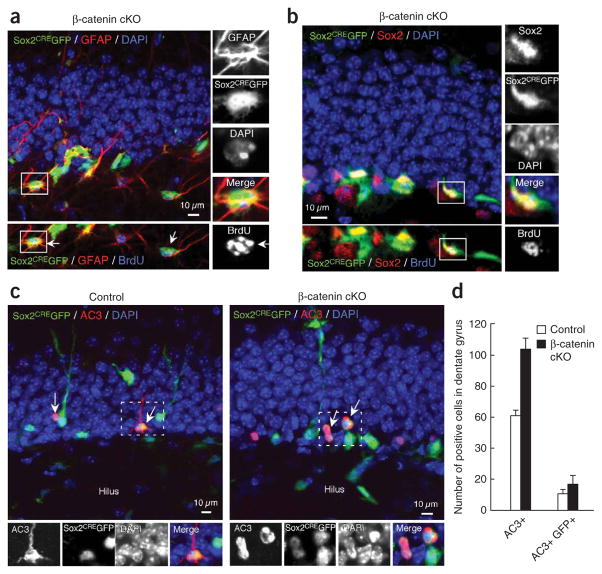
Figure 6.
Lineage tracing Sox2-positive NSCs in β-catenin cKO mice. Immunohistochemical analysis of the Sox2CREGFP cells. (a,b) Sox2-positive cells were able to give rise to GFAP-positive (red, a) and to Sox2-positive NSCs (red, b). Magnified images show triple immunohistochemistry of GFAP (a) or Sox2 (b), GFP (green) and BrdU (blue, lower panels). (c) Immunohistochemical analysis of apoptotic cells labeled by AC3 in β-catenin cKO mice. Representative image of AC3 (red), GFP (green) and DAPI (blue) in control mice (left) and β-catenin cKO mice (right). The GFP-positive cells colocalizing with the AC3-positive cells are indicated by white arrows. The GFP and AC3 double-positive cells in the white dotted square are magnified in the bottom panels. (d) Quantification of GFP-positive and AC3-positive cells in dentate gyrus of control and β-catenin cKO mice. The numbers of the AC3-positive and AC3 and GFP double-positive cells in the dentate gyrus of control mice (white bars) and β-catenin cKO mice (black bars) are plotted.
In contrast, there was a substantial decrease in Sox2CREGFP-positive cells that became mature neurons in β-catenin cKO mice (Fig. 5a). The percentage of NeuN and GFP double-positive cells was reduced by 85% relative to that of control mice (Fig. 5d). The number of Sox2CREGFP-positive cells that were also labeled by Prox-1, which labels both immature and mature neurons, was also reduced by 90% in β-catenin cKO mice compared with control mice (Supplementary Fig. 6).
To examine Sox2CREGFP-positive cells in the stem cell compartment, we stained the cells with GFAP, a marker of radial stem-like cells and astrocytes, and BrdU (Fig. 6a). The proportion of GFAP and GFP double-positive cells in β-catenin cKO mice was 4.4-fold higher than that in control mice (Fig. 5d). The Sox2-CREGFP-positive cells in β-catenin cKO mice were also positive for Sox2 and labeled with BrdU (Fig. 6b). There was no substantial change in the number of GFP and Sox2 double-positive cells (Fig. 5c). The proportion of GFP and Sox2 double-positive cells was 1.5-fold higher than that in control mice (Fig. 5d).
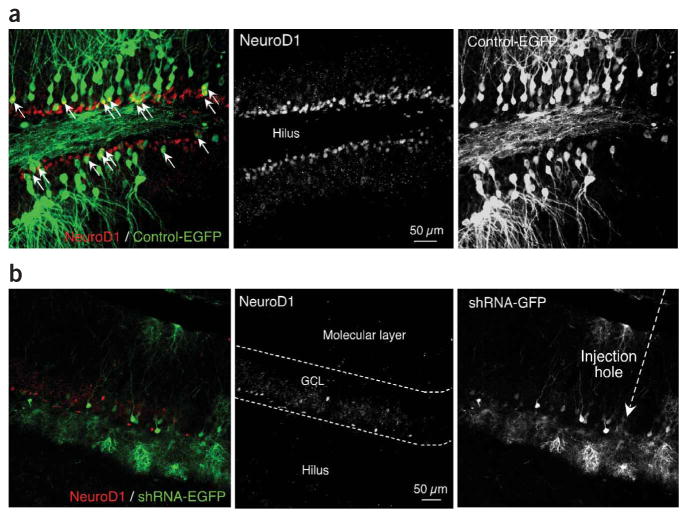
We observed similar results with a lentivirus approach in vivo. Cells that were positive for the lentivirus encoding β-catenin shRNA and GFP cells did not significantly colocalize with NeuroD1-positive cells (P < 0.001), whereas control lentivirus-GFP–positive cells colocalized with many NeuroD1-positive cells (Fig. 7 and Supplementary Fig. 7). Cells infected with a lentivirus expressing shRNA specific to β-catenin were positive for Sox2 and BrdU, similar to cells infected with a control lenti-virus expressing GFP (Supplementary Fig. 8). The shRNA-GFP–positive cells colocalized with GFAP-positive cells and Nestin-positive cells (Nestin is a radial glial cell marker) (Supplementary Fig. 9). These data suggest that the maintenance of the undifferentiated stem cell compartment and, to a large extent, astrocytic lineage cells and/or GFAP-positive radial stem-like cells remained intact with knockdown of β-catenin. Together, these data indicate that Sox2-positive cells cannot transition to immature and mature granule neurons when Wnt/β-catenin signaling is blocked, whereas the stem/progenitor cell compartment remains intact.
Figure 7.
Infection of lentivirus expressing β-catenin shRNA in vivo. To evaluate the effect of Wnt signaling in vivo, lentivirus expressing β-catenin shRNA or control lentivirus expressing only EGFP (lentivirus-GFP) was stereotactically injected into adult rat hippocampus. (a) Lentivirus-GFP in dentate gyrus. White arrows indicate the population of cells that were double positive for NeuroD1 (red) and EGFP (green). (b) Immunohistochemical analysis of the cells infected by the lentivirus encoding β-catenin shRNA and GFP. Cells expressing β-catenin shRNA and GFP (green) and NeuroD1-positive cells (red) are shown.
Wnt is important for survival of neuronal progenitor cells
The decrease of Sox2CREGFP-positive cells in β-catenin cKO mice could also be explained by a defect in the survival of neuronal progenitors. Thus, we examined the presence of dead and/or dying cells by activated caspase 3 (AC3) staining. We found more AC3-positive cells in dentate gyrus of β-catenin cKO mice than in control mice (Fig. 6c,d). Although we observed AC3-positive cells that colabeled with Sox2 (Supplementary Fig. 10), the total number of Sox2-positive cells remained unchanged in β-catenin cKO mice (Fig. 5c), suggesting that inhibition of Wnt/β-catenin signaling in Sox2-positive NSCs is important for the generation NeuroD1-positive cells from Sox2-positive NSCs, as well as for the survival of neuronal progenitors.
Finally, to examine whether Wnt/β-catenin–mediated neuronal differentiation is dependent on NeuroD1, we performed loss-of-function experiments in adult NSCs expressing siRNA specific to NeuroD1, as well as in subventricular zone and dentate gyrus neurospheres from 4-week-old Neurod1loxP/loxP mice (NeuroD1 cKO) mice15. The addition of Wnt3a ligand induced microtubule associated protein 2AB (Map2AB) expression in neurons in NSCs that were treated with of control random siRNA; Map2AB expression was completely suppressed with the introduction of NeuroD1 siRNA (Supplementary Fig. 11). Moreover, Wnt3a ligand treatment in NeuroD1 cKO neurospheres15 with control GFP lentivirus resulted in TUJ1-positive neurons after 2 d (Supplementary Fig. 12). However, in the presence of lentivirus expressing Cre, which caused the deletion of NeuroD1, Wnt3a stimulation failed to induce substantial numbers of TUJ1-positive neurons, which is consistent with the idea that NeuroD1 is essential for Wnt3a-mediated neuronal differentiation.
LINE-1 promoter activity in newborn and mature neurons
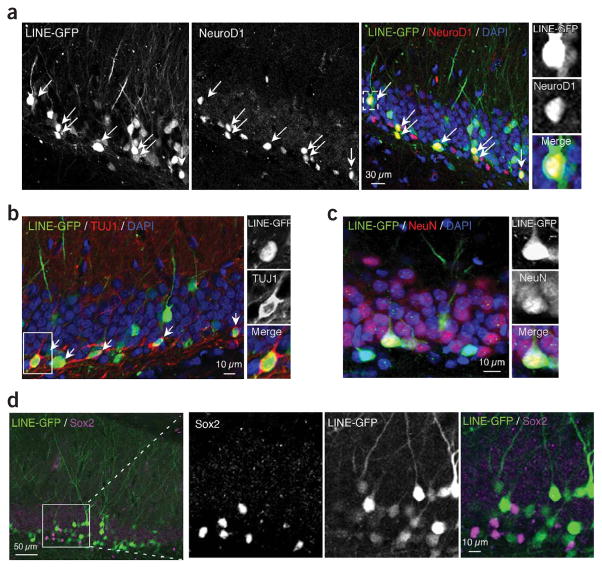
On the basis of our results, we hypothesized that LINE-1 sequences are active during adult neurogenesis. To evaluate LINE-1–based promoter activity in vivo, we prepared a lentiviral vector that encodes the LINE-1 ORF2 fragment fused to the egfp gene (LINE-GFP). LINE-GFP lentivirus was stereotactically injected into dentate gyrus of young adult rats. Notably, we observed LINE-GFP expression restricted to neurogenic areas, and GFP-positive cells colocalized with NeuroD1-positive neuronal progenitors and TUJ1-positive newborn granule neurons (Fig. 8a,b). Moreover, we observed a similar expression pattern of the 5′ UTR of LINE-GFP in TUJ1-positive newborn neurons in dentate gyrus (Supplementary Fig. 13). We also detected LINE-GFP–positive cells that migrated further into the granule cell layer, where mature neurons reside that were NeuN positive33 (Fig. 8c and Supplementary Fig. 14). LINE-GFP–positive cells were negative for Sox2 (Fig. 8d). These in vivo data are consistent with our in vitro results, which indicate that NeuroD1 and LINE-1 expression are specifically induced only when Sox2-positive NSCs transition to newborn neurons on Wnt/β-catenin activation.
Figure 8.
Activity of LINE-1 as a promoter in adult rat hippocampus. (a–d) We examined the activity and specificity of the LINE-1–based promoter in adult rat hippocampus. EGFP-expressing lentivirus, under the control of the LINE-1–based promoter, was stereotactically microinjected into the dentate gyrus of adult rats and the population of GFP-positive cells (green) was analyzed by immunohistochemistry using an antibody to NeuroD1 (red, a). Comparisons of the LINE-GFP population (green) to TUJ1 staining (red, b), NeuN staining (red, c) and Sox2 staining (magenta, d) are also shown.
DISCUSSION
We found an important function for canonical Wnt/β-catenin signaling in balancing self-renewal of NSCs and neuronal differentiation in adult dentate gyrus. Our results (as summarized in Supplementary Fig. 15) indicate that the Sox2 and TCF/LEF regulatory element in the Neurod1 promoter is critical for the transition from Sox2-mediated repression to Wnt/β-catenin-mediated activation, that the clear, dose-dependent activation of the Neurod1 promoter by the Wnt3a ligand is dependent on the Sox/LEF-binding site, that deletion of β-catenin leads to substantial loss of NeuroD1-positive cells, whereas the stem cell compartment remains intact in vivo, and that Wnt/β-catenin–mediated neuronal differentiation is dependent on NeuroD1, at least in vitro. Taken together, these findings indicate that the decreased adult neurogenesis observed in the Sox2–cre-gfp; Ctnnb1loxP/loxP mice is probably a result of a failure of neuronal lineage commitment from Sox2-positive NSCs and of survival of Sox2- and NeuroD1-positive progenitor cells/neuroblasts.
During embryonic development, neurogenins function as pro-neural proteins that activate transcription of Neurod1 through E-protein binding sites in its promoter28–30. However, the transcriptional/epigenetic mechanism that regulates NeuroD1 expression in the adult neurogenic niche is not clear. In the SGZ, hippocampal astrocyte–derived factors, such as Wnt proteins, signal to NSCs to promote adult neurogenesis. Among several Wnt proteins (Wnt3a, Wnt2a, Wnt5a, Wnt7a and Wnt8b)34–36, Wnt3a has a dominant role in CNS development, as the deletion of Wnt3a (Wnt3a−/− mice) results in the absence of dentate gyrus formation35. Furthermore, it has been reported that β-catenin is involved in the dendritic development of newborn neurons37. Both canonical4–8 and noncanonical37 Wnt/β-catenin signaling may contribute to the step-wise progression of adult hippocampal neurogenesis by removing Sox2 repression and turning on NeuroD1. Similar to a loss of Wnt/β-catenin signaling, NeuroD1 deficiency during hippocampal development leads to a complete loss of dentate gyrus formation in mice10,11. In adult stages, when the hippocampal formation is fully developed, conditional deletion of β-catenin and NeuroD1 (ref. 15) lead to a similar phenotype in dentate gyrus, that is, a decreased number of neuronal progenitors/newborn neurons, suggesting that the regulation of canonical Wnt/β-catenin signaling and Neurod1 gene expression are tightly linked and the functions of both β-catenin and NeuroD1 are indispensable for adult neurogenesis and for the survival of neuronal progenitors. Recently, it was shown that Wnt-mediated adult hippocampal neurogenesis contributed to learning and memory in rats38. Our data suggest that Wnt signaling activation might be a major environmental factor that is relayed to the NSC genome for neuronal lineage commitment to modulate behavior.
Our finding that the Wnt-mediated regulatory mechanism is required for the activation of NeuroD1 can be broadly extended to the regulation of LINE-1. Because retro-element sequences are scattered throughout the genome and contain Sox/LEF DNA regulatory elements, one possibility is that Sox/LEF-binding sites act as bidirectional promoters and cause nearby neuronal gene loci to become de-silenced and activated during adult neurogenesis. Thus, to explore the possibility that LINE-1 sequences containing Sox2 and TCF/LEF sites might confer cell type–specific regulation as described for NeuroD1, we searched for LINE-1 sequences proximal to the transcriptional start sites of known protein-coding genes in human, mouse and rat genomes. We were able to identify 79, 84 and 25 such LINE-1 elements within −6,000 and +1,000 base pairs of the target human, mouse and rat genes, respectively (Supplementary Tables 1–3). These bioinformatics analyses suggest that there is a global regulatory mechanism for controlling the activation/repression of neuronal gene expression that uses embedded retrotransposition sequences in the genome as a putative master regulatory pathway during adult neurogenesis. However, the causal relationship between LINE-1 sequences and transcriptional activation of nearby Sox/LEF-driven neuronal genes awaits future validation. Recently, it has been shown that environment is a robust stimulator of adult neurogenesis39, possibly through the activation of the Sox/LEF regulatory elements described here. Consistent with these data, we recently observed that voluntary exercise increased LINE-1 retrotransposition in dentate gyrus40. Our data provides a window into the molecular mechanism behind experience-dependent LINE-1 retrotransposition that may affect neuronal plasticity.
METHODS
Cell culture
Adult hippocampal NSCs were cultured as described27. Adult NSCs were cultured with Dulbecco’s modified Eagle’s medium/F-12 medium (Invitrogen) containing 20 ng mL−1 FGF-2 (Wako), 1% N2 supplement (Invitrogen), 1% antibiotic-antimycotic (Invitrogen), and 2 mM L-glutamine (Wako) in a 5% CO2 incubator at 37 °C. For neuronal differentiation, cells were cultured in N2 medium (Invitrogen) containing retinoic acid (1 μM, Sigma), forskolin (5 μM, Sigma) and KCl (40 mM, Wako). Dkk1 (500 ng mL−1, R&D Systems), Wnt3a (50 ng mL−1, R&D Systems) and 5 μM TDZD8 (4-benzyl-2-methyl-1,2,4-thiadiazolidine-3,5-dione, Calbiochem) were added to the medium to determine the effects of Wnt signaling. For the adult neurosphere culture from ND1 cKO mice (Supplementary Fig. 12), spheres were cultured on uncoated dishes. Lentivirus expressing GFP or Cre-GFP was introduced to the neurospheres. To examine the effect of Wnt signaling on Neurod1−/−cells (neurospheres), we added Wnt3a ligand (50 ng mL−1) for 2 d after the removal of FGF2 and epidermal growth factor 2.
Construction of plasmids, shRNA and siRNA
Five copies of the Sox/LEF-binding sites were fused upstream to the 200-bp cytomegalovirus (CMV) minimal promoter carrying a TATA box and linked to the luciferase gene (Sox/LEF-TATA). Five copies of the DNA binding sequence for Sox2 (Sox-TATA) and five copies of the DNA binding sequence for TCF/LEF (LEF-TATA) were also fused to the minimal promoter-driven luciferase. The mutant construct containing Neurod1 promoter–driven luciferase (NeuroD1 mutant luciferase in Fig. 2e) was designed to contain a mutation in the Sox/LEF site to abolish Sox2 and TCF/LEF binding (AAC AAA G sequence was exchanged to GCT AGC G). The Wnt3, DnWnt, Sox2, HDAC1 and β-catenin–expressing lentiviral vectors were constructed using CSC PW, a third-generation, self-inactivating lentiviral vector41. Each expression cassette was subcloned at the 3′ end of the CMV promoter on CSC SP PW. The shRNA for the Ctnnb1 gene was designed to target the sequence GCA ATC AGC TGG CCT GGT TTG, located in the last exon of the rat β-catenin transcript. It was constructed by connecting a shRNA sequence with a terminator under the murine U6 promoter. This shRNA cassette was subcloned into the TUHSA CS PW lentiviral vector, which contains cassettes of CMV-EGFP, with the ClaI and PmeI restriction sites, and the U6 promoter with the ClaI and HpaI sites. TUHSA CS PW lentiviral vector was derived from the original CSC SP PW plasmid. The knockdown effect of the Ctnnb1 shRNA was confirmed by western blot analysis using cell lysate from lentivirus-infected, GFP-positive adult hippocampal NSCs (Supplementary Fig. 2). The production of lentivirus has been described elsewhere42; the viral titers were greater than 1.5 × 104 transducing units ng−1, as determined by the p24 ELISA assay. The murine Neurod1 promoter was cloned by PCR from genomic DNA and inserted into CSC PW-Luci at the site of the CMV promoter using the restriction enzyme sites for ClaI and BamHI. Sox-TATA, Sox/LEF-TATA and LEF-TATA luciferase reporter plasmids included the binding sequences of the Sox transcription factor (AAC AAT Gtt tAA CAA TGa aaA ACA ATG ttt TAC AAT Gaa aAT ATC AAT G, where capital letters indicate binding sequences and lowercase letters indicate linker sequences), the Sox and LEF transcription factor (AAC AAA Gtt aAA CAA AGt ttA ACA AAG aaa AAC AAA Gta tAA CAA AG), and the LEF transcription factor (AAC AAA aaa AAC AAA ttt AAC AAA aaa AAC AAA ttt AAC AAA). Synthetic siRNAs targeting Neurog1, Neurog2, Ctnnb1 and Neurod1 rat mRNA were purchased from Ambion (Silencer Select siRNA). The siRNA-targeting Sox2 was custom-synthesized by Ambion (custom Select siRNA).
Immunofluorescence studies
Immunofluorescence studies were performed as described27. We used mouse monoclonal antibody to beta-tubulin III (TUJ1, 1:500, Promega), antibody to Sox2 (1:300, Chemicon), goat antibody to NeuroD (1:100, Santa Cruz Biotechnology), mouse antibody to nestin (BD Biosciences), guinea pig antibody to GFAP (1:500, Advanced Immunochemical), mouse antibody to NeuN (1:60, clone A10), mouse monoclonal antibody to Wnt3a (1:100, Abcam), goat antibody to Wnt3 (1:100, Everest), mouse monoclonal antibody to S100β (1:200, Abcam), rabbit monoclonal antibody to calretinin (1:100, Abcam), rabbit antibody to Ki67 (1:150, Abcam), rabbit antibody to doublecortin (1:100, Santa Cruz Biotechnology), rabbit antibody to RUNX3 (1:250, Abcam), rat antibody to BrdU (1:250, Abcam) and DAPI (Wako). To examine the proliferative status of NeuroD1-expressing cells (Fig. 1), we injected Fisher 344 rats (7–8-week-old rats) with BrdU (100 mg per kg) once a day for 1 week and used the brain sections for BrdU staining analysis. For the immunostaining analysis of AC3 (Fig. 6), additional pretreatment of brain sections was conducted15. All secondary antibodies were from Jackson ImmunoResearch. Images were analyzed using a Bio-Rad Radiance confocal imaging system or Carl Zeiss LSM confocal imaging system.
qRT-PCR assays
For qRT-PCR assays, total RNA was extracted from cells by Isogen (Nippon Gene). After DNase I treatment (TURBO DNA-free, Ambion), cDNA was synthesized from 1 μg of purified RNA by the SuperScript II First-Strand cDNA synthesis system (Invitrogen), according to the manufacturer’s instructions. qRT-PCR was performed with a real-time PCR machine (Bio-Rad). The reported results of the qRT-PCR assays are the averages of two independent RNA preparations. The PCR cycling parameters were 94 °C for 2 min, 40 cycles at 94 °C for 15 s, 60 °C for 20 s and 72 °C for 40 s. Data analysis was performed using the comparative threshold cycle value (Ct) method.
Luciferase assay
Adult NSCs were electroporated with reporter plasmids by a nucleofector device (Amaxa). Neurod1-luciferase in a pGL2-luci plasmid (or mutant Neurod1-luciferase) and renilla luciferase (R-luc) as an internal control were co-electroporated into adult NSCs. We measured luciferase activity 48 h after electroporation in 50 μL of lysis supernatant with the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s protocol. The luminescence signal was quantified with a luminometer (Lumant LB 9501). The ORF2 region and 5′ UTR of human LINE-1.3 were cloned and inserted into CSC PW (the same constructs were used for the EGFP reporter in Fig. 8) at the site of the CMV promoter and into the pGL2-luci plasmid.
ChIP and RT-PCR
ChIP was performed using a kit following the manufacturer’s protocol (Upstate). Chromatin from cell extracts (input) or immunoprecipitated with specific antibodies was amplified by PCR using primers designed to flank the Sox/LEF-binding site in the rat Neurod1 promoter and LINE-1. For primary antibodies, we used antibody to HDAC1 (rabbit, Upstate), antibody to acetyl-histone H4 (rabbit, Upstate), antibody to acetyl-histone H3 (rabbit, Upstate), antibody to dimethyl-histone H3 (Lys4, mouse, Upstate), antibody to dimethyl-histone H3 (Lys9, mouse, Upstate), antibody to Sox2 (rabbit, Chemicon) and antibody to β-catenin (rabbit, Cell Signaling). RT-PCR was performed using total RNA extracted from adult hippocampus NSCs. A total of 1 μg RNA was used for first-strand cDNA synthesis with SuperScript II (GibcoBRL). PCR primer sequences are available on request.
Bioinformatics analysis
The genomic sequences for rat (rn3), human (hg17) and mouse (mm5) and the species-specific annotation for known protein coding genes and LINEs (annotated with RepeatMasker) were obtained from the University of California Santa Cruz public genome database (http://genome.ucsc.edu). Candidate binding sites for Sox2 (WWC AAW G) and TCF/LEF (WWC AAA) were identified on either strand of genomic sequence (W for either A or T and N for either A, C, G or T). Promoters were defined as the regions 6,000 bases upstream and 1,000 bases downstream of the annotated transcriptional start sites of protein coding genes. Full-length (at least 6 kb) LINE elements containing TCF/LEF or Sox2 candidate sites located in promoters were retained. We identified 79, 84 and 25 such LINE elements in the human, mouse and rat genomes, respectively (Supplementary Tables 1–3).
Retrovirus injection into adult hippocampus
All animal procedures were performed in accordance with protocols approved by the Institutional Animal Care and Use Committee of the National Institute of Advanced Industrial Science and Technology. We used retrovirus expressing a Cre-GFP fusion protein under the control of the Sox2 promoter1,26 (Sox2CREGFP). It has been determined that Sox2 promoter (3.1 kb) on the retrovirus recapitulates endogenous Sox2 expression similarly in CNS1. Rosa26-egfp mice (Jackson Laboratory), harboring egfp and a loxP-flanked stop codon, were crossed with Ctnnb1loxP/loxP mice (Jackson Laboratory). The Ctnnb1loxP/loxP allele contains loxP sites flanking exons 2–6 and has been shown to be a conditional null allele32. Transduction of Cre recombinase under the Sox2 promoter deleted the stop codon to activate GFP expression in Sox2-positive cells (control mice). We injected Sox2CRE GFP retrovirus into Ctnnb1loxP/loxP mice that were crossed with a Rosa26-egfp reporter line and traced GFP-positive cells that had stem cell–specific deletion of Ctnnb1loxP/loxP (β-catenin cKO mice). The retrovirus (1.0–1.5 μL) was injected into either control (Rosa26-egfp) or Ctnnb1loxP/loxP; Rosa26-egfp mice (n = 6 per group). The retrovirus was stereotactically injected into the dentate gyrus of the hippocampus (anterioposterior −2.5 mm, lateral ±2.0 mm and dorsoventral −2.0 to 2.5 mm from Bregma) with a 26-gauge stainless microinjection needle at a rate of 0.5 μl min−1. The needle remained in place for an additional 2 min to facilitate delivery of the virus. Starting 2 weeks after the retrovirus injection, we injected BrdU (100 mg per kg) once a day for 2 weeks. We killed the mice 3 weeks post-surgery and perfused them with 4% paraformaldehyde solution. Brain sections were cut at a thickness of 30 μm with a microtome (ROM-380, Yamato). An immunohistochemical analysis was performed using a confocal microscope.
Lentivirus injection into adult rat hippocampus
Concentrated lentivirus (1.5 μl) was injected stereotactically into the dentate gyrus of the hippocampus (anterioposterior −3.5 mm, lateral ±2.5 mm and dorsoventral −3.5 mm from Bregma) of adult female CD (Sprague-Dawley [SD]) rats (6–8 weeks old, n = 6 per group; Fig. 5) with a 26-gauge stainless microinjection needle at a rate of 0.5 μl min−1. To assess LINE-1 promoter activity with lentivirus encoding the LINE-1 ORF2 fragment fused to gfp (LINE-GFP; Fig. 8), we stereotactically injected lentivirus aliquots injected into the dentate gyrus of young adult rats (7–8 weeks old, n = 6). The needle remained in place for an additional 2 min to facilitate delivery of the virus. We injected BrdU (100 mg per kg) once a day for 1 week starting 2 weeks after the initial injection of the virus. The rats were killed 3 weeks post-infection and perfused with 4% paraformaldehyde. Brain sections were cut at a thickness of 30 μm with a microtome (ROM-380, Yamato). An immunohistochemical analysis was performed using a confocal microscope.
Supplementary Material
Acknowledgments
We thank T. Ohtake for assistance with animal care and supporting our in vivo experiments. We thank M. Namihira and J. Kohyama for assistance in obtaining immunostaining data, E. Mosser for the constitutively active β-catenin construct, and G. Canettieri and M. Montminy for HDAC1. We thank A. Huynh for help with immunohistochemical analysis. We are grateful for the technical assistance of B. Miller and to M.L. Gage for editorial comments. T.K., M.W. and M.A. were supported by various grants from National Institute of Advanced Industrial Science and Technology. T.K. was partly supported by the Grant-in-Aid for Exploratory Research. F.H.G. was supported by grants from the US National Institutes of Health (MH082070) and the G. Harold and Leila Y. Mathers Charitable Foundation.
Footnotes
Note: Supplementary information is available on the Nature Neuroscience website.
AUTHOR CONTRIBUTIONS
T.K., J.H., A.M., K.N. and F.H.G. conceptualized and designed the study. T.K., J.H., A.M. and F.H.G. analyzed the data. T.K. conducted the experiments with assistance from M.W., J.H. and L.M. G.Y. and A.M. conducted the bioinformatics analysis. A.M. helped design the LINE-1 plasmid constructs, D.C.L. helped with the Wnt plasmid design and constructs, and M.W. designed the shRNA construct. M.A. contributed reagents and analytical tools. T.K. wrote the paper with comments from all of the authors.
Reprints and permissions information is available online at http://www.nature.com/reprintsandpermissions/.
References
- 1.Suh H, et al. In vivo fate analysis reveals the multipotent and self-renewal capacities of Sox2+ neural stem cells in the adult hippocampus. Cell Stem Cell. 2007;1:515–528. doi: 10.1016/j.stem.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Song H, Stevens CF, Gage FH. Astroglia induce neurogenesis from adult neural stem cells. Nature. 2002;417:39–44. doi: 10.1038/417039a. [DOI] [PubMed] [Google Scholar]
- 3.Barkho BZ, et al. Identification of astrocyte-expressed factors that modulate neural stem/progenitor cell differentiation. Stem Cells Dev. 2006;15:407–421. doi: 10.1089/scd.2006.15.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lie DC, et al. Wnt signaling regulates adult hippocampal neurogenesis. Nature. 2005;437:1370–1375. doi: 10.1038/nature04108. [DOI] [PubMed] [Google Scholar]
- 5.Galceran J, Miyashita-Lin EM, Devaney E, Rubenstein JL, Grosschedl R. Hippocampus development and generation of dentate gyrus granule cells is regulated by LEF1. Development. 2000;127:469–482. doi: 10.1242/dev.127.3.469. [DOI] [PubMed] [Google Scholar]
- 6.Machon O, van den Bout CJ, Backman M, Kemler R, Krauss S. Role of beta-catenin in the developing cortical and hippocampal neuroepithelium. Neuroscience. 2003;122:129–143. doi: 10.1016/s0306-4522(03)00519-0. [DOI] [PubMed] [Google Scholar]
- 7.Solberg N, Machon O, Krauss S. Effect of canonical Wnt inhibition in the neurogenic cortex, hippocampus and premigratory dentate gyrus progenitor pool. Dev Dyn. 2008;237:1799–1811. doi: 10.1002/dvdy.21586. [DOI] [PubMed] [Google Scholar]
- 8.Wexler EM, Geschwind DH, Palmer TD. Lithium regulates adult hippocampal progenitor development through canonical Wnt pathway activation. Mol Psychiatry. 2008;13:285–292. doi: 10.1038/sj.mp.4002093. [DOI] [PubMed] [Google Scholar]
- 9.Clevers H. Wnt/β-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 10.Miyata T, Maeda T, Lee JE. NeuroD is required for differentiation of the granule cells in the cerebellum and hippocampus. Genes Dev. 1999;13:1647–1652. doi: 10.1101/gad.13.13.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu M, et al. Loss of BETA2/NeuroD leads to malformation of the dentate gyrus and epilepsy. Proc Natl Acad Sci USA. 2000;97:865–870. doi: 10.1073/pnas.97.2.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tozuka Y, Fukuda S, Namba T, Seki T, Hisatsune T. GABAergic excitation promotes neuronal differentiation in adult hippocampal progenitor cells. Neuron. 2005;47:803–815. doi: 10.1016/j.neuron.2005.08.023. [DOI] [PubMed] [Google Scholar]
- 13.Deisseroth K, et al. Excitation-neurogenesis coupling in adult neural stem/progenitor cells. Neuron. 2004;42:535–552. doi: 10.1016/s0896-6273(04)00266-1. [DOI] [PubMed] [Google Scholar]
- 14.Hsieh J, Nakashima K, Kuwabara T, Mejia E, Gage FH. Histone deacetylase inhibition–mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc Natl Acad Sci USA. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao Z, et al. Neurod1 is essential for the survival and maturation of adult-born neurons. Nat Neurosci. 2009;12:1090–1092. doi: 10.1038/nn.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishimoto M, Fukushima A, Okuda A, Muramatsu M. The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol Cell Biol. 1999;19:5453–5465. doi: 10.1128/mcb.19.8.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferri AL, et al. Sox2 deficiency causes neurodegeneration and impaired neurogenesis in the adult mouse brain. Development. 2004;131:3805–3819. doi: 10.1242/dev.01204. [DOI] [PubMed] [Google Scholar]
- 18.Yuan H, Corbi N, Basilico C, Dailey L. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 1995;9:2635–2645. doi: 10.1101/gad.9.21.2635. [DOI] [PubMed] [Google Scholar]
- 19.Collignon J, et al. A comparison of the properties of Sox-3 with Sry and two related genes, Sox-1 and Sox-2. Development. 1996;122:509–520. doi: 10.1242/dev.122.2.509. [DOI] [PubMed] [Google Scholar]
- 20.Ambrosetti DC, Basilico C, Dailey L. Synergistic activation of the fibroblast growth factor 4 enhancer by Sox2 and Oct-3 depends on protein-protein interactions facilitated by a specific spatial arrangement of factor binding sites. Mol Cell Biol. 1997;17:6321–6329. doi: 10.1128/mcb.17.11.6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Avilion AA, et al. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003;17:126–140. doi: 10.1101/gad.224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bylund M, Andersson E, Novitch BG, Muhr J. Vertebrate neurogenesis is counteracted by Sox1–3 activity. Nat Neurosci. 2003;6:1162–1168. doi: 10.1038/nn1131. [DOI] [PubMed] [Google Scholar]
- 23.Graham V, Khudyakov J, Ellis P, Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- 24.Bani-Yaghoub M, et al. Role of Sox2 in the development of the mouse neocortex. Dev Biol. 2006;295:52–66. doi: 10.1016/j.ydbio.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 25.Muotri AR, et al. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 26.D’Amour KA, Gage FH. Genetic and functional differences between multipotent neural and pluripotent embryonic stem cells. Proc Natl Acad Sci USA. 2003;100:11866–11872. doi: 10.1073/pnas.1834200100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gage FH, et al. Survival and differentiation of adult neuronal progenitor cells transplanted to the adult brain. Proc Natl Acad Sci USA. 1995;92:11879–11883. doi: 10.1073/pnas.92.25.11879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma Q, Kintner C, Anderson DJ. Identification of neurogenin, a vertebrate neuronal determination gene. Cell. 1996;87:43–52. doi: 10.1016/s0092-8674(00)81321-5. [DOI] [PubMed] [Google Scholar]
- 29.Farah MH, et al. Generation of neurons by transient expression of neural bHLH proteins in mammalian cells. Development. 2000;127:693–702. doi: 10.1242/dev.127.4.693. [DOI] [PubMed] [Google Scholar]
- 30.Guillemot F. Vertebrate bHLH genes and the determination of neuronal fates. Exp Cell Res. 1999;253:357–364. doi: 10.1006/excr.1999.4717. [DOI] [PubMed] [Google Scholar]
- 31.Hsieh J, Gage FH. Chromatin remodeling in neural development and plasticity. Curr Opin Cell Biol. 2005;17:664–671. doi: 10.1016/j.ceb.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 32.Brault V, et al. Inactivation of the β-catenin gene by Wnt1-Cre–mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development. 2001;128:1253–1264. doi: 10.1242/dev.128.8.1253. [DOI] [PubMed] [Google Scholar]
- 33.Yang N, Zhang L, Zhang Y, Kazazian HH., Jr An important role for RUNX3 in human LINE-1 transcription and retrotransposition. Nucleic Acids Res. 2003;31:4929–4940. doi: 10.1093/nar/gkg663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grove EA, Tole S, Limon J, Yip L, Ragsdale CW. The hem of the embryonic cerebral cortex is defined by the expression of multiple Wnt genes and is compromised in Gli3-deficient mice. Development. 1998;125:2315–2325. doi: 10.1242/dev.125.12.2315. [DOI] [PubMed] [Google Scholar]
- 35.Lee SM, Tole S, Grove E, McMahon AP. A local Wnt-3a signal is required for development of the mammalian hippocampus. Development. 2000;127:457–467. doi: 10.1242/dev.127.3.457. [DOI] [PubMed] [Google Scholar]
- 36.Shimogori T, VanSant J, Paik E, Grove EA. Members of the Wnt, Fz and Frp gene families expressed in postnatal mouse cerebral cortex. J Comp Neurol. 2004;473:496–510. doi: 10.1002/cne.20135. [DOI] [PubMed] [Google Scholar]
- 37.Gao X, Arlotta P, Macklis JD, Chen J. Conditional knock-out of beta-catenin in postnatal-born dentate gyrus granule neurons results in dendritic malformation. J Neurosci. 2007;27:14317–14325. doi: 10.1523/JNEUROSCI.3206-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jessberger S, et al. Dentate gyrus–specific knockdown of adult neurogenesis impairs spatial and object recognition memory in adult rats. Learn Mem. 2009;16:147–154. doi: 10.1101/lm.1172609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Praag H, Shubert T, Zhao C, Gage FH. Exercise enhances learning and hippocampal neurogenesis in aged mice. J Neurosci. 2005;25:8680–8685. doi: 10.1523/JNEUROSCI.1731-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Muotri AR, Zhao C, Marchetto MCN, Gage FH. Environmental influence on L1 retrotransposons in the adult hippocampus. Hippocampus. doi: 10.1002/hipo.20564. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miyoshi H, Takahashi M, Gage FH, Verma IM. Stable and efficient gene transfer into the retina using an HIV-based lentiviral vector. Proc Natl Acad Sci USA. 1997;94:10319–10323. doi: 10.1073/pnas.94.19.10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pfeifer A, Brandon EP, Kootstra N, Gage FH, Verma IM. Delivery of the Cre recombinase by a self-deleting lentiviral vector: efficient gene targeting in vivo. Proc Natl Acad Sci USA. 2001;98:11450–11455. doi: 10.1073/pnas.201415498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.