Abstract
Plants use sugars as signaling molecules and possess mechanisms to detect and respond to changes in sugar availability, ranging from the level of secondary signaling molecules to altered gene transcription. G-protein-coupled pathways are involved in sugar signaling in plants. The Arabidopsis thaliana Regulator of G-protein Signaling protein 1 (AtRGS1) combines a receptor-like seven transmembrane domain with an RGS domain, interacts with the Arabidopsis Gα subunit (AtGPA1) in a D-glucose-regulated manner, and stimulates AtGPA1 GTPase activity. We determined that AtRGS1 interacts with additional components, genetically defined here, to serve as a plasma membrane sensor for D-glucose. This interaction between AtRGS1 and AtGPA1 involves, in part, the seven-transmembrane domain of AtRGS1.
Keywords: Arabidopsis, AtRGS1, glucose sensing, heterotrimeric G protein complex, Regulator of G Signaling protein 1
1. Introduction
Signal transduction pathways mediated by sugars play roles in virtually all aspects of life and development for most organisms [1]. For example, in Saccharomyces cerevisiae, a complex network of sugar signaling pathways has been characterized, involving at least four receptors for glucose. These include both intracellular receptors, such as hexokinase-1, and cell surface receptors, including the hexose transporter-like proteins Snf3 and Rgt2 and the G protein-coupled receptor Gpr1 [2]. Despite the wealth of knowledge on sugar signaling pathways in S. cerevisiae, relatively little is known about the apical signaling elements or downstream pathways involved in sugar signaling in multicellular organisms. Animals use G-protein signaling for taste perception of sugars; in humans, this is accomplished via the gustducin-coupled T1R2-T1R3 heterodimer, and recent evidence indicates that this G protein signaling network is also expressed in the gut, where it regulates expression of the Na+-dependent glucose co-transporter protein SGLT1 [3–5].
Like S. cerevisiae, Arabidopsis thaliana has a simple repertoire of G protein signaling elements, one canonical Gα subunit (AtGPA1), one Gβ subunit (AGB1), at least two Gγ subunits and one Regulator of G Signaling protein (RGS1), AtRGS1 [6]. The Arabidopsis heterotrimeric G protein complex has been implicated in an array, potentially a mélange, of plant physiologies [7] such as abscisic acid signaling [8–15], biotic and abiotic stress [16–22], germination and early development [23–25], and glucose signaling. The involvement of G-protein signaling pathways in the response of plants to changes in glucose availability has previously been suggested by the phenotypes of heterotrimeric G-protein signaling mutants in response to chronic treatment with high sugar concentrations, which inhibit seed germination and arrest growth of wild-type seedlings [26–34]. For example, Arabidopsis thaliana Gα subunit (AtGPA1)-null mutants are hypersensitive to glucose during germination and seedling development [30–33]. AtRGS1, comprised of a C-terminal RGS domain coupled to an N-terminal domain with a predicted seven transmembrane (7TM) topology [27], interacts with the AtGPA1 at the plasma membrane and functions as a GTPase-activating protein (GAP) for AtGPA1 [27,35,36]. Several lines of evidence also indicate the involvement of AtRGS1 in sugar-mediated regulatory pathways in Arabidopsis. In Atrgs1-null mutants, seed germination and seedling development are insensitive to D-glucose [28,29], while overexpression of AtRGS1 results in hypersensitivity to glucose during seedling growth [27,37]. Treatment with D-glucose also alters the interaction of AtRGS1 and AtGPA1 [36]. However, G protein signaling is poorly characterized in Arabidopsis and other plants relative to what is known from yeast and animals systems, and relatively little is known about processes lying downstream of AtGPA1.
Here, we conclude that AtRGS1, a putative extracellular receptor for D-glucose, together with the heterotrimeric G protein complex mediates the steady-state level of transcripts from a small set of sugar-regulated genes in a G protein-coupled signaling network.
2. Materials and methods
2.1 Arabidopsis
All experiments were conducted using Arabidopsis thaliana of the Columbia ecotype. The generation and characterization of the majority of Arabidopsis lines containing T-DNA insertions and transgenic alleles used in these studies are described in the literature [27,36,38–40]. Gene accession numbers are: AGB1, At4g34460; AtGPA1, At2g26300; AtRGS1, At3g26090; THF1, At220890
2.2. Cultivation of Arabidopsis seedlings for gene chip and real-time PCR analysis
Arabidopsis seeds were sterilized by sequential treatments with 70% ethanol + 0.05% Triton-X (15 minutes), 95% ethanol + 0.05% Triton-X (5 minutes) and 95% ethanol (5 minutes) followed by air-drying in a sterile hood. Roughly 200 seeds per sample were then transferred to 250-mL flasks containing 50 mL ½ MS (Murashige and Skoog) medium (pH 5.7) + 50 mM D-glucose. The flasks were incubated in the dark for 2 days at 4°C and were then transferred to a shaker at 22°C under constant low light conditions and incubated for 7 days. To sugar starve seedlings, the media was replaced with ½ MS medium containing no D-glucose and the seedlings were grown on a shaker in the dark for 2 days. Following sugar starvation, seedlings were removed from the dark and incubated under constant low light on a shaker with ½ MS media containing the indicated concentrations of D-glucose (0–300 mM) for the indicated time periods. Following this incubation, the seedlings were snap-frozen in liquid nitrogen. Cyclohexamide treatments were performed as described by Scherer et al [41], except seedlings were grown as described above. Briefly, seedlings were treated with cycloheximide for 1 hour before sugar treatment, and then throughout the sugar treatment. Concentrations of cyclohexamide used were 1 ug/ul and 10 ug/ul. These concentrations provided virtually identical results; therefore, the results were pooled for the final analysis. Experiments using 1 ug/ul cyclohexamide were performed twice, and experiments using 10 ug/ul were performed three times.
2.3 Bimolecular Fluorescence Complementation (BiFC)
The coding sequences of AtRGS1 and the mutant version AtRGS1(E320K) were cloned into BiFC vector pBatL-sYFP-N to generate RGS1-sYFP-N and RGS1(E320K)-sYFP-N vectors. The coding sequences of GPA1, PIP2A and p31 (AT3G01290) were cloned into BiFC vector pBatL-sYFP-C to generate GPA1-sYFP-C, PIP2A-sYFP-C and p31-sYFP-C vectors. All vectors were transformed into Agrobacteria stain GV3101 (pMP90). Overnight-grown Agrobacteria were resuspend in infiltration solution (10 mM MES pH5.7, 10 mM MgCl,150 μM acetosyringone) to OD=1.5 and incubated at room temperature for 4 hours. The indicated pair of Agrobacteria and Agrobacteria harboring p19 to suppress gene silencing [42] were mixed and used to infiltrate the leaves of 4–5 week-old Nicotiana benthamiana. Four days after infiltration, leaves were detached from plants and observed under an Olympus IX 81epi-fluorescence microscope. Images were captured by a cooled charge-coupled device camera (Photometrics Cascade digital camera, Roper Scientific).
2.4 Expression Arrays
Wild-type and Atrgs1–1 seedlings were grown to analyze expression profiles as in Scheible et al. [43] and Osuna et al. [44] except that after 7 days the seedlings were transferred to a fresh medium that contained no sugar, rather than nitrogen, and after an additional 2 days 15 or 100 mM glucose was added to the starved seedlings. Quality controls, RNA preparation, dye swaps, and replications are as described by Scheible, et al [43]. Measurements of carbohydrates showed that the seedlings were carbon depleted (data not shown). Plant material was harvested, RNA prepared and used for hybridization of Affymetrix ATH1 arrays, and the raw Affymetrix data (CEL files) processed using the RMA (log scale Robust Multi-array Analysis) software as in Scheible et al. [43]. RMA is based on the Quantile normalization method and has better precision than MicroArray Suite 5.0 (Affymetrix, Santa Clara, CA) and dCHIP (http://www.dchip.org/), especially for low expression values [45,46]. In addition, all signals called `not present' by the Affymetrix MASC software were excluded from the data (and are marked as `A' in the table in the supplementary material). The data were also visualized and figures produced using MapMan software [47]. A downloadable version for local application and a servlet version are available at http://gabi.rzpd.de/projects/MapMan/.
2.5 RNA extraction and cDNA synthesis
RNA was extracted from Arabidopsis seedlings by use of the RNeasy Plant Mini Kit (Qiagen, Valencia, CA) according to the manufacturer's instructions. cDNA was generated using SuperScript III Reverse Transcriptase (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Briefly, 3 μg total RNA with dNTPs (Invitrogen, 0.5 mM final concentration) and Oligo(dT)20 oligomers (Invitrogen, 2.5 μM final concentration) was incubated at 65°C for 5 min. First-strand cDNA synthesis was then performed by adding SuperScript III Reverse Transcriptase (Invitrogen, 200 U), DTT (Invitrogen, 5 mM final concentration), RNAseOut (Invitrogen, 2 U) and RNAse-free water to a final volume of 20 μl and incubating the samples at 50°C for 45 minutes. The reactions were terminated by incubation at 70°C for 15 minutes. Following first-strand cDNA synthesis, 1 μl RNAse H was added to the reactions, and the samples were incubated for 30 minutes at 37°C and stored at −80°C.
2.6 Real-time PCR technique and analysis
A 69-bp fragment of the At4g01080 gene (GenBank accession no. NM_116338) was amplified to quantitate transcript levels in seedlings exposed to different treatments. Real-time PCR reactions were assembled in a total volume of 50 μl using 25 μl of SYBR GREEN PCR Master Mix (Applied Biosystems, Foster City, CA), 2 μl of cDNA from the 20 μl first-strand cDNA synthesis reactions and primers specific for At4g01080 or the reference gene tubulin beta-4 chain (TUB4; At5g44340) at final concentrations of 0.2 pmol/μl. Reactions were performed in triplicate. Primer sequences for At4g01080 were 5'- GAA GAA CAA ATG GTG GGC TT -3' and 5'- ATG CAG ATG AGA GAC TGG ACA -3'; primer sequences for tubulin beta-4 chain were 5'- AGA GGT TGA CGA GCA GAT GA -3' and 5'- ACC AAT GAA AGT AGA CGC CA -3'.
Real-time PCR was performed in a DNA Engine Opticon 2 System (Bio-Rad, Hercules, CA) using Opticon Monitor 3.1 software with the following thermocycler program: 2 min of preincubation at 94°C followed by 40–45 cycles of 15 s at 94°C, 15 s at 55°C, and 15 s at 72°C. SYBR Green dye fluorescence was monitored at the end of the annealing phase. A melting curve from 65°C to 95°C was used to confirm the presence of single products. All amplification curves were baseline-adjusted by subtracting the lowest fluorescence signal measured in each well over all cycles and the average of the blank wells (global minimum baseline adjustment in the Opticon Monitor 3.1 software). The threshold was set manually to a position at which signal intensities were low but had significantly surpassed levels and begun to increase exponentially, and the number of cycles required to reach this value, CT, was determined for each sample.
2.7 Quantification of relative gene expression from real-time PCR data
A general mathematical model was used to determine the ratio of the expression of a gene following two different treatments by real-time PCR and was applied to expression of At4g01080 in different Arabidopsis genetic lines following different glucose treatments. For these calculations, tubulin beta-4 chain was used as a reference gene. The basic equation describing the ratio calculation based on real-time PCR amplification data is:
where Ct is the threshold cycle number, ΔCt target (treatment1-treatment2) is the difference in Ct values for the target gene (At4g01080) between the two treatments being compared, Δ Ct ref (treatment1-treatment2) is the difference in Ct values for the reference gene (tubulin beta-4 chain) between the same two treatments, Etarget is the PCR efficiency for the target gene (E = 1 corresponds to 100% efficiency) and Eref is the PCR efficiency for the reference gene. E is assumed to be independent of N in the particular amplification range and was calculated by the Opticon Monitor 3.1 software from the slope of a plot of Ct vs. log N0:
2.8 Statistical analysis
The statistical significance of changes in mRNA induction between groups was assessed using an unpaired Student's t-test. P-values <0.05 were considered to be significant.
3. Results and discussion
3.1 AtRGS1 mediates D-glucose regulation of expression of a limited set of genes
To investigate genetic and structural requisites of G protein- and AtRGS1-mediated sugar signaling in Arabidopsis, we first compared the D-glucose-induced gene expression profiles of wild-type and Atrgs1–2 null seedlings. Glucose-starved seedlings were treated with 100 mM mannitol or two concentrations of D-glucose for 3 hours. The arrays were normalized using the Robust Multi-array Analysis software [45,46] and all signals called absent by the MASC software were excluded from the analysis. The raw data sets for the various treatments are provided in supplementary data (S1) and deposited at http://www.ncbi.nlm.nih.gov.geo/ with the series number [upon notice of acceptance]. As expected, the profiles in Atrgs1-null seedlings were similar to wild-type plants, with regression coefficients of 0.999 and 0.992 in control comparisons of sugar-starved seedlings and of seedlings that received 100 mM mannitol as an osmotic control. A small number of genes showed strong responses to 100 mM mannitol in both genotypes, indicating they respond to mild water deficits, and were excluded from subsequent analyses. The regression decreased slightly in the presence of 15 and 100 mM glucose (0.987 and 0.986, respectively).
Addition of glucose leads to dramatic changes in the steady-state level of transcripts from many genes involved in central metabolism in wild-type and Atrgs1-null seedlings. Direct comparison of the expression profiles for this set of ca. 2000 genes in 100 mM glucose revealed small differences between the two genotypes (supplemental material S2). Ten genes were identified that showed a marked attenuation of the response to glucose in Atrgs1-null seedlings (highlighted in Supplemental Data (S1)). The specific transcript levels of ten of these are shown in supplemental material S3. These include five that encode putative myrosinase-binding proteins (At1g52000, At1g52040, At1g54020, At2g39330, At5g48850), a predicted receptor kinase (At1g35710), a MYB transcription factor (At1g56650), a trehalose 6-P phosphatase (At1g78090) and two proteins of unknown function (At4g01080, At5g48850).
Of the transcripts identified to be differentially regulated in wild-type and Atrgs1-null seedlings, At4g01080 showed the strongest increase in wild-type plants in response to 100 mM D-glucose treatment, little response in Atrgs1-null plants and no response in either line to treatment with mannitol (Figure S3), and was thus selected on the basis of these characteristics as a candidate for a marker of AtRGS1-mediated sugar sensing. The At4g01080 gene encodes a 442 amino acid protein of unknown function with a predicted molecular weight of 50687.3. The At4g01080 gene product is predicted by TMHMM 2.0 (http://www.cbs.dtu.dk/services/TMHMM-2.0), DAS-TMfilter (http://www.enzim.hu/DAS/DAS.html) and SOSUI (http://bp.nuap.nagoya-u.ac.jp/sosui/) to contain a transmembrane domain (residues 51–73) with a cytoplasmic N-terminus, and is predicted by Plant-PLoc (http://chou.med.harvard.edu/bioinf/plant/) and PSORT (http://psort.hgc.jp/) to localize to the plasma membrane or extracellularly [48–55]. The At4g01080 protein contains an InterPro DUF231 domain (residues 256–430), as well as domains similar to those found in leaf senescence-related proteins from Arabidopsis and rice [56,57].
3.2 D-glucose induction of At4g1080 is time, dose, and AtRGS1 dependent
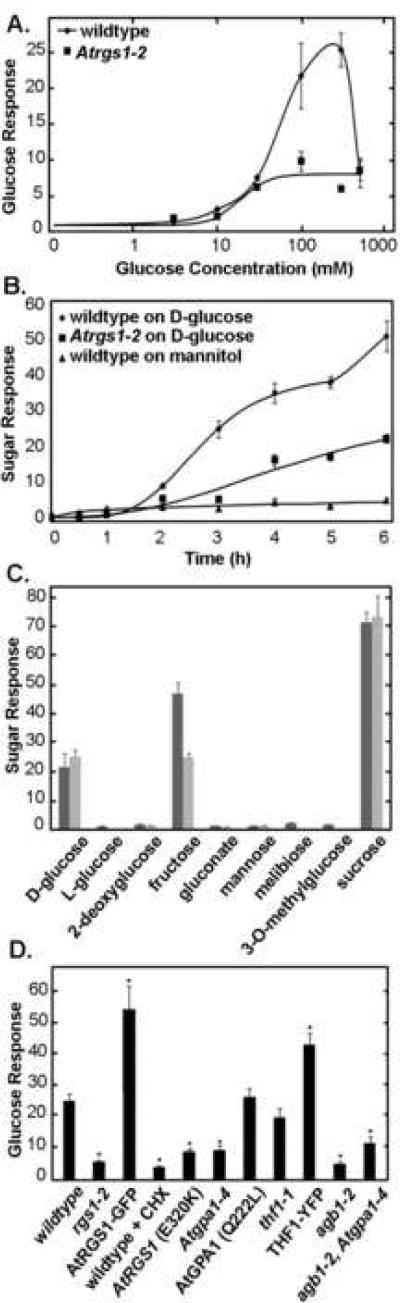
At4g01080 displayed a robust differential response to D-glucose between wild-type and Atrgs1-null plants (Figure 1A–B), with little or no response to treatment with mannitol (Figure 1B), confirming findings from the gene chip analysis. Above 10 mM glucose, the At4g01080 transcript level increased dramatically in wild-type plants, with the greatest increase over baseline levels observed in plants treated with 300 mM D-glucose (25.4-fold increase vs. control plants). Much smaller increases in At4g01080 transcript levels were observed in Atrgs1-null plants, where a roughly 6-fold increase was observed relative to untreated plants following incubation with 300 mM D-glucose. Three hundred millimolar glucose is commonly used in experiments linking altered sugar sensitivity to genotypes [28,31,32,40,58,59].
Fig. 1.
Induction of At4g01080 steady-state transcript level (Glucose/Sugar Response). (A) D-glucose dose-dependence of At4g01080 transcript increase by D-glucose. Wild-type and Atrgs1–2 7 day old seedlings were sugar-starved for 2 days and then treated with various concentrations of D-glucose for 3 hours. Wild-type is indicated here as Col-O and Atrgs1 null mutants indicated as rgs1–2. (B) Time dependency of At4g01080 transcript levels in response to treatment with D-glucose or mannitol. Wild-type and Atrgs1–2 7 day old seedlings were sugar-starved for 2 days and then treated with 300 mM D-glucose for varying time periods. (C) At4g01080 transcript level increase in response to a range of sugars and sugar analogues. At4g01080 transcript increase in response to treatment with various sugars and sugar analogues for 3 hours. Dark grey, 100 mM D-glucose; light grey, 300 mM D-glucose. (D) Regulation of At4g01080 transcript level in various genetic backgrounds in response to treatment with D-glucose. Treatment was 300 mM D-glucose for 3 h. rgs1–2: seedlings null for AtRGS1; RGS1-GFP: seedlings null for AtRGS1 and overexpressing an AtRGS1-GFP chimera; WT + CHX: wild-type seedlings treated with cycloheximide (1 or 10 μg/ml) for 1 hour before and during D-glucose treatment; RGS1-E320K: seedlings null for AtRGS1 over-expressing an AtRGS1 mutant containing mutation (E320K) that disrupts the interaction between AtRGS1 and AtGPA1; gpa1–4: seedlings null for AtGPA1; RGS-GFP + gpa1–4: seedlings null for AtRGS1 and AtGPA1 and over-expressing an AtRGS1-GFP chimera; GPA1 (Q222L): seedlings null for AtGPA1 overexpressing an AtGPA1 mutant containing mutation (Q222L), which results in a constitutively active form of the protein; thf1–1: seedlings null for THF1; THF1-YFP: seedlings over-expressing an THF1-YFP chimera; agb1–2: seedlings null for AGB1; agb1–2/gpa1–4: seedlings null for AGB1 and AtGPA1. * indicates values that are significantly different from wild-type plants (p < 0.01). (A–D) Bars or points represent means +/− S.E. After the indicated treatment, RNA was isolated from whole seedlings and used to generate cDNA using oligo dT primers as described in Materials and methods. These cDNA samples were then used for real-time PCR reactions with primers specific for the At4g01080 sequence and the TUB4 sequence (the reference) to determine the level of increase of the At4g01080 transcript level. Each mean is from at least three biological replicates with 3 internal replicates for each to assure precision. For the means presented in panel D, the number of replicates are: Col-O, eleven times1; rgs1–2, eight times; RGS1-GFP, six times; gpa1–4, three times; agb1–2, two times; agb1–2/gpa1–4, three times; GPA1(QL), two times; RGS1(E320K), six times; thf1–1, two times; cyclohexamide, five times.
At4g01080 transcript levels in wild-type plants increased substantially with incubations of 3 hours or longer. In Atrgs1-null plants, a 3-hour incubation with 300 mM D-glucose resulted in little change in At4g01080 mRNA levels above baseline; however, after 4 hours At4g01080 gene expression increased over the baseline levels in these plants (Figure 1B). The AtRGS1-independent increase in At4g01080 mRNA levels seen in Atrgs1-null plants at later time points is D-glucose-mediated, because treatment of wild-type plants with 300 mM mannitol as an osmotic control resulted in little increase in transcript level (Figure 1B).
At4g01080 is not a primary response gene because sugar induction of At4g01080 has a 2–3 hour lag period (Figure 1B) and because treatment with cycloheximide blocked D-glucose-mediated At4g01080 transcript level increase (Figure 1D). This is consistent with findings from Price et al. [60] demonstrating that gene induction by glucose requires protein translation on a global scale, while glucose gene repression is largely translation-independent.
3.3 AtRGS1-mediated At4g01080 transcription is sugar selective
Wild-type and Atrgs1-null seedlings displayed differential growth sensitivities to high concentrations of various sugars and sugar analogues that is dependent in part upon whether these molecules are able to be transported into plant cells, phosphorylated by hexokinases (HXKs) or metabolized [28]. In wild-type plants, monosaccharides (D-glucose, D-fructose) or a disaccharide (sucrose), which are transportable, metabolizable and phosphorylatable by HXKs, induced At4g01080 transcription (Figure 1C).
3.4 AtRGS1 regulates At4g01080 transcription in a dose-dependent manner
To better understand the signaling pathway involved in the AtRGS1-mediated transcriptional response to glucose, real-time PCR experiments were used to investigate the D-glucose-induced increase in At4g01080 transcript level in a number of G protein- and sugar-signaling-specific mutant genetic backgrounds. In agreement with the results from our gene chip analysis and from dose-response real-time-PCR experiments using wild-type and Atrgs1–2 plants, the induction of At4g01080 transcript level was significantly decreased in Atrgs1–2 seedlings relative to wild-type (Figure 1D; 6.0-fold induction for Atrgs1–2 vs. 25.4-fold induction for wild-type, p < 0.0001). In Atrgs1–2 plants over-expressing an AtRGS1-GFP construct (driven by a 35S cauliflower mosaic virus promoter and previously shown to rescue the Atrgs1–2 phenotype), At4g01080 transcript level was significantly increased relative to wild-type (54.9-fold induction, p = 0.0005), suggesting a dose-response effect for At4g01080 transcriptional or post-transcriptional regulation that is dependent upon the level of AtRGS1 protein expression.
3.5 At4g01080 transcript level regulation requires AtRGS1-AtGPA1 interaction, but does not require intrinsic GTPase activity
Since the best-described role for AtRGS1 is its function as a GAP for AtGPA1, we determined At4g01080 transcriptional levels following glucose treatment in Atrgs1–2 plants over-expressing an AtRGS1-GFP or AtRGS1 construct in which the AtRGS1 protein contains a mutation known to eliminate AtRGS1 GAP activity [36] and to eliminate interaction between AtRGS1 and AtGPA1 (Fig 1D, AtRGS1-E320K). The D-glucose-induced increase in At4g01080 transcript level was significantly decreased in these plants relative to wild-type (9.2-fold induction, p = 0.0002), suggesting a requirement for AtRGS1 GAP activity and/or an interaction of AtRGS1 with the Arabidopsis Gα subunit. In Atgpa1-null Arabidopsis seedlings (Atgpa1–4), the increase in At4g01080 transcript level was significantly less than in wild-type (9.7-fold induction, p = 0.0057); over-expression of AtRGS1 was unable to rescue this decrease in At4g01080 induction in Atgpa1–2 seedlings (9.6-fold induction, p = 0.0065). In Atgpa1–4 plants over-expressing a constitutively active form of AtGPA1 (AtGPA1-Q222L), At4g01080 transcript level was significantly increased over the levels observed in Atgpa1–4 plants (p = 0.0037) and similar to levels observed in wild-type plants (26.8-fold induction, p = 0.7787); thus, over-expression of AtGPA1-Q222L rescues the Atgpa1-null phenotype in our assay, consistent with previous findings that examined root growth phenotypes following chronic D-glucose exposure in these genotypes [32]. However, the AtGPA1-Q222L mutant lacks the intrinsic GTPase activity, leading to the conclusion that, while an interaction between AtRGS1 and AtGPA1 is critical for the At4g01080 transcript level increase, the intrinsic GAP activity AtRGS1, per se, is not critical for glucose induction of At4g01080 via AtGPA1. It does not preclude a role for the GAP function by AtRGS1 at later times in this signaling pathway.
To test this further we examined in vivo interaction between AtRGS1 and AtGPA1 using Bimolecular Fluorescence Complementation (BiFC) [61]. As shown in Figure 2, AtRGS1 and AtGPA1 –split YFP tagged proteins complement to reconstitute a fluorescent YFP. Interestingly, a tagged AtRGS1 (E320K) mutant also interacts with AtGPA1 using BiFC. Since this mutation has been shown to disrupt the interaction between AtGPA1 and the C-terminal RGS-box-containing- domain of AtRGS1, we conclude that the interaction occurs through the 7TM domain. It should be noted that BiFC is not a quantitative measure of interaction and that weak or strong transient interactions can drive stably-reconstituted YFP molecules [61].
Fig. 2.
Interaction between AtRGS1 and AtGPA1 through the 7-transmembrane (7TM) domain. Agrobacteria harboring RGS1-sYFP-N and GPA1-sYFP-C (A) and RGS1(E320K)-sYFP-N and GPA1-sYFP-C (B) were co-infiltrated into Nicotiana benthamiana leaves. Because it was previously shown that the E320K mutation in AtRGS1 disrupts interaction between the C-terminal-located RGS box and AtGPA1 [66], the in vivo interaction between AtGPA1 and AtRGS1 (E320K) shown here is likely occurring through the 7TM domain. The cell membrane proteins PIP2A (C) and p31 (D) were used as negative controls to monitor spontaneous re-association between N and C terminal halves of YFP. Because no fluorescence was visible, the images in C and D were taken at higher gain setting of camera than for A and B in order to visualize the cell outline. Bar=30 um.
3.6 The AtGPA1 interactor THF1 is involved indirectly in At4g01080 transcriptional regulation
RGS proteins attenuate Gα signaling via their GAP activities, but also can act as scaffolding proteins to bring together various components of a G-protein signaling complex [62,63]. Our earlier finding that AtGPA1 has rapid nucleotide exchange and is by default active at steady state [36] suggests an alternate form of regulation for the protein in Arabidopsis, possibly through selective localization via interaction with scaffolds such as the 7TM domain of AtRGS1. Furthermore, treatment with high concentrations of D-glucose promotes a transient change in conformation between AtGPA1 and AtRGS1, leading to increased FRET efficiency between fluorescently-labeled versions of these proteins [36]. A stable interaction between AtGPA1 and AtRGS1 via a scaffolding-like association where signaling is briefly allowed to proceed between the active Gα and its effector is consistent with the requirement of both AtRGS1 and AtGPA1 for glucose-enhanced At4g01080 transcript levels. The observed glucose-induced change in conformation between AtRGS1 and ATGPA1 is transient but this does not preclude the possibility that these two proteins are stably associated. We speculate that AtRGS1 would first promote signaling through a prebound AtGPA1 by facilitating the association of AtGPA1 with downstream partners. AtRGS1 would also deactivate AtGPA1 subunit by acting as a GAP protein to promote AtGPA1 GTP hydrolysis. The inactive AtGPA1 might remain associated with AtRGS1 during long-term treatments with glucose, albeit in a conformation that is not conducive to FRET [16], or that the scaffold-like docking site on AtRGS1 is only transiently available to AtGPA1 or its effector following glucose treatment. This would explain the opposing sugar sensitivity phenotypes displayed by Atrgs1-null and Atgpa1-null plants under conditions of chronic glucose exposure.
AtGPA1 associates with at least one other protein with a predicted scaffolding role, the plastid membrane protein THF1, which is itself regulated by D-glucose levels [32]. THF1 interacts with AtGPA1 in a nucleotide-independent manner at sites were plastids abut the plasma membrane. thf1-null mutants display variegated leaves and are hypersensitive to chronic exposure of glucose, while THF1-overexpressing plants are resistant to glucose [32,64]. Furthermore, THF1 protein levels are regulated by glucose, with high glucose concentrations leading to a proteasome-dependent degradation of the protein in roots [32]. The results observed here are consistent with the structure of AtRGS1 acting as a scaffold to facilitate an interaction between effector proteins and AtGPA1, perhaps newly released from its interaction with THF1 following D-glucose-mediated degradation of that protein. Equally plausibly, THF1 could be part of a glucose-mediated signaling complex with AtGPA1, aiding in or prolonging a transient scaffold-like association between AtGPA1 with AtRGS1 before THF1 is degraded. The hypersensitivity to chronic glucose treatment of both Atgpa1-null and thf1-null mutant plants would support the idea that THF1 acts to promote or prolong AtGPA1 signaling
To distinguish among these possibilities for the involvement of THF1 in AtRGS1-mediated glucose sensing, At4g01080 mRNA levels were assessed in thf1-null mutant plants before and after glucose treatment. In thf1–1 seedlings, there was a trend towards slightly higher basal level of At4g01080 mRNA compared to wild-type, but the difference was not significant (1.2-fold increase in thf1–1 vs. wild-type, p = 0.3827), suggesting that any increase in the pool of AtGPA1 available to interact with AtRGS1 resulting from deletion of THF1 had only a minor impact upon At4g01080 steady-state transcript levels under sugar-starved conditions. Increased availability of AtGPA1 to interact with AtRGS1 through sugar-mediated degradation of THF1, therefore, does not appear to be the primary mode of regulation for this glucose signaling pathway.
Instead, we speculate that THF1 may stabilize AtGPA1 interactions. Following glucose treatment of thf1–1 seedlings, the increase in At4g01080 transcription was less relative to wild-type, although the difference was again not considered significant by our criterion (Figure 1D; 20.1-fold induction, p = 0.3023). However, in plants over-expressing a THF1-GFP construct, At4g01080 mRNA levels were found to be significantly increased over wild-type levels following glucose treatment (43.6-fold induction, p = 0.0035). Thus, while not being absolutely required for glucose-mediated transcriptional regulation of At4g01080, THF1 does appear to play a role in this process. With regard to the scaffolding model for AtRGS1 proposed above, the minor decrease in At4g01080 mRNA levels in thf1–1 seedlings compared to wild-type following glucose treatment argues against THF1 recruitment of AtGPA1. The effects seen in THF1-YFP-overexpressing plants would instead suggest that THF1 promotes AtGPA1 signaling by enhancing a scaffold-like interaction between the active AtGPA1 and AtRGS1, either by inhibiting AtGPA1 deactivation via AtRGS1 GAP activity or by prolonging the availability of the docking site for AtGPA1 or its effector following glucose treatment.
3.7 The D-Glucose increase in At4g01080 steady-state transcript level requires AGB1
Another aspect of the mechanism of At4g01080 transcript up-regulation is that the process may require either the formation of a Gαβγ heterotrimer or a close association of an active Gα with the Gβγ dimer via a mechanism facilitated by AtRGS1. Based upon the in vitro rate constants observed for AtGPA1, which suggest that GTP hydrolysis rather than GDP/GTP exchange is the rate limiting step in the cycling between the active and inactive forms of the protein, it is predicted that more than 99% of AtGPA1 molecules would be present in the active form under steady state conditions [36]. Under such conditions, and in contrast to the case in metazoan systems, negative regulation of AtGPA1 through the GAP activity of AtRGS1 may be required to allow for the formation of appreciable amounts of αβγ heterotrimer. Therefore, in mutant Arabidopsis plants lacking AtRGS1 or AtGPA1, heterotrimer formation would not occur; thus, if normal regulation of At4g01080 transcripts requires the activity of the αβγ heterotrimer, a similar phenotype might be expected in both Atrgs1-and Atgpa1-null backgrounds. If either αβγ heterotrimer formation or the recruitment of Gα and the Gβγ dimer by AtRGS1 is required for regulation of At4g01080 gene expression, AGB1 expression would be necessary for this regulation, and agb1-null plants should display a phenotype for At4g01080 induction similar to that seen in the Atrgs1–2 and Atgpa1–4 mutants. In agb1–2 seedlings, the increase in At4g01080 transcript levels was attenuated relative to wild-type (Figure 1D; 5.4-fold induction, p = 0.0048). A similar phenotype was seen in Atgpa1–4, agb1–2 double mutant plants (11.9-fold induction, p = 0.0035).
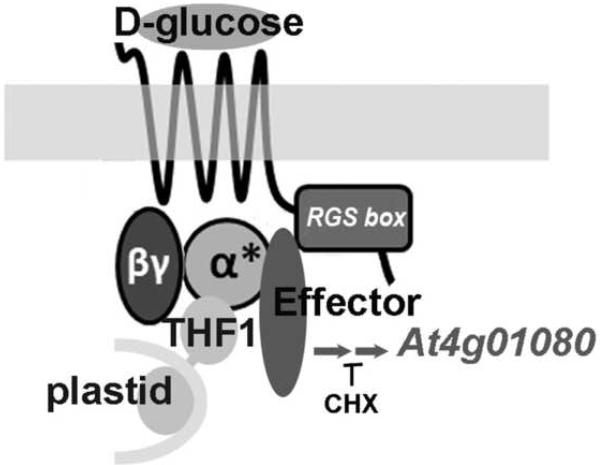
Taken together, these results demonstrate the involvement of AGB1 in glucose-mediated At4g01080 transcript level control, and lend support to the idea of signaling through the αβγ heterotrimer or through the combined signaling of Gα and the Gβγ dimer. The presence of a robust up-regulation of At4g01080 levels in plants with the constitutively active AtGPA1-Q222L mutant is still consistent with signaling through the heterotrimer as it has been shown that AtGPA1 Q222L remains a part of the heterotrimeric complex [65]. If signaling through both Gα and Gβ are important for glucose-induced At4g01080 gene induction while Gα GTPase is not necessary (as demonstrated by AtGPA1-Q222L), the critical role for AtRGS1 would again seem to be that of a networking protein, facilitating the interaction of Gα and the Gβγ dimer or, in the case that the Gαβγ heterotrimer does not dissociate, enhancing that interaction between the G protein heterotrimer and its downstream effector(s). In conclusion, the work here enables the assembly of some of the components of a novel glucose sensing complex at the plasma membrane (Figure 3).
Fig. 3.
A proposed physical model for a AtRGS1-G-protein sugar sensor based on the genetic data of Figure 1D. AtRGS1 is indicated by the 7-transmembrane protein containing the Regulator of G Signaling (RGS) motif in its carboxy-terminal cytoplasmic domain. The heterotrimeric G protein complex is associated with AtRGS1 and is represented by its G alpha (α) subunit and the G beta (β) and G gamma (γ) dimer. The activated form of a (α*) is known to be part of the heterotrimeric complex. A physical association between α and AtRGS1, between α and βγ, and between α* and THF1 have been shown previously. THF1 is a protein of the outer membrane of root cell plastids. The interaction interface has been mapped to the globular cytoplasmic domain on THF1. The effector has yet to be identified but is added here assuming that the effect of glucose activation of α* on gene transcription is not direct. CHX, cyclohexamide; At4g01080 encodes a plasma membrane protein of unknown function and the steady state level of its mRNA is used here as a rapid reporter of AtRGS1-mediated sugar sensing.
Structured summary.
MINT-6743118:
RGS1 (uniprotkb:Q8H1F2) and GPA1 (uniprotkb:P18064) physically interact (MI:0218) by bimolecular fluorescence complementation (MI:0809)
Supplementary Material
Acknowledgements
Work in A.M.J.'s lab on the Arabidopsis G proteins is supported by the NIGMS (GM065989-01), the DOE (DE-FG02-05ER15671), and the NSF (MCB-0718202 and MCB-0723515). Work in M.S.'s lab was supported by the Max Planck Society. We thank Ms. Jing Yang for her valuable technical assistance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Rolland F, Baena-Gonzalez E, Sheen J. Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu Rev Plant Biol. 2006;57:675–709. doi: 10.1146/annurev.arplant.57.032905.105441. [DOI] [PubMed] [Google Scholar]
- [2].Santangelo GM. Glucose signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2006;70:253–82. doi: 10.1128/MMBR.70.1.253-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Slone J, Daniels J, Amrein H. Sugar receptors in Drosophila. Curr Biol. 2007;17:1809–16. doi: 10.1016/j.cub.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Margolskee RF, et al. T1R3 and gustducin in gut sense sugars to regulate expression of Na+-glucose cotransporter 1. Proc Natl Acad Sci U S A. 2007;104:15075–80. doi: 10.1073/pnas.0706678104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Jang HJ, et al. Gut-expressed gustducin and taste receptors regulate secretion of glucagon-like peptide-1. Proc Natl Acad Sci U S A. 2007;104:15069–74. doi: 10.1073/pnas.0706890104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Temple BRS, Jones AM. The plant heterotrimeric G protein complex. Ann. Rev Plant Mol. Biol. 2007;58:249–266. doi: 10.1146/annurev.arplant.58.032806.103827. [DOI] [PubMed] [Google Scholar]
- [7].Perfus-Barbeoch L, Jones AM, Assmann SM. Plant heterotrimeric G protein function: insights from Arabidopsis and rice mutants. Current Opinion in Plant Biology. 2004;7:719–731. doi: 10.1016/j.pbi.2004.09.013. [DOI] [PubMed] [Google Scholar]
- [8].Chen J-G, Pandey S, Huang J, Alonso JM, Ecker JR, Assmann SM, Jones AM. GCR1 can act independently of heterotrimeric G-protein in response to brassinosteroids and gibberellins in Arabidopsis seed germination. Plant Physiol. 2004;135:907–915. doi: 10.1104/pp.104.038992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chen Y, Ji F, Xie H, Liang J, Zhang J. The regulator of G-protein signaling proteins involved in sugar and abscisic acid signaling in Arabidopsis seed germination. Plant Physiol. 2006;140:302–310. doi: 10.1104/pp.105.069872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Coursol S, Liu-Min F, Le Stunff H, Spiegel S, Gilroy S, Assmann SM. Sphingolipid signalling in Arabidopsis guard cells involves heterotrimeric G proteins. Nature. 2003;423:651–654. doi: 10.1038/nature01643. [DOI] [PubMed] [Google Scholar]
- [11].Fan L-M, Zhang W, Chen J-G, Taylor JP, Jones AM, Assmann SM. Abscisic acid regulation of guard-cell K + and anion channels in GÎ2- and RGS-deficient Arabidopsis lines. Proceedings of the National Academy of Sciences. 2008;105:8476–8481. doi: 10.1073/pnas.0800980105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Mishra G, Zhang W, Deng F, Zhao J, Wang X. A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science. 2006;312:264–266. doi: 10.1126/science.1123769. [DOI] [PubMed] [Google Scholar]
- [13].Pandey S, Assmann SM. The Arabidopsis putative G protein-coupled receptor GCR1 interacts with the G protein α subunit GPA1 and regulates abscisic acid signaling. Plant Cell. 2004;16:1616–1632. doi: 10.1105/tpc.020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pandey S, Chen J-G, Jones AM, Assmann SM. G-Protein complex mutants are hypersensitive to abscisic acid regulation of germination and postgermination development. Plant Physiol. 2006;141:243–256. doi: 10.1104/pp.106.079038. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [15].Wang X-Q, Ullah H, Jones AM, Assmann SM. G protein regulation of ion channels and abscisic acid signaling in Arabidopsis guard cells. Science. 2001;292:2070–2072. doi: 10.1126/science.1059046. [DOI] [PubMed] [Google Scholar]
- [16].Booker FL, Burkey KO, Overmyer K, Jones AM. Differential responses of G-protein Arabidopsis thaliana mutants to ozone. New Phytol. 2004;162:633–641. doi: 10.1111/j.1469-8137.2004.01081.x. [DOI] [PubMed] [Google Scholar]
- [17].Joo JH, Wang S, Chen JG, Jones AM, Fedoroff NV. Different signaling and cell death roles of heterotrimeric G Protein α and β subunits in the Arabidopsis oxidative stress response to ozone. Plant Cell. 2005;17:957–970. doi: 10.1105/tpc.104.029603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Llorente F, Alonso-Blanco C, Sanchez-Rodriguez C, Jorda L, Molina A. ERECTA receptor-like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina. The Plant Journal. 2005;43:165–180. doi: 10.1111/j.1365-313X.2005.02440.x. [DOI] [PubMed] [Google Scholar]
- [19].Trusov Y, Rookes JE, Chakravorty D, Armour D, Schenk PM, Botella JR. Heterotrimeric G proteins facilitate Arabidopsis resistance to necrotrophic pathogens and are involved in jasmonate signaling. Plant Physiol. 2006;140:210–220. doi: 10.1104/pp.105.069625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Trusov Y, et al. Heterotrimeric G protein γ subunits provide functional selectivity in Gβγ dimer signaling in Arabidopsis. Plant Cell. 2007;19:1235–1250. doi: 10.1105/tpc.107.050096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang S, Assmann SM, Fedoroff NV. Characterization of the Arabidopsis Heterotrimeric G Protein. J. Biol. Chem. 2008;283:13913–13922. doi: 10.1074/jbc.M801376200. [DOI] [PubMed] [Google Scholar]
- [22].Wang S, Narendra S, Fedoroff N. Heterotrimeric G protein signaling in the Arabidopsis unfolded protein response. PNAS. 2007;104:3817–3822. doi: 10.1073/pnas.0611735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Gao Y, Wang S, Asami T, Chen J-G. Loss-of-function mutations in the Arabidopsis heterotrimeric G-protein {alpha} subunit enhance the developmental defects of brassinosteroid signaling and biosynthesis mutants. Plant Cell Physiol. 2008;49:1013–1024. doi: 10.1093/pcp/pcn078. [DOI] [PubMed] [Google Scholar]
- [24].Ullah H, Chen J-G, Temple B, Boyes DC, Alonso JM, Davis KR, Ecker JR, Jones AM. The β subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes. Plant Cell. 2003;15:393–409. doi: 10.1105/tpc.006148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang HX, Perdue T, Weerasinghe R, Taylor JP, Cakmakci NG, Marzluff WF, Jones AM. A golgi hexose transporter is involved in heterotrimeric G protein regulated early development in Arabidopsis. Mol. Biol. Cell. 2006;17:4257–4269. doi: 10.1091/mbc.E06-01-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Trusov Y, et al. Heterotrimeric G protein gamma subunits provide functional selectivity in Gbetagamma dimer signaling in Arabidopsis. Plant Cell. 2007;19:1235–50. doi: 10.1105/tpc.107.050096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chen J-G, Willard FS, Huang J, Liang J, Chasse SA, Jones AM, Siderovski DP. A seven-transmembrane RGS rrotein that modulates plant cell proliferation. Science. 2003;301:1728–1731. doi: 10.1126/science.1087790. [DOI] [PubMed] [Google Scholar]
- [28].Chen JG, Jones AM. AtRGS1 function in Arabidopsis thaliana. Methods Enzymol. 2004;389:338–50. doi: 10.1016/S0076-6879(04)89020-7. [DOI] [PubMed] [Google Scholar]
- [29].Chen Y, Ji F, Xie H, Liang J, Zhang J. The regulator of G-protein signaling proteins involved in sugar and abscisic acid signaling in Arabidopsis seed germination. Plant Physiol. 2006;140:302–10. doi: 10.1104/pp.105.069872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ullah H, Chen JG, Young JC, Im KH, Sussman MR, Jones AM. Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science. 2001;292:2066–9. doi: 10.1126/science.1059040. [DOI] [PubMed] [Google Scholar]
- [31].Ullah H, Chen JG, Wang S, Jones AM. Role of a heterotrimeric G protein in regulation of Arabidopsis seed germination. Plant Physiol. 2002;129:897–907. doi: 10.1104/pp.005017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Huang J, Taylor JP, Chen JG, Uhrig JF, Schnell DJ, Nakagawa T, Korth KL, Jones AM. The plastid protein THYLAKOID FORMATION1 and the plasma membrane G-protein GPA1 interact in a novel sugar-signaling mechanism in Arabidopsis. Plant Cell. 2006;18:1226–38. doi: 10.1105/tpc.105.037259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pandey S, Chen JG, Jones AM, Assmann SM. G-protein complex mutants are hypersensitive to abscisic acid regulation of germination and postgermination development. Plant Physiol. 2006;141:243–56. doi: 10.1104/pp.106.079038. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [34].Wang HX, Weerasinghe RR, Perdue TD, Cakmakci NG, Taylor JP, Marzluff WF, Jones AM. A Golgi-localized hexose transporter is involved in heterotrimeric G protein-mediated early development in Arabidopsis. Mol Biol Cell. 2006;17:4257–69. doi: 10.1091/mbc.E06-01-0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Willard FS, Siderovski DP. Purification and in vitro functional analysis of the Arabidopsis thaliana regulator of G-protein signaling-1. Methods Enzymol. 2004;389:320–38. doi: 10.1016/S0076-6879(04)89019-0. [DOI] [PubMed] [Google Scholar]
- [36].Johnston CA, et al. GTPase acceleration as the rate-limiting step in Arabidopsis G proein-coupled sugar signaling. Proc. Nat. Acad. Sci. USA. 2007 doi: 10.1073/pnas.0704751104. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chen Y, Ji F, Xie H, Liang J. Overexpression of the regulator of G-protein signalling protein enhances ABA-mediated inhibition of root elongation and drought tolerance in Arabidopsis. J Exp Bot. 2006;57:2101–10. doi: 10.1093/jxb/erj167. [DOI] [PubMed] [Google Scholar]
- [38].Jones AM, Ecker JR, Chen JG. A reevaluation of the role of the heterotrimeric G protein in coupling light responses in Arabidopsis. Plant Physiol. 2003;131:1623–7. doi: 10.1104/pp.102.017624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ullah H, Chen JG, Temple B, Boyes DC, Alonso JM, Davis KR, Ecker JR, Jones AM. The beta-subunit of the Arabidopsis G protein negatively regulates auxin-induced cell division and affects multiple developmental processes. Plant Cell. 2003;15:393–409. doi: 10.1105/tpc.006148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Moore B, et al. Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science. 2003;300:332–6. doi: 10.1126/science.1080585. [DOI] [PubMed] [Google Scholar]
- [41].Scherer GFE, Zahn M, Callis J, Jones AM. A role for phospholipase A in auxin-regulated gene expression. FEBS Letters. 2007;581:4205–4211. doi: 10.1016/j.febslet.2007.07.059. [DOI] [PubMed] [Google Scholar]
- [42].Voinnet O, Rivas S, Mestre P, Baulcombe D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 2003;33:949–956. doi: 10.1046/j.1365-313x.2003.01676.x. [DOI] [PubMed] [Google Scholar]
- [43].Scheible W-R, et al. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 2004 doi: 10.1104/pp.104.047019. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Osuna D, et al. Temporal responses of transcripts, enzyme activities and metabolites after adding sucrose to carbon-deprived Arabidopsis seedlings. Plant J. 2007;49:463–91. doi: 10.1111/j.1365-313X.2006.02979.x. [DOI] [PubMed] [Google Scholar]
- [45].Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. mmaries of Affymetrix GeneChip probe level data. Nuc. Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- [47].Thimm O, et al. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004;37:914–939. doi: 10.1111/j.1365-313x.2004.02016.x. [DOI] [PubMed] [Google Scholar]
- [48].Cserzo M, Eisenhaber F, Eisenhaber B, Simon I. On filtering false positive transmembrane protein predictions. Protein Eng. 2002;15:745–52. doi: 10.1093/protein/15.9.745. [DOI] [PubMed] [Google Scholar]
- [49].Hirokawa T, Boon-Chieng S, Mitaku S. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 1998;14:378–9. doi: 10.1093/bioinformatics/14.4.378. [DOI] [PubMed] [Google Scholar]
- [50].Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–6. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- [51].Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–80. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- [52].Sonnhammer EL, von Heijne G, Krogh A. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol. 1998;6:175–82. [PubMed] [Google Scholar]
- [53].Chou KC, Shen HB. Large-scale plant protein subcellular location prediction. J Cell Biochem. 2007;100:665–78. doi: 10.1002/jcb.21096. [DOI] [PubMed] [Google Scholar]
- [54].Chou KC. Using amphiphilic pseudo amino acid composition to predict enzyme subfamily classes. Bioinformatics. 2005;21:10–19. doi: 10.1093/bioinformatics/bth466. [DOI] [PubMed] [Google Scholar]
- [55].Shen HB, Chou KC. Ensemble classifier for protein fold pattern recognition. Bioinformatics. 2006;22:1717–22. doi: 10.1093/bioinformatics/btl170. [DOI] [PubMed] [Google Scholar]
- [56].Zdobnov EM, Apweiler R. InterProScan--an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–8. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- [57].Servant F, Bru C, Carrere S, Courcelle E, Gouzy J, Peyruc D, Kahn D. ProDom: automated clustering of homologous domains. Brief Bioinform. 2002;3:246–51. doi: 10.1093/bib/3.3.246. [DOI] [PubMed] [Google Scholar]
- [58].Cho YH, Yoo SD, Sheen J. Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell. 2006;127:579–89. doi: 10.1016/j.cell.2006.09.028. [DOI] [PubMed] [Google Scholar]
- [59].Yanagisawa S, Yoo SD, Sheen J. Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature. 2003;425:521–5. doi: 10.1038/nature01984. [DOI] [PubMed] [Google Scholar]
- [60].Price J, Laxmi A, Martin SK, Jang JC. Global transcription profiling r eveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell. 2004;16:2128–50. doi: 10.1105/tpc.104.022616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Bhat RA, Lahaye T, Panstruga R. The visible touch: in planta visualization of protein-protein interactions by fluorophore-based methods. Plant Methods. 2006;2:12. doi: 10.1186/1746-4811-2-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Hepler JR. RGS protein and G protein interactions: a little help from their friends. Mol Pharmacol. 2003;64:547–9. doi: 10.1124/mol.64.3.547. [DOI] [PubMed] [Google Scholar]
- [63].Abramow-Newerly M, Roy AA, Nunn C, Chidiac P. RGS proteins have a signalling complex: interactions between RGS proteins and GPCRs, effectors, and auxiliary proteins. Cell Signal. 2006;18:579–91. doi: 10.1016/j.cellsig.2005.08.010. [DOI] [PubMed] [Google Scholar]
- [64].Wang Q, Sullivan RW, Kight A, Henry RL, Huang J, Jones AM, Korth KL. Deletion of the chloroplast-localized Thylakoid formation1 gene product in Arabidopsis leads to deficient thylakoid formation and variegated leaves. Plant Physiol. 2004;136:3594–604. doi: 10.1104/pp.104.049841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Adjobo-Hermans MJ, Goedhart J, Gadella TW., Jr. Plant G protein heterotrimers require dual lipidation motifs of Galpha and Ggamma and do not dissociate upon activation. J Cell Sci. 2006;119:5087–97. doi: 10.1242/jcs.03284. [DOI] [PubMed] [Google Scholar]
- [66].Johnston CA, et al. GTPase acceleration as the rate-limiting step in Arabidopsis G protein-coupled sugar signaling. Proceedings of the National Academy of Sciences. 2007;104:17317–17322. doi: 10.1073/pnas.0704751104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.