Abstract
Asymmetric divisions of radial glial progenitors produce self-renewing radial glia and differentiating cells simultaneously in the ventricular zone (VZ) of the developing neocortex. While differentiating cells leave the VZ to constitute the future neocortex, renewing radial glial progenitors stay in the VZ for subsequent divisions. The differential behaviour of progenitors and their differentiating progeny is essential for neocortical development; however, the mechanisms that ensure these behavioural differences are unclear. Here we show that asymmetric centrosome inheritance regulates the differential behaviour of renewing progenitors and their differentiating progeny. Centrosome duplication in dividing radial glial progenitors generates a pair of centrosomes with differently aged mother centrioles. During peak phases of neurogenesis, the centrosome retaining the old mother centriole stays in the VZ and is preferentially inherited by radial glial progenitors, whereas the centrosome containing the new mother centriole mostly leaves the VZ and is largely associated with differentiating cells. Removal of Ninein, a mature centriole-specific protein, disrupts the asymmetric segregation and inheritance of the centrosome and causes premature depletion of progenitors from the VZ. These results suggest that preferential inheritance of the centrosome with the mature older mother centriole is required for maintaining radial glial progenitors in the developing mammalian neocortex.
Radial glial cells constitute a major population of neural progenitor cells that occupy the proliferative ventricular zone (VZ) in the developing mammalian neocortex 1–3. In addition to their well-characterized function as a scaffold in supporting neuronal migration 4, radial glial cells display interkinetic nuclear oscillation and proliferate extensively at the luminal surface of the VZ (i.e. the VZ surface). During the peak phase of neurogenesis (around embryonic day 13 to 18, E13–E18, in mice), they predominantly undergo asymmetric division to self-renew while simultaneously giving rise either directly to a neuron, or to an intermediate progenitor cell (IPC) that subsequently divides symmetrically to produce neurons 5–8. While differentiating progeny progressively migrate away from the VZ to form the cortical plate (CP) – the future neocortex, renewing radial glial progenitors remain in the VZ for subsequent divisions. The distinct migratory behaviour of radial glial progenitors and their differentiating progeny is fundamental to the proper development of the mammalian neocortex; however, little is known about the basis of these behavioural differences.
Centrosomes, the main microtubule-organizing centres (MTOCs) in animal cells 9, play an important role in many cell processes, particularly during cell division 10 and cell migration 11–13. All normal animal cells initially inherit one centrosome, consisting of a pair of centrioles surrounded by an amorphous pericentriolar material (PCM). The two centrioles differ in their structure and function 9, 14, 15. The older “mother” centriole, which is formed at least one and a half generations earlier, possesses appendages/satellites that bear specific proteins, such as Cenexin/Odf2 16, 17 and Ninein 18–20, and anchor microtubules and support ciliogenesis 9, 21. In contrast, the younger “daughter” centriole, which is formed during the preceding S phase, lacks these structures. Full acquisition of appendages/satellites by the daughter centriole is not achieved until at least one and a half cell cycles later 22, 23. During each cell cycle, the centrosome replicates once in a semi-conservative manner 24, resulting in the formation of two centrosomes – one of which retains the original old mother centriole (i.e. the mother centrosome) while the other receives the new mother centriole (i.e. the daughter centrosome) 14, 15. This intrinsic asymmetry in the centrosome has recently been demonstrated to be important for proper spindle orientation during the division of male germline stem cells 25, 26 and neuroblasts 27, 28 in Drosophila, although female germline stem cells appear to divide normally in the absence of centrioles/centrosomes 29. These studies indicate a critical role for the differential behaviour of centrosomes with differently aged mother centrioles in asymmetric division of the progenitor/stem cells 30–33, although it remains unclear whether proper behaviour and development of the progenitor/stem cells and their differentiating daughter cells depend on centrosome asymmetry. Asymmetric division of radial glial progenitors accounts for nearly all neurogenesis in the developing mammalian neocortex 5–8. Three out of four autosomal recessive primary microcephaly (MCPH) genes identified so far encode centrosomal components 34, suggesting that proper neocortical neurogenesis and development entail a tight regulation of the centrosome 35, which is so far poorly understood. To address these issues, we investigated centrosome regulation during the peak phase of mammalian neocortical neurogenesis (Supplementary Fig. 1).
Centriole and centrosome asymmetry
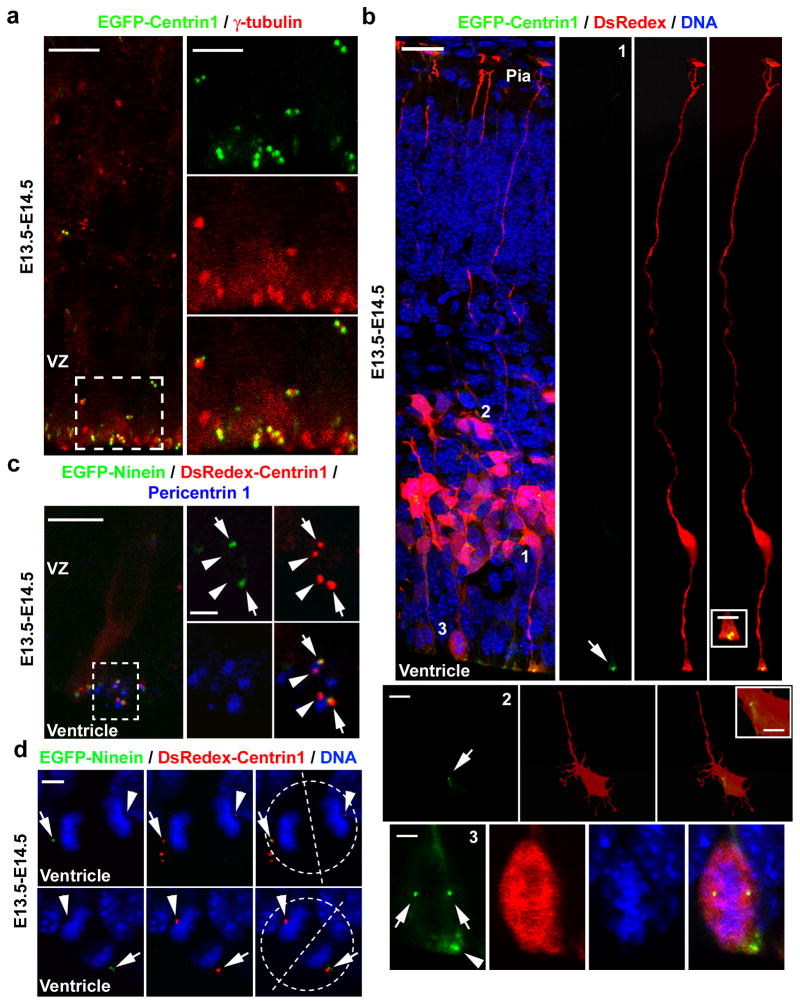
To examine centrosome behaviour, we introduced a plasmid encoding Centrin1, a central component of the centriole, fused with enhanced green fluorescent protein (EGFP-Centrin1) into the developing neocortex of E13.5 mouse embryos by in utero electroporation (Supplementary Fig. 2a). As expected, EGFP-Centrin1 formed pairs of dots that co-localized with γ-Tubulin, a centrosomal marker (Figure 1a), suggesting that transient expression of EGFP-Centrin1 reliably labels the two centrioles of individual centrosomes in the developing neocortex in vivo. Moreover, we observed that at the onset of peak neurogenesis (E13–E14), EGFP-Centrin1-labelled centrosomes were predominantly located at the VZ surface with a small subset located in the subventricular zone (SVZ) and the intermediate zone (IZ) (Fig. 1a and Supplementary Fig. 2b).
Figure 1. Centriole and centrosome asymmetry in the developing neocortex.
(a) Images of E14.5 cortices electroporated with EGFP-Centrin1 (green) at E13.5 (E13.5–E14.5) and immunostained for γ-Tubulin (red). (b) Images of cortices electroporated with EGFP-Centrin1 (green) and DsRedexpress (DsRedex, red) and counterstained with DAPI (blue). Arrows and the arrowhead indicate the centrosomes. (c) Images of cortices electroporated with EGFP-Ninein (green, arrows) and DsRedex-Centrin1 (red, arrows and arrowheads) and immunostained for Pericentrin1 (blue). (d) Images of dividing radial glial cells in late mitosis (broken circles) with condensed chromosomes (DAPI, blue) expressing EGFP-Ninein (green, arrows) and DsRedex-Centrin1 (red, arrows and arrowheads). Broken lines indicate the cleavage plane. Scale bars: 10 μm and 5 μm (a); 25 μm, 2.5 μm, 10 μm, 5 μm, and 5 μm (from top to bottom, b); 20 μm and 2 μm (c); 5 μm (d).
To identify the cell types harbouring EGFP-Centrin1-labelled centrosomes, we co-electroporated a plasmid encoding DsRedexpress (DsRedex), a red fluorescence protein that diffuses throughout cells and thereby reveals their morphology (Fig. 1b). We found that in bipolar radial glial progenitors in the VZ the centrosome was located in their ventricular endfeet at the VZ surface (Fig. 1b, cell 1) as previously suggested 36, 37, whereas in multipolar cells in the IZ and the SVZ the centrosome was harboured in their cell bodies (Fig. 1b, cell 2). Moreover, we observed dividing radial glial progenitors that possess a pair of centrosomes together with condensed chromosomes at the VZ surface (Fig. 1b, cell 3). Consistent with this differential centrosome localization between radial glial progenitors and their differentiating daughter cells, we observed a progressive increase in the appearance of EGFP-Centrin1-labelled centrosomes in the IZ and the CP as development proceeded, in addition to some that remained at the VZ surface (Supplementary Fig. 2 b and c). This gradual increase in centrosome localization in the IZ and the CP coincided with the production and migration of differentiating cells such as neurons to these regions during this period.
The distinct positioning of the centrosome in radial glial progenitors versus their differentiating progeny prompted us to ask whether the centrosomes in these two cell populations/types are different. To explore this, we electroporated a plasmid encoding Ninein, a mature centriole-specific protein that localizes to appendages/satellites 18, 19, fused with EGFP (EGFP-Ninein) together with a plasmid encoding Centrin1 fused to DsRedex (DsRedex-Centrin1) into the developing mouse neocortex at E13.5. As expected, both EGFP-Ninein and DsRedex-Centrin1 formed dot-like structures and co-localized to the centrosomes, especially those at the VZ surface, as identified by an antibody to the integral centrosomal protein Pericentrin1 (Fig. 1c). Interestingly, EGFP-Ninein was preferentially concentrated at one of the two centrioles marked by DsRedex-Centrin1 in individual centrosomes (Fig. 1c), suggesting that the two centrioles in interphase radial glial progenitors are not identical. Given that Ninein specifically associates with mature centrioles 18–20, these results indicate that the centriole with abundant EGFP-Ninein is the more mature mother centriole, whereas the one with little EGFP-Ninein is the less mature daughter centriole. A similar inequity in the recruitment of EGFP-Ninein by the duplicated centrosomes was also observed in dividing radial glial progenitors at the VZ surface (Fig. 1d), indicating that the duplicated centrosomes are not identical during late mitosis. The centrosome with abundant EGFP-Ninein is probably the centrosome that retains the mature old mother centriole and the centrosome with little EGFP-Ninein is likely the centrosome that bears the relatively immature new mother centriole.
Having found that the centrosomes in dividing radial glial progenitors exhibit asymmetry in their maturity, we next asked whether this centrosome asymmetry is related to the distinct behaviour of radial glial progenitors and their differentiating progeny in the developing neocortex during neurogenesis. To address this, we compared the relative distribution of centrosomes labelled by DsRedex-Centrin1 versus those labelled by EGFP-Ninein in the developing neocortex as development proceeded (Supplementary Fig. 3). Interestingly, while DsRedex-Centrin1-labelled centrosomes progressively occupied the IZ and the CP, where differentiating cells are situated, EGFP-Ninein-labelled centrosomes were mostly found in the VZ, where radial glial progenitors are located. Given that DsRedex-Centrin1 labels all centrosomes whereas EGFP-Ninein selectively labels mature centrosomes, these results point to an intriguing possibility that the duplicated centrosomes in dividing radial glial cells are differentially inherited depending on their age and maturity during neocortical neurogenesis. It is known that during each cell division, one centrosome retains the old mature mother centriole and the other bears the new less mature mother centriole14, 15, 22, 38. Thus, these results suggest that centrosomes with differently aged mother centrioles are differentially inherited by the two daughter cells of asymmetrically dividing radial glial progenitors.
In vivo pulse-chase labelling of centrosomes
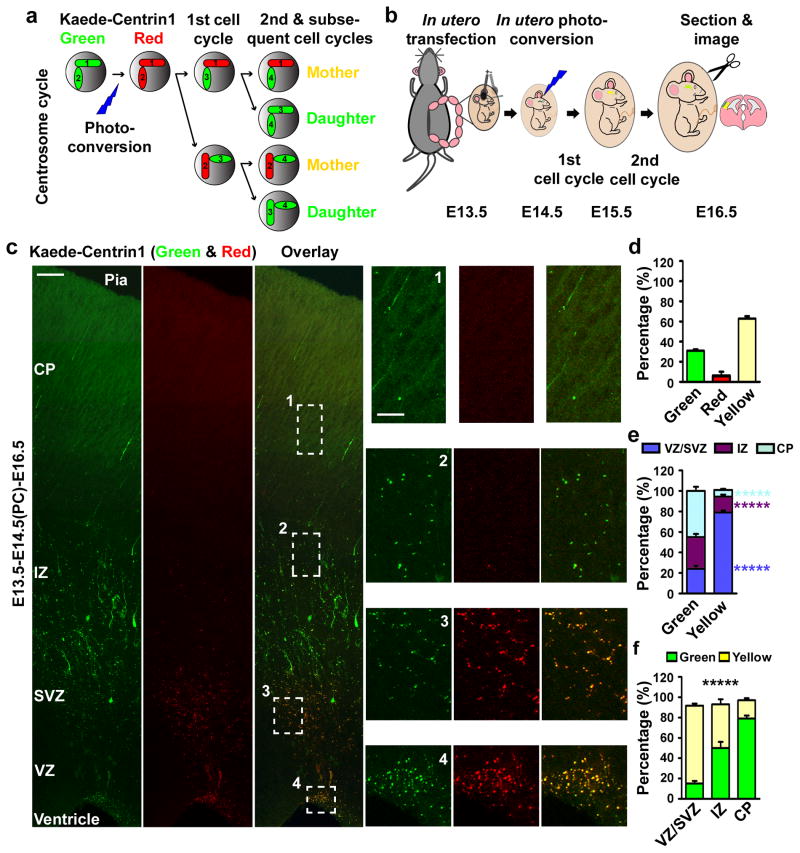
To test this, we first developed an assay to explicitly distinguish between the centrosome containing the old mother centriole and the centrosome containing the new mother centriole in the developing neocortex in vivo (Fig. 2a and b). The assay takes advantage of the photo-convertible fluorescent protein, Kaede 39, which changes from green to red fluorescence upon exposure to violet light. Centrioles in the developing neocortex were labelled by transient expression of Centrin1 fused with Kaede (Kaede-Centrin1). Photo-conversion was then performed to switch labelled centrioles from green to red fluorescence. This green-to-red fluorescence conversion of Kaede proteins is irreversible and the red protein is very stable 39, thus allowing long-term tracking of the existing photo-converted proteins and the structures with which they are associated. Moreover, all newly synthesized Kaede proteins are green fluorescent. It is known that centriole duplication requires new protein synthesis of Centrin 40. As a result, newly duplicated centrioles that contain newly synthesized Kaede-Centrin1 are green fluorescent, while previously existing centrioles are red fluorescent. Hence, in the first cell cycle after photo-conversion, both centrosomes contain a red fluorescent mother centriole and a green fluorescent daughter centriole. However, in the second and subsequent cell cycles, centrosomes with the new mother centriole contain only green fluorescent centrioles, whereas centrosomes retaining the original old mother centriole harbour both red and green fluorescent centrioles, thus distinguishing between centrosomes with differently aged mother centrioles (Fig. 2a).
Figure 2. Asymmetric segregation of centrosomes with differently aged mother centrioles.
(a, b) Strategy and experimental procedure for using Kaede-Centrin1 to distinguish between centrosomes with differently aged mother centrioles. (c) Images of E16.5 cortices electroporated with Kaede-Centrin1 at E13.5 and photo-converted (PC) at E14.5 (E13.5-E14.5(PC)-E16.5). Scale bars: 50 μm and 15 μm. (d–f) Quantifications of the percentage of labelled centrosomes that are green, red, or yellow fluorescent (d), the percentage of green or yellow fluorescent centrosomes that are located in different regions of the developing neocortex (e), and the percentage of labelled centrosomes located in different regions of the developing neocortex that are green or yellow fluorescent (f) (total 4,314 centrosomes from seven individual animals). Data are shown as mean±s.e.m.; *****, p<5e-5.
To carry out this assay in the developing neocortex in vivo, we developed an in utero photo-conversion procedure and combined it with in utero electroporation (Fig. 2b). Kaede-Centrin1, which localized specifically to the centrosomes (Supplementary Fig. 4), was introduced into the developing mouse neocortex at E13.5. One day later, i.e. E14.5, the forebrain of electroporated embryos was treated with a short exposure of violet light while still in the uterus, which effectively converted nearly all Kaede-Centrin1 proteins and their labelled centrosomes from green to red fluorescence (E13.5–E14.5(PC), Supplementary Fig. 5 a and b). The uterus was re-placed and the embryos continued to develop in vivo. The localization and inheritance of centrosomes were analyzed at different developmental stages thereafter.
We found that one day after photo-conversion (E13.5-E14.5(PC)-E15.5), around 95% of centrosomes contained both red and green fluorescent centrioles (indicated by yellow colour in the merged image) (Supplementary Fig. 5 c and d), consistent with the notion that the labelled cells have undergone one round of division and have duplicated their centrioles during the 24-hour period following photo-conversion. This was shown directly by imaging centrosomes at high magnification (Supplementary Fig. 6), revealing that each centriole was mostly only red or green fluorescence. This also demonstrates that there is little diffusion of Centrin proteins between duplicated centrioles, or between the centrioles and a cytoplasmic pool which was confirmed by fluorescence recovery after photo-bleaching (FRAP) experiments (Supplementary Fig. 7). Moreover, we found that more than 30% of centrosomes possessed only green fluorescent centrioles two days after photo-conversion (E13.5-E14.5(PC)-E16.5) (Fig. 2c and d). The appearance of the solely green fluorescence centrosomes 48 hours after photo-conversion indicates that the initially labelled radial glial progenitors have undergone two rounds of division during this period; this is consistent with the prior observation that the duration of the neocortical progenitor cell cycle is about 12 to 20 hours around this developmental stage 41. The ongoing division of labelled radial glial cells at a normal rate suggests that expression of Kaede-Centrin1 and the photo-conversion procedure had no effect on their cell cycle. In addition, no obvious DNA damage or cell death was induced by the photo-conversion treatment (Supplementary Fig. 8). Besides the green and yellow fluorescent centrosomes, we observed about 4% of solely red fluorescent centrosomes (Fig. 2d), indicating that a few labelled cells do not undergo cell division during this period.
Asymmetric segregation and inheritance of centrosomes
Having successfully distinguished the centrosomes with differently aged mother centrioles in the developing neocortex in vivo, we next examined their distribution to determine whether they are asymmetrically segregated. Remarkably, we found that more than 76% of centrosomes with the new mother centriole (i.e. only green fluorescent) were located in the IZ and the CP, whereas around 78% of centrosomes with the old mother centriole (i.e. both green and red fluorescent) were located in the VZ in addition to the SVZ (Fig. 2c, e and f). These results demonstrate that the centrosomes with differently aged mother centrioles are asymmetrically segregated in the developing neocortex during the peak phase of neurogenesis. It is worth noting that a small fraction of both green and red (i.e. yellow) fluorescent centrosomes was found in the IZ and the CP (Fig. 2e and f) and that these centrosomes likely originated from the first cell cycle after photo-conversion (Supplementary Fig. 5 c and d).
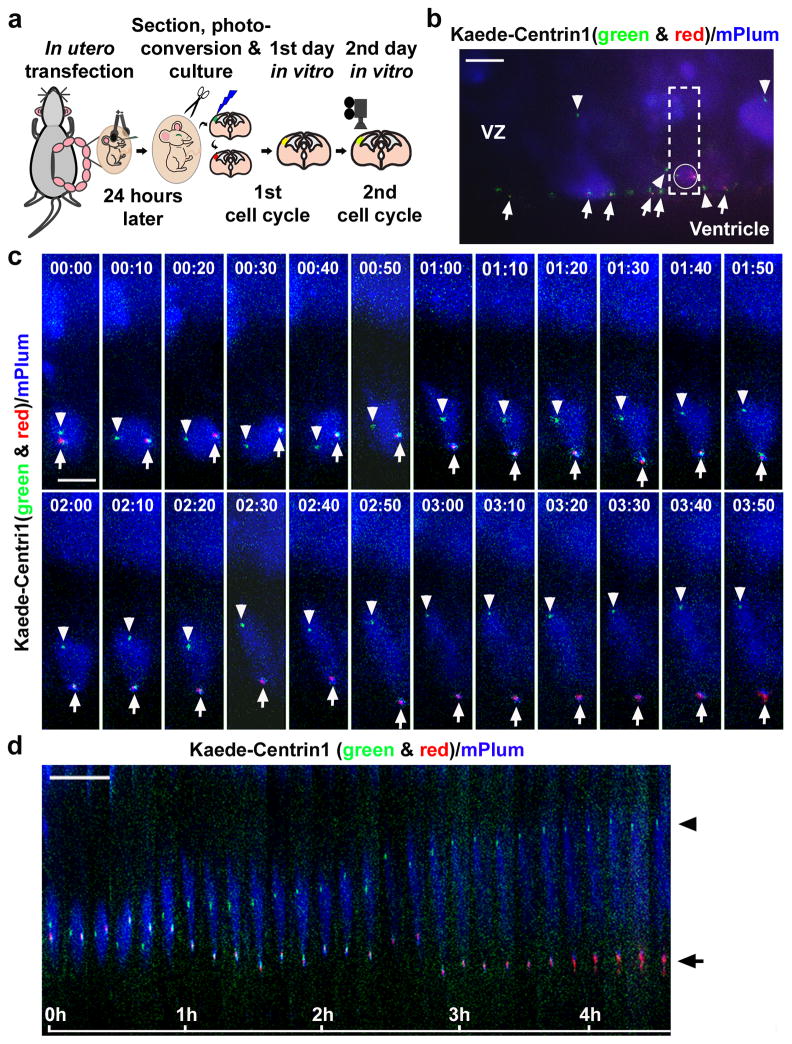
The asymmetric segregation of centrosomes suggests differential regulation of the duplicated centrosomes in dividing radial glial progenitors. To gather further evidence for this, we carried out time-lapse imaging experiments to monitor the behaviour of centrosomes with differently aged mother centrioles in dividing radial glial progenitors at the VZ surface in situ (Fig. 3a). Kaede-Centrin1 was introduced into radial glial cells together with mPlum, a far-red fluorescent protein, to label cell morphology. Around 24 hours later, cortical slices were prepared. Photo-conversion of the existing Kaede-Centrin1 proteins was then performed in individual slices, which were then cultured for another 24 hours before being subjected to time-lapse imaging. Labelled dividing radial glial cells with enlarged and rounded cell bodies possessing a pair of centrosomes at the VZ surface (Fig. 3b) were monitored at 10-minute intervals over a period of five to eight hours (Fig. 3c and d, Supplementary Video 1, and Supplementary Fig. 9). In six out of seven dividing radial glial cells that proceeded through mitosis at the VZ surface and reached the two-cell stage, the centrosome retaining the old mother centriole in both red and green fluorescence stayed at the VZ surface, whereas the centrosome containing the new mother centriole in solely green fluorescence migrated away from the VZ surface (Fig. 3c and d, Supplementary Video 1, and Supplementary Fig. 9). These results demonstrate that the centrosomes with differently aged mother centrioles in dividing radial glial progenitors exhibit distinct behaviour during the peak phase of neurogenesis.
Figure 3. Distinct behaviour of centrosomes with differently aged mother centrioles.
(a) Experimental procedure for time-lapse imaging analysis of centrosome behaviour. (b) Image of a cortical slice in culture expressing Kaede-Centrin1 (green and red) and mPlum (blue) prior to time-lapse imaging. Arrows indicate the centrosomes with the old mother centriole (both green and red fluorescent) and arrowheads indicate the centrosomes with the new mother centriole (green fluorescent only). The outlined region contains a dividing radial glial cell possessing a pair of centrosomes with differently aged mother centrioles (circled). (c, d) Time-lapse (c) and kymograph (d) images of the outlined region in b. The time is indicated at the top (c) or the bottom (d) of images. Arrows indicate centrosomes possessing the old mother centriole and arrowheads indicate centrosomes possessing the new mother centriole. Scale bars: 15 μm (b) and 10 μm (c, d).
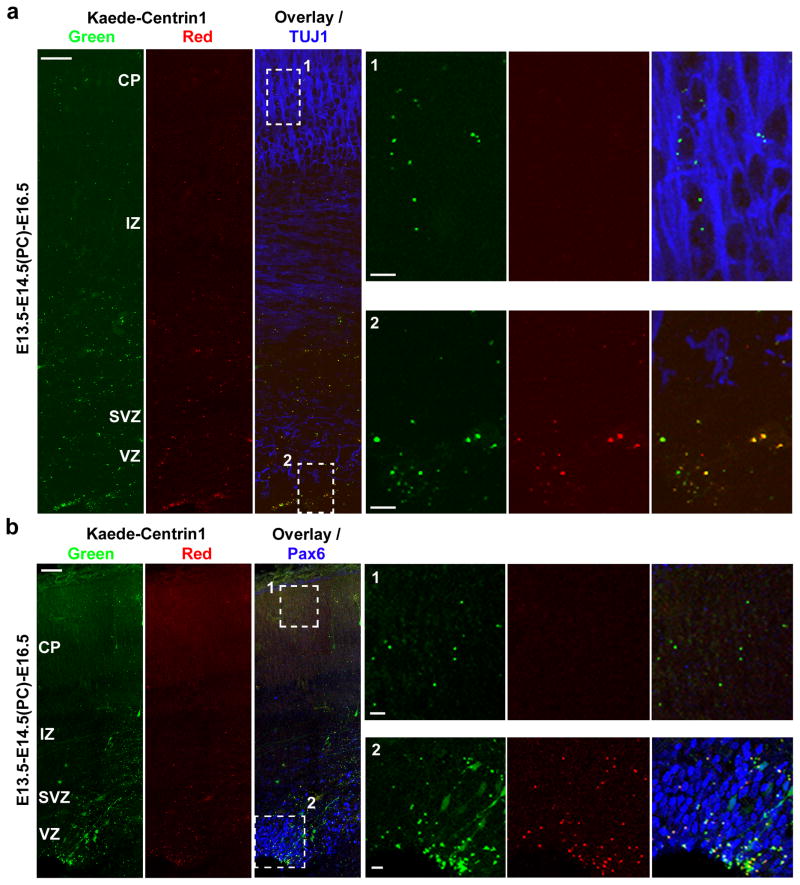
The distinct behaviour of the centrosomes suggests that they are differentially inherited by the two daughter cells embarking on different routes of fate specification and development. Based on their behaviour, we postulated that the centrosome with the new mother centriole is largely inherited by differentiating cells, such as neurons, while the centrosome with the old mother centriole that remains located at the VZ is mostly inherited by radial glial progenitors. Indeed, we found that two days after photo-conversion (E13.5-E14.5(PC)-E16.5) the centrosomes with the new mother centriole, marked by green fluorescence alone, were mostly associated with cells expressing TUJ1, a differentiating neuronal marker, in the CP and the IZ (Fig 4a). In contrast, the centrosomes that retained the old mother centriole in yellow, i.e. both green and red, fluorescence at the VZ were largely associated with cells expressing Pax6, a radial glial progenitor marker (Fig. 4b). These results show that the centrosomes with differently aged mother centrioles in dividing radial glial cells are asymmetrically inherited by the two daughter cells; while the renewing radial glial progenitor inherits the centrosome with the old mother centriole, the differentiating daughter cell inherits the centrosome with the new mother centriole.
Figure 4. Asymmetric inheritance of centrosomes with differently aged mother centrioles.
Images of cortices electroporated with Kaede-Centrin1, photo-converted, and immunostained for TUJ1 (a) or for Pax6 (b) (blue). High magnification images of the outlined regions are shown to the right. Scale bars: 50 μm, 10 μm and 10 μm.
Asymmetric centrosome inheritance maintains progenitors
Our data thus far show that centrosomes with differently aged mother centrioles are differentially inherited by the two daughter cells of asymmetrically dividing radial glial progenitors in the developing neocortex. We next tested whether the selective inheritance of the centrosome with the old mature mother centriole by radial glial progenitors is necessary for their maintenance at the VZ. Should this be the case, given that Ninein is an essential component of the appendage/satellite structures specific to the mature centriole, we predicted that removal of Ninein, which prevents centriole maturation 19, 42, would disrupt asymmetric segregation of centrosomes with differently aged mother centrioles and impair the maintenance of radial glial progenitors in the developing neocortex.
To test this, we developed short hairpin RNA (shRNA) sequences against Ninein that effectively suppressed its expression (Ninein shRNAs, Supplementary Fig. 10a). Consistent with our prediction, expression of Ninein shRNA, but not control shRNA, disrupted asymmetric segregation of centrosomes with differently aged mother centrioles labelled with Kaede-Centrin1 in the developing neocortex (Supplementary Fig. 11), suggesting that Ninein is necessary for centriole maturation thereby generating asymmetry between duplicated centrosomes. The presence of solely green fluorescent centrosomes in Ninein shRNA-expressing cortices indicates that centrosome duplication and segregation and cell division are not severely affected by removal of Ninein, as suggested previously 19, 42. More importantly, we found that removal of Ninein caused a premature depletion of cells from the VZ, where radial glial progenitors reside (Fig. 5a and b). This effect of Ninein shRNAs correlated with their efficacy in suppressing Ninein protein expression (Supplementary Fig. 10 a and b) and was rescued by a shRNA-insensitive Ninein plasmid (Supplementary Fig. 10c), suggesting that the effect of the Ninein shRNA is due to a specific depletion of the endogenous Ninein protein. A similar reduction in cells in the VZ was observed when Ninein expression was suppressed using small interfering RNA (siRNA) (Supplementary Fig. 10 d and e).
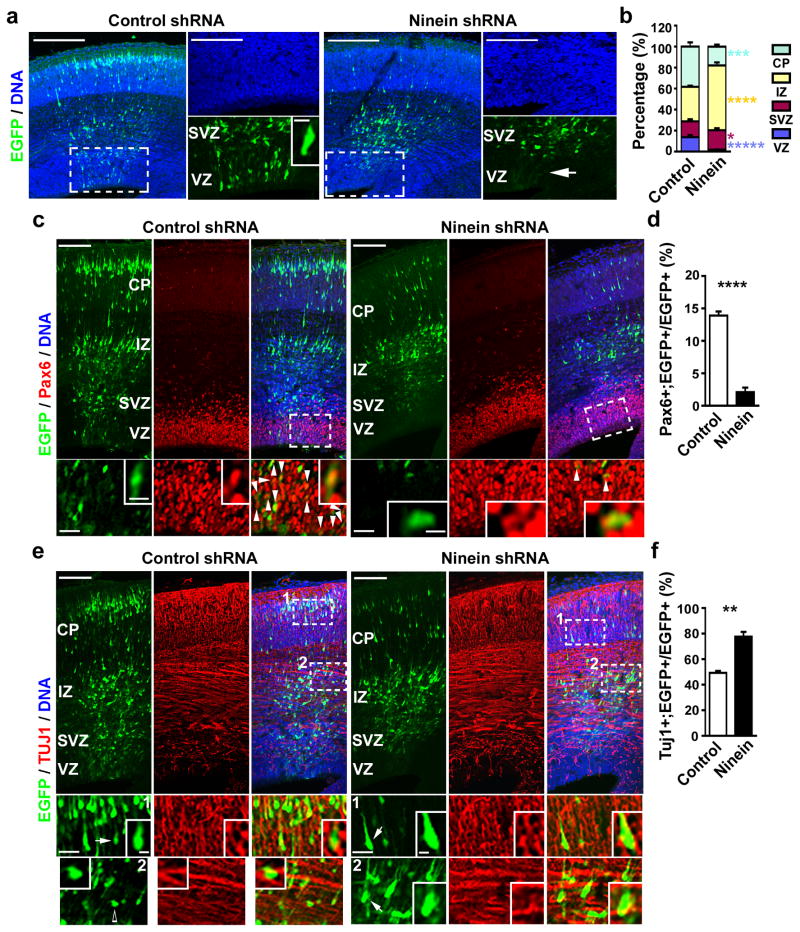
Figure 5. Preferential inheritance of the centrosome with the mature mother centriole maintains radial glial progenitors.
(a) Images of E16.5 cortices electroporated with either EGFP/Control (left, green) or EGFP/Ninein shRNA (right, green) at E13.5 and counterstained with DAPI (blue). (b) Quantification of the percentage of EGFP-expressing cells in different regions of the developing neocortex (Control shRNA: total 1,873 cells from five individual animals; Ninein shRNA: total 958 cells from five individual animals). (c, e) Images of E16.5 cortices electroporated with EGFP/Control (left, green) or EGFP/Ninein shRNA (right, green) at E13.5 and immunostained for Pax6 (c) or TUJ1 (e) (red). Arrowheads indicate EGFP-expressing cells positive for Pax6 (c). Arrows indicate EGFP-expressing cells that are positive for TUJ1 and the open arrow indicates an EGFP-expressing cell that is not positive for TUJ1 (e). (d, f) Quantification of the percentage of EGFP-expressing cells positive for Pax6 (d; Control shRNA: total 1,752 cells from five individual animals; Ninein shRNA: total 1,230 cells from five animals) or for TUJ1 (f; Control shRNA, total 1,383 cells from five individual animals; Ninein shRNA, total 1,247 cells from five individual animals). Data are shown as mean±s.e.m.; *, p<0.05; **, p<0.005; ***, p<0.001; ****, p<0.0005; *****, p<5e-5. Scale bars: 100 μm, 50 μm, and 10 μm (a); 100μm, 5 μm, and 25 μm (c); 100μm, 25 μm, and 5 μm (e).
To further characterize the extent to which removal of Ninein leads to a depletion of radial glial progenitors, we next examined the fate specification of cells expressing either control or Ninein shRNA (Fig. 5c–f). When compared with the control, expression of Ninein shRNA led to a drastic reduction in the percentage of cells positive for Pax6 and glutamate transporter (GLAST) (Fig. 5c and d; Supplementary Fig. 12), two radial glial progenitor markers, and a significant increase in the percentage of cells positive for TUJ1 (Fig. 5e and f), a differentiating neuronal marker. These results suggest that removal of Ninein leads to a depletion of radial glial progenitors and a concomitant increase in differentiating neurons. Consistent with this, we observed a significant reduction in phospho-Histone 3 (P-H3)-labelled mitotic cells at the VZ surface (Supplementary Fig. 13) and a drastic increase in cell cycle exit (Supplementary Fig. 14). No obvious change in the cleavage plane orientation of late stage mitotic cells at the VZ surface was observed (Supplementary Fig. 15).
Previous studies showed that the carboxyl-terminus of Ninein is responsible for its localization to the centriole and expression of this region displaces endogenous protein at the centriole 43. Interestingly, we found that, similar to removal of Ninein, expression of the carboxyl-terminus of Ninein (Ninein-Cter) led to a premature depletion of radial glial progenitor cells from the VZ (Supplementary Fig. 16), suggesting that centriolar Ninein is critical for maintaining radial glial progenitor cells in the VZ. Taken together, these results strongly suggest that preferential inheritance of a centrosome containing the mature mother centriole is required for the maintenance of radial glial progenitors in the proliferative VZ of the developing neocortex.
In summary, the results presented here suggest that the centrosomes with differently aged centrioles in asymmetrically dividing radial glial progenitors exhibit different behaviour and are differentially inherited by the two daughter cells during the peak phase of mammalian neocortical neurogenesis (Supplementary Fig. 1). While the centrosome with the less mature new mother centriole migrates away from the VZ surface and is largely inherited by differentiating cells, the centrosome with the more mature old mother centriole stays at the VZ surface and is predominantly inherited by renewing radial glial progenitors. Recently, asymmetric behaviour of centrosome has been observed during asymmetric division of Drosophila male germline stem cells (GSCs) and neuroblasts 25–28. Our findings suggest that this type of asymmetric centrosome regulation may be a general feature of asymmetric cell division across species 30–33. Furthermore, our findings provide new insight into centrosome regulation in the developing mammalian neocortex, which has been linked to the pathogenesis of human microcephaly 34, 44.
Centrosomes with differently aged mother centrioles differ in their protein composition and thereby in their biophysical properties, such as microtubule anchorage activity 9, 15 and the capability to mediate ciliogenesis 21, 23, 45. In this study, we found that Ninein, an appendage/satellite-specific protein required for centriole maturation, localized differently to the duplicated centrosomes in radial glial progenitors in late mitosis. Interestingly, another appendage/satellite-specific protein Cenexin/Odf2 was recently found to be asymmetrically localized to centrosomes in sister cells after mitosis; moreover, the cell receiving the more mature old mother centriole usually grew a primary cilium first 21. The asymmetric inheritance of centrosomes with distinct biophysical properties may thereby differentially regulate the behaviour and development of the daughter cells that receive them. For example, given that primary cilia play essential roles in a number of signal transduction pathways, including Sonic hedgehog (Shh) and platelet-derived growth factor (PDGF) signalling, the asynchrony in cilium formation could differentially influence the ability of the two daughter cells to respond to environmental signals and thereby their behaviour and fate specification. Also, the strong microtubule anchorage activity associated with the centrosome retaining the older mother centriole would facilitate its anchorage to a specific site (e.g. the VZ surface), thereby tethering the cell that inherits it. Indeed, we found that disruption of centriole maturation by removing Ninein not only impairs asymmetric segregation of centrosomes, but also depletes radial glial progenitors from the VZ, a proliferative niche in the developing mammalian neocortex. Aside from their participation in microtubule organization and ciliogenesis, centrosomes associate with messenger RNAs (mRNAs) 46 and membrane-bound organelles such as the Golgi and recycling endosomes and regulate protein degradation 47, 48, thereby raising the possibility that asymmetric centrosome inheritance might contribute to proper segregation of cell fate determinants to the two daughter cells of asymmetrically dividing progenitor/stem cells.
Methods summary
In utero electroporation and photo-conversion
In utero electroporation of the plasmids (e.g. EGFP-Centrin1) was performed as previously described 49, 50. For in utero photo-conversion, a similar surgical procedure was carried out as for in utero electroporation. The forebrain of the embryos that received electroporation was exposed to a brief (about three to five minutes) exposure of light at 350–400 nm while in the uterus. All procedures for animal handling and usage were approved by our institutional research animal resource centre (RARC).
Brain section, immunohistochemistry and imaging
Brains were fixed at the desired developmental stages and coronal sections were prepared using a vibratome (Leica Microsystems). Immunohistochemistry were performed as previously described 50. Images were acquired using a confocal laser scanning microscope (FV1000, Olympus) and analyzed using FluoView (Olympus), Volocity (Improvision) and Photoshop (Adobe Systems). Data were presented as mean and s.e.m. and statistical differences were determined using the nonparametric tests (Mann-Whitney-Wilcoxon test for two groups of data and Kruskal-Wallis test for three or more groups of data).
Cortical slice culture and time-lapse imaging
Cortical slice cultures were prepared and time-lapse imaging was acquired as previously described 12. Images were analyzed using MetaMorph (Molecular Devices) and Photoshop (Adobe Systems).
Methods
Plasmids and in utero electroporation and photo-conversion
Human Centrin1 cDNA was obtained by polymerase chain reaction (PCR) and cloned into the KpnI and BamHI sites of pEGFP-C1 (Clontech) to generate the EGFP-Centrin1 plasmid. Kaede and DsRedexpress (DsRedex) cDNAs were obtained by PCR and cloned into pEGFP-Centrin1-C1 to replace EGFP in generating the Kaede-Centrin1 and DsRedex-Centrin1 plasmids. The mouse Ninein (accession number AY515727) plasmid was kindly provided by Dr. Michel Bornens (CNRS/Institut Curie, France). Human Ninein cDNA (IMAGE ID 3090109, Open Biosystems) was cloned into the EcoRI and NotI sites of pCDNA3.1. Three shRNA sequences against mouse Ninein were designed as follows: shRNA-a (5′-GCAGAAGGCCAGCTGAGGT-3′), -b (GGCCGAGATCCGGCACTTG), -c (GCTTCAATTCAGACAATGG). All sense and anti-sense oligos were purchased from Sigma. Annealed oligos were cloned into the HpaI and XhoI sites of the Lentiviral vector pLL3.7, which contains a separate CMV promoter that drives expression of EGFP 50. In this study, mouse Ninein shRNA-c was primarily used after extensive characterization to demonstrate that it specifically suppressed Ninein protein expression and function. For siRNA experiments, synthetic oligos against Ninein and control were purchased from Santa Cruz Biotechnology (sc-61196). Human Ninein cDNA, which differs from mouse Ninein cDNA in the shRNA-c targeting region and is thereby insensitive to Ninein shRNA-c expression, was used for the rescue experiment. The amino-terminus (nucleotides 1–1120) and carboxyl-terminus (nucleotides 5623–6339) of mouse Ninein were amplified by PCR and cloned into the EcoRI and NotI sites of pCAG-IRES-EGFP. All plasmids were confirmed by sequencing.
In utero electroporation was performed as previously described 49, 50. In brief, a timed pregnant CD-1 mouse at 13.5 days of gestation (E13.5) or a rat at E16.5 was anesthetized, the uterine horns were exposed, and ~1 μl of plasmid DNA (1–3 μg/μl) mixed with Fast Green (Sigma,) was manually microinjected through the uterus into the lateral ventricle, using a bevelled and calibrated glass micropipette (Drummond Scientific). For electroporation, five 50 ms pulses of 40–50 mV with a 950 ms interval were delivered across the uterus with two 9-mm electrode paddles positioned on either side of the head (BTX, ECM830). For in utero photo-conversion, a similar surgical procedure was carried out as for in utero electroporation. The forebrain of the embryos that received electroporation was exposed to a brief (about three to five minutes) exposure of light at 350–400 nm while in the uterus. Throughout these surgical procedures, the uterus was constantly bathed with warm phosphate buffered saline (PBS, pH 7.4). After the procedure, the uterus was placed back in the abdominal cavity and the wound was surgically sutured. The animal was then placed in a 28°C recovery incubator under close monitoring until it recovered and resumed normal activity. All procedures for animal handling and usage were approved by our institutional research animal resource centre (RARC).
Brain sectioning and confocal imaging and analysis
Embryos were removed and transcardially perfused with ice-cold PBS (pH 7.4) followed by 4% paraformaldehyde (PFA) in PBS (pH 7.4). For cell cycle exit analysis, electroporated embryos were exposed to bromodeoxyuridine (BrdU, ~50–100 mg/kg body weight) for 24 hours prior to sacrifice. Brains were dissected out and coronal sections were prepared using a vibratome (Leica Microsystems). For immunohistochemistry, sections were incubated for one hour at room temperature in a blocking solution (10% normal goat or donkey serum as appropriate, 0.1% Triton X-100, and 0.2% gelatin in PBS), followed by incubation with the primary antibodies overnight at 4°C. Sections were then washed in 0.1% Triton X-100 in PBS and incubated with the appropriate secondary antibody for one to two hours at room temperature.
The primary antibodies used were: rabbit polyclonal anti-γ-Tubulin (Sigma, 1:500), mouse monoclonal anti-Pericentrin1 (BD Biosciences, 1:1000), mouse monoclonal anti-β-III Tubulin (clone TUJ1) (Covance, 1:500), rabbit polyclonal anti-Pax6 (Covance, 1:500), rabbit polyclonal anti-Tbr2 (Millipore/Chemicon, 1:500), rat monoclonal anti-BrdU (Abcam, 1:400), mouse polyclonal anti-Ki67 (Novus Biological, 1:200), rabbit polyclonal anti-GLAST (Invitrogen, 1:400), rabbit polyclonal anti-phospho-Histone 3 (Millipore/Upstate, 1:1000), mouse monoclonal anti-phospho-H2AX (Millipore/Upstate, 1:250) and rabbit polyclonal anti-cleaved Caspase 3 (Cell Signalling Technology, 1:250). Secondary antibodies used were: goat or donkey anti-mouse or anti-rabbit Alexa-546 and Alexa-647 conjugated antibodies (Invitrogen/Molecular Probes, 1:500). DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen/Molecular Probes). Images were acquired with an Olympus FV1000 confocal microscope, and analyzed with FluoView (Olympus), Volocity (Improvision), and Photoshop (Adobe Systems).
Data are presented as mean ± standard error of the mean (s.e.m.) and nonparametric tests (Mann-Whitney-Wilcoxon test for two groups of data and Kruskal-Wallis test for three or more groups of data) were used for statistical significance estimation.
Cortical slice culture and time-lapse imaging
Cortical slice cultures were prepared and time-lapse imaging was acquired as previously described 12. About 12 hours after in utero electroporation, embryos were removed and the brain was extracted into ice-cold artificial cerebro-spinal fluid (ACSF) containing (in mM): 125 NaCl, 5 KCl, 1.25 NaH2PO4, 1 MgSO4, 2 CaCl2, 25 NaHCO3 and 20 glucose; pH 7.4, 310 mOsm/L. Brains were embedded in 4% low-melting agarose in ACSF and sectioned at 400 μm using a vibratome (Leica microsystems). Brain slices that contained Kaede-Centrin1- and mPlum- expressing cells were transferred onto a slice culture insert (Millicell) in a glass-bottom petri dish (MatTek Corporation) with culture medium containing (by volume): 66% BME, 25% Hanks, 5% FBS, 1% N-2, 1% Penicillin/Streptomycin/Glutamine (Invitrogen/GIBCO) and 0.66% D-(+)-glucose (Sigma). Cultures were maintained in a humidified incubator at 37°C with constant 5% CO2 supply. Twenty-four hours later, petri dishes with slice cultures were transferred to an inverted microscope DMIRB (Leica) with a 10X objective and the slices were exposed to brief (~12 seconds) epifluorescent illumination using a DAPI filter. The slices were returned to the incubator and cultured for another 24 hours. Time-lapse images of dividing radial glial cells expressing Kaede-Centrin1 and mPlum were acquired using a 40X objective lens and a CoolSnap HQ camera (Roper Scientific) every 10 minutes for about five to eight hours. Images were analyzed using MetaMorph (Molecular Devices) and Photoshop (Adobe Systems).
Supplementary Material
Acknowledgments
We thank Drs. A. Hall, A. Joyner, K. V. Anderson, J. Kaltschmidt, B. Tsou, Y. Chin and L. A. McDowell for comments on the manuscript, the anonymous referees for helpful suggestions, members of the Shi laboratory for discussions, Dr. A. Hadjantonakis for human Centrin1 cDNA, Dr. M. Bornens for EGFP-Ninein (mouse) and Ninein truncation mutant plasmid, Dr. Y.-R. Hong for EGFP-Ninein (human) plasmid, Dr. A. Miyawaki for pCS2+-Kaede plasmid, Drs. H. Zhong, K. Svoboda and R. Tsien for DsRedexpress and mPlum cDNA constructs. We thank Drs. C. T. Anderson and T. Stearns for sharing unpublished data. This work is supported by grants from March of Dimes Birth Defects Foundation, Whitehall Foundation, DANA Foundation, Autism Speaks Foundation, Klingenstein Foundation, NARSAD (to S.-H.S.), and NIH (to S.-H.S. and R.B.V.).
Footnotes
Author Contributions
X.W. and S.-H.S. conceived the project. X.W. performed most of the experiments. J.-W.T., W.-N.L. and R.B.V. contributed to the time-lapse imaging experiment and J.H.I. contributed to the characterization of Kaede-Centrin1 co-localization and in utero photo-conversion procedure. X. W. and S.-H. S. analyzed data, interpreted results and wrote the manuscript. All authors edited the manuscript.
References
- 1.Miyata T, Kawaguchi A, Okano H, Ogawa M. Asymmetric inheritance of radial glial fibers by cortical neurons. Neuron. 2001;31:727–41. doi: 10.1016/s0896-6273(01)00420-2. [DOI] [PubMed] [Google Scholar]
- 2.Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–20. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- 3.Malatesta P, Hartfuss E, Gotz M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals a neuronal lineage. Development. 2000;127:5253–63. doi: 10.1242/dev.127.24.5253. [DOI] [PubMed] [Google Scholar]
- 4.Rakic P. Elusive radial glial cells: historical and evolutionary perspective. Glia. 2003;43:19–32. doi: 10.1002/glia.10244. [DOI] [PubMed] [Google Scholar]
- 5.Miyata T, et al. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development. 2004;131:3133–45. doi: 10.1242/dev.01173. [DOI] [PubMed] [Google Scholar]
- 6.Noctor SC, Martinez-Cerdeno V, Ivic L, Kriegstein AR. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 2004;7:136–44. doi: 10.1038/nn1172. [DOI] [PubMed] [Google Scholar]
- 7.Chenn A, McConnell SK. Cleavage orientation and the asymmetric inheritance of Notch1 immunoreactivity in mammalian neurogenesis. Cell. 1995;82:631–41. doi: 10.1016/0092-8674(95)90035-7. [DOI] [PubMed] [Google Scholar]
- 8.Noctor SC, Martinez-Cerdeno V, Kriegstein AR. Distinct behaviors of neural stem and progenitor cells underlie cortical neurogenesis. J Comp Neurol. 2008;508:28–44. doi: 10.1002/cne.21669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol. 2002;14:25–34. doi: 10.1016/s0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- 10.Doxsey S, McCollum D, Theurkauf W. Centrosomes in cellular regulation. Annu Rev Cell Dev Biol. 2005;21:411–34. doi: 10.1146/annurev.cellbio.21.122303.120418. [DOI] [PubMed] [Google Scholar]
- 11.Xie Z, et al. Cep120 and TACCs control interkinetic nuclear migration and the neural progenitor pool. Neuron. 2007;56:79–93. doi: 10.1016/j.neuron.2007.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsai JW, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat Neurosci. 2007;10:970–9. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- 13.Solecki DJ, Model L, Gaetz J, Kapoor TM, Hatten ME. Par6alpha signaling controls glial-guided neuronal migration. Nat Neurosci. 2004;7:1195–203. doi: 10.1038/nn1332. [DOI] [PubMed] [Google Scholar]
- 14.Meraldi P, Nigg EA. The centrosome cycle. FEBS Lett. 2002;521:9–13. doi: 10.1016/s0014-5793(02)02865-x. [DOI] [PubMed] [Google Scholar]
- 15.Delattre M, Gonczy P. The arithmetic of centrosome biogenesis. J Cell Sci. 2004;117:1619–30. doi: 10.1242/jcs.01128. [DOI] [PubMed] [Google Scholar]
- 16.Lange BM, Gull K. A molecular marker for centriole maturation in the mammalian cell cycle. J Cell Biol. 1995;130:919–27. doi: 10.1083/jcb.130.4.919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakagawa Y, Yamane Y, Okanoue T, Tsukita S, Tsukita S. Outer dense fiber 2 is a widespread centrosome scaffold component preferentially associated with mother centrioles: its identification from isolated centrosomes. Mol Biol Cell. 2001;12:1687–97. doi: 10.1091/mbc.12.6.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouckson-Castaing V, et al. Molecular characterisation of ninein, a new coiled-coil protein of the centrosome. J Cell Sci. 1996;109 (Pt 1):179–90. doi: 10.1242/jcs.109.1.179. [DOI] [PubMed] [Google Scholar]
- 19.Ou YY, Mack GJ, Zhang M, Rattner JB. CEP110 and ninein are located in a specific domain of the centrosome associated with centrosome maturation. J Cell Sci. 2002;115:1825–35. doi: 10.1242/jcs.115.9.1825. [DOI] [PubMed] [Google Scholar]
- 20.Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol. 2000;149:317–30. doi: 10.1083/jcb.149.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson TC, TS Centriole Age Underlies Asynchronous Primary Cilium Growth in Mammalian Cells. Curr Biol. 2009 doi: 10.1016/j.cub.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chretien D, Buendia B, Fuller SD, Karsenti E. Reconstruction of the centrosome cycle from cryoelectron micrographs. J Struct Biol. 1997;120:117–33. doi: 10.1006/jsbi.1997.3928. [DOI] [PubMed] [Google Scholar]
- 23.Vorobjev IA, Chentsov Yu S. Centrioles in the cell cycle. I Epithelial cells. J Cell Biol. 1982;93:938–49. doi: 10.1083/jcb.93.3.938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442:947–51. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- 25.Cheng J, et al. Centrosome misorientation reduces stem cell division during ageing. Nature. 2008;456:599–604. doi: 10.1038/nature07386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamashita YM, Mahowald AP, Perlin JR, Fuller MT. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 2007;315:518–21. doi: 10.1126/science.1134910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rebollo E, et al. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev Cell. 2007;12:467–74. doi: 10.1016/j.devcel.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 28.Rusan NM, Peifer M. A role for a novel centrosome cycle in asymmetric cell division. J Cell Biol. 2007;177:13–20. doi: 10.1083/jcb.200612140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stevens NR, Raposo AA, Basto R, St Johnston D, Raff JW. From stem cell to embryo without centrioles. Curr Biol. 2007;17:1498–503. doi: 10.1016/j.cub.2007.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cabernard C, Doe CQ. Stem cell self-renewal: centrosomes on the move. Curr Biol. 2007;17:R465–7. doi: 10.1016/j.cub.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 31.Spradling AC, Zheng Y. Developmental biology. The mother of all stem cells? Science. 2007;315:469–70. doi: 10.1126/science.1138237. [DOI] [PubMed] [Google Scholar]
- 32.Yamashita YM, Fuller MT. Asymmetric centrosome behavior and the mechanisms of stem cell division. J Cell Biol. 2008;180:261–6. doi: 10.1083/jcb.200707083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonzalez C. Spindle orientation, asymmetric division and tumour suppression in Drosophila stem cells. Nat Rev Genet. 2007;8:462–72. doi: 10.1038/nrg2103. [DOI] [PubMed] [Google Scholar]
- 34.Cox J, Jackson AP, Bond J, Woods CG. What primary microcephaly can tell us about brain growth. Trends Mol Med. 2006;12:358–66. doi: 10.1016/j.molmed.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 35.Higginbotham HR, Gleeson JG. The centrosome in neuronal development. Trends Neurosci. 2007;30:276–83. doi: 10.1016/j.tins.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 36.Hinds JW, Ruffett TL. Cell proliferation in the neural tube: an electron microscopic and golgi analysis in the mouse cerebral vesicle. Z Zellforsch Mikrosk Anat. 1971;115:226–64. doi: 10.1007/BF00391127. [DOI] [PubMed] [Google Scholar]
- 37.Chenn A, Zhang YA, Chang BT, McConnell SK. Intrinsic polarity of mammalian neuroepithelial cells. Mol Cell Neurosci. 1998;11:183–93. doi: 10.1006/mcne.1998.0680. [DOI] [PubMed] [Google Scholar]
- 38.Bornens M, Piel M. Centrosome inheritance: birthright or the privilege of maturity? Curr Biol. 2002;12:R71–3. doi: 10.1016/s0960-9822(01)00678-9. [DOI] [PubMed] [Google Scholar]
- 39.Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci U S A. 2002;99:12651–6. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salisbury JL, Suino KM, Busby R, Springett M. Centrin-2 is required for centriole duplication in mammalian cells. Curr Biol. 2002;12:1287–92. doi: 10.1016/s0960-9822(02)01019-9. [DOI] [PubMed] [Google Scholar]
- 41.Cai L, Hayes NL, Nowakowski RS. Local homogeneity of cell cycle length in developing mouse cortex. J Neurosci. 1997;17:2079–87. doi: 10.1523/JNEUROSCI.17-06-02079.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mogensen MM, Malik A, Piel M, Bouckson-Castaing V, Bornens M. Microtubule minus-end anchorage at centrosomal and non-centrosomal sites: the role of ninein. J Cell Sci. 2000;113 (Pt 17):3013–23. doi: 10.1242/jcs.113.17.3013. [DOI] [PubMed] [Google Scholar]
- 43.Delgehyr N, Sillibourne J, Bornens M. Microtubule nucleation and anchoring at the centrosome are independent processes linked by ninein function. J Cell Sci. 2005;118:1565–75. doi: 10.1242/jcs.02302. [DOI] [PubMed] [Google Scholar]
- 44.Bond J, et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet. 2005;37:353–5. doi: 10.1038/ng1539. [DOI] [PubMed] [Google Scholar]
- 45.Preble AM, Giddings TM, Jr, Dutcher SK. Basal bodies and centrioles: their function and structure. Curr Top Dev Biol. 2000;49:207–33. doi: 10.1016/s0070-2153(99)49010-6. [DOI] [PubMed] [Google Scholar]
- 46.Lambert JD, Nagy LM. Asymmetric inheritance of centrosomally localized mRNAs during embryonic cleavages. Nature. 2002;420:682–6. doi: 10.1038/nature01241. [DOI] [PubMed] [Google Scholar]
- 47.Wigley WC, et al. Dynamic association of proteasomal machinery with the centrosome. J Cell Biol. 1999;145:481–90. doi: 10.1083/jcb.145.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fuentealba LC, Eivers E, Geissert D, Taelman V, De Robertis EM. Asymmetric mitosis: Unequal segregation of proteins destined for degradation. Proc Natl Acad Sci U S A. 2008;105:7732–7. doi: 10.1073/pnas.0803027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tabata H, Nakajima K. Efficient in utero gene transfer system to the developing mouse brain using electroporation: visualization of neuronal migration in the developing cortex. Neuroscience. 2001;103:865–72. doi: 10.1016/s0306-4522(01)00016-1. [DOI] [PubMed] [Google Scholar]
- 50.Bultje RS, et al. Mammalian Par3 regulates progenitor cell asymmetric division via notch signaling in the developing neocortex. Neuron. 2009;63:189–202. doi: 10.1016/j.neuron.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.