Abstract
Mass spectrometry based structural proteomics approaches for probing protein structures are increasingly gaining in popularity. The potential for such studies is limited because of the lack of analytical techniques for the automated interpretation of resulting data. In this paper, a suite of algorithms called ProtMapMS is developed, integrated, and implemented specifically for the comprehensive automatic analysis of mass spectrometry data obtained for protein structure studies using covalent labeling. The functions include data format conversion, mass spectrum interpretation, detection and verification of all peptide species, confirmation of the modified peptide products, and quantification of the extent of peptide modification. The results thus obtained provide valuable data for use in combination with computational approaches for protein structure modeling. The structures of both monomeric and hexameric forms of insulin were investigated by oxidative protein footprinting followed by high-resolution mass spectrometry. The resultant data was analyzed both manually and using ProtMapMS without any manual intervention. The results obtained using the two methods were found to be in close agreement and overall were consistent with predictions from the crystallographic structure.
1 Introduction
Structural Proteomics is the large scale study of the structural description of proteins and their higher order complexes present in a given cell. It holds special significance since cellular behavior and disease are functions of the interactions between macromolecular complexes involved in cellular biological transactions. Important questions in structural proteomics involve elucidating the structure of these multicomponent assemblies, including their sub components and their assembly, and relating their structure to function. Mass spectrometry (MS) based approaches are proving to be indispensable for addressing these questions.1 These approaches include chemical cross-linking with mass spectrometry,2 hydrogen-deuterium exchange,3 and covalent labelling.4-7 Chemical cross-linking utilizes cross-linking reagents in order to covalently tether interacting proteins in a multi-protein complex. The identification of the cross-linked species from the mass spectral analysis provides insights into the interfaces of the protein complex. Hydrogen-deuterium exchange methods are based upon the principle that some of the backbone hydrogen atoms in proteins exchange positions with the deuterium atoms from the surrounding deuterium oxide solution, which can be detected using mass spectrometry. In addition, transiently unfolding segments of the protein can exchange. The rate of exchange of hydrogens is a function of the protein structure and solvent accessibility, providing information about the protein dynamics in solution.
In the process of covalent labelling by means of protein footprinting, the protein surface is typically exposed to hydroxyl radicals resulting in the oxidation of protein. 8 Multiple methods are used to produce the hydroxyl radicals, these include transition metal-mediated chemical methods (Fenton Chemistry),9 laser dissociation of hydrogen peroxide,10 and radiolysis of water by X-rays or gamma rays.8,11 The hydroxyl radicals react with the side chains of specific surface residues giving rise to specific modification products, minimally impacting the globular protein structure. The modified protein is subjected to enzymatic digestion, followed by liquid chromatography coupled mass spectrometry (LC-MS). The readouts from oxidized residues in the mass spectrometric analysis provide information for mapping the protein surface and defining the solvent accessibility for residues of interest.12-15 The process is repeated for the same protein, when it is present as a part of a macromolecular complex or when bound to ligands of any kind. Investigation into the differential extent of oxidation of a protein in the two forms, for example isolated protein versus protein-ligand complex, provides information on the structural changes introduced during ligand binding and protein-protein interactions. Owing to the similarity of hydroxyl radicals to water molecules, they are quite suitable to act as solvent accessibility probes. Their high reactivity together with well defined chemical selectivity makes them ideal tools for biomolecular structural examination. Previous studies report the following order of reactivity for the oxidation of amino acids and their efficiency of detection by mass spectrometry:Cys>Met>Trp>Tyr>Phe>Cystine>His>Leu, Ile>Arg, Lys, Val>Ser, Thr, Pro>Gln, Glu>Asp, Asn>Ala>Gly.8,16-18 The oxidatively labeled products of various amino acids are shown in Table 1.
Table 1.
Primary oxidation products and resulting mass changes for various amino acid side chains subjected to radiolytic modification and detected by mass spectrometry 17
| Residue | Side chain modifications and associated mass changes |
|---|---|
| Cys | sulfonic acid (+48), sulfonic acid (+32), hydroxy (-16) |
| cystine | sulfonic acid, sulfonic acid |
| Met | sulfoxide (+16), sulfone (+32), aldehyde (-32) |
| Trp | hydroxy- (+16, +32, +48, etc.), pyrrol ring-open (+32, etc.) |
| Tyr | hydroxy- (+16, +32, etc.) |
| Phe | hydroxy- (+16, +32, etc.) |
| His | oxo- (+16), ring-open (-22, -10, +5) |
| Leu | hydroxy- (+16), carbonyl (+14) |
| Ile | hydroxy- (+16), carbonyl (+14) |
| Val | hydroxy- (+16), carbonyl (+14) |
| Pro | hydroxy- (+16), carbonyl (+14) |
| Arg | deguanidination (-43), hydroxy- (+16), carbonyl (+14) |
| Lys | hydroxy- (+16), carbonyl (+14) |
| Glu | decarboxylation (-30), hydroxy- (+16), carbonyl (+14) |
| Gln | hydroxy- (+16), carbonyl (+14) |
| Asp | decarboxylation (-30), hydroxy- (+16) |
| Asn | hydroxy- (+16) |
| Ser | hydroxy- (+16), carbonyl (-2- or +16-H2O) |
| Thr | hydroxy- (+16), carbonyl (-2- or +16-H2O) |
| Ala | hydroxy- (+16) |
2 Goals
Structural mass spectrometry techniques have evolved and been improved such that these studies are now carried out routinely. The interpretation of high volumes of data resulting from such experiments represents the biggest bottleneck for the overall experiment, thus, limiting their potential. Manual interpretation of the experimental data is tedious, time consuming, and prone to human error. Although progress has been made for the semi-automation of data processing for hydrogen-deuterium exchange19-22 and chemical crosslinking23 experiments, currently there is a lack of analytical tools that are specifically tailored to meet the needs of covalent labelling experiments. As seen in Table 1, a typical hydroxyl radical-mediated covalent labeling experiment leads to multiple oxidation states of various amino acid side chains,17,18 leading to a challenging task for data analysis. Established database searching methods such as Mascot 24 and SEQUEST25 are not well suited to handle the large number of combinations of possible side-chain oxidation states as generated from hydroxyl radical oxidation(Table 1). Although MyriMatch, 26 InsPecT,27 and ByOnic28,29 offer improved database searching allowing for a large number of modifications, a number of manual labor intensive steps are required followed by the search in order to extract the relevant quantitative information with respect to oxidation extent for peptides of interest. Moreover, relationship information between retention time in the chromatographic column of unoxidized and oxidized forms of peptide is largely ignored in such searches although these data are very valuable to the analysis. We describe an integrated suite of algorithms, implemented as a software package called ProtMapMS, aimed at the automated mass spectrum analysis especially targeted towards protein structure studies using protein footprinting techniques. The goals of the project are - rapid identification and verification of all peptide species, localizing the site of modification of the oxidized peptide products using multistage mass spectrometry data, and quantification of the extent of peptide modification as a function of exposure time to hydroxyl radicals. ProtMapMS is available for academic use free of charge through a licensing agreement.
3 Approaches
The project is subdivided into the following modules: Data format conversion, Mass spectrum interpretation, Extracting single ion chromatograms, and Generation of dosage response curves. Each of the modules is discussed below:
3.1 Data format conversion
The original format of a mass spectrum collected on commercial instruments is proprietary; this needs to be converted into a readable format in order to be analyzed by other methods. The RAW spectrum is first converted into an mzXML30 format which is followed by a conversion into text format for each of the MS scans, using Proteowizard31 utilities called msconvert and msaccess respectively based upon tools from Institute of Systems Biology.32 ProtMapMS is able to accept Thermo Fisher RAW data files and common open representation mzXML format. 30 The resulting output contains extensive information about the retention time, comprehensive m/z and spectral intensity values from MS1(output from the first stage of an MS experiment), and MS2(output from the second stage of an MS experiment) experiments in an accessible text format.
3.2 Mass Spectral Interpretation
The goal of this step is to detect and confirm the identity of the peptides from the protein(s) of interest so that the confirmed species can be further subjected to quantitative characterization subsequently. In the first three steps, the unmodified peptides are assigned, in the fourth step, the oxidatively modified forms are evaluated. It consists of the following steps:
Generation of the theoretical m/z values of interest For a given protein or a protein complex, a list of theoretical monoisotopic and average mass values of peptides is generated based upon the protein sequence(s), protease used (allowed enzymes are Trypsin, Chymotrypsin, Glu-C, Lys-C, AspN, non-specific digestion, and Custom Enzyme digestion allowing for digestion at user-specified residues), missed cleavages permitted, and allowable fixed modification(s) (e.g., alkylation of Cysteines). For each of these average masses, a theoretical isotopic distribution is generated using Mercury33 algorithm to determine the location and relative intensities of various isotopes. The isotopic mass values are further converted into m/z domain corresponding to all the isotopes greater than 60% of the most abundant isotope for each of the user-specified range of charge states (1-4, typically).
Matching the theoretical m/z against precursor ion m/z values The next step is to detect the peptides observed in the spectrum. This is done by comparing the theoretical m/z list obtained in the previous step against the experimentally observed precursor ion m/z values which were subjected to MS2 analysis during the experiment. Only the matches within a user specified upper error limit (typically, <10 parts-per-million (ppm) for Fourier Transform MS data) are considered to be “possible” matches, which are subjected to structural confirmation in the next step.
- Assignments Confirmation The assignments made above are confirmed by the analysis of MS2 information. This step helps resolve cases when there can be multiple possible assignments for a given precursor ion. The observed MS2 spectra are compared against their theoretical counterparts for the likely candidates with a user-specified maximum permissible error range (typically 0.3 Daltons for MS2 stage in an LTQ-FT instrument), and the candidate with the best match is identified as the true match for the observed mass. This is done by first generating the theoretical MS2 spectra for each of the candidates peptides, representing the uniformally abundant ions formed by backbone cleavage typically observed in a CID (Collision Induced Dissociation) spectrum. This is followed by calculating the cross-correlation coefficient between the experimental spectrum S, and theoretical MS2 spectrum, T, as follows:34
A p-value is calculated for testing the hypothesis of no correlation. Each p-value is the probability of getting a correlation as large as the observed value by random chance, when the true correlation is zero. The p-value is computed by transforming the correlation to create a Student’s T statistic using a standard function “corrcoef” in Matlab (R2008b). 35 Note that in a typical footprinting experiment, the identity of the protein is known a priori, so that there are only a limited number of possible assignments for an observed precursor ion. This is in contrast to the situation of typical database search engines such as Sequest, Mascot, and ByOnics that encounter a more difficult problem of assigning the protein identity from a very high number of possible sequences within the protein database.(1) Detecting oxidatively labeled peptides In a typical protein footprinting experiment, the protein(s) under study is gently exposed to the hydroxyl radicals such that only a very small fraction of the protein gets oxidatively labeled. Hence, the oxidative counterparts of only the peptides detected in unoxidized form are selected as candidates for further search. This helps in narrowing down the search space to relevant species, reduce computational time, and decrease the rate of false positives. This feature is especially useful when the number of possible modifications being considered is as large as 44 as in the present case. 36-39 However, there can be an unlikely although possible situation where the peptide is completely oxidized and no unoxidized version of the peptide is available. Such special cases can be detected by the quality control experiment that is usually carried out for understanding the unexposed sample prior to the whole footprinting experiment. In such cases, the observed persistent oxidation can be treated as a fixed modification (a case similar to the alkylation of Cysteines), and the remaining analysis can be carried out as usual. Oxidatively labeled peptides are detected by repeating the above steps 1 through 3 for peptides containing a single oxidative modification. Special care is taken to localize the site of modification by examination of the surrounding ions during the “Assignments Confirmation” step. This is particularly useful for cases there are multiple possible sites for an oxidative modification to occur within the same peptide. If a peptide is observed to be oxidized, the difference between the peak retention times of oxidized and unoxidized forms is utilized for further confirmation of the assignments of such species. A figure representing labeled MS2 spectrum indicating the ion assignments is saved for each of the peptide assignments so that it be retrieved at a later time for visual inspection if the need arises. If a particular residue on a peptide is confirmed to be oxidized during this process, it is added to the list called “Mods List” containing modified residues detected along with the type of modification. This step is followed by searching for two oxidative modifications per peptide using the above steps 1 through 3, with the allowable modifications selected from “Mods List”. It is rare to observe more than one oxidative modifications per peptide in a typical hydroxyl radical-mediated protein footprinting experiment since special care is taken to suppress the secondary oxidation and the total modification extent is limited.36
3.3 Quantitative Characterization
A dose response curve is a plot that indicates the fraction of unmodified peptide as a function of the exposure time of the peptide to overall radical dose by means of X-ray radiation. After the identification of peptides that are observed in both the modified and unmodified forms, the corresponding selected ion chromatogram (SIC) is extracted for each of the identified species. User is allowed to specify a window in terms of ppm around the mass value of interest for extracting the SIC, as opposed to using a constant width around m/z irrespective of its value. This ensures that the SIC window is uniform in mass domain. Area for each of the peaks representing the different forms of a peptide is computed using curve integration. The abundance of each peptide form is calculated as the integrated area under the peak in the single ion chromatogram. The fraction of the unmodified peptide (UF) is computed from the sum total of all the modified forms and unmodified peptide peak areas as follows:
| (2) |
where U represents the peak area under SIC for the unmodified peptide, Mi represents the peak area under the SIC for the ith modified form of the peptide, and n represents the total number of observed oxidatively labeled forms. A dose response curve is generated by plotting the value of UF for each time of exposure. The resulting curve typically obeys the following pseudo first order reaction:
| (3) |
where y(t) is the fraction of the unmodified peptide UF(equation 2), k is the rate constant, t is the exposure time in seconds.
4 Results and Discussion
Insulin, a metabolism-regulating hormone, exists as a monomer or a dimer form depending upon the solution conditions, it assembles to form hexamers in the presence of zinc. The structures of both monomeric and hexameric forms of insulin were investigated by oxidative protein footprinting followed by high-resolution mass spectrometry.
Human insulin monomer and hexamer proteins were prepared in cacodylic acid sodium salt trihydrate buffer (pH 7.4) at a concentration of 5 μ M. A volume of 5 μ L of the prepared samples were exposed to the X-28C X-ray white beam at the National Synchrotron Light Source, Brookhaven National Laboratory for 0, 8, 15 and 20 ms at ambient temperature. 15,40,41 After exposure, protein samples were reduced and alkylated and subjected to proteolysis by modified trypsin (Promega) at an enzyme-to-protein ratio of 1:20 wt/wt at 37° C overnight. The digest mixtures (~ 1 pmol) were loaded onto a 300 μm (internal diameter) x 5 mm C18, PepMap reverse phase trapping column to pre-concentrate and wash away excess salts using a nano HPLC UltiMate -3000 (Dionex, CA) column switching technique. The reverse phase separation was performed on a 75 Âţm (internal diameter) x 15 cm C18, PepMap column using the nano HPLC UltiMate -3000 platform (Dionex, CA). Proteolytic peptides eluting from the column with a gradient of acetonitrile of 2% per min were directed to a LTQ-FT mass spectrometer (Thermo Fisher Scientific, CA) equipped with a nano-spray ion source and with the needle voltage of 2.4 kV. All mass spectra were acquired in data dependent experiments. These experiments were set up such as MS and tandem MS spectra were acquired in positive ion mode, with the following acquisition cycle: a full scan recorded in the FT analyzer at resolution R=100000 followed by MS/MS of the eight most-intense peptide ions in the LTQ analyzer. The dose-response curves were obtained by plotting the fraction unmodified for each peptide as a function of exposure time. The resulting spectra were used for testing ProtMapMS for overall analysis.
A list of m/z values corresponding to the abundant isotopes was generated with masses obtained by theoretically digesting the insulin sequences using custom enzyme digestion (allowing for cleavages after residues Arg, Lys, and His) for charge states ranging from 1-4, and was compared against the experimentally observed precursor ion m/z values. In addition to cleaving after the expected Arg and Lys residues, Trypsin has also been observed to cleave after the residue His in the protein sequence. This observation led us to use the custom enzyme setting to allow for observing peptides resulting from all expected cleavage sites. All the matches within 10 ppm were retained and subjected to MS2 verification. The observed MS2 spectra were compared against their theoretical counterparts for the likely candidates, and the candidate with the best match is identified as the true match for the observed mass as discussed in section 3. A sample output of monoisotopic mass assignments from a mass spectrum of insulin exposed to hydroxyl radicals analyzed using this approach is depicted in Table 2. For example, first row in the table indicates that the theoretical monoisotopic mass (MI Mass) of 858.4276 matches the experimentally observed mass of doubly charged B-chain peptide 23-29 within an error of 1 ppm. The peptide, observed in the mass spectrum at an m/z value of 430.2222 with a charge state of 2, elutes at 22.10 minutes in the liquid chromatography column. Rows 2 through 6 in the table represent the various site-specific oxidative modifications observed on the peptide.
Table 2.
Sample output of mass assignments for human insulin, SAA and and EAA columns indicate the residue identities and positions of the start and end amino acid residues of the peptides representing the theoretical monoisotopic mass (MI Mass), observed within a certain error (Err), specified in parts-per-million (ppm). Retention time in LC column, m/z location of isotopic cluster in the spectrum, the charge state value (Z), and modifications, if any, on the identified peptide
| SAA | EAA | MI Mass | Err | Retention Time | m/z | Z | Modifications |
|---|---|---|---|---|---|---|---|
| G23 | K29 | 858.4276 | -1 | 21.25 | 430.2222 | 2 | |
| G23 | K29 | 872.4069 | 0 | 21.65 | 437.2113 | 2 | P28+14 |
| G23 | K29 | 874.4225 | 1 | 20.51 | 438.2186 | 2 | F24+16 |
| G23 | K29 | 874.4225 | 1 | 19.52 | 438.2186 | 2 | F25+16 |
| G23 | K29 | 874.4225 | 1 | 21.15 | 438.2186 | 2 | Y26+16 |
| G23 | K29 | 890.4174 | -1 | 18.41 | 446.2168 | 2 | F25+32 |
Figure 1 illustrates the liquid chromatographic elution profile for human insulin monomer peptide 23-29 identified in Table 2. The four plots in Figure 1 represent the chromatographic elution plots for doubly charged insulin B-chain peptide 23-29 for an X-ray exposure time of 0, 8, 15, and 20 ms. The unmodified form of the peptide is indicated by cyan (m/z=430.22) color. The oxidatively modified forms are magnified by a factor of 18 for easier viewing and are represented by blue (m/z value of 437.21, P28 oxidation (+14 Da)), green (m/z=438.22, single oxidation of F24, F25, and Y26 (+16 Da)), and red (m/z location of 446.22, double oxidation (+32 Da)) colors respectively (See also Table 2). When the protein is unexposed to any hydroxyl radicals by means of X-ray exposure, none of the modified forms the peptide is observed as expected (Figure 1). As the X-ray exposure time to the protein increases, the relative intensities of the modified forms increase as seen in Figure 1. This is because of the fact that the peptide has increased opportunity to react with hydroxyl radicals. As expected, the oxidized peptides elute at a slightly different time than their unoxidized counterpart due to the change in hydrophobicity with the addition of polar oxygen atoms. Typically, depending upon the identity of the amino acid residue being modified, the oxidatively labeled peptide elutes a few seconds to four minutes earlier than its unmodified counterpart.42 In contrast, owing to a different chemistry,17 oxidatively modified products of Arginine, Histidine, and Proline have been recently observed to migrate after the unmodified form of the peptide in the liquid chromatography column. For example, the modified products for peptide segment 23-29 that incorporated one oxygen atom represents five isomeric forms of the peptide molecule (Figure 1, green SIC peaks labeled as A-E). Tandem MS indicates that the peaks A, B, and C represent F25+16 form of the peptide, while peaks D and E represent a mixture of F24+16 and F25+16 isomeric forms, while Y26+16 is the predominant species representing peak F. Note that the F25 residue of the protein is highly solvent accessible (Table 3), which could potentially allow for multiple possibilities for the site of oxidation on the F25 ring. We speculate that the varying chromatographic elution times in peaks A-C might represent different isoforms of the F25+16 modified peptide, with each isoform containing oxidative group at a different position on the F25 ring. Such variations in behavior for ortho-, para-, or meta-substituted phenols is well precedented.43
Figure 1.
Chromatographic elution plots for doubly charged human insulin B-Chain peptide 23-29. The unmodified form is shown in cyan, while the modified forms (magnified by a factor of 18) are shown in blue (P28+14), green (mixture of F24+16, F25+16, and Y26+16), and red (F25+32).
Table 3.
Oxidation table for Insulin Monomer and Hexamer: Detected trypsin digested peptides, oxidized residues determined using mass spectrometry (Col 2 and 6), solvent accessibility area as obtained from X-Ray crystallography (Col 3 and 7), rate constants evaluated manually (Cols 4 and 8) and automatically using ProtMapMS (Cols 5 and 9)
| Peptide Sequence |
Insulin Monomer | Insulin Hexamer | ||||||
|---|---|---|---|---|---|---|---|---|
| Oxid. Res. |
SA (Å) | Rate Const (s-1) (Manual) |
Rate Const (s-1) (ProtMapMS) |
Oxid. Res. |
SA (Å) | Rate Const (s-1) (Manual) |
Rate Const (s-1) (ProtMapMS) |
|
| FVNQHLCGSH (1-10, B-Chain) |
F1 | 123.6 | 1.41±0.14 | 1.36±0.16 | F1 | 11.1 | 0.40±0.02 | 0.46±0.06 |
| H5 C7 H10 |
91.6 44.5 119.3 |
14.89±1.42 | 13.07±0.92 | H5 C7 H10 |
92.0 49.7 54.0 |
7.64±0.44 | 6.67±0.41 | |
| LVEALYLVCGER (11-22, B-Chain) |
Y16 L17 R22 |
137.3 117.2 124.1 |
8.59±0.57 | 5.64±0.55 | Y16 L17 R22 |
37.1 9.4 104.9 |
1.81±0.11 | 1.53±0.24 |
| GFFYTPK (23-29, B-Chain) |
F24 F25 Y26 P28 |
30.9 140.1 66.1 52.1 |
9.33±0.12 | 8.14±0.3 | F24 F25 Y26 P28 |
0.0 55.8 9.2 15.9 |
5.11±0.11 | 4.61±0.16 |
| GIVEQCCTSI CSLYQLENYCN (1-21, A-Chain) |
C7 T8 I10 L13 Y14 Y19 |
38.9 90.0 93.0 44.7 137.6 62.5 |
56.4±7.6 | 47.5±8.6 | C7 T8 I10 — Y14 Y19 |
24.0 89.9 77.1 1.1 110.3 62.5 |
32.7±1.8 | 29.8±5.3 |
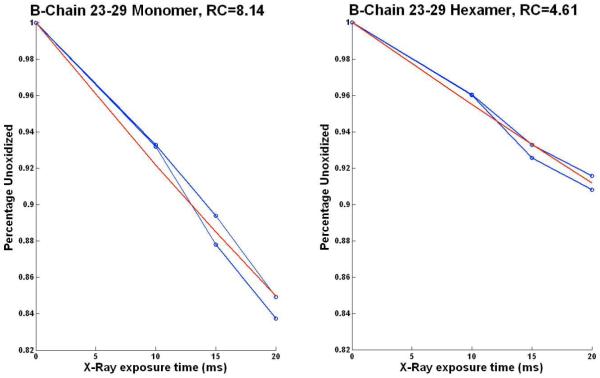
A total of sixteen independent mass spectrometry experiments were carried out (samples exposed to hydroxyl radicals for 0, 8, 15, 20 ms; two sets each for insulin monomer and hexamer) and fitted globally to provide the oxidation rate constants using nonlinear regression. The data obeys a pseudo first-order reaction at suitable irradiation times as seen in Figure 2. ProtMapMS is able to automatically generate such time resolved dose response plots for each of the observed oxidatively modified peptides. Typically, the m/z extraction window is 10 ppm wide and centered at the experimental m/z value so that window width varies with m/z value. The rate constants calculated using this method are indicative of the solvent accessibility and reactivity of the peptide residues. For example, Figure 2 shows the dose response curves for the insulin monomer (left) and hexamer (right) peptide obtained using two independent experiments (blue lines), with each experiment representing four time intervals corresponding to an exposure time of 0, 8, 15, and 20 ms. The experimental curves for insulin monomer and hexamer fit to the theoretical curves (red lines) with a rate constant of 8.14 s-1 and 4.61 s-1 respectively. Higher rate constant for the monomeric form indicates that the oxidized residues have higher solvent accessibility than in the case of hexameric form. Such measures have proved to be very useful for providing atomic structural models when used in conjunction with computational methods such as protein-protein docking and homology modeling.13,44
Figure 2.
Dose Response plots for human insulin B-Chain peptide 23-29. The experimental curves (blue) from two sets of experiments fit to the first order theoretical curves (red) corresponding to a rate constant of 8.14 for monomeric form versus 4.61 for the hexamer. Lower rate constant for the hexamer indicates the reduced solvent accessibility for the peptide.
At longer irradiation times, with the accumulation of multiple oxidative modifications, the biomolecule may vary from its native configuration. This introduces non-linearity in the dose response plots at longer time points due to conformational variation of the reactive site under study. Thus, special consideration is given to selecting appropriate irradiation times to avoid such a phenomenon. However, if such a case does occur in practice, ProtMapMS provides the user with an option to select the data points of interest and reject the outliers, and it recalculates the rate constant based upon the new selection.
Table 3 compares the rate constants obtained manually and automatically using ProtMapMS for tryptic peptides of Insulin monomer and hexamer. Column 1 indicates the sequence and position of the observed peptide, columns 2 and 6 represent the residues found to be oxidized using mass spectrometric analysis of monomeric and hexameric form respectively, Solvent Accessibility (SA, represented in Å) for the observed oxidized residues is indicated as determined by their crystallographic structures. The rate constant values obtained for each of the peptides evaluated using ProtMapMS are shown in columns 5 and 9 for monomer and hexamer respectively, while those calculated using manual analysis are represented by columns 4 and 8 respectively.
In order to test the accuracy of the results obtained using ProtMapMS, the data was also manually examined as follows: The mass spectra were searched using Bioworks 3.3.1 software (Thermo Scientific, CA) in multiple steps against a database comprising only the sequence of human insulin allowing for a total of 28 oxidative modifications. Since Bioworks 3.3.1 allows for only a limited number of modifications, multiple searches were required, with each search allowing for a different set of oxidative modifications. The results from each search were compiled together manually. As a result of this search, 17 unique side chain oxidation products were observed. Four tryptic peptides comprised of 1-10, 11-22 and 23-29 segments of insulin B-chain, and 1-21 segment of insulin A-chain covering 99% of human insulin sequence were detected. All four peptides were observed to be oxidized in the human insulin monomer and hexamer. The peak area for m/z ion signals of these peptides and their radiolytic products were calculated by integration of the intensity values of these ions signals that were manually extracted from the total ion current chromatogram (TIC). The extent of modification was manually calculated for each peptide as the ratio of the area under the ion signal for unmodified peptide to the sum of the areas under the ion signals for unmodified peptide and their radiolytic products at each time of exposure.
As shown in Table 3, ProtMapMS was also able to detect the tryptic peptides 1-10, 11-22, 23-29 segments of B-chain, and 1-21 segment of A-chain. In addition, all the 17 side chain oxidation products that were observed in manual analysis were also observed, and the oxidation of these residues was confirmed by tandem MS analysis. Although the rates of oxidation measured by the two methods were similar, we did not expect the two oxidation rate constants to be identical. This is due to the differences in the parameters used in the two approaches. The manual analysis employed visual examination and subjective judgement by the user for selecting the parameters while ProtMapMS uses optimized and specific parameters across the whole analysis. For example, the manual analysis selected the first isotope for SIC extraction, doubly charged state for each peptide, fixed m/z window of 0.02 Daltons for SIC extraction, and used visual inspection for selecting the retention time window for SIC integration. On the other hand, ProtMapMS uses optimized parameters such as the most abundant isotope for SIC extraction, charge state representing the highest current intensity, an m/z window of 10 ppm so that it varies with the absolute value of m/z and allows for better specificity, and a retention time window of six minutes for SIC integration around the peak retention time for each of the species to account for the all the isoforms of modified peptides. Nevertheless, despite the variation in the parameters, the rate constants using the two approaches are comparable as seen in Table 3.
The Solvent Accessibility, SA (in Å)), of human insulin monomer and hexamer was calculated in order to verify the primary target residues that were likely to be modified using VADAR, 45 and is shown in columns 3 and 7 for insulin monomer and hexamer respectively. All of the four peptides within the B-chain of human insulin hexamer exhibited significant decrease in the oxidation compared with the case for human insulin monomer. B-Chain peptide 1-10 experienced a drop in the rate constant by a factor of 1.9, while the rate constants for peptides 11-22 and 23-29 dropped by 4.7 and 1.8 times respectively in the insulin hexamer form. Because the solvent accessibility of the side chain residues on the surface of the protein correlates with the reaction rates, 15,16 these observations suggest that the reactive side chains that showed decrease in the extent of oxidation are protected in the human insulin hexamer compared to the human insulin monomer. These results are consistent with the crystallographic data for human insulin monomer and hexamer. Specifically, the crystal structure revealed dramatic decreases in the solvent accessibility of the amino acid side chains of insulin upon hexamer formation including F1 in the segment 1-10, Y16 and L17 in the segment 11-22, and F24, Y26 and P28 in the segment 23-29 of the B-chain from 5 to 30 fold. In addition, residues C7 and L13 in the segment 1-21 of the insulin A-chain showed a decrease in the solvent accessibility, which is reflected in the drop in the rate constant by 1.5 times for the peptide upon hexamer formation. Moreover, residue L13 of A-chain was observed to be oxidized only in monomeric form, we failed to detect its oxidative state in the hexamer. However, residue F24 of B-chain was observed to be oxidized in the hexamer, although it is not solvent accessible as observed in the crystallographic form. This may suggest a different structural conformations of the protein in the crystal and the solution form.
An interesting observation revealed through the automated analysis using ProtMapMS was that the oxidative modification on H5 and H10 of B-chain peptide 1-10 were observed only for the doubly charged peptide, while no such modifications were observed on its triply charged counterpart. This could possibly be due to the fact that the oxidative group attached to the Histidine side chains interferes with the ability of the residue to retain the positive charge. Moreover, there was strong experimental evidence for an hydroxy-oxidative modification only on residue F1 for the peptide. This enabled us to calculate the rate constant for the individual residue F1 from the triply charged peptide as shown in Table 3. The solvent accessibility area for F1 residue drops by 11 times in going from monomeric to hexameric state, the rate constant for the residue drops by 3 times. Overall, the substantial decrease in the oxidation of various amino acids indicates that the structural reorganization associated with the insulin hexamer formation is extensive.
ProtMapMS provides a comprehensive solution for all data analysis needs for covalent labeling experiments. Dose response curves and chromatographic elution plots for each of the oxidatively modified peptides were generated indicating the different oxidative states of the peptide as a function of time, which was previously not possible. The manual analysis of the protein spectra using semi-automated methods including database search tools took about 50 man hours of an expert footprinter for processing the data, while the same results were obtained using ProtMapMS in an hour, followed by an hour of manual validation of the automated results. This illustrates that ProtMapMS has significantly reduced one of the largest bottlenecks in protein footprinting experiments. Automated analysis ensures that consistent comparison can be made across different experiments using the same parameter values. This eliminates the possibility of human bias due to subjectivity and human error in the interpretation. In addition, it is easier to change the parameter settings and observe the affect of changing various parameters such as SIC extraction window, charge state, allowed modifications etc., across different experiments. As the spectrum complexity rises for larger biomolecules/macromolecular complexes, it becomes extremely difficult to perform manual analysis. On the other hand, ProtMapMS scales very well with the complexity of the data. Owing to the technological advancements in the computational power, no significant increase in computational time is expected in going from simple to complex set of data. As the data interpretation becomes easier, ProtMapMS provides the possibility of extending the scope of utilization of footprinting technique to a wider audience by eliminating the need for expert judgement for data analysis.
The proposed computational approach is generic and can be extended to structural mass spectrometry using any kind of covalent labeling. As it becomes easier to interpret the data resulting from covalent labeling experiments, this approach can be used to probe large proteins, macromolecular interfaces, conformational changes and biomolecular dynamics. Such experimental data can be used in conjunction with computational structure modeling techniques such as comparative modeling and threading. These theoretical models in the absence of support from experimental data lack reliability, especially when appropriate templates are not available in the case of ab-initio modeling. Hybrid approaches representing a combination of computational modeling and experimental methods such as hydrogen-deuterium exchange and covalent labelling are increasingly gaining interest to combine the benefits from both the approaches. 46,47 Specifically, the results from experimental analysis provide explicit constraints, which determine the surface accessibility or burial of particular residues, that can be incorporated for refining computational structure prediction results, greatly reducing the model space to be considered. Footprinting data has been successfully used for RNA structure prediction.48 There is a great potential for similar progress for protein structure prediction.
5 Conclusions
Structural mass spectrometry provides valuable insights into macromolecular structures and interactions. Data analysis presents itself as the biggest bottleneck for such experiments till date. A computational framework and algorithms have been developed and integrated in the form of ProtMapMS for the automated analysis of mass spectrometry data aimed specifically at such studies. ProtMapMS modules include data format conversion, mass spectrum interpretation, liquid chromatography data extraction, assignment and verification of all the observed masses, identification of the oxidized peptide products, verification of the site of modification, quantitatively characterizing the oxidatively labeled peptides, and generating the dose response curves for each oxidative peptide. Human insulin samples both in monomeric and hexameric forms were exposed to synchrotron X-ray beam for intervals ranging from 0 to 20 milliseconds. A total of four independent mass spectrometry experiments representing tryptic peptides of hydroxyl radical exposed human insulin (two each of insulin monomeric and hexameric form) were carried out. The resultant data was analyzed both manually and using ProtMapMS without any manual intervention. Both manual and automated analysis revealed 21 unique side chain oxidation products of the protein. Dose response plot of the modified peptides was constructed automatically to assist in determining the rate constant. The rate constants obtained for peptides from insulin hexamer were significantly smaller than the case of monomeric state, indicating that the structural reorganization associated with the insulin hexamer formation is extensive. The results obtained using ProtMapMS and manual analysis were found to be in close agreement and were overall were consistent with the predictions from the crystallographic structure. Automated analysis reduced the data interpretation time from weeks to couple of hours. Results from this approach provide extremely valuable set of constraints that can be used for validation purposes in computational modeling for protein structure determination. The generic nature of the approach allows it to be easily extended for hydrogen/deuterium exchange studies and chemical cross-linking experiments.
Acknowledgement
This work was supported by NIH grants P41-EB-01979 and U54-GM-074945. We thank Prof Keiji Takamoto, Prof Peter B. O’Connor, Dr Amisha Kamal, and Dr Xiaojing Zheng for scientific discussions, Jennifer Burgoyne, Dr Sayan Gupta, and Dr Serguei Ilchenko for experimental assistance. ProtMapMS is available through a free license for academic use. For further questions regarding the availability of ProtMapMS, please contact the authors.
References
- (1).Aebersold R, Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- (2).Back JW, de Jong L, Muijsers AO, de Koster CG. J Mol Biol. 2003;331:303–313. doi: 10.1016/s0022-2836(03)00721-6. [DOI] [PubMed] [Google Scholar]
- (3).Wales TE, Engen JR. Mass Spectrom. Rev. 2006;25:158–70. doi: 10.1002/mas.20064. [DOI] [PubMed] [Google Scholar]
- (4).Suckau D, Mak M, Przybylski M. Proc Natl Acad Sci USA. 1992;89:5630–5634. doi: 10.1073/pnas.89.12.5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Sheshberadaran H, Payne LG. Proc Natl Acad Sci USA. 1988;85:1–5. doi: 10.1073/pnas.85.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Sharp JS, Becker JM, Hettich RL. Analytical Chemistry. 2004;76:672–683. doi: 10.1021/ac0302004. [DOI] [PubMed] [Google Scholar]
- (7).Watson C, Janik I, Zhuang T, Charva£tova£ O, Woods RJ, Sharp JS. Analytical Chemistry. 2009;81:2496–2505. doi: 10.1021/ac802252y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Maleknia SD, Brenowitz M, Chance MR. Anal Chem. 1999;71:3965–3973. doi: 10.1021/ac990500e. [DOI] [PubMed] [Google Scholar]
- (9).Sharp JS, Becker JM, Hettich RL. Anal Biochem. 2003;313:216–225. doi: 10.1016/s0003-2697(02)00612-7. [DOI] [PubMed] [Google Scholar]
- (10).Hambly DM, Gross ML. Journal of the American Society for Mass Spectrometry. 2005;16:2057–2063. doi: 10.1016/j.jasms.2005.09.008. [DOI] [PubMed] [Google Scholar]
- (11).Sclavi B, Sullivan M, Chance MR, Brenowitz M, Woodson SA. Science. 1998;279:1940–1943. doi: 10.1126/science.279.5358.1940. [DOI] [PubMed] [Google Scholar]
- (12).Kiselar JG, Mahaffy R, Pollard TD, Almo SC, Chance MR. Proc Natl Acad Sci USA. 2007;104:1552–1557. doi: 10.1073/pnas.0605380104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Takamoto K, Kamal JKA, Chance MR. Structure. 2007;15:39–51. doi: 10.1016/j.str.2006.11.005. [DOI] [PubMed] [Google Scholar]
- (14).Sharp JS, Tomer KB. Biophysical Journal. 2007;92:1682–1692. doi: 10.1529/biophysj.106.099093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Guan J-Q, Almo SC, Chance MR. Accounts of Chemical Research. 2004;37:221–229. doi: 10.1021/ar0302235. [DOI] [PubMed] [Google Scholar]
- (16).Kiselar JG, Maleknia SD, Sullivan M, Downard KM, Chance MR. Int J Radiat Biol. 2002;78:101–114. doi: 10.1080/09553000110094805. [DOI] [PubMed] [Google Scholar]
- (17).Xu G, Chance MR. Chemical Reviews. 2007;107:3514–3543. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- (18).Takamoto K, Chance MR. Annu Rev Biophys Biomol Struct. 2006;35:251–276. doi: 10.1146/annurev.biophys.35.040405.102050. [DOI] [PubMed] [Google Scholar]
- (19).Weis DD, Engen JR, Kass IJ. J Am Soc Mass Spectrom. 2006;17:1700–1703. doi: 10.1016/j.jasms.2006.07.025. [DOI] [PubMed] [Google Scholar]
- (20).Pascal BD, Chalmers MJ, Busby SA, Griffin PR. Journal of the American Society for Mass Spectrometry. 2009;20:601–610. doi: 10.1016/j.jasms.2008.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Slysz G, Baker C, Bozsa B, Dang A, Percy A, Bennett M, Schriemer D. BMC Bioinformatics. 2009;10:162. doi: 10.1186/1471-2105-10-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Lou X, Kirchner M, Renard B, Koethe U, Voss B, Graf C, Steen J, Steen H, Mayer M, Hamprecht F. HeXicon: Fully Automated HX-MS Data Analysis with Complete Deuteration Distribution Estimation”; Proceedings of 56th American Society of Mass Spectrometry conference on Mass Spectrometry; 2008. [Google Scholar]
- (23).Chen T, Jaffe JD, Church GM. J Comput Biol. 2001;8:571–583. doi: 10.1089/106652701753307494. [DOI] [PubMed] [Google Scholar]
- (24).Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- (25).Eng JK, McCormack AL, Yates JR. Journal of the American Society for Mass Spectrometry. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- (26).Tabb DL, Fernando CG, Chambers MC. Journal of Proteome Research. 2007;6:654–661. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Tanner S, Shu H, Frank A, Wang L-C, Zandi E, Mumby M, Pevzner PA, Bafna V. Analytical Chemistry. 2005;77:4626–4639. doi: 10.1021/ac050102d. [DOI] [PubMed] [Google Scholar]
- (28).Bern M, Cai Y, Goldberg D. Analytical Chemistry. 2007;79:1393–1400. doi: 10.1021/ac0617013. [DOI] [PubMed] [Google Scholar]
- (29).Charvátová O, Foley BL, Bern MW, Sharp JS, Orlando R, Woods RJ. Journal of the American Society for Mass Spectrometry. 2008;19:1692–1705. doi: 10.1016/j.jasms.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Pedrioli PGA, et al. Nat Biotechnol. 2004;22:1459–1466. doi: 10.1038/nbt1031. [DOI] [PubMed] [Google Scholar]
- (31).Kessner D, Chambers M, Burke R, Agus D, Mallick P. Bioinformatics. 2008;24:2534–2536. doi: 10.1093/bioinformatics/btn323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Seattle Proteome Center - Proteomics Tools. http://tools.proteomecenter.org/software.php.
- (33).Rockwood AL. Anal. Chem. 1996;68:2027–2030. doi: 10.1021/ac951158i. [DOI] [PubMed] [Google Scholar]
- (34).Rodgers JL, Nicewander WA. The American Statistician. 1988;42:59–66. [Google Scholar]
- (35).Biometrika. 1908;6:1–25. STUDENT. [Google Scholar]
- (36).Xu G, Kiselar J, He Q, Chance MR. Anal Chem. 2005;77:3029–3037. doi: 10.1021/ac048282z. [DOI] [PubMed] [Google Scholar]
- (37).Xu G, Chance MR. Analytical Chemistry. 2005;77:4549–4555. doi: 10.1021/ac050299+. [DOI] [PubMed] [Google Scholar]
- (38).Xu G, Chance MR. Analytical Chemistry. 2004;76:1213–1221. doi: 10.1021/ac035422g. [DOI] [PubMed] [Google Scholar]
- (39).Xu G, Takamoto K, Chance MR. Anal Chem. 2003;75:6995–7007. doi: 10.1021/ac035104h. [DOI] [PubMed] [Google Scholar]
- (40).Sclavi B, Woodson S, Sullivan M, Chance M, Brenowitz M. Methods Enzymology. 1998;275:379–402. doi: 10.1016/s0076-6879(98)95050-9. [DOI] [PubMed] [Google Scholar]
- (41).Y RC, B S, M S, L DM, A WS, R CM, M B. Methods Enzymology. 2000;317:353–368. doi: 10.1016/s0076-6879(00)17024-7. [DOI] [PubMed] [Google Scholar]
- (42).Kiselar JG, Janmey PA, Almo SC, Chance MR. Mol Cell Proteomics. 2003;2:1120–1132. doi: 10.1074/mcp.M300068-MCP200. [DOI] [PubMed] [Google Scholar]
- (43).Dowle CJ, Malyan AP, Matheson AM. The Analyst. 1990;115:105. [Google Scholar]
- (44).Kamal JKA, Benchaar SA, Takamoto K, Reisler E, Chance MR. Proc Natl Acad Sci USA. 2007;104:7910–7915. doi: 10.1073/pnas.0611283104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Willard L, Ranjan A, Zhang H, Monzavi H, Boyko RF, Sykes BD, Wishart DS. Nucl. Acids Res. 2003;31:3316–3319. doi: 10.1093/nar/gkg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Pantazatos D, Kim JS, Klock HE, Stevens RC, Wilson IA, Lesley SA, Woods VL. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:751–756. doi: 10.1073/pnas.0307204101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Zhu MM, Rempel DL, Du Z, Gross ML. Journal of the American Chemical Society. 2003;125:5252–5253. doi: 10.1021/ja029460d. [DOI] [PubMed] [Google Scholar]
- (48).Sclavi B, Sullivan M, Chance MR, Brenowitz M, Woodson SA. Science. 1998;279:1940–1943. doi: 10.1126/science.279.5358.1940. [DOI] [PubMed] [Google Scholar]