Abstract
Oncogenes influence nutrient metabolism and nutrient dependence. The oncogene c-Myc stimulates glutamine metabolism and renders cells dependent on glutamine to sustain viability (“glutamine addiction”), suggesting that treatments targeting glutamine metabolism might selectively kill c-Myc-transformed tumor cells. However, many current or proposed cancer therapies interfere with the metabolism of glucose, not glutamine. Here we studied how c-Myc-transformed cells maintained viability when glucose metabolism was impaired. In SF188 glioblastoma cells, glucose deprivation did not affect net glutamine utilization but elicited a switch in the pathways used to deliver glutamine carbon to the tricarboxylic acid cycle, with a large increase in the activity of glutamate dehydrogenase (GDH). The effect on GDH resulted from the loss of glycolysis, because it could be mimicked with the glycolytic inhibitor 2-deoxyglucose and reversed with a pyruvate analog. Furthermore, inhibition of Akt signaling, which facilitates glycolysis, increased GDH activity and over-expression of Akt suppressed it, suggesting that Akt indirectly regulates GDH through its effects on glucose metabolism. Suppression of GDH activity with RNA interference or an inhibitor showed that the enzyme is dispensable in cells able to metabolize glucose, but is required for cells to survive impairments of glycolysis brought about by glucose deprivation, 2-deoxyglucose or Akt inhibition. Thus, inhibition of GDH converted these glutamine addicted cells to glucose addiction. The findings emphasize the integration of glucose metabolism, glutamine metabolism and oncogenic signaling in glioblastoma cells, and suggest that exploiting compensatory pathways of glutamine metabolism can improve the efficacy of cancer treatments that impair glucose utilization.
Keywords: Glioblastoma, metabolism, glutamine, glucose, Akt
Introduction
Tumor cells display enhanced, autonomous, or otherwise unusual metabolic activities compared to differentiated cells (1). Since Warburg's experiments in the 1920s, the metabolic idiosyncrasies of tumors have been proposed to hold therapeutic opportunities in cancer, and many studies have focused on identifying pathways supporting tumor cell survival and growth. So far, enhanced rates of glucose and glutamine utilization are best established as hallmarks of tumor metabolism (2-4). The central roles of these nutrients derive from their abundance in vivo, their use as bioenergetic substrates, and especially from the fact that they provide precursors for the synthesis of lipids, proteins and nucleic acids. Metabolic flux analysis in proliferating glioblastoma cells demonstrated that the coordinated metabolism of glucose and glutamine enabled a metabolic platform supporting both bioenergetics and biosynthesis (5). These observations suggest that a better understanding of the relationships between glucose and glutamine metabolism will lead to novel approaches to curb tumor growth.
Consistent with the importance of glucose and glutamine for tumor growth, many studies have demonstrated that tumor suppressors and oncogenes influence the metabolism of these nutrients. The phosphatidylinositol 3′-kinase (PI3K)/Akt pathway, which is enhanced in a large fraction of human tumors, positively regulates glucose uptake, glycolysis, and the use of glucose as a macromolecular precursor during cell growth (6, 7). The oncogene c-Myc facilitates glutamine metabolism by increasing the expression of surface transporters and enzymes (8, 9). In glioblastoma cells with enhanced c-Myc activity, Akt inhibition decreases glycolysis without affecting glutamine consumption, while suppression of c-Myc reduced the uptake and mitochondrial processing of glutamine (9). Thus, while many tumor cells exhibit high rates of glucose and glutamine consumption, distinct oncogenic pathways regulate discrete aspects of the metabolic phenotype.
A surprising consequence of transformation is that it can limit metabolic flexibility in vitro. For example, Akt not only increases glucose metabolism, but also increases sensitivity to glucose withdrawal (10). This effect is due to an impaired ability to activate β-oxidation, which normally supports bioenergetics during glucose starvation (10, 11). Similarly, cells with enhanced c-Myc activity cannot survive glutamine withdrawal because they cannot maintain pools of tricarboxylic acid (TCA) cycle intermediates in the absence of glutamine (9, 12). These nutrient “addiction” studies suggest that targeting intermediary metabolism will be selectively toxic to cancer cells, with therapeutic specificity defined by the tumor genotype. This might be particularly useful in glucose addiction, because many current cancer therapies alter tumor glucose uptake and are correlated with reduced signal on 18fluorodeoxyglucose positron emission tomography (18FDG-PET) (13).
Importantly, however, tumor cells may retain the capacity for metabolic compensation in vivo to survive periods of diminished glucose metabolism. Recent studies have documented that a decrease in 18FDG-PET signal during cancer therapy does not necessarily predict histopathological resolution or improved patient outcome (14, 15). These studies imply that glycolytic tumors survive stress by activating alternative metabolic pathways, and that defining and targeting these pathways will improve cancer treatment. Given the complementary roles of glucose and glutamine in intermediary metabolism, we studied how glutamine-addicted glioblastoma cells survived glucose deprivation.
Materials and Methods
Reagents
Methyl-pyruvate (CH3-Pyr), dimethyl-α-ketoglutarate (dm-αKG), epigallocatechin gallate (EGCG) and 2-deoxyglucose (2-DG) were from Sigma. The isozyme selective (Akt 1/2) inhibitor Akti VIII was from Calbiochem. Isotopes were obtained from Isotec (L-[3-13C]-glutamine) or Cambridge Isotope Laboratories (L-[α-15N]-glutamine; L-[γ-15N]-glutamine).
Cell culture, metabolism and viability experiments
SF188 cells (UCSF Brain Tumor Registry) were passaged in culture as described (5). These experiments used an SF188-derived cell line over-expressing human Bcl-xL previously used to study metabolism during Akt inhibition (9). The mouse astrocyte cell lines were described previously (16). For all experiments involving nutrient depletion or isotopically-enriched nutrients, Dulbecco's Modified Eagle's Medium was prepared from powder lacking glucose, L-glutamine, phenol red, sodium pyruvate and sodium bicarbonate (Sigma). This was supplemented with 10% dialyzed fetal calf serum (Hyclone), sodium bicarbonate (42.5 mM), HEPES (25 mM), Penicillin/Streptomycin (10 U and 10 μg/mL, respectively), and glucose/glutamine as indicated. To measure metabolic rates, one million cells were plated into 6-cm dishes and cultured until 90% confluent. At time 0, the cells were rinsed in PBS, fed with 2 mL of the test medium and cultured. End-point experiments proceeded for 6-8 hours, then the medium was collected and analyzed for metabolite abundance. Rates were based on the change in metabolite abundance from the start medium. Ammonia was measured with a spectrophotometric assay (Megazyme). Glucose, lactate, glutamine and glutamate were measured with a chemical analyzer (NOVA Biomedical), and other amino acids were measured by HPLC. For viability measurements, the culture medium and trypsinized cells were combined, stained with Trypan Blue and counted with a hemacytometer. Phase contrast images were obtained with a Nikon Eclipse TE300 microscope and processed with MetaMorph (Universal Imaging).
RNA interference
siRNAs were obtained from Ambion, Inc, reconstituted in water to 20 μM and transfected using Effectene (Qiagen). Manufacturer protocols were used to transfect 200 picomoles of each siRNA into one million cells at a final siRNA concentration of 40 nM. Glucose withdrawal experiments were performed on the 7th day after transfection.
Western Blotting
Whole-cell lysates were prepared in RIPA buffer and quantified using the BCA Protein Assay (Thermo Scientific). Protein was separated by SDS-PAGE, transferred to nitrocellulose, and probed with monoclonal antibodies against GDH (Novus Biologicals) or β-actin (Sigma). Detection was performed using SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific) and a UVP BioImaging System.
13C nuclear magnetic resonance (NMR)
Cells were cultured in three 15-cm dishes until 90% confluent, then rinsed and overlaid with 25 mL of medium containing 2 mM L-[3-13C]-glutamine, with or without 10 mM unlabeled glucose. After 7 hours, the medium was removed and the cells were rinsed in PBS. The dishes were placed at −80°C for 10 minutes to halt metabolic activity. Frozen cells were homogenized by sonication in 4 mL of 6% ice-cold perchloric acid. After centrifuging to remove debris, the acid-soluble material was neutralized with 8 N potassium hydroxide and centrifuged. The supernatant was lyophilized, reconstituted in D2O and analyzed in a 5-mm NMR tube. NMR analysis was performed on a Bruker Advance™ 400 wide bore 9.4T instrument equipped with a 5-mm 1H/13C dual probe. 13C spectra were recorded under approximately fully relaxed conditions and broadband proton decoupling. Carbon spectra were acquired under the following conditions: pulse flip angle 45°; repetition time 1.5 seconds; spectral width 25 KHz; number of data points 64,000; number of scans approximately 25,000. Free induction decays were zero-filled to 131,072 points and apodized with exponential multiplication. Spectra were analyzed using NMR Utility Transform Software (Acorn NMR) and the relative abundances of glutamate C4 and C5 signals were determined by normalizing areas to natural abundance HEPES resonances.
Gas chromatography/mass spectrometry (GC/MS)
Enrichment in NH4+ was determined using a modification of a published method (17). Briefly, NH4+ from 50-100 μL of culture medium was converted to aminobutyrate in the presence of oxobutyrate, purified glutamate dehydrogenase, and NADH (Sigma) for 30 minutes at 37°C. The sample was applied to an ion exchange column (AG 50W-X4, BioRad), washed with water, and eluted with 2 mL of 4 N ammonium hydroxide. To determine enrichment in alanine, culture medium was subjected to the same column purification. Standards containing known ratios of unlabeled/15N-labeled NH4+ and alanine were prepared using the same method. Samples were evaporated and derivatized in 100 μL of Tri-Sil (Thermo Scientific), then analyzed by GC/MS with an Agilent 6890N GC coupled to an Agilent 5973 Mass Selective Detector. The oven temperature was ramped from 70°C to 150°C by 5°C/minute, then by 10°C/minute to 325°C. The following ion ratios were measured: alanine m/z 116 (unenriched) and 117 (enriched); and aminobutyrate m/z 130 (unenriched) and 131 (enriched). The atom percent excess (A.P.E.) of each product was calculated by comparing the ion ratio to the standard curve.
Results
Glucose withdrawal stimulates mitochondrial glutamine metabolism
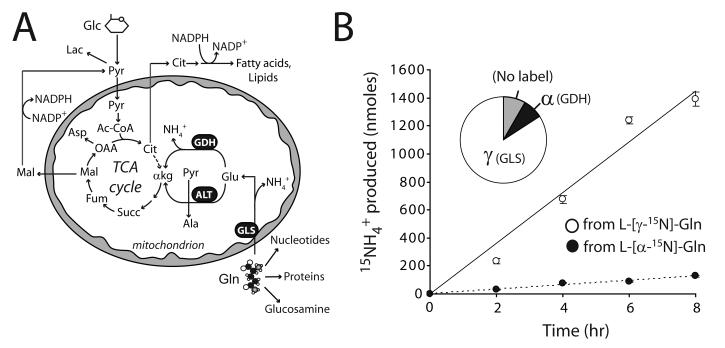
We previously determined that SF188 glioblastoma cells metabolized carbon from glutamine in the TCA cycle and secreted ammonia in the process (5). To understand how the cells converted glutamine into the TCA cycle intermediate α-ketoglutarate (α-KG), we compared the activities of glutaminase (GLS) and GDH. These two enzymes generate ammonia from the γ and α nitrogens of glutamine, respectively, and their sequential activity produces α-KG (Fig. 1A). We cultured cells with glutamine that was labeled with 15N in either the γ- or α-position, and followed production of 15NH4+ by GC/MS (Fig. 1B). After 8 hours, more than 90% of the ammonia in the medium was derived from these two nitrogens (Fig. 1B). More than half (58±1%) of the glutamine consumed by the cells was metabolized by GLS, and thus this enzyme outpaced all other glutamine-consuming systems combined. Furthermore, the GDH flux was approximately 10% of the GLS flux, consistent with the known contribution of other glutamate-consuming enzymes (e.g. alanine aminotransferase, aspartate aminotransferase) under glucose-replete conditions (5).
Figure 1. Glutamine metabolism in glioblastoma cells.
A, Glutamine (Gln) is a precursor for synthesis of nucleotides, proteins and glucosamine. It can also be metabolized in the mitochondria, providing carbon to the TCA cycle as α-ketoglutarate (α-kg). Downstream metabolism converts glutamine carbon into oxaloacetate (OAA) and/or acetyl-CoA (Ac-CoA), although the latter is predominantly formed from glucose. B, Glioblastoma cells were cultured in medium supplemented with unlabeled glucose and either L-[α-15N]-glutamine or L-[γ-15N]-glutamine. Glutaminase (GLS) and glutamate dehydrogenase (GDH) activities were followed by measuring transfer of 15N to 15NH4+. The time course shows the average and SD at each time point for three parallel cultures. The pie graph shows relative contributions of the γ and α nitrogens of glutamine to the total NH4+ pool after 8 hours. The small amount of unlabeled ammonium (gray wedge) mostly resulted from metabolism of unlabeled glutamate by GDH. Other abbreviations: Glc, glucose; Pyr, pyruvate; Lac, lactate; Cit, citrate; Glu, glutamate; Succ, succinate; Fum, fumarate; Mal, malate; Asp, aspartate; NH4+, ammonium; ALT, alanine aminotransferase.
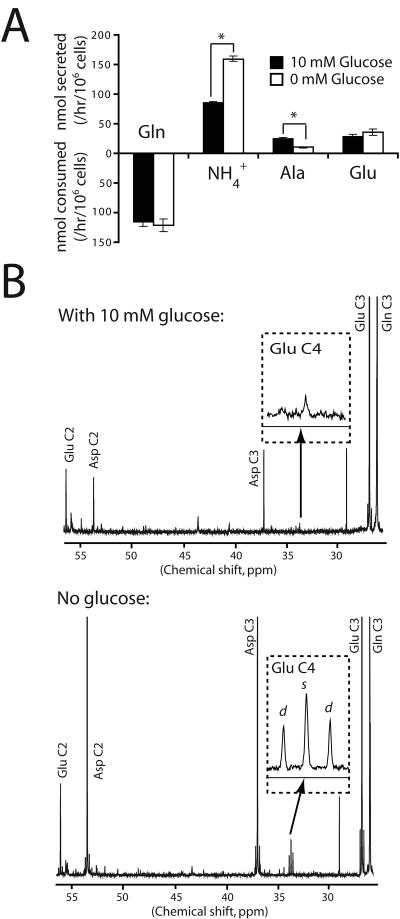
Because these cells normally use carbon from both glucose and glutamine to operate the TCA cycle (Fig. 1A), we tested whether glucose availability influenced glutamine metabolism. Cells were cultured in 10 mM or 0 mM glucose and the medium was analyzed to measure the consumption of glutamine and the secretion of by-products of glutamine catabolism (ammonia, alanine and glutamate). Under both conditions, the cells consumed glutamine at a rate at least 10-fold higher than any other amino acid, and alanine and glutamate were by far the most rapidly secreted amino acids (Supplementary Table 1). ATP levels were maintained over the time course (Supplementary Fig. 1). Surprisingly, glucose deprivation did not affect the net utilization of glutamine or of any other amino acid, suggesting that the cells did not enhance amino acid consumption rates to compensate for the loss of glucose (Supplementary Table 1 and Fig. 2A). However, glucose deprivation doubled total ammoniagenesis (Fig. 2A), implying an increased allocation of glutamine into ammonia-generating mitochondrial reactions.
Figure 2. Glucose withdrawal increases mitochondrial glutamine metabolism.
A, Glutamine-dependent metabolic rates in the presence and absence of glucose. Averages and SDs of three independent cultures are shown. *: p<0.005. B, 13C NMR spectra of metabolites from cells cultured in medium containing L-[3-13C]-glutamine plus or minus unlabeled glucose. The inset shows an enlargement of the glutamate C4 resonance at 34 ppm. The doublet (d) corresponds to glutamate labeled both at C4 (from acetyl-CoA) and at C3 (from glutamine-derived OAA), and the singlet (s) corresponds to glutamate molecules labeled at C4 but not C3. The experiment was performed twice for each condition. Abbreviation: Ala, alanine.
Mitochondrial metabolism of glutamine follows two divergent pathways (Fig. 1A). In one, glutamine carbon is used to produce OAA in the TCA cycle. In the other, glutamine-derived malate leaves the mitochondria to be used as a minor source of pyruvate and acetyl-CoA, both of which are predominantly formed from glucose carbon (5). To determine how glucose withdrawal affected these pathways, we cultured cells in L-[3-13C]-glutamine in the presence or absence of 10 mM glucose and analyzed intracellular metabolites by 13C NMR. In the presence of glucose, glutamine enriched the OAA pool as shown by the labeling in aspartate carbons 2 and 3 (Fig. 2B, top). Glutamine was only a minor source of acetyl-CoA, as shown by the minimal labeling in glutamate carbon 4, which arises from acetyl-CoA (Fig. 2B, top). When glucose was withdrawn, glutamine not only enriched the OAA pool but also contributed carbon to pyruvate/acetyl-CoA, because the labeling in glutamate carbon 4 (C4) increased 5-fold (Fig. 2B, bottom). Labeling in glutamate C5, also derived from acetyl-CoA, was similarly increased (data not shown). To confirm increased enrichment of the pyruvate pool, we measured 13C content in secreted lactate. In the presence of glucose, less than 5% of the lactate was derived from glutamine, but this percentage gradually increased as the glucose concentration in the medium declined (Supplementary Fig. 2A). However, the total abundance of 13C-labeled lactate fell during glucose deprivation (Supplementary Fig. 2B), as would be expected if more of the glutamine-derived pyruvate was converted to acetyl-CoA and oxidized in the TCA cycle. Thus, these cells use glutamine to produce OAA, pyruvate and acetyl-CoA, and to maintain TCA cycle function in the absence of glucose.
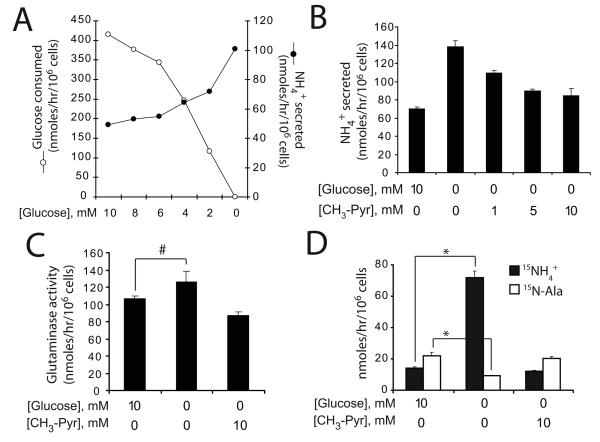
Glucose withdrawal stimulates GDH activity
We next studied the mechanism by which glucose deprivation increased total ammoniagenesis. Glucose withdrawal caused a gradual increase in ammonia production (Fig. 3A). This could be reversed with CH3-Pyr (Fig. 3B), a pyruvate analog metabolized in the mitochondria (18), suggesting that the increased ammonia production resulted from reduced delivery of pyruvate to the mitochondria. Surprisingly, glucose deprivation only marginally increased GLS activity (Fig 3C). Instead, the excess ammonia was due to a large increase in GDH activity, which was completely reversed by CH3-Pyr (Fig. 3D). The opposite trend was observed for alanine secretion (Fig. 3D). Thus, glucose withdrawal caused a twelve-fold increase in the ratio of GDH to ALT activity. This effect was not specific for glioblastoma cells, because withdrawal of glucose from mouse embryonic fibroblasts also increased ammonia production and GDH activity (Supplementary Fig 3).
Figure 3. Glucose withdrawal activates glutamate dehydrogenase.
A, Cells were cultured in decreasing glucose concentrations, and the rates of glucose consumption and ammonia production were measured. B, Total ammoniagenesis was measured in cells cultured with or without glucose, and with increasing concentrations of methyl-pyruvate (CH3-Pyr). The average and SD of three independent cultures are shown. C, Glutaminase activity was measured by following transfer of 15N from L-[γ-15N]-glutamine to 15NH4+ in the presence or absence of glucose and CH3-Pyr. The average and SD of three independent cultures are shown. #: p<0.05. D, Cells were cultured in L-[α-15N]-glutamine and the accumulation of 15NH4+ (GDH activity) and of 15N-alanine (ALT activity) was measured in the presence or absence of glucose and CH3-Pyr. The average and SD of three independent cultures are shown. *: p<0.0005
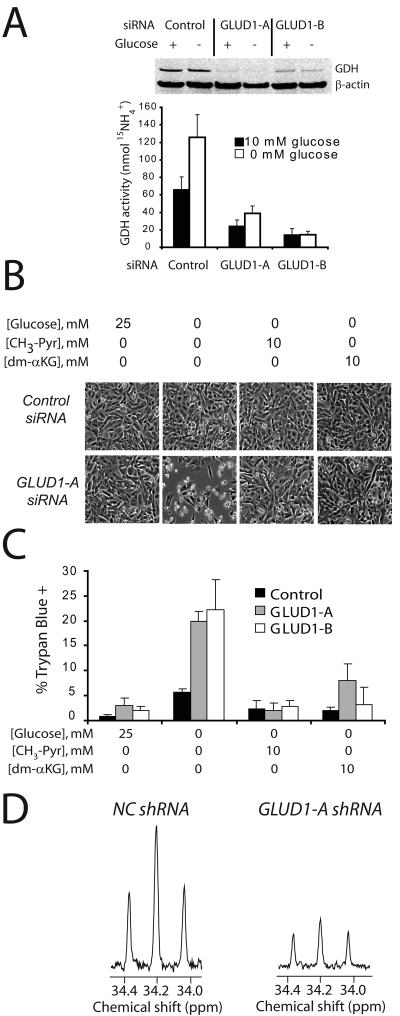
Loss of GDH activity limits the use of glutamine carbon in the TCA cycle and sensitizes glioblastoma cells to glucose withdrawal
The abrupt increase in GDH activity during glucose withdrawal raised the possibility that GDH compensates for glucose deprivation and allows cells to maintain viability. To test this, we used siRNAs targeting the two GDH isoforms, encoded by the genes GLUD1 and GLUD2. An siRNA directed against GLUD2 did not affect GDH abundance (data not shown) whereas two siRNAs directed against the GLUD1 transcript reduced GDH protein abundance and enzyme activity (Fig. 4A). Cells containing either of these siRNAs were indistinguishable from cells transfected with a control siRNA when glucose was present, but they lost viability in the absence of glucose for 24 hours (Fig. 4B,C). CH3-Pyr completely rescued viability of the glucose-deprived cells, as did dm-αKG an analog of α-KG (Fig. 4B,C), which like CH3-Pyr suppressed GDH activity (Supplementary Figure 4). To determine the effect of GDH knockdown on the TCA cycle, we generated a cell line with stable shRNA-mediated suppression of GDH (Supplementary Figure 5A,B). Compared to cells expressing a control shRNA, GDH knockdown cells lost viability in the absence of glucose unless supplemented with CH3-Pyr (Supplementary Fig. 5C). We cultured these cells in L-[3-13C]-glutamine and analyzed intracellular metabolites with 13C NMR. Suppression of GDH activity impaired the conversion of glutamine to acetyl-CoA, as shown by decreased labeling in glutamate C4 (Fig. 4D). This effect was not due to global metabolic defects, since labeling in aspartate was similar between the two cell lines (Supplementary Fig. 5D).
Figure 4. Glioblastoma cells require GDH to survive glucose deprivation.
A, Cells were transfected with a control siRNA or siRNAs directed against the GLUD1 transcript. The effect on GDH protein abundance (top) and flux (bottom) was determined in the presence and absence of glucose. The average and SD of three independent cultures are shown. B-C, Cells transfected with the control or GLUD1 siRNAs were subjected to glucose withdrawal, and effects on morphology and viability were determined. The metabolites CH3-Pyr and dm-αKG were tested for their ability to rescue viability in glucose-deprived cells. The average and SD of three independent cultures are shown for each condition. D, Glucose-deprived cells expressing a control (NC) shRNA or an shRNA against the GLUD1 transcript (GLUD1-A) were cultured with L-[3-13C]-Gln. Labeling in carbon 4 of glutamate was analyzed by 13C NMR.
Inhibition of glycolysis or Akt signaling stimulates GDH activity
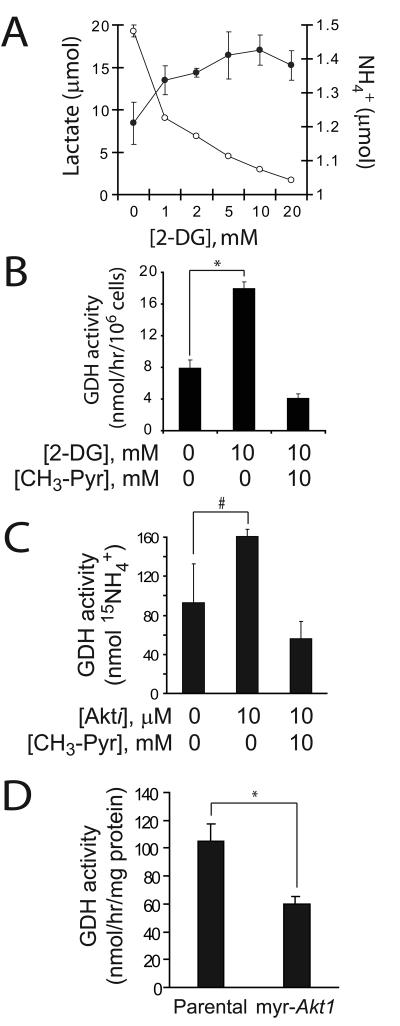
We next tested whether inhibitors of glycolysis could influence GDH activity. First, we treated cells with 2-deoxyglucose (2-DG), a competitive hexokinase inhibitor. Addition of 2-DG suppressed lactate production and increased ammoniagenesis (Fig. 5A). The drug also increased GDH activity, and this effect was reversed by CH3-Pyr (Fig. 5B). Next, we treated cells with an Akt inhibitor previously reported to reduce glycolysis in these cells (9). Similar to the effects of glucose withdrawal, the Akt inhibitor suppressed alanine secretion and mildly increased GLS activity (data not shown), while GDH activity was significantly enhanced (Fig. 5C). Addition of CH3-Pyr suppressed GDH activity during treatment with the Akt inhibitor (Fig. 5C). Furthermore, mouse astrocytes expressing a constitutively active Akt allele displayed chronically suppressed GDH activity compared to parental cells (Fig. 5D). Together, these data show that pharmacological impairment of glycolysis enhances GDH activity, and that Akt suppresses GDH activity through its effects on glucose metabolism.
Figure 5. Inhibition of glycolysis or Akt signaling activates GDH.
A, Glioblastoma cells were cultured in increasing concentrations of 2-deoxyglucose (2-DG) and the release of lactate (○) and NH4+ (●) was measured. B, Cells were cultured in 4 mM L-[α-15N]-glutamine and the accumulation of 15NH4+ was measured in the presence or absence of 2-DG and CH3-Pyr. C, Cells were cultured with an Akt inhibitor previously shown to suppress glycolysis in these cells (9). The effect on GDH activity in the absence and presence of CH3-Pyr was determined. D, GDH activity was compared between immortalized mouse astrocytes lacking or containing a constitutively-active Akt allele (myr-Akt1). In all experiments, the average and SD of three independent cultures are shown for each condition. #:p<0.05; *: p<0.005.
A GDH inhibitor sensitizes glioblastoma cells to drugs that suppress glycolysis
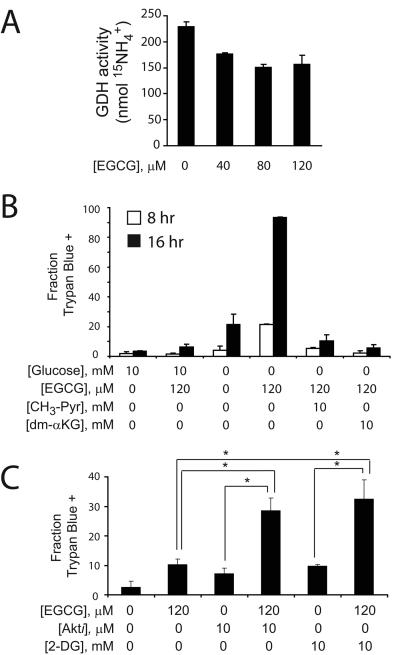
We next tested whether we could recapitulate the effects of GDH knockdown using epigallocatechin gallate (EGCG), an abundant green tea polyphenol that inhibits GDH in vitro (19). EGCG blunted maximal GDH activity in glucose-deprived cells (Fig. 6A). Like the effect of GDH knockdown, EGCG enhanced cell death in the absence of glucose, and addition of either CH3-Pyr or dm-αKG rescued viability (Fig. 6B). Finally, to test whether EGCG could enhance the effect of drugs that limit glucose metabolism, we cultured cells in the presence of glucose with EGCG and either the Akt inhibitor or 2-DG. EGCG increased the effect of both these inhibitors on cell death (Fig. 6C).
Figure 6. A GDH inhibitor sensitizes glioblastoma cells to glucose withdrawal and to inhibitors of glycolysis and Akt signaling.
A, The effect of EGCG on GDH activity was determined in glucose-deprived cells. B, Glioblastoma cells were cultured in the presence or absence of glucose, EGCG, CH3-Pyr and dm-αKG. The effects of these treatments, alone or in combination, were determined on cell viability at 8 and 16 hours. C, Glioblastoma cells were cultured in the presence or absence of EGCG, an Akt inhibitor or 2-DG, alone or in combination. After 24 hours, viability was determined by Trypan blue staining. In all experiments, the average and SD of three independent cultures are shown for each condition. *: p≤0.005.
Discussion
This decade has seen renewed interest in understanding the metabolic activities that support tumor cell survival and growth. The abundance of data showing that mutations in oncogenes and tumors suppressors influence glucose metabolism, coupled with the utility of 18FDG-PET, have raised the expectation that metabolic therapies directed against glycolysis will be effective in cancer. However, recent studies showing imperfect correlation between improvement on 18FDG-PET and patient survival have tempered this hope. Such studies suggest that tumors are capable of surviving periods of reduced glucose metabolism by using alternative metabolic pathways. Finding and targeting these survival pathways may ultimately improve the long-term efficacy of treatments that interfere with tumor glucose metabolism. Here we identified such a survival pathway, catalyzed by GDH, in a highly glycolytic gliobastoma cell line.
The emerging picture of tumor cell metabolism is that a complex network links signal transduction with the metabolism of glucose, glutamine and other nutrients. Understanding this network will enhance efforts to develop metabolically-targeted cancer therapies. This work emphasizes how distinct signaling pathways can influence glucose and glutamine metabolism in the same cells. In the glioblastoma cells we studied, Akt facilitates glucose metabolism but exerts no effect on total glutamine consumption (9). Rather, c-Myc stimulates glutamine metabolism by increasing the expression of amino acid transporters and glutaminase, resulting in increased glutamine catabolism and increased sensitivity to glutamine withdrawal (8, 9, 12). c-Myc activates a metabolic program in which glucose-replete cells require transamination reactions to produce α-KG (9). Thus, while Akt directs the delivery of glucose-derived acetyl-CoA to the TCA cycle, c-Myc directs the delivery of glutamine-derived α-KG and ultimately OAA for anaplerosis and cell growth. Glucose-replete cells can tolerate suppression of GDH because there are other sources of both α-KG and acetyl-CoA.
The current work adds to this picture by describing an alternative pathway that is activated in glucose-starved cells. The new pathway involves a large increase in GDH activity compared to ALT, which is the most active transaminase when glucose is present (5). Unlike transamination reactions, GDH has the advantage of delivering α-KG to the TCA cycle without the expenditure of a keto-acid, which could otherwise be oxidized to supplement ATP production. For example, the cells we studied displayed aspartate aminotransferase activity during glucose withdrawal, as shown by the transfer of 13C from glutamine to aspartate (Fig. 2B). This reaction converts glutamate to α-KG, but consumes OAA in the process. Thus, the pathway serves as a “mini-cycle” that generates aspartate but does not produce OAA for traditional TCA cycling (20). The concomitant induction of GDH would yield net OAA and therefore support ongoing citrate synthesis and function of the TCA cycle. Moreover, under these conditions glutamine metabolism also generated acetyl-CoA for the TCA cycle in a GDH-dependent manner. This may explain how the cells maintained their ATP content in the absence of glycolysis (Supplementary Fig. 1).
The effects of glucose withdrawal on both ALT and GDH were reversed by CH3-Pyr (Fig. 3D). Presumably this molecule stimulates ALT by supplying a large intracellular pyruvate pool for transamination. Since both ALT and GDH consume glutamate, competition between the two enzymes might contribute to CH3-Pyr's negative effect on GDH. However, the large discrepancy between GDH suppression (~60 nmol/hr/million cells) and ALT induction (only ~10 nmol/hr/million cells) suggest the involvement of additional mechanisms.
GDH has not received much attention in cancer cell metabolism, probably because its activity is suppressed during robust glycolysis in vitro. GDH is a widely-expressed homohexameric enzyme localized to the mitochondrial matrix, where it coordinates carbon and nitrogen metabolism. It determines the rate of oxidative degradation of glutamate and other nonessential amino acids, and thus is uniquely poised to respond to glucose deprivation. Regulation of GDH activity is extremely complex and involves allosteric effects, post-translational modifications and other levels of control (21, 22). Suppression of GDH activity by glucose is not restricted to glioblastoma, since we also observed it in mouse embryonic fibroblasts (Supplementary Fig. 3), and another study observed it in myeloma and hybridoma cells (23). The mechanism for GDH activation during glucose withdrawal is independent of changes in mRNA and protein levels (data not shown and Fig. 4A). Because the forward reaction (oxidative deamination of glutamate) uses NAD+ as a cofactor, it is possible that the low cellular NAD+/NADH ratio of highly glycolytic cells holds GDH activity in check. A similar phenomenon has been observed in brain slices, where glucose deprivation increased the cytoplasmic and mitochondrial NAD+/NADH ratio and ammoniagenesis (24). It is significant that in the cells studied here, there was no toxicity associated with the increased production of ammonia when glucose was withdrawn. Rather, the additional GDH activity sustained cell viability. GDH serves a similar compensatory role in plants. Mutant strains of Arabidopsis lacking GDH activity grew normally in carbon-replete conditions, but could not survive carbon starvation induced by prolonged growth in the dark. Viability was restored to the mutants simply by providing an exogenous carbohydrate source (25). Thus, we conclude that de-repression of GDH is necessary for some cells to adapt to and survive low-glucose conditions.
It is not known to what extent the metabolism of cultured tumor cells parallels tumor metabolism in vivo, but evidence suggests that many core metabolic activities are shared between settings. First, 18FDG-PET and 1H magnetic resonance spectroscopy reveal robust glucose consumption and lactate production in aggressive tumors. Second, expression studies have shown that enzymes involved in glucose and glutamine metabolism are abundantly expressed in tumor tissue and in some cases predict patient outcome (26-29). Third, inhibition of some of these enzymes impairs tumor growth in animal models (30-33). Fourth, some mechanisms of metabolic compensation, particularly autophagy, allow cells to survive nutrient stress in vitro and in vivo (34-36). Likewise, GDH inhibition may increase the efficacy of treatments that interfere with glucose metabolism. There are many such treatments already in use that could benefit from concomitant blockade of GDH. Alkylators and other DNA damaging agents suppress glycolysis in vitro and in vivo through PARP-dependent depletion of cytoplasmic NAD (37-39). The kinase inhibitors imatinib and rapamycin inhibit glycolysis in vitro by reversing the effects of oncogenic signaling pathways on glucose metabolism, and they suppress 18FDG-PET signal in some tumors (15, 40-42).
A major challenge in cancer therapeutics is to develop strategies that antagonize tumor cell survival without causing dose-limiting toxicity. Previous efforts to inhibit tumor glutamine metabolism were complicated by nausea, mucositis and pancytopenia (43). In this regard, tea polyphenols are intriguing because they are consumed in large quantities by millions of people worldwide and because a number of epidemiological studies have demonstrated the benefits of green tea in preventing the initiation or progression of cancer (44). EGCG in particular has a number of pharmacological properties that could independently suppress tumor cell growth (45). The relevance of EGCG's activity as a GDH inhibitor is unknown, although this has been proposed as a mechanism to explain the protective effect of green tea against diabetes (19). In the glioblastoma cells we studied, the effect of EGCG perfectly mimicked GDH knockdown in that it was enhanced during decreased glucose metabolism and reversed by providing GDH-dependent nutrients. Thus it will be interesting to test whether genetic or pharmacological impairment of GDH activity can suppress tumor growth or enhance the effect of conventional cancer therapies.
Supplementary Material
Acknowledgments
We thank Andrew Mullen for experimental assistance, Suzanne Wehrli for NMR spectroscopy and Dinesh Rakhejah and Vivian Jones of the Children's Medical Center Metabolics Laboratory for HPLC. We thank Charles Stanley, Changhong Li, Russell Jones, Aron Jaffe and Julian Lum for discussing the data and manuscript.
This work was supported by grants from the NIDDK (DK072565) and the American Cancer Society (ACS-IRG-02-196) to RJD.
Footnotes
The authors deny conflicts of interest.
References
- 1.Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–7. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- 2.Kim JW, Dang CV. Cancer's molecular sweet tooth and the Warburg effect. Cancer Res. 2006;66:8927–30. doi: 10.1158/0008-5472.CAN-06-1501. [DOI] [PubMed] [Google Scholar]
- 3.Medina MA. Glutamine and cancer. J Nutr. 2001;131:2539S–42S. doi: 10.1093/jn/131.9.2539S. discussion 50S-1S. [DOI] [PubMed] [Google Scholar]
- 4.DeBerardinis RJ, Sayed N, Ditsworth D, Thompson CB. Brick by brick: metabolism and tumor cell growth. Curr Opin Genet Dev. 2008;18:54–61. doi: 10.1016/j.gde.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeBerardinis RJ, Mancuso A, Daikhin E, et al. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–50. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elstrom RL, Bauer DE, Buzzai M, et al. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64:3892–9. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 7.Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24:6314–22. doi: 10.1038/sj.onc.1208773. [DOI] [PubMed] [Google Scholar]
- 8.Gao P, Tchernyshyov I, Chang TC, et al. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009 doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wise DR, DeBerardinis RJ, Mancuso A, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–7. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buzzai M, Bauer DE, Jones RG, et al. The glucose dependence of Akt-transformed cells can be reversed by pharmacologic activation of fatty acid beta-oxidation. Oncogene. 2005;24:4165–73. doi: 10.1038/sj.onc.1208622. [DOI] [PubMed] [Google Scholar]
- 11.DeBerardinis RJ, Lum JJ, Thompson CB. Phosphatidylinositol 3-kinase-dependent modulation of carnitine palmitoyltransferase 1A expression regulates lipid metabolism during hematopoietic cell growth. J Biol Chem. 2006;281:37372–80. doi: 10.1074/jbc.M608372200. [DOI] [PubMed] [Google Scholar]
- 12.Yuneva M, Zamboni N, Oefner P, Sachidanandam R, Lazebnik Y. Deficiency in glutamine but not glucose induces MYC-dependent apoptosis in human cells. J Cell Biol. 2007;178:93–105. doi: 10.1083/jcb.200703099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ben-Haim S, Ell P. 18F-FDG PET and PET/CT in the evaluation of cancer treatment response. J Nucl Med. 2009;50:88–99. doi: 10.2967/jnumed.108.054205. [DOI] [PubMed] [Google Scholar]
- 14.Tan MC, Linehan DC, Hawkins WG, Siegel BA, Strasberg SM. Chemotherapy-induced normalization of FDG uptake by colorectal liver metastases does not usually indicate complete pathologic response. J Gastrointest Surg. 2007;11:1112–9. doi: 10.1007/s11605-007-0218-8. [DOI] [PubMed] [Google Scholar]
- 15.Ma WW, Jacene H, Song D, et al. [18F]Fluorodeoxyglucose Positron Emission Tomography Correlates With Akt Pathway Activity but Is Not Predictive of Clinical Outcome During mTOR Inhibitor Therapy. J Clin Oncol. 2009 doi: 10.1200/JCO.2008.18.8383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de la Iglesia N, Konopka G, Puram SV, et al. Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway. Genes Dev. 2008;22:449–62. doi: 10.1101/gad.1606508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brosnan JT, Brosnan ME, Yudkoff M, et al. Alanine metabolism in the perfused rat liver. Studies with (15)N. J Biol Chem. 2001;276:31876–82. doi: 10.1074/jbc.M103890200. [DOI] [PubMed] [Google Scholar]
- 18.Jijakli H, Nadi AB, Cook L, Best L, Sener A, Malaisse WJ. Insulinotropic action of methyl pyruvate: enzymatic and metabolic aspects. Arch Biochem Biophys. 1996;335:245–57. doi: 10.1006/abbi.1996.0505. [DOI] [PubMed] [Google Scholar]
- 19.Li C, Allen A, Kwagh J, et al. Green tea polyphenols modulate insulin secretion by inhibiting glutamate dehydrogenase. J Biol Chem. 2006;281:10214–21. doi: 10.1074/jbc.M512792200. [DOI] [PubMed] [Google Scholar]
- 20.Yudkoff M, Nelson D, Daikhin Y, Erecinska M. Tricarboxylic acid cycle in rat brain synaptosomes. Fluxes and interactions with aspartate aminotransferase and malate/aspartate shuttle. J Biol Chem. 1994;269:27414–20. [PubMed] [Google Scholar]
- 21.Smith TJ, Stanley CA. Untangling the glutamate dehydrogenase allosteric nightmare. Trends Biochem Sci. 2008;33:557–64. doi: 10.1016/j.tibs.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 22.Haigis MC, Mostoslavsky R, Haigis KM, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–54. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 23.Martinelle K, Doverskog M, Jacobsson U, Chapman BE, Kuchel PW, Haggstrom L. Elevated glutamate dehydrogenase flux in glucose-deprived hybridoma and myeloma cells: evidence from 1H/15N NMR. Biotechnol Bioeng. 1998;60:508–17. doi: 10.1002/(sici)1097-0290(19981120)60:4<508::aid-bit13>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 24.Gibson G, Blass JP. A relation between (NAD+)/(NADH) potentials and glucose utilization in rat brain slices. J Biol Chem. 1976;251:4127–30. [PubMed] [Google Scholar]
- 25.Miyashita Y, Good AG. NAD(H)-dependent glutamate dehydrogenase is essential for the survival of Arabidopsis thaliana during dark-induced carbon starvation. J Exp Bot. 2008;59:667–80. doi: 10.1093/jxb/erm340. [DOI] [PubMed] [Google Scholar]
- 26.Altenberg B, Greulich KO. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics. 2004;84:1014–20. doi: 10.1016/j.ygeno.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 27.Langbein S, Zerilli M, Zur Hausen A, et al. Expression of transketolase TKTL1 predicts colon and urothelial cancer patient survival: Warburg effect reinterpreted. Br J Cancer. 2006;94:578–85. doi: 10.1038/sj.bjc.6602962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aledo JC, Segura JA, Medina MA, Alonso FJ, Nunez de Castro I, Marquez J. Phosphate-activated glutaminase expression during tumor development. FEBS Lett. 1994;341:39–42. doi: 10.1016/0014-5793(94)80236-x. [DOI] [PubMed] [Google Scholar]
- 29.Unwin RD, Craven RA, Harnden P, et al. Proteomic changes in renal cancer and co-ordinate demonstration of both the glycolytic and mitochondrial aspects of the Warburg effect. Proteomics. 2003;3:1620–32. doi: 10.1002/pmic.200300464. [DOI] [PubMed] [Google Scholar]
- 30.Lobo C, Ruiz-Bellido MA, Aledo JC, Marquez J, Nunez De Castro I, Alonso FJ. Inhibition of glutaminase expression by antisense mRNA decreases growth and tumourigenicity of tumour cells. Biochem J. 2000;348(Pt 2):257–61. [PMC free article] [PubMed] [Google Scholar]
- 31.Fantin VR, St-Pierre J, Leder P. Attenuation of LDH-A expression uncovers a link between glycolysis, mitochondrial physiology, and tumor maintenance. Cancer Cell. 2006;9:425–34. doi: 10.1016/j.ccr.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 32.Christofk HR, Vander Heiden MG, Harris MH, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–3. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 33.Hatzivassiliou G, Zhao F, Bauer DE, et al. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–21. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 34.Lum JJ, Bauer DE, Kong M, et al. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–48. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- 35.Degenhardt K, Mathew R, Beaudoin B, et al. Autophagy promotes tumor cell survival and restricts necrosis, inflammation, and tumorigenesis. Cancer Cell. 2006;10:51–64. doi: 10.1016/j.ccr.2006.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amaravadi RK, Yu D, Lum JJ, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–36. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams SN, Anthony ML, Brindle KM. Induction of apoptosis in two mammalian cell lines results in increased levels of fructose-1,6-bisphosphate and CDP-choline as determined by 31P MRS. Magn Reson Med. 1998;40:411–20. doi: 10.1002/mrm.1910400311. [DOI] [PubMed] [Google Scholar]
- 38.Zong WX, Ditsworth D, Bauer DE, Wang ZQ, Thompson CB. Alkylating DNA damage stimulates a regulated form of necrotic cell death. Genes Dev. 2004;18:1272–82. doi: 10.1101/gad.1199904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Day SE, Kettunen MI, Gallagher FA, et al. Detecting tumor response to treatment using hyperpolarized 13C magnetic resonance imaging and spectroscopy. Nat Med. 2007;13:1382–7. doi: 10.1038/nm1650. [DOI] [PubMed] [Google Scholar]
- 40.Wei LH, Su H, Hildebrandt IJ, Phelps ME, Czernin J, Weber WA. Changes in tumor metabolism as readout for Mammalian target of rapamycin kinase inhibition by rapamycin in glioblastoma. Clin Cancer Res. 2008;14:3416–26. doi: 10.1158/1078-0432.CCR-07-1824. [DOI] [PubMed] [Google Scholar]
- 41.Gottschalk S, Anderson N, Hainz C, Eckhardt SG, Serkova NJ. Imatinib (STI571)-mediated changes in glucose metabolism in human leukemia BCR-ABL-positive cells. Clin Cancer Res. 2004;10:6661–8. doi: 10.1158/1078-0432.CCR-04-0039. [DOI] [PubMed] [Google Scholar]
- 42.Basu S, Mohandas KM, Peshwe H, Asopa R, Vyawahare M. FDG-PET and PET/CT in the clinical management of gastrointestinal stromal tumor. Nucl Med Commun. 2008;29:1026–39. doi: 10.1097/MNM.0b013e328313bbe7. [DOI] [PubMed] [Google Scholar]
- 43.Souba WW. Glutamine and cancer. Ann Surg. 1993;218:715–28. doi: 10.1097/00000658-199312000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Khan N, Mukhtar H. Multitargeted therapy of cancer by green tea polyphenols. Cancer Lett. 2008;269:269–80. doi: 10.1016/j.canlet.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ju J, Lu G, Lambert JD, Yang CS. Inhibition of carcinogenesis by tea constituents. Semin Cancer Biol. 2007;17:395–402. doi: 10.1016/j.semcancer.2007.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.