Abstract
The discovery of hepcidin as a key regulator of iron homeostasis has advanced our current knowledge of this field. Liver-derived hepcidin peptide is secreted in response to iron and inflammation and interacts with the iron export protein ferroportin. This review summarizes recent advances discussed at the Symposium. A particular focus is on molecular interactions between hepcidin and ferroportin, the regulation of hepcidin expression by iron and inflammation, and emerging methods to measure serum hepcidin in human populations.
Introduction
Our understanding of iron metabolism has advanced dramatically in the past few years, mainly as a result of the discovery of hepcidin, a key regulator of whole-body iron homeostasis. Hepcidin is a peptide hormone produced primarily by the liver and secreted into the circulation. Its synthesis increases in response to iron and inflammation and decreases in response to erythropoiesis. Hepcidin regulates systemic iron metabolism by interacting with its receptor ferroportin, a transmembrane iron-export protein. Ferroportin is abundantly expressed on the cell surface of reticuloendothelial (RE)7 macrophages (i.e., resident macrophages of the liver, spleen, and bone marrow) and on the basolateral membrane of duodenal enterocytes. These 2 cell types are the main suppliers of iron to the plasma. By recycling iron from scenescent erythrocytes, RE macrophages release 20–25 mg of iron into the plasma per day. Enterocytes add another ∼1–2 mg of iron to the plasma through the absorption of dietary iron. Hepcidin inhibits iron release at both of these sites by binding to cell-surface ferroportin and causing its internalization and subsequent degradation. Hepcidin can therefore be considered as a negative regulator of iron absorption and recycling.
The aim of the 2008 symposium “Hepcidin Regulation of Iron Transport” was to report recent advances regarding the molecular interactions between hepcidin and ferroportin, the regulation of hepcidin expression by iron, and the roles of hepcidin and ferroportin in iron overload disorders, inflammation, and infection. Methods of measuring hepcidin in human populations were also discussed. The symposium was cosponsored by the American Society for Nutrition and the American Physiological Society. Support for the symposium was generously provided by Intrinsic LifeSciences and Xenon Pharmaceuticals. Participating speakers included Drs. Elizabeta Nemeth (UCLA), Ivana DeDomenico (University of Utah), Janet Hunt (USDA-ARS Grand Forks Human Nutrition Research Center), David Haile (UTHSCSA), and Marianne Wessling-Resnick (HSPH). This review was prepared by the Symposium organizers and cochairs, Drs. Jamie Collins (currently at the University of Florida-Gainesville), Mitch Knutson (UF-Gainesville) and Marianne Wessling-Resnick (HSPH).
Hepcidin in iron-overload disorders
Many iron-overload disorders in humans, such as hemochromatosis disorders and genetic anemias, are the result of perturbed regulation of hepcidin expression in the liver, ultimately leading to abnormally low hepcidin production in the setting of increased body iron levels. Hepcidin is regulated by several molecules including HFE (product of a high-iron gene), hemojuvelin (HJV), and transferrin receptor 2 (TfR2), as described in more detail below (see summary in Table 1 and Figure 1). Two point mutations in the HFE gene are a principal cause of hereditary hemochromatosis (HH), which is characterized by iron accumulation causing organ and tissue damage. In HH, hepcidin expression is inappropriately low despite systemic iron overload (1), suggesting that HFE is required for normal hepcidin expression. Mutations in HFE that are common in HH disrupt interactions of HFE with the iron-sensing machinery, leading to decreased production of hepcidin.
TABLE 1.
Proteins involved with hepcidin regulation of iron transport
| Protein | Properties | Role in hepcidin regulation of iron transport |
|---|---|---|
| Ferroportin | Transmembrane protein | Hepcidin receptor and iron exporter |
| HFE | Single point mutation responsible for most iron overload | Mutations in HFE are associated with low hepcidin production; precise function unknown |
| Transferrin receptor 2 (TfR2) | Homologous to TfR1; abundantly expressed in liver | May sense plasma iron level through HFE |
| BMP6 | Member of the TGFβ superfamily of cytokines; expression is induced by iron | Binds to BMP receptors I and II, inducing phosphorylation of SMAD proteins |
| HJV | Membrane-bound and soluble protein | Serves as a BMP coreceptor to activate SMAD |
| SMADs | Cellular mediators of transcriptional activation | Activated SMAD induce transcription of hepcidin |
| JAK | Involved in a signaling pathway that regulates ferroportin trafficking; transcriptional response | May phosphorylate ferroportin; activates STAT31 in response to IL-6 |
| STAT3 | Cellular mediator of transcriptional activation | Activated STAT3 induces transcription of hepcidin |
STAT3, signal transducer and activator of transcription 3.
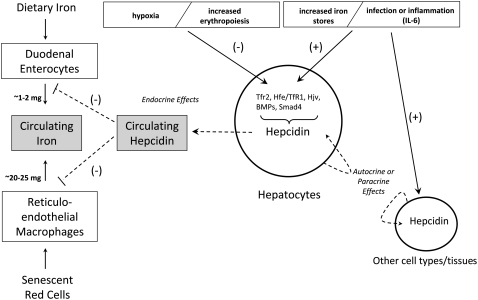
FIGURE 1 .
Regulation of body iron homeostasis by hepcidin. Hepcidin is secreted primarily by hepatocytes into the circulation, where it functions to inhibit iron absorption in the proximal small intestine and iron release from RE macrophages by binding to its receptor ferroportin and causing its internalization and degradation. Hepcidin gene expression is down-regulated by low tissue oxygen tension and by increased erythropoietic demand and up-regulated by increased body iron stores and infection or inflammation. Up-regulation of hepcidin gene expression during infection and inflammation is in part mediated by IL-6. Signals emanating from these physiological effectors are transduced into the nucleus to regulate hepcidin gene transcription via interaction of the hepcidin gene promoter with several proteins including TfR2, a complex of HFE with TfR1, HJV, SMAD4, and BMP. Hepcidin produced by hepatocytes may also have autocrine and/or paracrine effects on various cell types in the liver, and hepcidin produced by other cell types and tissues may have similar effects. Hepcidin produced by nonhepatic tissues contributes little to circulating hepcidin levels but may have important local effects.
Other forms of hemochromatosis are caused by mutations in the TFR2 and hemojuvelin (HJV) genes. Homozygous disruption of the TFR2 gene leads to a more severe form of hemochromatosis in which urinary hepcidin levels are very low (2,3). Juvenile hemochromatosis, which manifests much earlier in life, is caused by mutations in the HJV gene or the hepcidin gene itself (4). Patients with this disorder have essentially undetectable urinary hepcidin levels (5). Regulation of hepcidin expression is also perturbed in chronic anemias with hemolysis or increased erythropoiesis in which iron overload and maldistribution occur (6). Serum hepcidin levels are severely decreased in these patients despite systemic iron overload (7). These findings suggest that the effects of anemia, particularly in the setting of enhanced red blood cell production, override the influence of increased body iron levels on hepcidin gene expression. Low hepcidin levels may contribute to the iron overload seen in these patients.
Another important player in the regulation of body iron levels is the iron exporter ferroportin. Ferroportin is the receptor through which hepcidin exerts its functional effects on serum iron levels. Interestingly, inactivating mutations in ferroportin also lead to a form of inherited hemochromatosis called “ferroportin disease” (8,9). Apparently, no other iron exporters can compensate for its loss of function.
Synthesis and regulation of hepcidin expression
As denoted by its name, hepcidin is produced primarily by hepatocytes. Other tissues and cells have been shown to express hepcidin, although to a much smaller extent. The human liver synthesizes hepcidin initially as an 84-amino-acid preprohepcidin. The first 24 amino acids of preprohepcidin contain an endoplasmic reticulum targeting signal that is cleaved to produce prohepcidin. Further enzymatic cleavage of prohepcidin produces the bioactive 25-amino-acid hepcidin. Bioactive hepcidin is released into plasma and excreted into the urine. Urinary hepcidin levels increase markedly after acute administration of an oral iron supplement (10) and are elevated 50- to 1000-fold in patients with secondary iron overload (11).
How iron increases the expression of hepcidin is an active area of study. Studies in primary hepatocytes show that hepcidin responds to iron only when it is bound to its plasma transport protein transferrin (12). In vivo, the hepcidin response to dietary iron is proportional to the increase in the amount of iron carried on plasma transferrin (i.e., transferrin saturation). These observations suggest that the liver uses transferrin saturation as an iron sensor. At the cellular level, iron sensing appears to involve transferrin receptor 2 (TfR2) because liver-specific disruption of TfR2 in mice markedly diminished hepcidin expression despite hepatic iron overload and substantially elevated transferrin saturation (13). HFE also appears to be required for iron sensing because patients with mutations in HFE do not produce hepcidin in response to oral iron as do normal individuals (1). In vivo studies suggest that HFE induces hepcidin expression only when it is not in complex with TfR1 (14). However, in vitro studies showing that HFE and TfR2 interact suggest that these 2 molecules work together to sense iron (15,16). It has been proposed that HFE uncouples from TfR1 to form the HFE/TfR2 complex that enables up-regulation of hepcidin expression to decrease dietary iron uptake and reduce iron recycling from macrophages in response to transferrin saturation (14).
Iron sensing is also mediated through the bone morphogenic protein (BMP)/SMAD [proteins that are homologs of both the Drosophila protein mothers against decapentaplegic (MAD) and the C. elegans protein SMA] signaling pathway. BMP represent a class of secreted ligands that regulate cell proliferation and differentiation. Binding of BMP ligands to cell-surface BMP receptors causes the phosphorylation of SMAD proteins that translocate to the nucleus to induce transcription of specific genes, including hepcidin. The unexpected link between the BMP/SMAD signaling pathway and hepcidin expression was first made in studies of mice lacking SMAD4 in the liver. Like TfR2-deficient animals, SMAD4-null mice developed hepatic iron overload with markedly reduced hepcidin expression (17). Moreover, these mice were unable to produce hepcidin in response to iron injection, thus demonstrating that the BMP/SMAD4 pathway is required to sense iron. BMP have been also shown to bind to HJV (16,18), the protein mutated in juvenile hemochromatosis. HJV exists bound to the cell surface or in a circulating soluble form. Cell-surface HJV binds to BMP and stimulates BMP signaling and hepcidin synthesis, whereas the soluble form antagonizes this effect. Overall, the transcriptional regulation of hepcidin expression by iron appears increasingly complex, involving the orchestration of a number of proteins. Models integrating all of these proteins have been proposed (16,19).
Measurement of hepcidin in human populations and correlation with markers of body iron stores
Several methods have recently been developed to measure hepcidin levels in humans. These include assays developed to detect preprohepcidin (by mRNA expression detected from liver biopsy samples), prohepcidin, and hepcidin-25 (the biologically active form of hepcidin) in serum and urine. The utility of these assays has been the subject of debate in the scientific community. One assay was used by Hadley et al. (20) in an attempt to correlate prohepcidin expression with iron absorption in healthy women volunteers. These authors had previously demonstrated that there was a strong correlation between nonheme iron absorption and serum ferritin levels (21) in that individuals with low serum ferritin absorbed more iron than those with high serum ferritin (in the absence of detectable inflammation). Using a commercial immunosorbent assay, these authors demonstrated that there was no significant correlation between iron absorption in healthy women and prohepcidin, although there was a significant correlation between serum prohepcidin and serum ferritin (20). Another group confirmed that serum prohepcidin did not correlate with iron absorption in a study of healthy men, some of whom were heterozygous for mutations of the HFE gene (22). Serum prohepcidin did not correlate with serum ferritin in this study (22).
Many of these recently developed hepcidin assays are cumbersome to perform and/or require extensive research instrumentation, and their availability to the clinical research community has been limited. Some recent studies of physiologically relevant hepcidin in humans have relied on a human urinary assay (10) in which hepcidin is extracted by cation-exchange chromatography and is detected by an immunodot chemiluminescent assay. Hepcidin is normalized to urinary creatinine to account for varying dilution of urine samples. Although this assay has proven useful, it is laborious, and its accuracy depends on the unproven supposition that urinary hepcidin correlates with circulating serum hepcidin. More recently, hepcidin assays based on mass spectroscopic techniques have been developed (3,23–25), but these assays are semiquantitative and depend on expensive scientific instrumentation.
Very recently, as revealed at the Experimental Biology symposium, a competitive enzyme-linked immunosorbent assay for human hepcidin has been developed (26). This assay utilizes a hepcidin antibody previously developed (10) and biotinylated hepcidin-25 as a tracer. These investigators have demonstrated that the assay can be performed in a high-throughput format in 96-well plates, and that it accurately and reproducibly detects physiological and pathological changes in serum or urine hepcidin levels. This assay may thus prove to be a superior method for the detection of hepcidin in human samples and should allow the thorough characterization of hepcidin levels in various human pathologies and in normal populations.
Molecular interactions of hepcidin with ferroportin
When hepcidin binds to ferroportin at the cell surface, the 2 proteins are internalized and degraded in lysosomes (27). The molecular signals involved in ferroportin internalization and degradation have recently been identified. Cell culture studies show that hepcidin binding to ferroportin at the cell surface initiates the rapid phosphorylation of 1 of 2 adjacent tyrosine residues located on a putative cytosolic region of ferroportin (28). After phosphorylation, the hepcidin-ferroportin complex is internalized through clathrin-coated pits. Once internalized, the phosphates are removed, and ferroportin becomes ubiquitinated, which directs the protein through the multivesicular body and subsequently to the lysosome for ultimate degradation. Defective ferroportin phosphorylation may contribute to iron-overload disorders because 2 human ferroportin mutations, N144H and Q182H, were found to display no or delayed phosphorylation, consistent with the insensitivity of these mutants to hepcidin-induced degradation. Preliminary studies indicate that phosphorylation of ferroportin is mediated by Janus kinase 2 (JAK2). Current efforts are directed at identifying which amino acids of ferroportin bind hepcidin, i.e., the hepcidin-binding domain.
The role of hepcidin during inflammation and infection
The ability of hepcidin to down-regulate ferroportin from the cell surface not only provides a molecular explanation for the regulation of iron homeostasis but also helps to explain the coordinated response producing the hypoferremia of inflammation and infection. Nemeth et al. (10) have shown that the cytokine interleukin (IL)-6 is necessary and sufficient for induction of hepcidin during inflammation, establishing that the iron regulatory peptide plays a key role in the anemia of chronic diseases. Induction by IL-6 involves signal transduction by JAK kinase regulation of Signal Activator of Transcription (29,30), adding to the complexity of transcriptional responses dictating hepcidin regulation. The integration of the signaling responses for appropriate expression of hepcidin is a key area of research that warrants further study (16,19).
Although production by the liver most likely accounts for the majority of hepcidin in the systemic circulation, the regulatory peptide can be synthesized in other tissues including heart (31,32), kidney (33), adipose tissue (34), spinal cord (35), myeloid cells (36), splenic and alveolar macrophages (37), and monocytes (38). These observations raise key questions about the function of hepcidin in local interactions. In particular, the coexpression of hepcidin with ferroportin in macrophages, neutrophils, heart, and retina raises the possibility for autocrine regulation in these settings. Autocrine biosynthesis of hepcidin has been shown to increase iron retention and to down-regulate ferroportin expression in monocytes and macrophages (37–39). Although infection and inflammation are known to induce liver expression of hepcidin, autocrine synthesis can more selectively activate the appropriate cell type and response. In fact, there is evidence that hepcidin may interact with ferroportin of macrophages and intestinal epithelial cells quite differently (40), suggesting that autocrine responses to locally produced hepcidin can help fine-tune the regulation of iron metabolism and response to infection.
It has long been known that iron is essential for both host and pathogen, leading to a rational basis for the hypoferremia of infection. However, although down-regulation of ferroportin by hepcidin would restrict systemic iron necessary for pathogen growth, cells expressing ferroportin would be particularly vulnerable to infection because of iron retention. This idea has recently been supported by studies of flatiron mice, a strain heterozygous for loss-of-function ferroportin mutations (41). Compared with control macrophages, flatiron macrophages show increased growth of Chlamydia and Legionella (42). These data are consistent with observations that demonstrate that ferroportin levels can also influence growth of Salmonella (43,44). Paradoxically, autocrine regulation of hepcidin expression by myeloid cells (36) and retinal cells (45) has been shown to respond to bacterial pathogens through toll-like receptor 4. Concomitant with this effect, expression of ferroportin declines. Sow et al. (46) have suggested that hepcidin may have a direct antimicrobial activity under these circumstances. However, new data discussed at the Experimental Biology symposium suggest that manipulation of intracellular iron pools through hepcidin-ferroportin regulation may also alter macrophage-mediated immune responses involving cytokine production (44,47) as well as nitric oxide production (48). These observations bolster the view that autocrine regulation of hepcidin synthesis in response to infection may play a key role in innate immunity that goes well beyond systemic iron withdrawal to actually modulate local inflammatory effects.
Conclusions and future directions
This article summarizes key findings from the EB2008 symposium on hepcidin and the regulation of iron homeostasis. A more comprehensive review of this topic can be found in a recent article by Nemeth and Ganz (6). It is now widely accepted that hepcidin is the key regulator of overall body iron homeostasis. Historically, several regulators of iron homeostasis were recognized that controlled iron flux in response to reductions in tissue oxygen levels, changes in body iron stores, alterations in erythropoiesis, and inflammation and infection. Recent findings have demonstrated that hepcidin gene expression is regulated by all these physiological changes, thus providing evidence that it is the long-sought iron regulatory hormone. It has also become apparent that hepcidin regulation occurs predominantly at the transcriptional level and that mRNA expression levels correlate with circulating protein levels. Pathophysiological alterations in certain individuals and in mouse models have revealed a number of players in the transcriptional regulation of hepcidin gene expression, many of which signal via the BMP/SMAD pathway.
Several important areas for future research exist in the hepcidin/iron field. A better understanding of the precise interaction between ferroportin and hepcidin would pave the way for the development of drugs that could mimic or block hepcidin activity and, in doing so, alter iron homeostasis. A better understanding of the transcriptional regulatory networks that signal to the hepcidin gene, particularly in response to iron and inflammation, will allow researchers to more fully appreciate the exquisite control of body iron levels, which often go awry in various pathological conditions. Furthermore, the development of a useful and accessible clinical assay is important to be able to assess alterations in hepcidin expression in various populations. It is our hope that the new assay developed by Ganz et al. will fit these criteria and become widely accessible to the research community. And last, although methods for the production of recombinant hepcidin have been reported in the scientific literature, more work needs to be done in this area so as to be able to provide pure hepcidin ultimately for clinical trials in humans with iron-overload disorders.
In conclusion, the last decade has seen an explosion of our knowledge of iron homeostasis, and the next 10 years will undoubtedly provide even more information that will benefit clinicians in their management of patients with pathological perturbations of body iron homeostasis.
Published as a supplement to The Journal of Nutrition. Presented as part of the symposium “Hepcidin Regulation of Iron Transport” given at the 2008 Experimental Biology meeting on April 6, 2008, in San Diego, CA. The symposium was sponsored by the American Society for Nutrition and the American Physiological Society Gastrointestinal and Liver Physiology Section and supported by educational grants from Xenon Pharmaceuticals and Intrinsic Life Sciences. The symposium was chaired by Marianne Wessling-Resnick, Mitchell Knutson, and Jamie Collins.
Supported by grants from the NIH to J.F.C. (NIDDK R01-DK 074867), M.W.-R. (NIDDK R01-DK064750, NIDA R21-DA025573, and NIEHS R01-ES014638), and to M.D.K. (K01-DK065064). The views expressed in this document are solely those of the authors and do not necessarily reflect those of NIDDK, NIEHS, or NIDA.
Author disclosures: J. F. Collins, M. Wessling-Resnick, and M. D. Knutson, no conflicts of interest.
Abbreviations used: BMP, bone morphogenic protein; HFE, high-iron gene; HH, hereditary hemochromatosis; HJV, hemojuvelin; IL, interleukin; JAK, Janus kinase; RE, reticuloendothelial; SMAD, proteins that are homologs of both the Drosophila protein mothers against decapentaplegic (MAD) and the C. elegans protein SMA; TfR1 or 2, transferrin receptor 1 or 2.
References
- 1.Piperno A, Girelli D, Nemeth E, Trombini P, Bozzini C, Poggiali E, Phung Y, Ganz T, Camaschella C. Blunted hepcidin response to oral iron challenge in HFE-related hemochromatosis. Blood. 2007;110:4096–100. [DOI] [PubMed] [Google Scholar]
- 2.Nemeth E, Roetto A, Garozzo G, Ganz T, Camaschella C. Hepcidin is decreased in TFR2 hemochromatosis. Blood. 2005;105:1803–6. [DOI] [PubMed] [Google Scholar]
- 3.Kemna E, Tjalsma H, Laarakkers C, Nemeth E, Willems H, Swinkels D. Novel urine hepcidin assay by mass spectrometry. Blood. 2005;106:3268–70. [DOI] [PubMed] [Google Scholar]
- 4.Roetto A, Papanikolaou G, Politou M, Alberti F, Girelli D, Christakis J, Loukopoulos D, Camaschella C. Mutant antimicrobial peptide hepcidin is associated with severe juvenile hemochromatosis. Nat Genet. 2003;33:21–2. [DOI] [PubMed] [Google Scholar]
- 5.Papanikolaou G, Samuels ME, Ludwig EH, MacDonald ML, Franchini PL, Dube MP, Andres L, MacFarlane J, Sakellaropoulos N, et al. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nat Genet. 2004;36:77–82. [DOI] [PubMed] [Google Scholar]
- 6.Nemeth E, Ganz T. Regulation of iron metabolism by hepcidin. Annu Rev Nutr. 2006;26:323–42. [DOI] [PubMed] [Google Scholar]
- 7.Papanikolaou G, Tzilianos M, Christakis JI, Bogdanos D, Tsimirika K, MacFarlane J, Goldberg YP, Sakellaropoulos N, Ganz T, Nemeth E. Hepcidin in iron overload disorders. Blood. 2005;105:4103–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montosi G, Donovan A, Totaro A, Garuti C, Pignatti E, Cassanelli S, Trenor CC, Gasparini P, Andrews NC, Pietrangelo A. Autosomal-dominant hemochromatosis is associated with a mutation in the ferroportin (SLC11A3) gene. J Clin Invest. 2001;108:619–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pietrangelo A. The ferroportin disease. Blood Cells Mol Dis. 2004;32:131–8. [DOI] [PubMed] [Google Scholar]
- 10.Nemeth E, Rivera S, Gabayan V, Keller C, Taudorf S, Pedersen BK, Ganz T. IL-6 mediates hypoferremia of inflammation by inducing the synthesis of the iron regulatory hormone hepcidin. J Clin Invest. 2004;113:1271–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nemeth E, Valore EV, Territo M, Schiller G, Lichtenstein A, Ganz T. Hepcidin, a putative mediator of anemia of inflammation, is a type II acute-phase protein. Blood. 2003;101:2461–3. [DOI] [PubMed] [Google Scholar]
- 12.Lin L, Valore EV, Nemeth E, Goodnough JB, Gabayan V, Ganz T. Iron transferrin regulates hepcidin synthesis in primary hepatocyte culture through hemojuvelin and BMP2/4. Blood. 2007;110:2182–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wallace DF, Summerville L, Subramaniam VN. Targeted disruption of the hepatic transferrin receptor 2 gene in mice leads to iron overload. Gastroenterology. 2007;132:301–10. [DOI] [PubMed] [Google Scholar]
- 14.Schmidt PJ, Toran PT, Giannetti AM, Bjorkman PJ, Andrews NC. The transferrin receptor modulates Hfe-dependent regulation of hepcidin expression. Cell Metab. 2008;7:205–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goswami T, Andrews NC. Hereditary hemochromatosis protein, HFE, interaction with transferrin receptor 2 suggests a molecular mechanism for mammalian iron sensing. J Biol Chem. 2006;281:28494–8. [DOI] [PubMed] [Google Scholar]
- 16.Nemeth E. Iron regulation and erythropoiesis. Curr Opin Hematol. 2008;15:169–75. [DOI] [PubMed] [Google Scholar]
- 17.Wang RH, Li C, Xu X, Zheng Y, Xiao C, Zerfas P, Cooperman S, Eckhaus M, Rouault T, et al. A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell Metab. 2005;2:399–409. [DOI] [PubMed] [Google Scholar]
- 18.Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, Samad TA, Campagna JA, Chung RT, Schneyer AL, et al. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet. 2006;38:531–9. [DOI] [PubMed] [Google Scholar]
- 19.De Domenico I, Ward DM, Kaplan J. Hepcidin regulation: ironing out the details. J Clin Invest. 2007;117:1755–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hadley KB, Johnson LK, Hunt JR. Iron absorption by healthy women is not associated with either serum or urinary prohepcidin. Am J Clin Nutr. 2006;84:150–5. [DOI] [PubMed] [Google Scholar]
- 21.Hunt JR, Roughead ZK. Adaptation of iron absorption in men consuming diets with high or low iron bioavailability. Am J Clin Nutr. 2000;71:94–102. [DOI] [PubMed] [Google Scholar]
- 22.Roe MA, Spinks C, Heath AL, Harvey LJ, Foxall R, Wimperis J, Wolf C, Fairweather-Tait SJ. Serum prohepcidin concentration: no association with iron absorption in healthy men; and no relationship with iron status in men carrying HFE mutations, hereditary haemochromatosis patients undergoing phlebotomy treatment, or pregnant women. Br J Nutr. 2007;97:544–9. [DOI] [PubMed] [Google Scholar]
- 23.Tomosugi N, Kawabata H, Wakatabe R, Higuchi M, Yamaya H, Umehara H, Ishikawa I. Detection of serum hepcidin in renal failure and inflammation by using ProteinChip System. Blood. 2006;108:1381–7. [DOI] [PubMed] [Google Scholar]
- 24.Kemna EH, Tjalsma H, Podust VN, Swinkels DW. Mass spectrometry-based hepcidin measurements in serum and urine: analytical aspects and clinical implications. Clin Chem. 2007;53:620–8. [DOI] [PubMed] [Google Scholar]
- 25.Murphy AT, Witcher DR, Luan P, Wroblewski VJ. Quantitation of hepcidin from human and mouse serum using liquid chromatography tandem mass spectrometry. Blood. 2007;110:1048–54. [DOI] [PubMed] [Google Scholar]
- 26.Ganz T, Olbina G, Girelli D, Nemeth E, Westerman M. Immunoassay for human serum hepcidin. Blood. 2008; prepublished online Aug 8; doi:10.1182/blood-2008–02–139915. [DOI] [PubMed]
- 27.Nemeth E, Tuttle MS, Powelson J, Vaughn MB, Donovan A, Ward DM, Ganz T, Kaplan J. Hepcidin regulates cellular iron efflux by binding to ferroportin and inducing its internalization. Science. 2004;306:2090–3. [DOI] [PubMed] [Google Scholar]
- 28.De Domenico I, Ward DM, Langelier C, Vaughn MB, Nemeth E, Sundquist WI, Ganz T, Musci G, Kaplan J. The molecular mechanism of hepcidin-mediated ferroportin down-regulation. Mol Biol Cell. 2007;18:2569–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wrighting DM, Andrews NC. Interleukin-6 induces hepcidin expression through STAT3. Blood. 2006;108:3204–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verga Falzacappa MV, Vujic Spasic M, Kessler R, Stolte J, Hentze MW, Muckenthaler MU. STAT3 mediates hepatic hepcidin expression and its inflammatory stimulation. Blood. 2007;109:353–8. [DOI] [PubMed] [Google Scholar]
- 31.Qian ZM, Chang YZ, Leung G, Du JR, Zhu L, Wang Q, Niu L, Xu YJ, Yang L, et al. Expression of ferroportin1, hephaestin and ceruloplasmin in rat heart. Biochim Biophys Acta. 2007;1772:527–32. [DOI] [PubMed] [Google Scholar]
- 32.Merle U, Fein E, Gehrke SG, Stremmel W, Kulaksiz H. The iron regulatory peptide hepcidin is expressed in the heart and regulated by hypoxia and inflammation. Endocrinology. 2007;148:2663–8. [DOI] [PubMed] [Google Scholar]
- 33.Kulaksiz H, Theilig F, Bachmann S, Gehrke SG, Rost D, Janetzko A, Cetin Y, Stremmel W. The iron-regulatory peptide hormone hepcidin: expression and cellular localization in the mammalian kidney. J Endocrinol. 2005;184:361–70. [DOI] [PubMed] [Google Scholar]
- 34.Bekri S, Gual P, Anty R, Luciani N, Dahman M, Ramesh B, Iannelli A, Staccini-Myx A, Casanova D, et al. Increased adipose tissue expression of hepcidin in severe obesity is independent from diabetes and NASH. Gastroenterology. 2006;131:788–96. [DOI] [PubMed] [Google Scholar]
- 35.Park CH, Valore EV, Waring AJ, Ganz T. Hepcidin, a urinary antimicrobial peptide synthesized in the liver. J Biol Chem. 2001;276:7806–10. [DOI] [PubMed] [Google Scholar]
- 36.Peyssonnaux C, Zinkernagel AS, Datta V, Lauth X, Johnson RS, Nizet V. TLR4-dependent hepcidin expression by myeloid cells in response to bacterial pathogens. Blood. 2006;107:3727–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu XB, Nguyen NB, Marquess KD, Yang F, Haile DJ. Regulation of hepcidin and ferroportin expression by lipopolysaccharide in splenic macrophages. Blood Cells Mol Dis. 2005;35:47–56. [DOI] [PubMed] [Google Scholar]
- 38.Theurl I, Theurl M, Seifert M, Mair S, Nairz M, Rumpold H, Zoller H, Bellmann-Weiler R, Niederegger H, et al. Autocrine formation of hepcidin induces iron retention in human monocytes. Blood. 2008;111:2392–9. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen NB, Callaghan KD, Ghio AJ, Haile DJ, Yang F. Hepcidin expression and iron transport in alveolar macrophages. Am J Physiol Lung Cell Mol Physiol. 2006;291:L417–25. [DOI] [PubMed] [Google Scholar]
- 40.Chaston T, Chung B, Mascarenhas M, Marks J, Patel B, Srai SK, Sharp P. Evidence for differential effects of hepcidin in macrophages and intestinal epithelial cells. Gut. 2008;57:374–82. [DOI] [PubMed] [Google Scholar]
- 41.Zohn IE, De Domenico I, Pollock A, Ward DM, Goodman JF, Liang X, Sanchez AJ, Niswander L, Kaplan J. The flatiron mutation in mouse ferroportin acts as a dominant negative to cause ferroportin disease. Blood. 2007;109:4174–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paradkar P, De Domenico I, Durchfort N, Zohn I, Kaplan J, Ward DM. Iron-depletion limits intracellular bacterial growth in macrophages. Blood. 2008;112:866–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chlosta S, Fishman DS, Harrington L, Johnson EE, Knutson MD, Wessling-Resnick M, Cherayil BJ. The iron efflux protein ferroportin regulates the intracellular growth of Salmonella enterica. Infect Immun. 2006;74:3065–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nairz M, Theurl I, Ludwiczek S, Theurl M, Mair SM, Fritsche G, Weiss G. The co-ordinated regulation of iron homeostasis in murine macrophages limits the availability of iron for intracellular Salmonella typhimurium. Cell Microbiol. 2007;9:2126–40. [DOI] [PubMed] [Google Scholar]
- 45.Gnana-Prakasam JP, Martin PM, Mysona BA, Roon P, Smith SB, Ganapathy V. Hepcidin expression in mouse retina and its regulation via lipopolysaccharide/Toll-like receptor-4 pathway independent of Hfe. Biochem J. 2008;411:79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sow FB, Florence WC, Satoskar AR, Schlesinger LS, Zwilling BS, Lafuse WP. Expression and localization of hepcidin in macrophages: a role in host defense against tuberculosis. J Leukoc Biol. 2007;82:934–45. [DOI] [PubMed] [Google Scholar]
- 47.Wang L, Johnson EE, Shi H, Walker W, Wessling-Resnick M, Cherayil BJ. Attenuated inflammation in hemochromatosis reveals a role for iron in translational control of macrophage cytokine expression. J Immunol. 2008;181:2723–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Johnson EE, Murray M, Wessling-Resnick M. Ferroportin modulates macrophage-mediated immune responses. FASEB J. 2008;22:692.5. [Google Scholar]