Abstract
Insulin induces and dietary n-3 PUFAs suppress hepatic de novo lipogenesis by controlling sterol-regulatory element binding protein-1 nuclear abundance (nSREBP-1). Our goal was to define the mechanisms involved in this regulatory process. Insulin treatment of rat primary hepatocytes rapidly augments nSREBP-1 and mRNASREBP-1c while suppressing mRNAInsig-2 but not mRNAInsig-1. These events are preceded by rapid but transient increases in Akt and Erk phosphorylation. Removal of insulin from hepatocytes leads to a rapid decline in nSREBP-1 [half-time (T1/2) ~ 10 h] that is abrogated by inhibitors of 26S proteasomal degradation. 22:6,n-3, the major n-3 PUFA accumulating in livers of fish oil-fed rats, suppresses hepatocyte levels of nSREBP-1, mRNASREBP-1c, and mRNAInsig-2 but modestly and transiently induces mRNAInsig-1. More importantly, 22:6,n-3 accelerates the disappearance of hepatocyte nSREBP-1 (T1/2 ~ 4 h) through a 26S proteasome-dependent process. 22:6,n-3 has minimal effects on microsomal SREBP-1 and sterol-regulatory element binding protein cleavage-activating protein or nuclear SREBP-2. 22:6,n-3 transiently inhibits insulin-induced Akt phosphorylation but induces Erk phosphorylation. Inhibitors of Erk phosphorylation, but not overexpressed constitutively active Akt, rapidly attenuate 22:6,n-3 suppression of nSREBP-1. Thus, 22:6,n-3 suppresses hepatocyte nSREBP-1 through 26S proteasome- and Erk-dependent pathways. These studies reveal a novel mechanism for n-3 PUFA regulation of hepatocyte nSREBP-1 and lipid metabolism.—Botolin, D., Y. Wang, B. Christian, and D. B. Jump. Docosahexaneoic acid (22:6,n-3) regulates rat hepatocyte SREBP-1 nuclear abundance by Erk- and 26S proteasome-dependent pathways.
Supplementary key words: sterol regulatory element binding protein-1, Insig-1, Insig-2
Sterol-regulatory element binding proteins (SREBP-1a, SREBP-1c, and SREBP-2) are basic helix-loop-helix-leucine zipper transcription factors that play a central role in controlling the transcription of genes involved in cholesterol and fatty acid synthesis (1). The principal mechanism for SREBP regulation of gene transcription involves the control of its nuclear abundance (nSREBP). nSREBP is regulated by two posttranslational mechanisms, proteolytic processing (1) and 26S proteasomal degradation (2). All SREBPs are synthesized as precursors (pSREBP; ~ 125 kDa) tethered to the endoplasmic reticulum (ER) and escorted to the Golgi complex by sterol-regulatory element binding protein cleavage-activating protein (SCAP) for proteolytic processing. nSREBP is transported to the nucleus via importin-β (3), where it binds sterol-regulatory elements in promoters of specific genes, recruits coactivators to the promoter, and stimulates gene transcription (4). Phosphorylation and ubiquitination of nSREBP targets nSREBP for 26S proteasomal degradation (5). Sterols regulate nSREBP levels by controlling the proteolytic processing step, not 26S proteasomal degradation. Instead, sterols induce the ER-resident proteins Insig-1 and Insig-2 to bind SCAP, which retains the SCAP-SREBP complex in the ER, preventing its cleavage to nSREBP (6). This is the molecular basis for the cholesterol suppression of nuclear SREBP-2 abundance and the suppression of cholesterol synthesis.
Although SREBP-1c and SREBP-2 are structurally similar, their regulation in the liver by nutrients and hormones and during postnatal development is quite different. SREBP-1c, but not SREBP-2, is induced by insulin and liver X receptor (LXR) agonists (7, 8). Both insulin and LXR agonists stimulate de novo lipogenesis. Oxysterol-activated LXR/retinoid X receptor heterodimers bind DR-4 regulatory elements on the SREBP-1c promoter and induce SREBP-1c gene transcription (8, 9). Insulin induction of SREBP-1c gene transcription has been correlated with phosphatidylinositol 3-kinase (PI3K) activation and Akt phosphorylation (10–12). Insulin also induces changes in Insig-1, Insig-2a, and Insig-2b mRNA abundance (13, 14), implicating effects of insulin on SREBP processing. However, the linkage between the insulin-regulated signaling pathways and the control of nSREBP-1c remains poorly defined.
PUFAs suppress nSREBP-1 but not nSREBP-2. The decline in nSREBP-1 accounts for the PUFA-mediated suppression of de novo lipogenesis (15, 16). The mechanism for the suppression of nSREBP-1 is complex and has been attributed to the inhibition of SREBP-1c gene transcription, enhanced mRNASREBP-1 degradation, and inhibition of SREBP-1 proteolytic processing (16–25). In some established cell lines, PUFAs interfere with oxysterol-activated LXRα receptors (26, 27). However, in primary hepatocytes and rat liver, PUFAs do not interfere with LXR-regulated gene expression (28).
Our goal is to identify pathways regulated by insulin and n-3 PUFAs that control nSREBP-1. We will show that insulin and n-3 PUFAs rapidly control nSREBP-1 through posttranslational mechanisms. Moreover, both insulin and n-3 PUFAs affect the phosphorylation status of Akt and Erk, which impacts nSREBP-1. Our studies reveal a previously unrecognized mechanism by which n-3 PUFAs control nSREBP-1.
MATERIALS AND METHODS
Animals and primary hepatocytes
All procedures for the use and care of animals for laboratory research have been approved by the All University Committee for Animal Use and Care at Michigan State University.
Feeding study
Male Sprague-Dawley rats were acclimated to meal-feeding a high-carbohydrate (glucose) diet (ICN Biochemicals, Aurora, OH) supplemented with olive oil (Pompiean, Baltimore, MD) at 10% (w/w) for 7 days. The meal began at 8 AM and ended at noon. After the acclimation period, rats were either maintained on the olive oil diet or switched to a high-carbohydrate diet supplemented with fish oil (Dyets, Inc., Bethlehem, PA) at 10% (w/w) (29, 30). Animals were maintained on the olive oil or fish oil for 7 days. Two hours after completion of the final meal, animals were euthanized for tissue collection.
Primary hepatocytes
Rats were maintained on Harlan-Teklad laboratory chow (8640) and water ad libitum. Rat primary hepatocytes were prepared from Teklad chow-fed (ad libitum) male Sprague-Dawley rats, cultured on BioCoat (type 1 collagen) plates (Becton-Dickinson, Bedford, MA), and treated with insulin (Invitrogen, Carlsbad, CA) or fatty acids (Nu-Chek Prep, Elysian, MN) as described previously (28).
Quantitation of hepatic fatty acid composition
Total lipids were extracted from liver in chloroform-methanol (2:1) plus 1 mM butylated hydroxytoluene (30). 7-Nonadecenoic acid (19:1) was added as a recovery standard at the time of extraction. Protein (Bio-Rad, Hercules, CA) was measured in extracts after the initial homogenization step. Total lipids were saponified, fractionated, and quantified by reverse-phase HPLC using a YMC J-Sphere (ODS-H80) column and a sigmoidal gradient starting at 86.5% acetonitrile + acetic acid (0.1%) and ending at 100% acetonitrile + acetic acid (0.1%) over 50 min with a flow rate of 1.0 ml/min using a Waters 600 controller. Fatty acids were detected using both ultraviolet light absorbance at 192 nm (Waters model 2487) and evaporative light scatter (Waters model 2420). Fatty acid composition and structures were confirmed at the Michigan State University mass spectrometry facility by GC-MS (www.bch.msu.edu/facilities/massspec/index.html). Fatty acid standards for reverse-phase HPLC were obtained from Nu-Chek Prep.
RNA extraction and real-time PCR
RNA was extracted from primary hepatocytes (30) and used as a template for real-time PCR. Specific primers for each gene (see below) were designed using Primer Express software (Applied Biosystems, Foster City, CA). First-strand cDNA was synthesized using SuperScript II RNase H– Reverse Transcriptase (Invitrogen). Synthesized cDNA was mixed with 2× SYBR Green PCR Master Mix (Applied Biosystems) and various sets of gene-specific forward and reverse primers and subjected to real-time PCR quantification using the ABI PRISM 7700 Sequence Detection System (Applied Biosystems). All reactions were performed in triplicate. The relative amounts of mRNAs were calculated using the comparative threshold cycle method. Cyclophilin was used as a control, and all results were normalized to the abundance of cyclophilin mRNA. Primers used for real-time PCR are as follows: SREBP-1c forward, TGGACTACTAGTGTTGGCCTGCTT; SREBP-1c reverse, ATCCAGGTCAGCTTGTTTGCGATG; Insig-1 forward, TGCAGATCCAGCGGAATGT; Insig-1 reverse, CCAGG-CGGAGGAGAAGATG; Insig-2a forward, GACGGATGTGTTGAA-GGATTTCT; Insig-2a reverse, TGGACTGAAGCAGACCAATGTC; Insig-2b forward, CCGGCAGAGCTCAGGATTT; Insig-2b reverse, AACTGTGGACTGAAGCAGACCAA; cyclophilin forward, TGGA-TGGCAAGCATGTGGTCTTTG; cyclophilin reverse, CTTCTTGC-TGGTCTTGCCATTCCT.
Immunoblotting
Extracts of primary hepatocytes were prepared as described previously (30, 31). Proteins (50–100 μg) extracted from cytosolic, microsomal, and nuclear fractions were separated electrophoretically by SDS-PAGE (NuPAGE 4–12% polyacrylamide Bis-Tris; Invitrogen) and transferred to nitrocellulose membranes. Membranes were incubated with antibodies for SREBP-1 [IgG-2A4 (31) and sc-13551], SREBP-2 [IgG-7D4 (31) and sc-5603], SCAP (sc-9675), Akt1 (sc-1618), pAkt1/2/3-Ser473 (sc-7985), ERK1 (sc-94), and phosphor-ERK (sc-7383) (Santa Cruz Biotechnology, San Cruz, CA). Anti-mouse and anti-rabbit secondary antibodies were obtained from Bio-Rad; anti-goat antibodies were obtained from Santa Cruz Biotechnology. The SuperSignal West Pico chemiluminescence kit (Pierce) detection system was used.
Recombinant adenovirus infection
Recombinant adenovirus expressing luciferase, kinase-dead Akt (Adv-Akt-KD), and constitutively active Akt (Adv-Akt-CA) were obtained from C. Rhodes (Pacific Northwest Research Institute, Seattle, WA) (32). Confluent primary hepatocytes were infected (10 plaque-forming units/cell) and harvested for evaluation of microsomal and nuclear SREBP-1 abundance.
Statistical analysis
Statistical analysis used Student’s t-test and ANOVA plus post hoc Tukey’s test (http://faculty.vassar.edu/lowry/VassarStats.html).
RESULTS
Effect of dietary fat on hepatic lipid composition
Compared with olive oil-fed rats, rats fed n-3 PUFA (fish oil)-enriched diets have lower levels of nSREBP-1 and SREBP-1 target genes (28). The mol% of n-3 PUFAs in the fish oil diet is as follows: 18:3,n-3, 1.8%; 20:5,n-3, 13%; 22:5,n-3, 1.7%; and 22:6,n-3, 16.3%. Analysis of the hepatic fatty acid composition of animals fed these diets indicates that fish oil feeding leads to no significant change in the total amount of lipid in the liver but significant changes in the type of fatty acids (Table 1). Despite the nearly equal levels of 20:5,n-3 and 22:6,n-3 in the diet, 22:6,n-3 accumulates in livers of fish oil-fed rats. Other n-3 PUFAs in the fish oil diet (i.e., 18:3,n-3, 20:5,n-3, and 22:5,n-3) are likely elongated, desaturated, and β-oxidized in the peroxisome to form 22:6,n-3, the end product of n-3 PUFA synthesis (33). Hepatic enrichment of 22:6,n-3 occurs in neutral lipids (triglycerides and cholesteryl esters), phospholipids, and nonesterified fatty acids (data not shown).
TABLE 1.
Effect of olive oil and fish oil feeding on hepatic fatty acid composition
| Fatty Acid | Olive Oil | Fish Oil | Mass Change | P |
|---|---|---|---|---|
| nmol/mg protein | ||||
| 16:0 | 527 ± 218 | 391 ± 128 | −136 | 0.21 |
| 18:0 | 55 ± 24 | 19 ± 13 | −36 | 0.041a |
| 18:1,n-9 | 224 ± 95 | 60 ± 18 | −164 | 0.021a |
| 18:2,n-6 | 127 ± 45 | 66 ± 2 | −61 | 0.05a |
| 18:3,n-3 | 8 ± 5 | 16 ± 2 | +8 | 0.026a |
| 18:3,n-6 | 69 ± 5 | 5 ± 1 | −4 | 0.17 |
| 20:3,n-6 | 12 ± 8 | 3 ± 0.3 | −9 | 0.047 |
| 20:3,n-9 | 26 ± 13 | 0.5 ± 0.1 | −25.5 | 0.012a |
| 20:4,n-6 | 204 ± 106 | 115 ± 15 | −89 | 0.11 |
| 20:5,n-3 | 5 ± 3 | 75 ± 15 | +70 | 0.0007a |
| 22:5,n-3 | 5 ± 3 | 25 ± 8 | +20 | 0.009a |
| 22:6,n-3 | 236 ± 129 | 568 ± 150 | +332 | 0.022a |
| Total | 1,437 ± 621 | 1,341 ± 333 | −96 | 0.414 |
| % of total | ||||
| 18:3,n-3 | 0.6 | 1.2 | ||
| 20:5,n-3 | 0.4 | 5.6 | ||
| 22:5,n-3 | 0.4 | 1.9 | ||
| 22:6,n-3 | 16.4 | 42.4 | ||
Male Sprague-Dawley rats were meal fed for 7 days (see Materials and Methods). The meal consisted of a high-carbohydrate diet supplemented with either olive oil or fish oil at 10% (w/w) (28). Total hepatic lipid was extracted, saponified, and quantified (see Materials and Methods). Results were obtained from three separate animals per group (mean ± SD). Statistical analysis used Student’s t-test, and one-tailed P values were calculated (http://faculty.vassar.edu/lowry/VassarStats.html).
P values ≤ 0.05 reflect significant differences between the means of the olive oil- and fish oil-fed groups.
Differential effects of n-3 PUFAs on rat hepatic SREBP-1 regulation
Because 22:6,n-3 accumulates in livers of fish oil-fed rats, we examined the effect of 22:6,n-3 on pSREBP-1 and nSREBP-1 in primary hepatocytes. Increasing the dose of 22:6,n-3 to 250 μM suppressed both precursor and nuclear forms of SREBP-1. However, nSREBP-1 was more sensitive to 22:6,n-3 suppression than microsomal SREBP-1, pSREBP-1 (Fig. 1A). We next compared the effect of 20:5,n-3 and 22:6,n-3 on SREBP-1 abundance. At the 100 μM dose, both fatty acids suppressed microsomal pSREBP-1 equally, 25–40% (Fig. 1B, C). 20:5,n-3 and 22:6,n-3 suppressed nSREBP-1 by 43% and 76%, respectively. 22:6,n-3 was ~ 2-fold more effective than 20:5,n-3 at suppressing nSREBP-1.
Fig. 1.
Effects of n-3 PUFAs on sterol-regulatory element binding protein-1 (SREBP-1) abundance in primary hepatocytes. Primary hepatocytes were incubated overnight in Williams E medium + 20 mM lactate + 10 nM DEX with no insulin or serum. The next day, cells were treated with 10 nM insulin and 25 mM glucose in the absence and presence of n-3 PUFAs with BSA (fatty acid/BSA = 5). A: Primary hepatocytes were treated with and without varying concentrations of 22:6,n-3. Cells were harvested after 24 h for isolation of microsomal and nuclear proteins for the measurement of precursor SREBP-1 (pSREBP-1; solid line) and nuclear SREBP-1 (nSREBP-1; dashed line) by immunoblotting (see Materials and Methods). The antibody recognizes both SREBP-1a and SREBP-1c. Results are presented as percentage of control after treatment with fatty acids and are representative of two separate experiments. B: Cells were treated with or without 100 μM 20:5,n-3 or 22:6,n-3 for 24 h. Microsomal and nuclear protein was extracted for measurement of pSREBP-1 and nSREBP-1 by immunoblotting (see Materials and Methods). Veh, vehicle. C: Results of five separate experiments were quantified, presented as arbitrary density units (means ± SD), and evaluated using ANOVA plus post hoc Tukey’s honestly significant difference test (http://faculty.vassar.edu/lowry/VassarStats.html). * P < 0.05, vehicle versus 22:6,n-3. DEX, dexamethasone.
Rapid effects of insulin and 22:6,n-3 on hepatic nSREBP-1c
Because 22:6,n-3 accumulates in livers of fish oil-fed rats (Table 1) and is more potent than 20:5,n-3 at suppressing nSREBP-1 (Fig. 1), 22:6,n-3 was used to examine the time course of n-3 PUFA effects on the regulation of nSREBP-1, pSREBP-1, and mRNASREBP-1c in rat primary hepatocytes. Cells incubated overnight in serum-free Williams E medium containing no insulin decreased nSREBP-1 by ~ 80% (Fig. 2A), with minimal (~ 10%) effect on pSREBP-1 (Fig. 2B). The addition of insulin (10 nM) to the culture medium induced nSREBP-1, pSREBP-1, and mRNASREBP-1c (Fig. 2A–C) 6-, 1.2-, and 5-fold after 24 h. 22:6,n-3 rapidly and significantly attenuated the insulin induction of SREBP-1 nuclear protein (Fig. 2A) but modestly suppressed microsomal SREBP-1 (Fig. 2B). 22:6,n-3 had no effect on nSREBP-2 (Fig. 2A, inset).
Fig. 2.
Time course of insulin (Ins) and 22:6,n-3 regulation of pSREBP-1 and nSREBP-1 abundance in rat primary hepatocytes. Primary rat hepatocytes were maintained overnight in Williams E medium + 20 mM lactate + 10 nM DEX with no serum or insulin. The next morning, cells were switched to medium supplemented with 25 mM glucose and 10 nM insulin in the absence and presence of 100 μM 22:6,n-3. Cells were harvested at the times indicated for extraction of nuclear (A) and microsomal (B) proteins for measurement of SREBP-1 by immunoblotting. Insets in A and B are representative immunoblots of SREBP-1 and SREBP-2 after 24 h of treatment. Immunoblots (A, B) were quantified, and the results are normalized to the level of SREBP-1 expressed in livers of 90 day old male rats (mean ± SD; n = 3). Veh, vehicle. C: Primary hepatocytes were incubated overnight as described above. The next day, cells were switched to medium supplemented with 25 mM glucose and 10 nM insulin in the absence or presence of 20:5,n-3 or 22:6,n-3 at 100 μM. At the times indicated, cells were harvested for RNA extraction and measurement of SREBP-1c and cyclophilin mRNA by real-time PCR (see Materials and Methods). Results are expressed as fold change in SREBP1c/cyclophilin (mean ± SD; n = 6).
Both 20:5,n-3 and 22:6,n-3 suppressed the insulin induction of mRNASREBP-1c. Although 20:5,n-3 effects on mRNASREBP-1c appear transient, 22:6,n-3 has a sustained suppressive effect on mRNASREBP-1c. The fact that 22:6,n-3 also decreased the abundance of mRNASREBP-1c by ~ 70% over 24 h suggests enhanced mRNA decay. 20:5,n-3 also promoted a decline in mRNASREBP-1c at 6 h, but by 24 h mRNASREBP-1c levels increased. Thus, 22:6,n-3 has a sustained suppressive effect on mRNASREBP-1c, whereas the 20:5,n-3 effect on mRNASREBP-1c is transient.
Together, these studies indicate that insulin rapidly induces nSREBP-1 and that 22:6,n-3 rapidly inhibits this response (Fig. 2). Neither factor had a major effect on microsomal SREBP-1 (Fig. 2B) or nuclear SREBP-2 (Fig. 2A). Although both insulin and 22:6,n-3 affected mRNASREBP-1c (Fig. 2C), the absence of a parallel change in microsomal SREBP-1 suggests that both insulin and 22:6,n-3 act rapidly at a posttranslational level to control nSREBP-1.
Effects of insulin and 22:6,n-3 on SCAP, Insig-1, and Insig-2
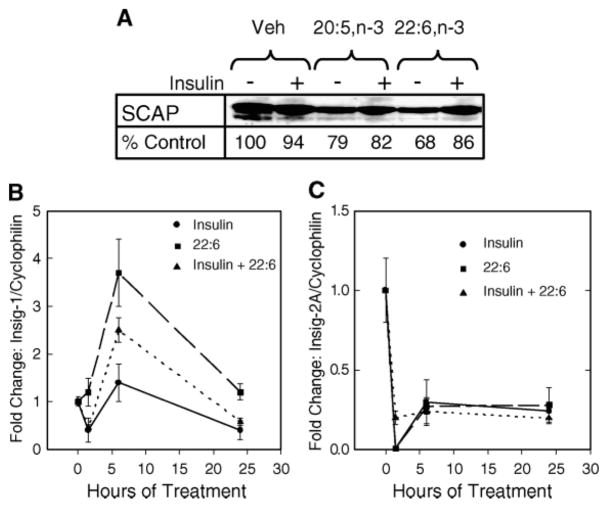
nSREBP is regulated by two opposing posttranslational mechanisms, SREBP processing (1) and 26S proteasomal degradation (2). We examined the effects of 22:6,n-3 on components involved in SREBP processing (e.g., SCAP, Insig-1, and Insig-2).
Microsomal SCAP levels remained marginally affected by insulin or n-3 PUFAs (Fig. 3A). Insig-1 mRNA abundance was modestly increased by insulin. 22:6,n-3 transiently induced Insig-1 mRNA in the presence and absence of insulin (Fig. 3B). Insig-2A mRNA was rapidly suppressed by insulin and 22:6,n-3 (Fig. 3C). Insig-2B levels were unaffected by insulin or 22:6,n-3 (data not shown).
Fig. 3.
Effects of n-3 PUFAs on sterol-regulatory element binding protein cleavage-activating protein (SCAP), Insig-1, and Insig-2 expression. A: Primary rat hepatocytes were treated with and without insulin and 20:5,n-3 or 22:6,n-3 for 24 h. After treatment, microsomal protein was isolated for analysis of SCAP protein by immunoblotting. The level of SCAP protein was quantified, and the results are shown as percentage of control and are representative of two separate studies. Veh, vehicle. B, C: Total RNA was extracted from primary hepatocytes treated as described for Fig. 2C. Levels of Insig-1 (B), Insig-2 (C), and cyclophilin mRNA were measured by real-time PCR. Results are presented as fold change in Insig/cyclophilin and are representative of three separate studies with triplicate samples/treatment (mean ± SD). Solid line-circles, insulin-treated cells; dashed line-squares, 22:6,n-3-treated cells; dotted line-triangles, insulin- and 22:6,n-3-treated cells.
The effects of insulin on Insig-1 and Insig-2 mRNA abundance confirm earlier reports (13). 22:6,n-3 effects on Insig-1 and Insig-2 expression suggest that n-3 PUFAs change Insig subtype expression in the liver. However, the effect of 22:6,n-3 + insulin on Insig-1 expression is modest and transient and may be insufficient to affect SREBP-1 processing. Finding that both insulin and 22:6,n-3 suppressed Insig-2 mRNA abundance is inconsistent with a role of Insig-2 in the 22:6,n-3 regulation of nSREBP-1. The outcome of these studies suggests that 22:6,n-3 may have minimal effects on SREBP-1 processing.
Effect of 26S proteasome inhibitors on insulin and 22:6,n-3 control of nSREBP-1
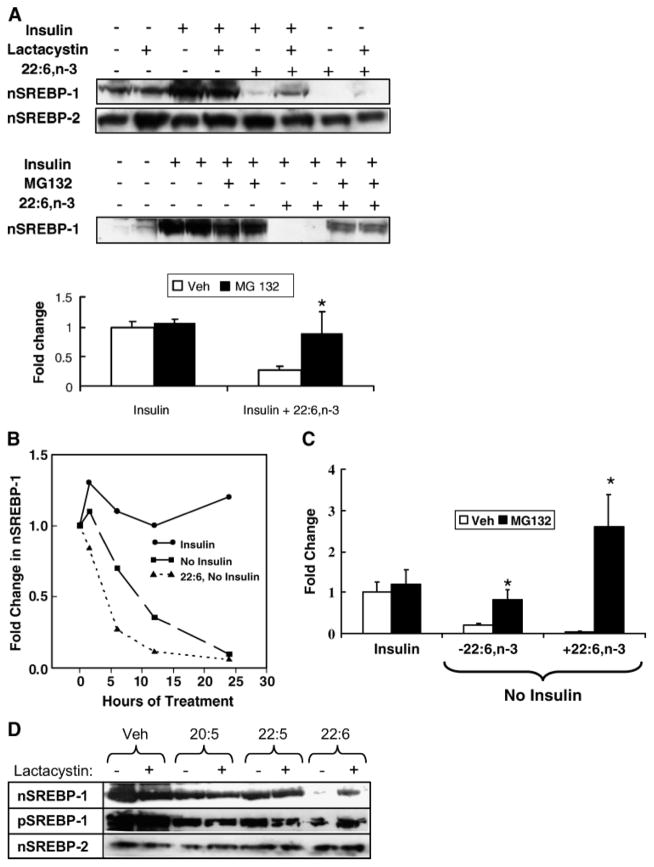
SREBP-1 and SREBP-2 are ubiquitinated and degraded by a 26S proteasome-dependent pathway (2, 5). To determine whether either insulin or 22:6,n-3 affects SREBP-1 26S proteasomal degradation, primary hepatocytes incubated overnight in serum- and insulin-free medium were treated with insulin or insulin + 22:6,n-3 in the absence and presence of the 26S proteasome inhibitors MG132 and lactacystin (Fig. 4A).
Fig. 4.
Effect of 26S proteasome inhibitors on insulin and 22:6,n-3 regulation of SREBP-1. A: Primary rat hepatocytes were maintained overnight in Williams E medium containing 10 nM DEX and 20 mM lactate but no serum, insulin, or fatty acids. The next morning, cells were switched to medium supplemented with 25 mM glucose in the absence or presence of insulin (10 nM), 22:6,n-3 (100 μM), lactacystin (0.5 μM), or MG132 (10 μM). Lactacystin and MG132 are inhibitors of 26S proteasomal degradation of proteins. In the MG132 experiments, cells were harvested after 12 h of treatment. In the lactacystin studies, cells were harvested after 6 h of treatment. Microsomal and nuclear proteins were assayed for SREBP-1 and SREBP-2 by immunoblotting. Representative immunoblots illustrate the effects of insulin, 22:6,n-3, and lactacystin (upper blot) on nSREBP-1 and nSREBP-2 or MG132 (lower blot) on nSREBP-1. In the absence of insulin or 22:6,n-3, MG132 has no detectable effect on nSREBP-1 abundance. Results of the MG132 experiment are quantified and presented as fold change in nSREBP-1 induced by insulin or insulin + 22:6,n-3 (mean ± SD; n = 4). Veh, vehicle. Results were evaluated by Student’s t-test. * P < 0.001 for vehicle- versus MG132-treated cells. B: Time course of the disappearance of SREBP-1 from hepatocyte nuclei. Primary rat hepatocytes were maintained overnight in Williams E medium containing 25 mM glucose, 10 nM DEX, and 10 nM insulin to induce nSREBP-1. The next morning, the medium was changed to Williams E medium containing 10 nM DEX and 25 mM glucose with insulin, without insulin, or without insulin but with 100 μM 22:6,n-3. Cells were harvested at 1.5, 6, 12, and 24 h for isolation of nuclear proteins. Levels of nSREBP-1 were quantified by immunoblotting. C: Primary rat hepatocytes were maintained overnight in Williams E medium containing 10 nM DEX, 25 mM glucose, and 10 nM insulin to induce nSREBP-1. The next morning, the medium was changed to Williams E medium containing 10 nM DEX and 25 mM glucose with insulin, without insulin, or without insulin but with 100 μM 22:6,n-3. All three groups received vehicle or MG132 at 10 μM. Twelve hours later, cells were harvested for measurement of nSREBP-1 by immunoblotting. Results are expressed as fold change from control (insulin-treated with no MG132) (mean ± SD; n = 3). Results were evaluated by Student’s t-test. * P < 0.01, vehicle- versus MG132-treated cells. D: Cells were treated with insulin in the absence or presence of n-3 PUFAs (100 μM) for 24 h in the absence or presence of lactacystin. Nuclear and microsomal proteins were isolated and assayed for nSREBP-1, pSREBP-1, and nSREBP-2 by immunoblotting. The results are representative of two separate studies.
Lactacystin had no effect on basal or insulin-induced (4-fold) nSREBP-1 (Fig. 4A, graph). Treatment of hepatocytes with insulin + 22:6,n-3 significantly (>90%) suppressed nSREBP-1. Coincubation with lactacystin or MG132 significantly attenuated the 22:6,n-3 effect on nSREBP-1 abundance (Fig. 4A). Incubating primary hepatocytes with 22:6,n-3 alone reduced SREBP-1 to undetectable levels; inclusion of lactacystin restored SREBP-1 to a detectable level. These results suggest that 22:6,n-3 regulates nSREBP-1 through a 26S proteasome-dependent process.
To examine this process differently, primary hepatocytes were treated with insulin overnight to induce nSREBP-1. Cells were maintained for an additional 24 h with insulin or the insulin was withdrawn; 22:6,n-3 was added to one group of cells in which insulin was withdrawn (Fig. 4B). Maintaining insulin treatment for an additional 24 h had little effect on nSREBP-1. However, removal of insulin led to a rapid decline in SREBP-1 from the nucleus [approximate half-time (~ T1/2) = 10 h]. Addition of 22:6,n-3 to the minus-insulin group accelerated the loss of nSREBP-1 (~ T1/2 ≤ 4 h).
This study was repeated with the addition of MG132, and the cells were harvested 12 h after insulin removal (Fig. 4C). Removal of insulin resulted in an ~ 80% decrease in nSREBP-1 after 12 h; addition of 22:6,n-3 suppressed nSREBP-1 to a 96% loss. Addition of MG132 totally prevented the loss of nSREBP-1 in cells receiving no insulin in the absence and presence of 22:6,n-3.
Lactacystin (Fig. 4D) or MG132 (data not shown) had no consistent effect on pSREBP-1 or nSREBP-2. Moreover, the suppression of nSREBP-1 by 20:5,n-3 and 22:5,n-3 was not affected by lactacystin. Thus, the action of 26S proteasome inhibitors (lactacystin and MG132) on nSREBP-1 is specific for 22:6,n-3. The outcome of these results support the notion that 22:6,n-3 regulates nSREBP-1 through a 26S proteasome-dependent mechanism.
Akt is a target for both insulin and 22:6,n-3 regulation
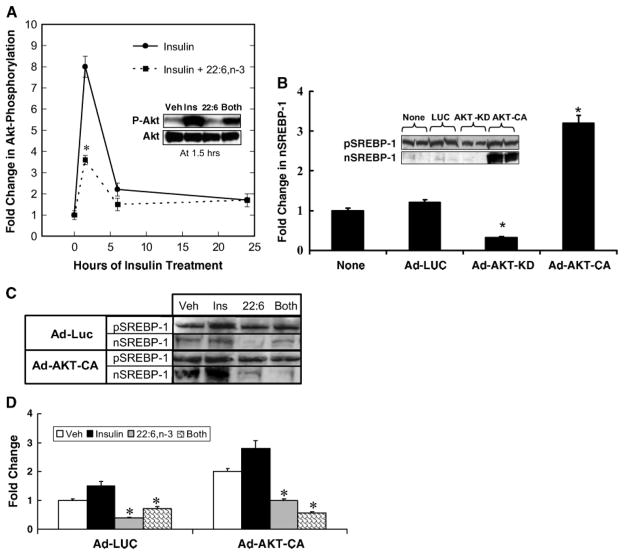
Next, we focused on determining which signal transduction mechanisms were involved in the regulation of nSREBP-1. Insulin activation of PI3K and Akt are established routes for the insulin control of nSREBP-1 (10, 12). We confirmed the involvement of PI3K by using the inhibitor LY294002 to block the insulin-mediated induction of nSREBP-1 in primary rat hepatocytes (data not shown). The effect of insulin and 22:6,n-3 on Akt phosphorylation (Ser473) was examined in primary hepatocytes (Fig. 5A).
Fig. 5.
Role of Akt in regulating nSREBP-1. A: Time course of insulin induction of Akt phosphorylation. Primary rat hepatocytes were maintained overnight in Williams E medium containing 10 nM DEX and 20 mM lactate, but with no serum, insulin, or fatty acids. The next morning, cells were switched to medium supplemented with 25 mM glucose and 10 nM insulin or 25 mM glucose, 10 nM insulin, and 100 μM 22:6,n-3. Cells were harvested at the times indicated for extraction of total hepatic proteins for measurement of total Akt and phospho-Akt (P-Akt) by immunoblotting. Results are presented as fold change in Akt phosphorylation and are normalized to the level of phospho-Akt in cells before insulin or 22:6,n-3 treatment (mean ± SD; n = 3). Results were evaluated by Student’s t-test. * P < 0.001, insulin + 22:6,n-3- versus insulin-treated cells. The inset shows a representative immunoblot for P-Akt and total Akt at 1.5 h of treatment. Ins, insulin; Veh, vehicle. B: Primary hepatocytes were either not infected (None) or infected with recombinant adenovirus expressing luciferase (Ad-LUC; control), a kinase-dead Akt (Adv-Akt-KD), or a constitutively active Akt (Adv-Akt-CA). Cells were maintained in Williams E medium containing 10 nM DEX with no serum or insulin. Twenty-four hours after infection, cells were harvested for analysis of pSREBP-1 and nSREBP-1 by immunoblotting (inset). Levels of pSREBP-1 and nSREBP-1 were quantified by immunoblotting. Results are expressed as fold change in nSREBP-1 and are normalized to levels of nSREBP-1 in noninfected cells (mean ± SD; n = 3). Results were evaluated by Student’s t-test. * P < 0.01, Ad-Akt-KD- or AD-Akt-CA- versus Ad-LUC-infected cells. C: Primary hepatocytes were infected with recombinant adenovirus expressing luciferase (Ad-LUC) or constitutively active Akt (Adv-Akt-CA). Twenty-four hours after infection, cells were treated with BSA (Veh), insulin (10 nM) + BSA, 22:6,n-3 (100 μM) + BSA, or a combination of insulin and 22:6,n-3 (Both). Six hours after treatment, cells were harvested for measurement of nSREBP-1 by immunoblotting. D: Results of four separate studies are quantified and normalized to the level of nSREBP-1 in vehicle-treated Ad-LUC-infected cells. Results are expressed as fold change (mean ± SD; n = 4). Results were evaluated by Student’s t-test. * P < 0.001, 22:6,n-3- versus insulin-treated cells.
Removal of insulin from the medium overnight decreases Akt phosphorylation, whereas insulin addition induces hepatocyte Akt phosphorylation 8-fold within 1.5 h. Six hours after insulin treatment, Akt phosphorylation was ~ 2-fold above basal values. 22:6,n-3 significantly attenuated (50%) the insulin-stimulated Akt phosphorylation at 1.5 h but had no significant effect on Akt phosphorylation after 6 h. The immunoblot (Fig. 5A, inset) illustrates the effect of insulin, 22:6,n-3, and combination treatment on Akt phosphorylation at 1.5 h. Total Akt protein levels remained unaffected by insulin or 22:6,n-3 treatment. Note that cells treated with 22:6,n-3 alone had no effect on Akt phosphorylation. Thus, 22:6,n-3 affects only insulin-stimulated Akt phosphorylation.
Because insulin-induced Akt phosphorylation was attenuated by 22:6,n-3, we determined whether overexpression of a constitutively active form of Akt would affect the 22:6,n-3 control of nSREBP-1 in primary hepatocytes. In a preliminary study, primary hepatocytes were infected with recombinant adenovirus expressing luciferase (Ad-Luc, as a control), Akt kinase-dead form (Adv-Akt-KD), or constitutively active Akt (Adv-Akt-CA) (Fig. 5B). The conditions of infection resulted in a ≥10-fold increase in Akt-CA and Akt-KD expression (data not shown). Cells received no insulin treatment. Compared with uninfected cells, Ad-Luc and Akt-KD had little effect on pSREBP-1. Akt-KD suppressed nSREBP-1 by ~ 50%. Akt-CA significantly induced nSREBP-1c (3-fold) with little effect on pSREBP-1. Thus, changes in Akt activity affect nSREBP-1 through a posttranslational mechanism.
To determine whether overexpressed Akt (Adv-Akt-CA) could abrogate 22:6,n-3 control of SREBP-1, primary hepatocytes were infected with Ad-Luc and Adv-Akt-CA and treated with insulin, 22:6,n-3, or insulin + 22:6,n-3 for 6 h (Fig. 5C, D). Microsomal pSREBP-1 remained unaffected by insulin or 22:6,n-3 in Ad-Luc- or Adv-Akt-CA-infected hepatocytes (Fig. 5C). Adv-Akt-CA infection induced nSREBP-1 nearly 3-fold, and insulin treatment of both Ad-Luc and Adv-Akt-CA had modest effects on nSREBP-1. More importantly, 22:6,n-3 treatment of hepatocytes, in the absence or presence of insulin, significantly suppressed nSREBP-1 in both Ad-Luc- and Adv-Akt-CA-treated cells. Overexpressed Akt-CA failed to abrogate the 22:6,n-3 effect on nSREBP-1. Thus, the inhibitory effect of 22:6,n-3 on Akt phosphorylation (Fig. 5A) is not linked to the 22:6,n-3-mediated suppression of nSREBP-1 (Fig. 5D).
Effects of insulin and 22:6,n-3 on Erk phosphorylation
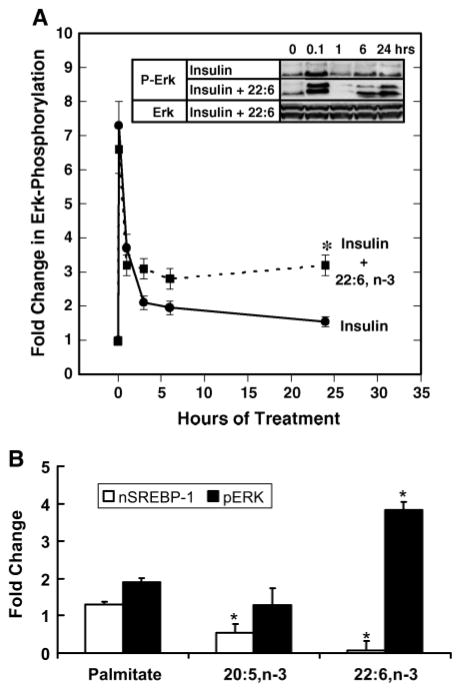
Insulin regulation of mitogen-activated protein kinase signaling pathways is well established (14, 34). Insulin treatment of primary rat hepatocytes rapidly, but transiently, increased Erk phosphorylation (Fig. 6A). This biphasic response has been described by other investigators (14). The combination of 22:6,n-3 and insulin had no effect on the early induction of Erk phosphorylation, suggesting that 22:6,n-3 does not interfere with insulin-stimulated Erk phosphorylation. However, 24 h after initiating treatment, Erk phosphorylation was sustained at a level significantly above that of the insulin-treated group. Insulin and 22:6,n-3 had no effect on total Erk protein abundance. Treatment of primary hepatocytes with 22:6, n-3 alone also increased Erk phosphorylation after 3 h (data not shown). Repeating this study with palmitate, 20:5,n-3, or 22:6,n-3 (Fig. 6B) indicated that after 24 h of treatment, only 22:6,n-3 significantly induced Erk phosphorylation. Increased Erk phosphorylation correlates with robust suppression of nSREBP-1.
Fig. 6.
Role of Erk in regulating nSREBP-1. A: Primary rat hepatocytes were maintained overnight in Williams E medium containing 20 mM lactate and 10 nM DEX with no serum or insulin. Cells were switched to medium supplemented with either 10 nM insulin (solid line) or insulin + 22:6,n-3 (dotted line). Cells were harvested at the times indicated for measurement of Erk phosphorylation by immunoblotting. The inset shows representative immunoblots of the effects of insulin and 22:6,n-3 on total Erk (Erk) and Erk phosphorylation (P-Erk). Results are normalized to the level of Erk phosphorylation in untreated primary hepatocytes and are shown as fold change in Erk phosphorylation (mean ± SD; n = 4). Results were evaluated by Student’s t-test. * P < 0.01, insulin + 22:6,n-3-versus insulin-treated cells. B: Effect of palmitate, 20:5,n-3, and 22:6,n-3 on nSREBP-1 (white bars) and Erk phosphorylation (black bars). Rat primary hepatocytes were maintained overnight in Williams E medium containing 20 mM lactate and 10 nM DEX with no insulin or serum. Cells were switched to medium supplemented with 25 mM glucose, 10 nM insulin, and 20 μM BSA in the absence or presence of fatty acids at 100 μM plus BSA (20 μM). Cells were harvested 24 h later for nSREBP-1 and Erk phosphorylation (pERK) by immunoblotting. Total Erk protein was unaffected by these treatments (not shown). Results are normalized to the level of nSREBP-1 or Erk phosphorylation in untreated cells (mean ± SD; n = 4). Results were evaluated by Student’s t-test. * P < 0.01, vehicle- versus fatty acid-treated cells.
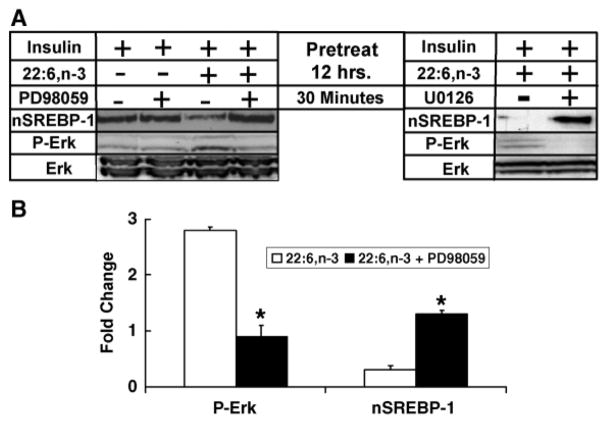
To determine whether Erk phosphorylation is linked to the regulation of nSREBP-1, cells were treated overnight with 10 nM insulin in the absence and presence of 100 μM 22:6,n-3 (Fig. 7). Overnight insulin treatment induced nSREBP-1 but had no effect on Erk phosphorylation. Overnight treatment with insulin + 22:6,n-3 suppressed nSREBP-1 and induced Erk phosphorylation. The two groups of hepatocytes were treated with vehicle or MEK [mitogen-activated protein (MAP) kinase kinase] inhibitors of Erk phosphorylation, 10 nM PD98059 or 10 nM U0126. After 30 min of treatment, hepatocytes were harvested for analysis of nSREBP-1 and Erk phosphorylation. The results using PD98059 are quantified in Fig. 7 (graph). Treatment of primary hepatocytes with PD98059 or U0106 effectively suppressed 22:6,n-3-induced Erk phosphorylation. More surprisingly, these MEK inhibitors (PD98059 and U0106) significantly induced nSREBP-1 in cells treated with insulin +22:6,n-3 overnight. These results indicate that chronic suppression of nSREBP-1 by 22:6,n-3 is rapidly (within 30 min) reversed by treatment with MEK inhibitors (PD98059 or U0106) of Erk phosphorylation.
Fig. 7.
The MEK inhibitors PD98059 and U0126 rapidly abrogate 22:6,n-3 suppression of nSREBP-1. A: Primary hepatocytes were maintained overnight in Williams E medium containing 20 mM lactate, 10 nM insulin, 10 nM DEX, and 20 μM BSA with or without 100 μM 22:6,n-3. The next morning, the medium was replaced with Williams E medium containing 25 mM glucose, 10 nM insulin, and 10 nM DEX in the absence and presence of 22:6,n-3 (100 μM) or 10 μM PD98059 or U0126. After 30 min of treatment, cells were harvested for protein isolation and immunoblotting for nSREBP-1 and phospho-Erk (P-Erk) or Erk total protein (Erk). B: Immunoblots from the PD98059 study were quantified for the effects of 22:6,n-3 on Erk phosphorylation (P-Erk) and nSREBP-1. White bars, 22:6,n-3; black bars, 22:6,n-3 + PD98059. Results are from four independent studies and are normalized to the level of P-Erk and nSREBP-1 in cells receiving no 22:6,n-3 or PD98059 (mean ± SD; n = 4). Results were evaluated by Student’s t-test. * P < 0.01 for 22:6,n-3- versus 22:6,n-3 + PD98059-treated cells.
The p38 inhibitor, SB203580, had no effect on Erk phosphorylation or nSREBP-1. The Jnk inhibitor, SP600125, was found unsuitable for our studies because it stimulated Erk phosphorylation. Together, these results indicate that 22:6,n-3 regulates nSREBP-1 through an Erk-dependent mechanism.
DISCUSSION
Insulin and n-3 PUFAs regulate hepatic de novo lipogenesis, at least in part, by controlling nSREBP-1 (1, 16, 35). Our goal was to define the mechanisms involved in this regulatory process. These studies have revealed a novel pathway by which one n-3 PUFA, namely 22:6,n-3, rapidly controls nSREBP-1. The evidence presented in this report supports the concept that 22:6,n-3 accelerates the degradation of nSREBP-1 by a 26S proteasome-dependent pathway while having little impact on microsomal SREBP-1 or nSREBP-2 (Figs. 1–4). 22:6,n-3 is the major n-3 PUFA accumulating in livers of fish oil-fed animals (Table 1). Moreover, 22:6,n-3 is the end product of n-3 PUFA synthesis from the essential fatty acid 18:3,n-3 (33). As such, 22:6,n-3 is a feedback inhibitor of its own synthesis. Fish oil feeding suppresses the expression of key hepatic enzymes in this pathway, specifically Elovl-5 and Δ5 and Δ6 desaturases, and activates peroxisomal β-oxidation for the degradation of ≥C22 PUFAs through a peroxisome proliferator-activated receptor α-dependent mechanism (29, 36). This regulatory scheme is analogous to the cholesterol control of its own synthesis by SREBP-2 (1), but with an important distinction. Cholesterol regulates the proteolytic conversion of pSREBP-2 to the nuclear form with little impact on 26S proteasomal degradation or SREBP-2 gene expression (1, 5). In contrast, 22:6,n-3 suppresses nSREBP-1 by accelerating 26S proteasome-dependent degradation of nSREBP-1 (Fig. 4). Like 20:5,n-3, 22:6,n-3 also controls nSREBP-1 through effects on SREBP-1c promoter activity (unpublished observation) and mRNASREBP-1 abundance (Fig. 2C). However, 22:6,n-3 is the only n-3 PUFA controlling nSREBP-1c through a 26S proteasome-dependent mechanism (Fig. 4D).
SREBP-1a, -1c, and -2 are ubiquitinated and degraded by the 26S proteasome (2, 5, 37). The signal initiating SREBP degradation involves the phosphorylation of Thr426 and Ser430 in the 424TLTTPPPSD motif in SREBP-1a and of Thr432 and Ser436 in the 430LMSPPASD motif of SREBP-2 (2). These sites correspond to Thr393 and Ser399 in SREBP-1c. Phosphorylation of the TLTTPPPSD motif by Gsk3β promotes binding of the ubiquitin ligase, SCFFbw7, which targets SREBP for 26S proteasomal degradation. Inhibition of Gsk3β activity by LiCl (38) or insulin (2) promotes the accumulation of SREBP. Insulin inhibits Gsk3β activity by increasing the Akt-mediated phosphorylation of Gsk3β at Ser9.
Our studies support the concept that insulin rapidly controls nSREBP-1 by regulating SREBP-1 degradation. Overexpression of kinase-dead Akt suppresses nSREBP-1, whereas constitutively active Akt induces nSREBP-1 in the absence of insulin. Overexpression of these Akt variants has little effect on microsomal SREBP-1 abundance (Fig. 5B). Moreover, insulin induction of Akt phosphorylation (Fig. 5A) precedes the increase in nSREBP-1 with minimal effects on pSREBP-1 (Fig. 2). Removal of insulin from hepatocytes promotes a rapid decline in nSREBP-1 (T1/2 ~ 10 h) (Fig. 4B) that is abrogated by 26S proteasome inhibitors (Fig. 4C). Together, these results suggest that insulin acts rapidly to induce nSREBP-1 by inhibiting 26S proteasomal degradation. Insulin effects on SREBP-1 promoter activity (22), mRNASREBP-1, and microsomal SREBP-1 abundance (Fig. 2) may be secondary to the primary effects of insulin on SREBP-1 phosphorylation and its proteasomal degradation.
22:6,n-3 suppresses insulin-induced nSREBP-1 with little impact on pSREBP-1 or nSREBP-2 (Figs. 1, 2, 4D). The transient inhibition of 22:6,n-3 on insulin-induced Akt phosphorylation (Fig. 5A) might be expected to enhance Gsk3β-mediated phosphorylation of SREBP-1 and promote SREBP-1 degradation in the proteasome. However, over-expression of constitutively active Akt failed to abrogate the 22:6,n-3-mediated suppression of nSREBP-1 (Fig. 5C, D). Thus, 22:6,n-3 suppression of Akt phosphorylation is not the sole route for 22:6,n3 control of nSREBP-1.
Insulin also rapidly, but transiently, induces Erk phosphorylation (Fig. 6) (14). Like Akt phosphorylation, insulin induction of Erk phosphorylation precedes changes in nSREBP-1 (Figs. 2, 6). 22:6,n-3 does not impair the early phase of insulin-induced Erk phosphorylation but sustains increased Erk phosphorylation after 6 h (Fig. 6A). Enhanced Erk phosphorylation correlates with lower levels of nSREBP-1 in hepatocytes (Fig. 6) and in liver (unpublished observation). MEK inhibitors (PD95098 and U0126) rapidly (within 30 min) attenuate Erk phosphorylation and induce nSREBP-1 (Fig. 7). The rapidity of the effect of MEK inhibitors on nSREBP-1 supports a role of the MEK/Erk pathway in controlling nSREBP-1.
SREBP-1a is phosphorylated by Erk in vitro at Ser117, and mutation of this site impairs insulin-stimulated regulation of the LDL receptor promoter by SREBP-1a (39). Ser117 corresponds to Ser92 in SREBP-1c. Other Erk sites have been identified in SREBP-1c at Ser39, Ser73, and Thr395 (http://scansite.mit.edu/). Based on in silico analysis, Thr395, which is located within the ubiquitin ligase binding motif of SREBP-1 (TLTTPPPSD), may be phosphorylated by both Gsk3β and Erk. Erk phosphorylation of this site may induce ubiquitin ligase binding and target SREBP-1 to the 26S proteasome. Interestingly, 430LMSPPASD in SREBP-2 is also phosphorylated by Gsk3β and binds SCFFbw7 (2), but it is not an Erk phosphorylation site (www.scansite.mit.edu). This sequence difference may account for the selective effect of 22:6,n-3 on SREBP-1 26S proteasomal degradation. Clearly, more studies are required to evaluate the role that the 393TLTPPPSD motif plays in the 22:6,n-3-mediated control of 26S proteasomal degradation of SREBP-1.
An important outcome of our study was finding that not all n-3 PUFAs regulate nSREBP-1 at the level of the 26S proteasome. 18:3,n-3 is a weak regulator of mRNASREBP-1 and nSREBP-1 (data not shown). Although both 20:5,n-3 and 22:6,n-3 suppress mRNASREBP-1, the 20:5,n-3 effect is transient (Fig. 2C). Both 20:5,n-3 (21) and 22:6,n-3 (unpublished observation) suppress SREBP-1c promoter activity. Thus, the decline in mRNASREBP-1 is likely attributable to the inhibition of SREBP-1c gene transcription and enhanced mRNASREBP-1 decay. The SREBP-1c promoter contains a sterol-regulatory element, and SREBP-1c promoter activity is induced by increased nSREBP-1 (8, 9). We suggest that 22:6,n-3-mediated activation of 26S proteasomal degradation of nSREBP-1 may be an antecedent mechanism for the 22:6,n-3 control of SREBP-1c gene transcription. However, 26S proteasomal degradation of nSREBP-1 cannot explain the n-3 PUFA control of mRNASREBP-1 degradation. PUFA-regulated signaling pathways controlling mRNASREBP-1 degradation remain unresolved.
Inhibitors of 26S proteasomal regulation are able to abrogate much, but not all, of the suppressive effect of 22:6,n-3 on nSREBP-1 (Fig. 4A, C). Effects of 22:6,n-3 on mRNASREBP-1 abundance as well as the modest effects on mRNAInsig-1 and microsomal SCAP levels (Fig. 3) may represent additional mechanisms contributing to the 22:6,n-3 control of nSREBP-1 (Fig. 4). Unlike cholesterol control of SREBP-2 (1), 22:6,n-3 acts at multiple levels to control nSREBP-1.
In summary, our studies have identified a novel mechanism by which n-3 PUFAs suppress nuclear levels of SREBP-1 in rat primary hepatocytes. 22:6,n-3, the main n-3 PUFA accumulating in livers of fish oil-fed rats, accelerates the loss of SREBP-1 from hepatocyte nuclei through 26S proteasome- and Erk-dependent pathways. Inhibition of these pathways abrogates the 22:6,n-3 effect on nuclear SREBP-1 abundance. The outcome of these studies has generated new unanswered questions. In particular, the mechanisms for 22:6,n-3 control of MEK activity, Erk phosphorylation, and 26S proteasomal degradation of nSREBP-1 remain unresolved. Because insulin and 22:6,n-3 act rapidly to control nSREBP-1, it will be important to determine whether the effects of insulin and n-3 PUFAs on SREBP-1c promoter activity (21, 25) are attributable to antecedent posttranslational mechanisms controlling nSREBP-1.
Acknowledgments
The authors thank Christopher J. Rhodes (Pacific Northwest Research Institute, Seattle, WA) for generously providing the recombinant adenovirus expressing luciferase and Akt. The authors also thank Drs. Julia Busik, Weiqin Chen, Kathleen Gallo, and Karl Olson for helpful suggestions during the course of these studies. This project was supported by the National Institutes of Health (Grant DK-43220), the National Research Initiative of the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service (Grant 2003-35200-13400), and the Michigan Agriculture Experiment Station.
Abbreviations
- ER
endoplasmic reticulum
- LXR
liver X receptor
- MEK
mitogen-activated protein kinase kinase
- PI3K
phosphatidylinositol 3-kinase
- SCAP
sterol-regulatory element binding protein cleavage-activating protein
- SREBP
sterol-regulatory element binding protein
- T1/2
half-time
References
- 1.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sundqvist A, Bengoechea-Alonso MT, Ye X, Lukiyanchuk V, Jin J, Harper JW, Ericsson J. Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCGFBW7. Cell Metabolism. 2005;1:379–391. doi: 10.1016/j.cmet.2005.04.010. [DOI] [PubMed] [Google Scholar]
- 3.Nagoshi E, Imamoto N, Sato R, Yoneda Y. Nuclear import of sterol regulatory element-binding protein-2, a basic helix-loop-helix-leucine zipper (bHLH-Zip)-containing transcription factor, occurs through the direct interaction of importin beta with HLH-Zip. Mol Biol Cell. 1999;10:2221–2233. doi: 10.1091/mbc.10.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett MK, Toth JI, Osborne TF. Selective association of sterol regulatory element binding protein isoforms with target promoters in vivo. J Biol Chem. 2004;279:37360–37367. doi: 10.1074/jbc.M404693200. [DOI] [PubMed] [Google Scholar]
- 5.Hirano Y, Yoshida M, Shimizu M, Sato R. Direct demonstration of rapid degradation of nuclear sterol regulatory element-binding proteins by the ubiquitin-proteasome pathway. J Biol Chem. 2001;276:36431–36437. doi: 10.1074/jbc.M105200200. [DOI] [PubMed] [Google Scholar]
- 6.Adams CM, Reitz J, De Brabander JK, Feramisco JD, Li L, Brown MS, Goldstein JL. Cholesterol and 25-hydroxycholesterol inhibit activation of SREBPs by different mechanisms, both involving SCAP and Insigs. J Biol Chem. 2004;279:52772–52780. doi: 10.1074/jbc.M410302200. [DOI] [PubMed] [Google Scholar]
- 7.Shimomura I, Bashmakov Y, Ikemoto S, Horton JD, Brown MS, Goldstein JL. Insulin selectively increases SREBP-1c mRNA in the livers of rats with streptozotocin-induced diabetes. Proc Natl Acad Sci USA. 1999;96:13656–13661. doi: 10.1073/pnas.96.24.13656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schultz JR, Tu H, Luk A, Repa JJ, Medina JC, Li L, Schwendner S, Wang S, Thoolen M, Mangelsdorf DJ, et al. Role of LXRs in control of lipogenesis. Genes Dev. 2000;14:2831–2838. doi: 10.1101/gad.850400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro JM, Shimomura I, Shan B, Brown MS, Goldstein JL, Mangelsdorf DJ. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleischmann M, Iynedjian PB. Regulation of sterol regulatory-element binding protein 1 gene expression in liver: role of insulin and protein kinase B/cAkt. Biochem J. 2000;349:13–17. doi: 10.1042/0264-6021:3490013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ono H, Shimano H, Katagiri H, Yahagi N, Sakoda H, Onishi Y, Anai M, Ogihara T, Fujishiro M, Viana AYI, et al. Hepatic AKT activation induces marked hypoglycemia, hepatomegaly and hypertriglyceridemia with sterol regulatory element binding protein involvement. Diabetes. 2003;52:2905–2913. doi: 10.2337/diabetes.52.12.2905. [DOI] [PubMed] [Google Scholar]
- 12.Ribaux PG, Iynedjian PB. Analysis of the role of protein kinase B (cAKT) in insulin-dependent induction of glucokinase and sterol regulatory element-binding protein 1 (SREBP1) mRNAs in hepatocytes. Biochem J. 2003;376:697–705. doi: 10.1042/BJ20031287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yabe D, Komuro R, Liang G, Goldstein JL, Brown MS. Liver-specific mRNA for Insig-2 down-regulated by insulin: implications for fatty acid synthesis. Proc Natl Acad Sci USA. 2003;100:3155–3160. doi: 10.1073/pnas.0130116100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keeton AB, Bortoff KD, Franklin JL, Messina JL. Blockade of rapid versus prolonged ERK1/2 activation has differential effects on insulin-induced gene expression. Endocrinology. 2005;146:2716–2725. doi: 10.1210/en.2004-1662. [DOI] [PubMed] [Google Scholar]
- 15.Jump DB. Dietary polyunsaturated fatty acids and regulation of gene transcription. Curr Opin Lipidol. 2002;13:155–164. doi: 10.1097/00041433-200204000-00007. [DOI] [PubMed] [Google Scholar]
- 16.Jump DB. Fatty acid regulation of gene transcription. Crit Rev Clin Lab Sci. 2004;41:41–78. doi: 10.1080/10408360490278341. [DOI] [PubMed] [Google Scholar]
- 17.Xu J, Nakamura MT, Cho HP, Clarke SD. Sterol regulatory element binding protein-1 expression is suppressed by dietary polyunsaturated fatty acids. A mechanism for the coordinate suppression of lipogenic genes by polyunsaturated fats. J Biol Chem. 1999;274:23577–23583. doi: 10.1074/jbc.274.33.23577. [DOI] [PubMed] [Google Scholar]
- 18.Hannah VC, Ou J, Luong A, Goldstein JL, Brown MS. Unsaturated fatty acids down-regulate srebp isoforms 1a and 1c by two mechanisms in HEK-293 cells. J Biol Chem. 2001;276:4365–4372. doi: 10.1074/jbc.M007273200. [DOI] [PubMed] [Google Scholar]
- 19.Mater MK, Thelen AP, Pan DA, Jump DB. Sterol response element-binding protein 1c (SREBP1c) is involved in the polyunsaturated fatty acid suppression of hepatic S14 gene transcription. J Biol Chem. 1999;274:32725–32732. doi: 10.1074/jbc.274.46.32725. [DOI] [PubMed] [Google Scholar]
- 20.Xu J, Teran-Garcia M, Park JH, Nakamura MT, Clarke SD. Polyunsaturated fatty acids suppress hepatic sterol regulatory element-binding protein-1 expression by accelerating transcript decay. J Biol Chem. 2001;276:9800–9807. doi: 10.1074/jbc.M008973200. [DOI] [PubMed] [Google Scholar]
- 21.Deng X, Cagen LM, Wilcox HG, Park EA, Raghow R, Elam MB. Regulation of the rat SREBP-1c promoter in primary rat hepatocytes. Biochem Biophys Res Commun. 2002;290:256–262. doi: 10.1006/bbrc.2001.6148. [DOI] [PubMed] [Google Scholar]
- 22.Cagen LM, Deng X, Wilcox HG, Park EA, Raghow R, Elam MB. Insulin activates the rat sterol-regulatory-element-binding protein 1c (SREBP-1c) promoter through the combinatorial actions of SREBP, LXR, Sp-1 and NF-Y cis-acting elements. Biochem J. 2005;385:207–216. doi: 10.1042/BJ20040162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Worgall TS, Sturley SL, Seo T, Osborne TF, Deckelbaum RJ. Polyunsaturated fatty acids decrease expression of promoters with sterol regulatory elements by decreasing levels of mature sterol regulatory element-binding protein. J Biol Chem. 1998;273:25537–25540. doi: 10.1074/jbc.273.40.25537. [DOI] [PubMed] [Google Scholar]
- 24.Chen G, Liang G, Ou J, Goldstein JL, Brown MS. Central role for liver X receptor in insulin-mediated activation of Srebp-1c transcription and stimulation of fatty acid synthesis in liver. Proc Natl Acad Sci USA. 2004;101:11245–11250. doi: 10.1073/pnas.0404297101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakatani T, Katsumata A, Miura S, Kamei Y, Ezaki O. Effects of fish oil feeding and fasting on LXRalpha/RXRalpha binding to LXRE in the SREBP-1c promoter of mouse liver. Biochim Biophys Acta. 2005;1736:77–86. doi: 10.1016/j.bbalip.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 26.Ou J, Tu H, Shan B, Luk A, DeBose-Boyd RA, Bashmakov Y, Goldstein JL, Brown MS. Unsaturated fatty acids inhibit transcription of the sterol regulatory element-binding protein-1c (SREBP-1c) gene by antagonizing ligand-dependent activation of the LXR. Proc Natl Acad Sci USA. 2001;98:6027–6032. doi: 10.1073/pnas.111138698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pawar A, Xu J, Jerks E, Mangelsdorf DJ, Jump DB. Fatty acid regulation of liver X receptors (LXR) and peroxisome proliferator-activated receptor alpha (PPARalpha) in HEK293 cells. J Biol Chem. 2002;277:39243–39250. doi: 10.1074/jbc.M206170200. [DOI] [PubMed] [Google Scholar]
- 28.Pawar A, Botolin D, Mangelsdorf DJ, Jump DB. The role of liver X receptor-alpha (LXR-alpha) in the fatty acid regulation of hepatic gene expression. J Biol Chem. 2003;278:40736–40743. doi: 10.1074/jbc.M307973200. [DOI] [PubMed] [Google Scholar]
- 29.Pan DA, Mater MK, Thelen AP, Peters JM, Gonzalez FJ, Jump DB. Evidence against the peroxisome proliferator-activated receptor alpha (PPARalpha) as the mediator for polyunsaturated fatty acid suppression of hepatic L-pyruvate kinase gene transcription. J Lipid Res. 2000;41:742–751. [PubMed] [Google Scholar]
- 30.Wang Y, Botolin D, Christian B, Busik C, Xu J, Jump DB. Tissue-specific, nutritional and developmental regulation of rat fatty acid elongases. J Lipid Res. 2005;46:706–715. doi: 10.1194/jlr.M400335-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Botolin D, Jump DB. Selective proteolytic processing of rat hepatic sterol regulatory element binding protein-1 (SREBP-1) and SREBP-2 during postnatal development. J Biol Chem. 2003;278:6959–6962. doi: 10.1074/jbc.M212846200. [DOI] [PubMed] [Google Scholar]
- 32.Wrede CE, Dickson LM, Lingohr MK, Briaud I, Rhodes CJ. Protein kinase B/Akt prevents fatty acid-induced apoptosis in pancreatic beta-cells (INS-1) J Biol Chem. 2002;277:49676–49684. doi: 10.1074/jbc.M208756200. [DOI] [PubMed] [Google Scholar]
- 33.Sprecher H. Metabolism of highly unsaturated n-3 and n-6 fatty acids. Biochim Biophys Acta. 2000;1486:219–231. doi: 10.1016/s1388-1981(00)00077-9. [DOI] [PubMed] [Google Scholar]
- 34.Keeton AB, Bortoff KD, Bennett WL, Franklin JL, Venable DY, Messina JL. Insulin-regulated expression of Egr-1 and Krox20: dependence on ERK1/2 and interaction with p38 and PI3-kinase pathways. Endocrinology. 2003;144:5402–5410. doi: 10.1210/en.2003-0592. [DOI] [PubMed] [Google Scholar]
- 35.Pegorier JP, Le May C, Girard J. Control of gene expression by fatty acids. J Nutr. 2004;134(Suppl):2444–2449. doi: 10.1093/jn/134.9.2444S. [DOI] [PubMed] [Google Scholar]
- 36.Ren B, Thelen AP, Peters JM, Gonzalez FJ, Jump DB. Polyunsaturated fatty acid suppression of hepatic fatty acid synthase and S14 gene expression does not require peroxisome proliferator-activated receptor alpha. J Biol Chem. 1997;272:26827–26832. doi: 10.1074/jbc.272.43.26827. [DOI] [PubMed] [Google Scholar]
- 37.Sundqvist A, Ericsson J. Transcription-dependent degradation controls the stability of the SREBP family of transcription factors. Proc Natl Acad Sci USA. 2003;100:13833–13838. doi: 10.1073/pnas.2335135100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim KH, Song MJ, Yoo EJ, Choe SS, Park SD, Kim JB. Regulatory role of GSK3 for transcriptional activity of ADD1/SREBP1c. J Biol Chem. 2004;279:51999–52006. doi: 10.1074/jbc.M405522200. [DOI] [PubMed] [Google Scholar]
- 39.Roth G, Kotzka J, Kremer L, Lehr S, Lohaus C, Meyer HE, Krone W, Muller-Wieland D. MAP kinases Erk1/2 phosphorylate sterol regulatory element-binding protein (SREBP)-1a at serine 117 in vitro. J Biol Chem. 2000;275:33302–33307. doi: 10.1074/jbc.M005425200. [DOI] [PubMed] [Google Scholar]