Abstract
Background
Variation in LDL-cholesterol (LDL-C) among individuals is a complex genetic trait involving multiple genes and gene-environment interactions.
Methods and Results
In a genome-wide association study (GWAS) to identify genetic variants influencing LDL-C in an isolated population from Kosrae, we observed associations for SNPs in the gene encoding HMG-CoA reductase (HMGCR). Three of these SNPs (rs7703051, rs12654264 and rs3846663) met the statistical threshold of genome-wide significance when combined with data from the Diabetes Genetics Initiative GWAS. We followed up the association results and identified a functional SNP in intron13 (rs3846662), which was in linkage disequilibrium with the SNPs of genome-wide significance and affected alternative splicing of HMGCR mRNA. In vitro studies in human lymphoblastoid cells demonstrated that homozygosity for the rs3846662 minor allele was associated with up to 2.2-fold lower expression of alternatively spliced HMGCR mRNA lacking exon13 and minigene transfection assays confirmed that allele status at rs3846662 directly modulated alternative splicing of HMGCR exon13 (42.9±3.9 vs. 63.7±1.0 %Δexon13/total HMGCR mRNA, p=0.02). Further, the alternative splice variant could not restore HMGCR activity when expressed in HMGCR deficient UT-2 cells.
Conclusion
We identified variants in HMGCR that are associated with LDL-C across populations and affect alternative splicing of HMGCR exon13.
Keywords: HMG-CoA reductase, SNP, genome-wide association study, LDL-C, alternative splicing
Elevated levels of LDL-cholesterol (LDL-C) are a primary risk factor for atherosclerotic cardiovascular disease, the major cause of morbidity and mortality in industrialized countries today 1. Variation in LDL-C among individuals is a complex genetic trait, involving multiple genes and significant gene- environment interactions 2. Candidate gene and linkage studies have identified some of the genetic factors contributing to the population variance in plasma lipoprotein levels 3, 4, but these factors only explain a small fraction of the heritability, suggesting that additional variants influencing lipid levels remain to be identified.
Our group has previously used candidate gene 5 and linkage 6 approaches to identify genetic loci affecting plasma lipid and lipoprotein levels in a cohort from the Island of Kosrae, Federated States of Micronesia, a genetic isolate with significant founder effects and a high prevalence of traits related to the metabolic syndrome 7.
Recently, genome-wide association studies (GWAS) have been shown to be successful in gene discovery for complex traits and offer a new approach to identify common genetic variants with modest effects 8. Using Affymetrix gene chip 500k arrays, we have performed a GWAS in ~2400 Kosraens for LDL-C and other metabolic traits (Lowe et al, manuscript in preparation). The strongest association for LDL-C was found for two SNPs on chromosome 19q13 near APOE, a candidate gene with known, common coding polymorphisms 9. The second best hits for LDL-C were SNPs that mapped to the HMG-CoA reductase (HMGCR) gene, the rate-limiting enzyme in cholesterol biosynthesis and the target of LDL-C lowering statin drugs. During the preparation of this manuscript, four GWAS for LDL-C in Caucasian cohorts were published 10–13 and one of them 13 included an association between the same SNPs in HMGCR and plasma LDL-C in Caucasians.
Identifying a strong and replicable association signal in a GWAS is just the first step in elucidating the specific genetic variants involved in predisposing individuals to complex traits. In most cases, the underlying causal variant is not directly captured on the SNP array. Rather, one or more SNPs on the array are acting as a proxy for a functional non-genotyped SNP with which it is in linkage disequilibrium. Fine-mapping, re-sequencing and hypothesis-driven approaches have been proposed to unearth the actual causal variants.
In this manuscript, we report associations of SNPs in HMGCR with LDL-C in Kosraens that, in combination with similar findings from studies in Caucasians, indicate that the same genetic variants at HMGCR contribute to differences in LDL-C across populations. To follow-up the association signal, we have implemented a hypothesis driven strategy and performed in vitro studies to identify a functional variant in intron13 of HMGCR that affects alternative splicing of exon13.
Methods
An expanded methods section is provided in the supplemental Methods (please see http://atvb.ahajournals.org).
Kosrae Study Subjects and GWAS
A genome-wide association study for LDL-C was carried out in 2346 people of the Island of Kosrae, Federated States of Micronesia using SNPs from the Affymetrix 500k platform. Details of this study will be described elsewhere (Lowe et al, manuscript in preparation). All participants in the study provided written informed consent and IRB approval was obtained from all participating institutions.
We used publicly available data from Saxena et al. 14 to validate our findings. The p-values of the Kosrae and DGI study analyses were combined using Fisher's method 15 to quantify the overall evidence for association. We set 5×10−8 as threshold for genome-wide statistical significance of the combined p-values.
Cell lines
Lymphoblastoid cell lines from 18 Caucasian CEU individuals of the HapMap collection were obtained from the Coriell Institute for Medical Research. These individuals were homozygous for the major (n=9) or minor allele (n=9) at rs3846662 and the three proxy SNPs from the Affymetrix 500k array (rs3846663, rs7703051 and rs12654264). For HMGCR mRNA expression studies cells were seeded at a density of 250 000 cells/ml medium.
UT-2 cells (a gift from Drs. Russell DeBose-Boyd and Michael Brown, UT Southwestern Medical Center) are mutant chinese hamster ovary (CHO) cells that lack HMG-CoA reductase and require mevalonate for growth. Stock cultures of UT-2 cells were grown in F12:MEM, 10% FCS, 0.2 mM mevalonate and growth experiments were carried out as described 16.
Quantitative Real-Time PCR
RNA was extracted using TRIzol reagent (Invitrogen) and cDNA was synthesized using SuperScript III (Invitrogen). Quantitative RT-PCR was performed in an ABI PRISM 7700 Sequence Detector (Applied Biosystems). Specific primers and probes for full-length HMGCR, Δexon13 HMGCR and β-actin mRNA were selected to span exon junctions to avoid co-amplification of genomic DNA. mRNA expression levels were normalized to β-actin as a housekeeping gene.
Minigene Constructs
Minigenes were used to assess the influence of SNP rs3846662 on alternative splicing of HMGCR exon13. PCR amplified genomic DNA for both alleles (rs3846662/A and rs3846662/G), containing exons12–14, internal introns and parts of the flanking introns, was cloned into the exon-trapping vector pSPL3 (kindly provided by Drs. Woohyun Yoon and David B. Goldstein, Duke University). Identities of the minigenes were confirmed by DNA sequencing. To confirm the causal polymorphism of the observed effects in the minigene systems, we converted SNP rs3846662 in the major form construct (allele A) to the minor form (allele G) by site directed mutagenesis (QuikChange Lightning Kit, Strategene).
Cell Transfection
HEK293 cells were transfected with HMGCR pSPL3 minigenes (A, G and A→G) and empty pSPL3 vector (negative control) using FuGene6 reagent (Roche Applied Bioscience). After 24h, RNA was isolated and reverse transcribed using the SA2 Primer (5’-ATCTCAGTGGTATTTGTGAGC-3’), corresponding to a transcribed exonic sequence in the pSPL3 vector and thus allowing analysis of only vector-specific HMGCR transcripts. HMGCR splicing pattern was analyzed by Real-Time PCR as described above.
Stable expressing cell lines
The open reading frames of HMGCR full-length and Δexon13 mRNA were PCR amplified and cloned into the pcDNA3.1 expression vector (Invitrogen). UT-2 cells with stable expression of human full-length (UT-2+FL) or Δexon13 HMGCR (UT-2+ex13) were generated by G418 selection of FuGene6 transfected UT-2cells.
Other assays
Cell protein was measured using the BioRad DC kit. HMG-CoA reductase activity was measured in detergent solubilized cell extracts as described 17 except that mevalonolactone was separated by ion exchange chromatography 18.
Results
Common variants in HMGCR are associated with plasma LDL-C across populations
We performed genome-wide association analysis for LDL-C using 2346 individuals from the Micronesian Island of Kosrae. The strongest association was found for SNPs on chromosome 19q13 in the APOE/C1/C4/C2 gene cluster (rs4420638, p= 1.89×10−7). The effect of the APOE polymorphisms on LDL-C is well established in many ethnicities and our result thus implicates that the same genetic factors are important in the Kosraen population. The second best locus for LDL-C mapped to a region on chromosome 5q13 containing the HMGCR gene (Table1). This gene encodes HMG-CoA reductase, the rate-limiting enzyme in cholesterol biosynthesis and thus represents an interesting candidate gene with high plausibility. The difference in LDL-C between homozygotes at this locus was 0.30 mmol/L (11.6 mg/dL) and the fraction of the population variance for LDL-C explained by this locus was 2.1%.
Table 1.
Primary associations and meta-analysis results for LDL-C
| Kosrae | DGI | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SNP | Chr | Position | Locus | A1/A2 | MAF | p | β | A1/A2 | MAF | p | β | p-comb |
| rs11957260 | 5 | 74644488 | hmgcr | T/C | 0.41 | 1.17×10−5 | 0.18 | T/C | 0.43 | 5.05×10−3 | 0.08 | 5.91×10−8 |
| rs7703051 | 5 | 74661243 | hmgcr | A/C | 0.41 | 1.75×10−5 | 0.18 | A/C | 0.41 | 5.86×10−4 | 0.10 | 1.03×10−8 |
| rs12654264 | 5 | 74684359 | hmgcr | T/A | 0.42 | 2.40×10−5 | 0.18 | T/A | 0.41 | 4.09×10−4 | 0.10 | 9.82×10−9 |
| rs3846663 | 5 | 74691482 | hmgcr | T/C | 0.39 | 1.28×10−6 | 0.20 | T/C | 0.41 | 5.71×10−4 | 0.10 | 7.31×10−10 |
SNP: dbSNP ID, Chr: chromosome, Position: NCBI build 36, Locus: closest gene, A1/A2: allele 1(major)/ allele2 (minor), MAF: minor allele frequency, p: p-value, β: beta-coefficient represents the proportion of 1 s.d. change in standardized LDL-C residual (mean=0 , s.d =1) per copy of the allele modelled (additive model), p-comb combined p-value of Kosrae and DGI studies using Fisher’s method
However, the results did not surpass genome-wide significance when the Bonferroni correction was applied for multiple testing (most associated SNP rs3846663: p=1.28×10−6). To validate our findings in an independent cohort, we combined data from our analysis with public data for LDL-C of the Diabetes Genetics Initiative (DGI) GWAS. Since Kosraens (Micronesian) and DGI participants (European Caucasians) are of different ancestry, we first investigated linkage disequilibrium (LD) patterns in a 1 Mb region around the locus. As shown in supplemental Fig.I, pairwise LD (r2) was symmetrical between Caucasian HapMap samples and Micronesians from Kosrae.
Combining p-values across both studies, we validated multiple SNPs at the HMGCR locus, three of which surpassed a genome-wide significance of p<5 ×10−8 (rs7703051, rs12654264 and rs3846663) (Table 1 and supplemental Fig.II). A regional association plot for the combined results showed a peak of association signal over a 47 kb region containing the HMGCR gene (Fig.II). In both study populations, the minor allele frequencies were comparable and the minor alleles were associated with an increase in LDL-C.
Figure 2.
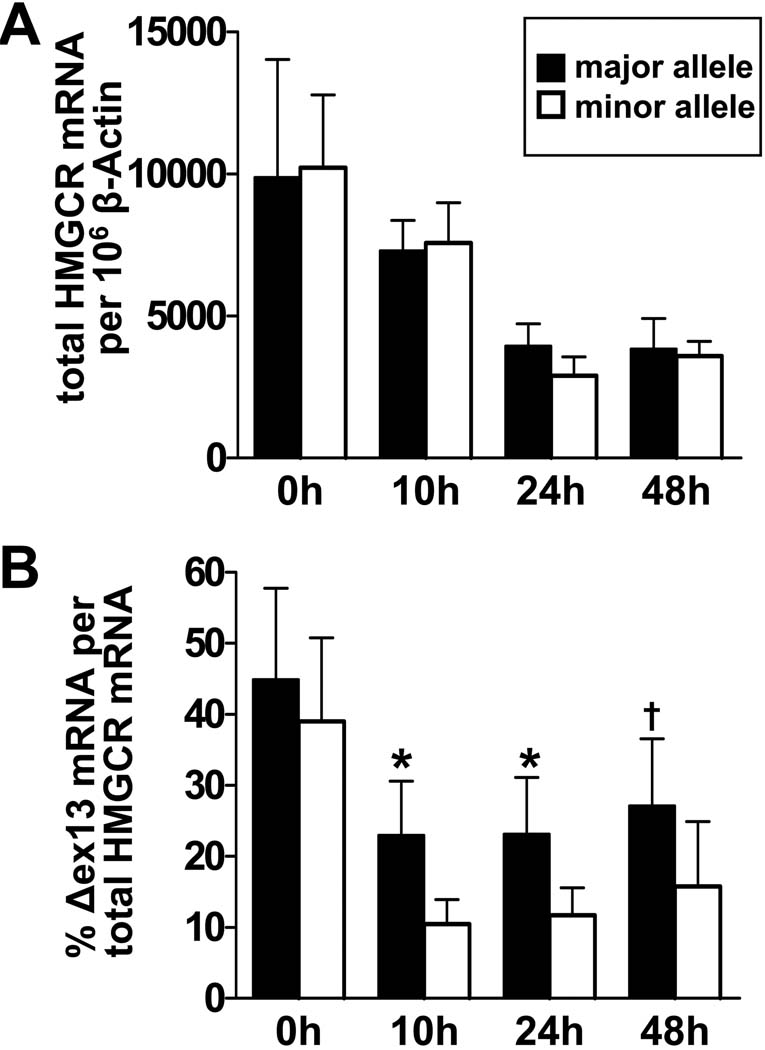
HMGCR mRNA expression in lymphoblastoid cells of major and minor allele homozygotes at rs3846662 (n=9 per genotype). (A) Total HMGCR mRNA expression normalized to β-actin over 48h. (B) Alternatively spliced HMGCR mRNA (Δex13 mRNA) as percentage of total HMGCR mRNA expression. *p<0.01 and †p<0.05 between major and minor allele samples.
We also combined the p-values of the association analysis results for plasma total cholesterol from the Kosrae and DGI studies and found genome-wide significance for the same three SNPs at the HMGCR locus (supplemental Table I).
Association between SNP rs3846662 and alternative splicing of HMGCR exon13 in human lymphoblastoid cell lines
To follow-up the association results, we next aimed to discover functional variants at the HMGCR locus and study their molecular mode of action. We used existing resequencing data of the region containing the entire HMGCR gene from 23 Caucasians - estimated to have >99% power to detect variants with a minor allele frequency of >5% 19- to identify candidate functional SNPs. Under our hypothesis driven model we focused on SNPs that would have strong potential for changing HMGCR function or levels.
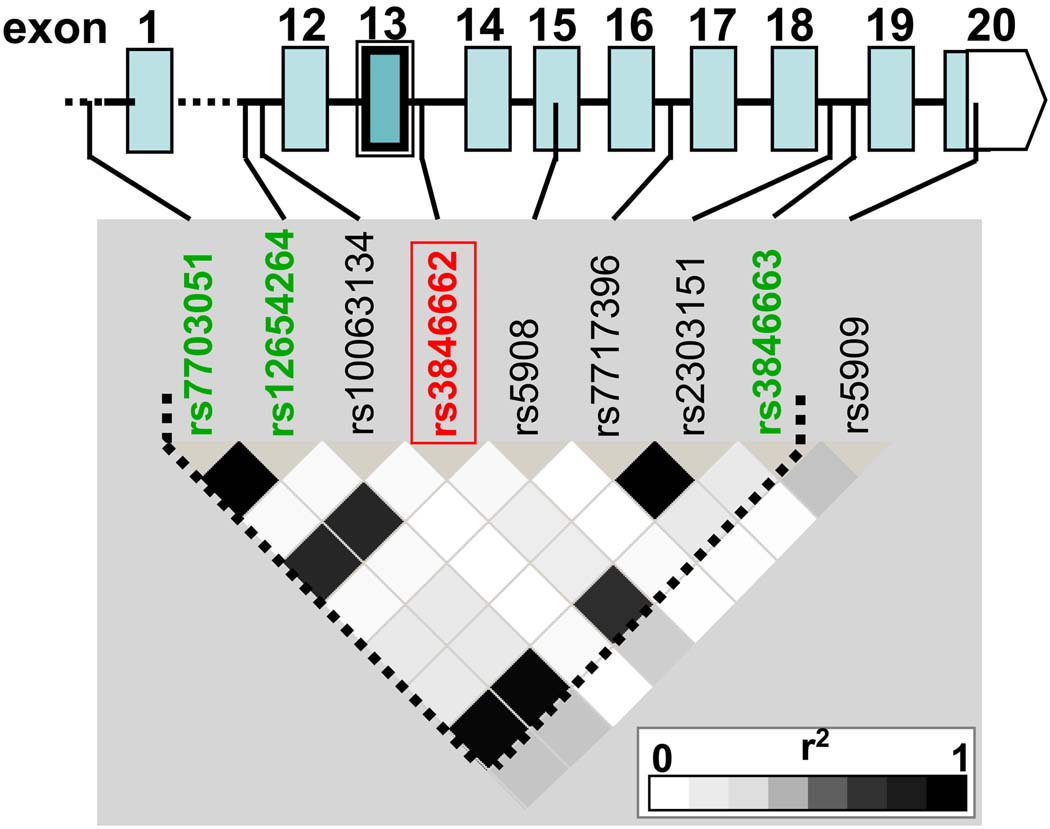
All associated SNPs of the Kosrae and DGI studies were non-coding variants and in LD (r2 0.81–1), suggesting they represent the same association. The lack of LD (r2≤0.02) between these SNPs and the only known non-synonymous SNP in HMGCR, rs5908 (I638V) in exon15, suggested that the association was not due to this protein coding mutation (Fig.1). Since the existence of a second HMGCR mRNA transcript resulting from alternative splicing had been reported in humans 20, we looked for SNPs in the vicinity of exon-intron borders. We detected that SNP rs3846662 was located 47bp downstream of exon13 and in LD with the genotyped variants of genome-wide significance (r2:0.82–0.93, Fig.1).We hypothesized that this intronic SNP may be functional and modulate the splicing efficiency of exon13.
Figure 1.
LD plot (r2 values) in HapMap CEU individuals for SNPs at the HMGCR gene locus demonstrating LD between Affymetrix 500k SNPs with genome-wide significance (green) and the functional intronic SNP rs3846662 (red). Coding exons are shown as blue boxes. Exon13, which is subject to alternative splicing is double framed.
To analyze whether rs3846662 was associated with HMGCR splicing efficiency we obtained lymphoblastoid cell lines (LCLs) from Caucasian CEU individuals of the HapMap collection, who were either homozygous for the major (rs3846662/AA) or minor allele (rs3846662/GG). Likewise, these individuals were also homozygous for the three proxy SNPs with genome-wide significance in the association meta-analysis (rs7703051, rs12654264 and rs3846663), where these SNPs denote a haplotype. LCLs (n=9 per genotype) were seeded in medium supplemented with 10% fetal calf serum and mRNA was harvested at various time points (0h, 10h, 24h and 48h) for expression analysis. We first analyzed if total HMGCR mRNA expression differed between both groups. As shown in Fig.2A, we did not detect significant differences in total HMGCR mRNA levels between the two groups at any time point, indicating that total HMGCR mRNA expression is not influenced by allele status at these SNPs.
We went on to determine the amounts of full-length and alternatively spliced HMGCR (Δexon13) mRNA separately. Δexon13 HMGCR mRNA was detectable in all samples and showed significant variation along the time course (Fig.2B). We observed a distinct decrease in the percentage of Δexon13 HMGCR mRNA in both genotype groups over the first 10h. However, the decrease in percentage of Δexon13 HMGCR mRNA was significantly less pronounced in LCLs from homozygotes for the rs3846662 major allele. Hence, the percentage of Δexon13 HMGCR mRNA per total HMGCR mRNA was significantly higher in homozygotes for the major allele as compared to homozygotes for the minor allele at 10h, 24h and 48h (10h: 23.0±7.6 vs. 10.4±3.4, 24h: 23.1±7.9 vs. 11.6±3.8, 48h: 27.1±9.4 vs. 15.7±9.1, % Δexon13/total HMGCR mRNA, major vs. minor allele, Fig.2B). In contrast to the other timepoints, the difference in Δexon13 mRNA expression at 0h did not reach significance, possibly attributable to regulatory mechanisms that first need to be initiated during this stage of acute adaption.
Similar results were obtained when we studied expression patterns in LCLs which were incubated in medium supplemented with 10% lipoprotein deficient serum (supplemental Fig.III).
Expression pattern of full-length and Δexon13 HMGCR mRNA in human tissues
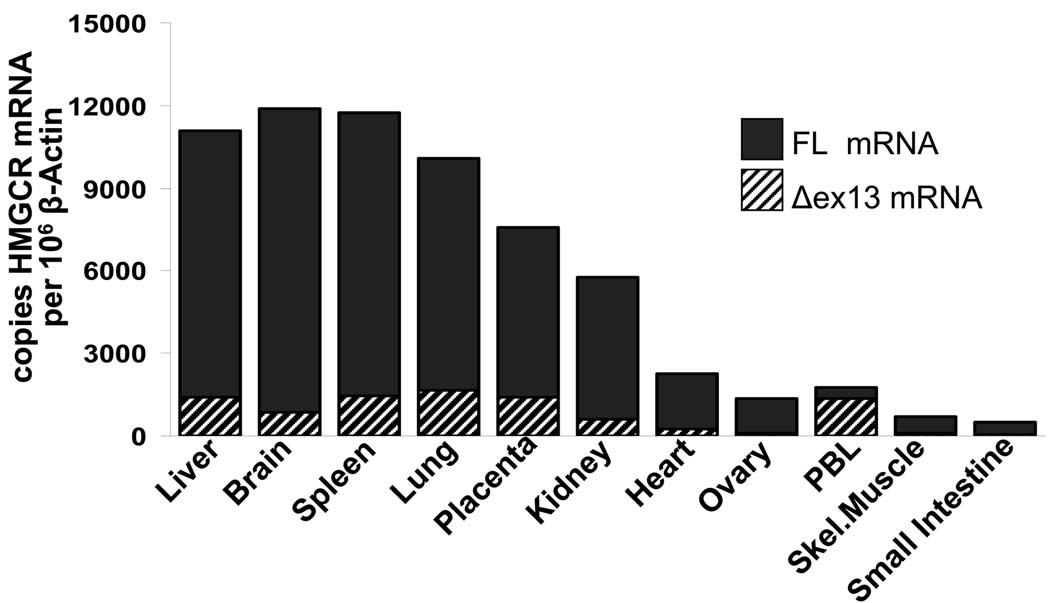
We next quantified the expression levels of both HMGCR transcripts in vivo, using cDNA samples from various human tissues. Both variants were expressed in cDNA pools from human liver, brain, spleen, lung, placenta, kidney, heart, ovary, peripheral blood leukocytes, skeletal muscle and small intestine, but their relative amounts differed significantly. The percentage of Δexon13 HMGCR mRNA per total HMGCR mRNA varied between 7 and 18%, with the exception of peripheral blood leukocytes. In peripheral blood leukocytes, Δexon13 HMGCR mRNA accounted for 79% of total HMGCR mRNA transcripts (Fig.3).
Figure 3.
Absolute amounts of full-length and Δexon13 HMGCR mRNA expression in cDNA pools of various human tissues (Clontech). HMGCR transcripts were normalized to β-actin as housekeeping gene.
SNP rs3846662 modulates alternative splicing of exon13 in HMGCR minigene constructs
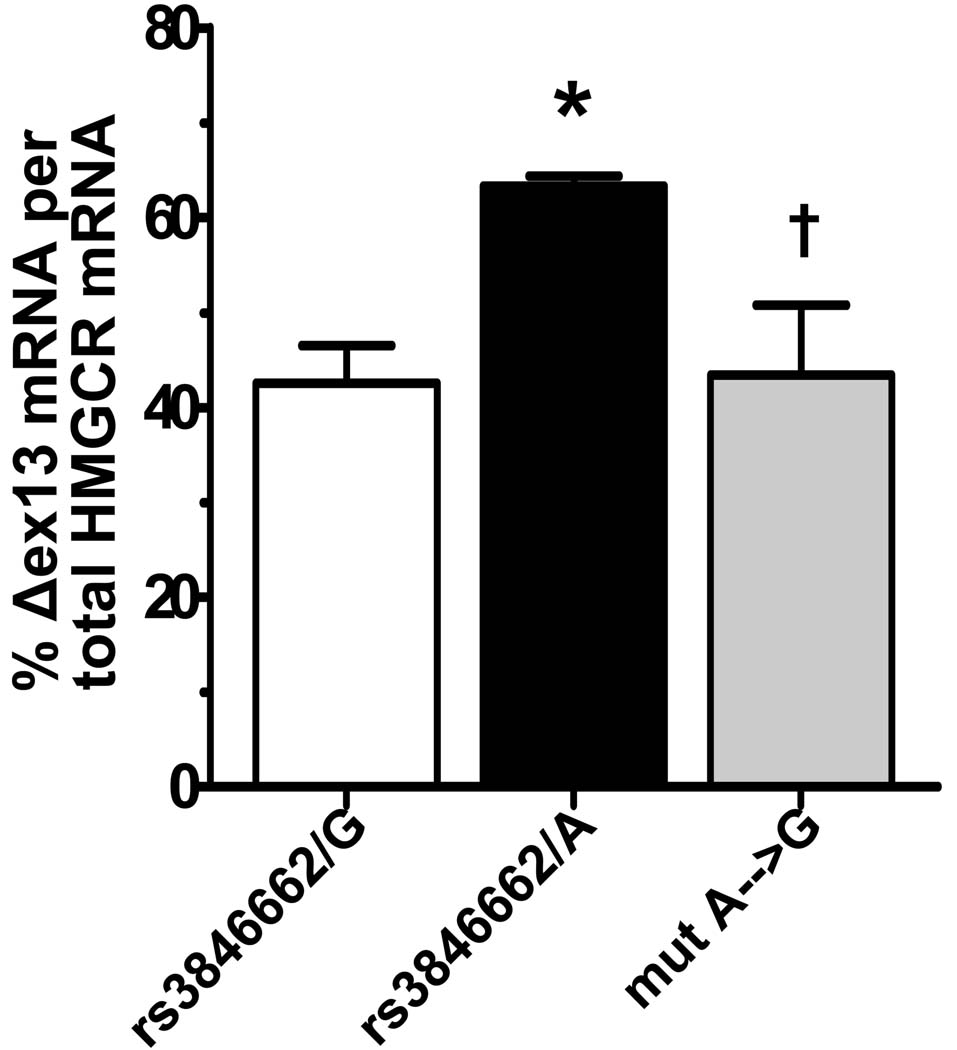
Our studies in LCLs demonstrated that the amount of Δexon13 HMGCR mRNA was associated with allele status at SNP rs3846662. Therefore, we aimed to specifically evaluate the functionality of SNP rs3846662 in splicing efficiency. We created exon-trapping vectors containing the genomic DNA sequence of HMGCR from intron12 to intron14 of rs3846662/AA (major allele) and rs3846662/GG (minor allele) individuals, respectively and transfected them into HEK293 cells.
In accordance with our previous results in human LCLs, we found significantly lower levels for Δexon13 HMGCR mRNA in cells transfected with the minor allele minigene (rs3846662/G) as compared to cells transfected with the major allele minigene (rs3846662/A) (Fig.4). The difference in exon13 splicing efficiency between the two minigenes was 20.8% (42.9±3.9 vs. 63.7±1.0 %Δexon13 HMGCR mRNA/total HMGCR mRNA, p=0.02). This difference in splicing efficiency was abolished when we transfected a construct in which we had used site directed mutagenesis to convert rs3846662/A to the minor G allele (Fig.4), further corroborating that allelic variants at rs3846662 directly modulate the efficiency of HMGCR exon13 splicing.
Figure 4.
HMGCR minigene splicing in transfected HEK cells. Alternatively spliced RNA (Δex13) from minigene constructs is expressed as percentage of total HMGCR minigene RNA. rs3846662/G: minor allele minigene, rs3846662/A: major allele minigene, mut A→G: major allele minigene after site directed mutagenesis, *p<0.05 rs3846662/G vs rs3846662/A; †p<0.05 rs3846662/A vs mut A→G
Full-length and Δexon13 HMGCR expression in stable transfected UT-2 cells
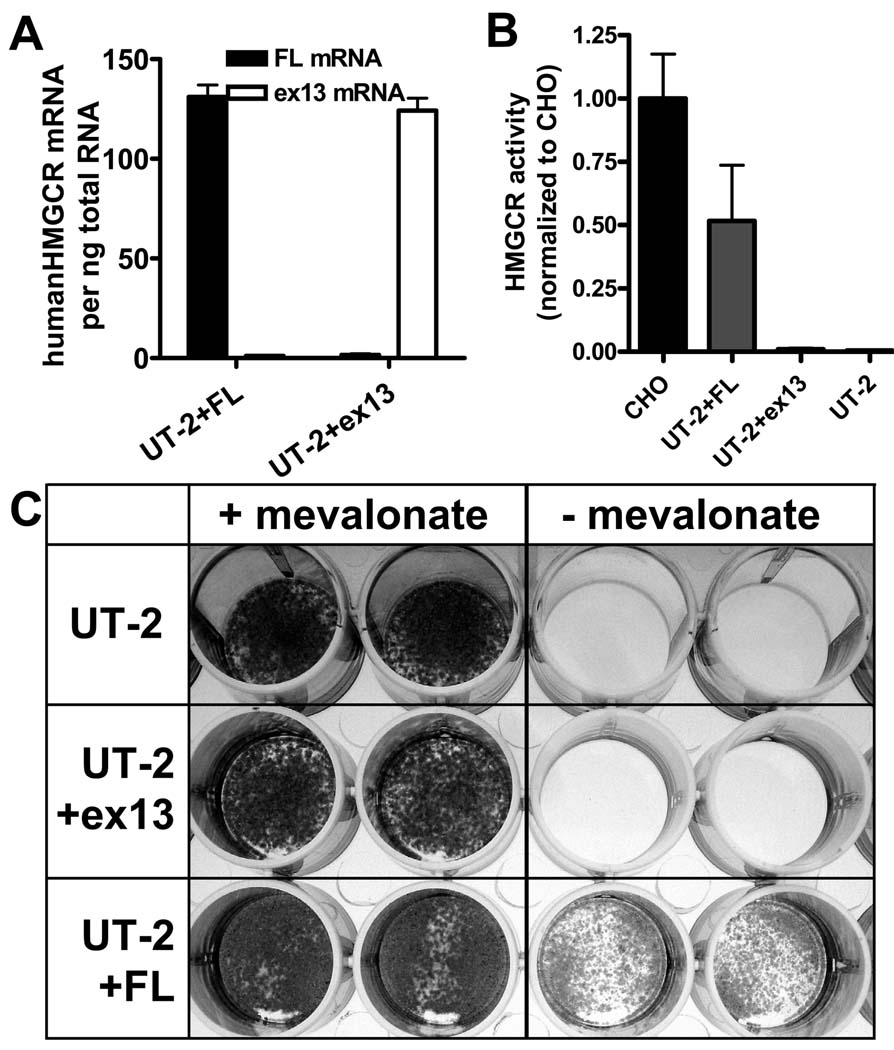
Alternative splicing of HMGCR mRNA leads to an in-frame deletion of 53 amino acids in the catalytic domain of the protein. To investigate the effect of this deletion on enzyme activity, we stably expressed human full-length (UT-2+FL) and Δexon13 (UT-2+ex13) HMGCR variants at comparable levels in UT-2 cells (Fig.5A), a CHO cell-line that lacks HMGCR activity and requires exogenous mevalonate for growth 16. UT-2+FL cells displayed 51% HMGCR enzyme activity of wild-type CHO cells, whereas UT-2+ex13 cells lacked enzyme activity and were indistinguishable from control UT-2 cells (Fig.5B). Further, UT-2+FL cells grew in the absence of mevalonate, whereas UT-2+ex13 and parental UT-2 cells died without mevalonate supplementation (Fig.5C), suggesting that the Δexon13 HMGCR variant is unable to restore enzyme activity in these cells.
Figure 5.
Characterization of UT-2 cells with stable expression of human full-length (UT-2+FL) and Δexon13 (UT-2+ex13) HMGCR: (A) HMGCR mRNA expression; (B) HMGCR activity normalized to wild-type CHO cells; (C) Growth characteristics of UT-2, UT-2+ex13 and UT-2+FL cells in the presence and absence of 0.2 mM mevalonate
Discussion
We identified variants in the HMGCR gene that were among our top hits for LDL-C in a GWAS in a population from the Island of Kosrae. We then conducted in vitro studies to follow-up the association signals and to identify a functional variant at the HMGCR locus. We present evidence that a common intronic SNP (rs384662) that is in linkage disequilibrium with the variants typed in the genome scan modulates alternative splicing of HMGCR mRNA. The resulting splice variant could not restore enzyme activity when expressed in HMGCR deficient UT-2 cells.
HMG-CoA reductase is a key enzyme in cholesterol homeostasis and catalyzes the rate limiting step in cholesterol biosynthesis 21. In contrast to other well known determinants of cholesterol homeostasis, e.g. LDL-receptor or Apolipoprotein E, associations between variants in HMGCR and LDL-C have only recently emerged in the context of GWAS. As in the Kosrae study, the initial results of the DGI GWAS in 2758 Caucasians supported associations between SNPs in HMGCR and LDL-C, but did not meet the statistical threshold of genome-wide significance by themselves (best associated SNP rs12654264: p=4.09×10−4) 13. In this study, genome-wide significance was clearly established for HMGCR SNP rs12654264 after validation in three additional Caucasian cohorts, resulting in a combined p-value of 1×10−20 in a total of ~18000 subjects 13. However in a separate study, the associations between SNPs in HMGCR and LDL-C that were observed in the DGI study were not strengthened by a meta analysis approach, consisting of the DGI and two other Caucasian GWAS (best associated SNP rs3846663: p=2.79×10−4 ) 12. This discrepancy might be attributable to some source of heterogeneity, e.g. differences in sample ascertainment or the impact of non-additive interactions with other genetic variants or unaccounted environmental exposures 22, 23. Combining the association results from the Kosrae and DGI studies revealed three variants in LD (r2>0.81) with genome-wide significance at the HMGCR locus, including the two SNPs mentioned above (rs12654264, rs3846663) and SNP rs7703051. Our data obtained in the Kosrae isolate thereby adds important evidence about the generalizibility of genetic associations at the HMGCR locus, demonstrating that these associations also extend to other ancestries. Interestingly, two pharmacogenetic studies investigating if genetic variants in HMGCR influence response to statin therapy demonstrated that common SNP haplotypes in HMGCR contribute to variation in statin response 24, 25. These haplotypes included the SNPs that were associated with plasma LDL-C in the Kosrae and DGI studies and it is possible that the same underlying mechanisms contribute to variation in LDL-C levels and variation in statin response.
A major aspect of our study was to follow-up the findings from the GWAS and to identify the putative functional variant at the HMGCR locus. To address this question we used human lymphoblastoid cells from the HapMap CEU collection which have previously been established as a suitable model to study the regulation of cholesterol biosynthesis in normal subjects and subjects with genetic abnormalities in lipid metabolism 26. Our efforts were facilitated by a near complete inventory (99%) of all common (>5% minor allele frequency) regional sequence variations, resulting from resequencing of the complete HMGCR locus in 23 Caucasians 19. Since the only known common coding SNP in HMGCR (rs5908, I638V) is not in LD with any of the genotyped SNPs, we consider it to be unlikely that this variant is responsible for the association signal. Likewise, since we did not detect significant differences in total HMGCR mRNA expression, we consider it to be unlikely that the causal SNP is located in a regulatory element affecting HMGCR transcription. On the other hand, we provide mutually supportive evidence that a common intronic variant (rs3846662) in LD with the genotyped variants is functional and alters the efficiency of HMGCR exon13 alternative splicing: We could demonstrate that (1.) expression levels of alternatively spliced Δexon13 HMGCR mRNA were significantly lower in lymphoblastoid cells from homozygotes for the rs3846662 minor allele and (2.) allele status at rs3846662 directly modulated alternative splicing of HMGCR mRNA in minigene constructs. Further, alternative splicing of HMGCR appeared to be regulated and was present in vivo, as we could detect Δexon13 HMGCR mRNA in all eleven human tissues that we studied.
HMGCR mRNA lacking exon13 was described in a survey of alternative pre-mRNA splicing by Johnson et al 20, however its function and the underlying mechanisms remain unknown. The regulation of gene splicing in mammalians involves both cis- and trans-factors, which are composed of auxiliary element sequences in the pre-mRNA, known as splicing enhancers and silencers 27 and cellular splicing factors which include several protein families 28. The most likely explanation for the observed differences in HMGCR mRNA splicing between major and minor allele homozygotes at rs3846662 is that this SNP is located in a binding motif for a splice auxiliary protein and allele status changes the binding affinity of this protein. Homozygosity for the major allele at rs3846662 increased the proportion of HMGCR mRNA lacking exon13. Skipping of exon 13 (159 bp) does not change the reading frame and the resulting protein lacks 53 amino acids in the catalytic domain. When we stably expressed both HMGCR variants in CHO cells deficient of endogenous HMGCR activity, the Δexon13 variant appeared to be non-functional and was not able to restore cell growth in the absence of mevalonate. At present, we can only speculate about the exact underlying mechanisms of this observation. Exon13 encodes parts of the catalytic domain and it contains the highly conserved sequence element ENVIGX3I/LP which is thought to mediate dimerization of the enzyme’s monomers 29. Thus, deletion of exon13 could potentially impact the stability of the enzyme, since experiments in which monomeric soluble proteins were fused to the HMGCR membrane domains illustrated that the protein was degraded faster when it was smaller than tetrameric 30. In addition, exon13 contains the E559 residue which is located at the front of the active site and was proposed to directly participate in the reduction of HMG-CoA by serving as a proton donor to mevaldehyde 29. Therefore, alternative splicing of HMGCR appears to result in altered enzymatic activity and could also lead to more rapid degradation of the protein. A decrease in HMGCR activity would lead to lower cellular cholesterol synthesis and subsequently a counter-regulatory increase of cholesterol uptake from the plasma via the LDL-receptor pathway to maintain intracellular cholesterol homeostasis. In accordance with this hypothesis the allele at rs3846662 that was causing higher levels of Δexon13 HMGCR mRNA in our in vitro studies was sharing a haplotype with the alleles that were associated with lower LDL-C in the genome-wide association studies. HMG-CoA reductase activity is subject to multivalent control on transcriptional and post-transcriptional levels and alternative splicing may be an additional regulatory mechanism.
Modulation of alternatively spliced HMGCR mRNA levels could be of pharmacologic interest with regard to response to statin therapy or as target for antisense-mediated exon skipping. Recently, an antisense oligonucleotide (AON)-mediated skipping approach related to lowering plasma cholesterol levels was applied by Khoo et al 31. In their study, AON-mediated exon27 skipping of the Apolipoprotein B transcript specifically lowered the amount of functional ApoB100 protein, while maintaining ApoB48 levels 31.
Therefore, identification of specific factors that regulate HMGCR alternative splicing and elucidating the underlying mechanism may lead to a better understanding of its impact on regulating cellular cholesterol homeostasis and plasma cholesterol levels.
Supplementary Material
Acknowledgements
Ralph Burkhardt is a fellow of the Deutsche Forschungsgemeinschaft (DFG Bu2263/1-1)
Footnotes
Disclaimer: The manuscript and its contents are confidential, intended for journal review purposes only, and not to be further disclosed.
Author Disclosures
Ralph Burkhardt: No disclosures
Eimear E Kenny: No disclosures
Jennifer K Lowe: No disclosures
Andrew Birkeland: No disclosures
Rebecca Josowitz: No disclosures
Martha Noel: No disclosures
Jacqueline Salit: No disclosures
Julian B Maller: No disclosures
Itsik Pe'er: No disclosures
Mark J Daly: No disclosures
David Altshuler: No disclosures
Markus Stoffel: No disclosures
Jeffrey M. Friedman: No disclosures
Jan L. Breslow: No disclosures
Addendum:
While this manuscript was under review another study reported that alternative splicing of HMG-CoA Reductase exon13 is associated with plasma LDL-C response to simvastatin (Medina MW et al, Circulation. 2008 Jul 22;118(4):355-62), further supporting the functional significance of HMGCR alternative splicing.
References
- 1.Kuulasmaa K, Tunstall-Pedoe H, Dobson A, Fortmann S, Sans S, Tolonen H, Evans A, Ferrario M, Tuomilehto J. Estimation of contribution of changes in classic risk factors to trends in coronary-event rates across the WHO MONICA Project populations. Lancet. 2000;355:675–687. doi: 10.1016/s0140-6736(99)11180-2. [DOI] [PubMed] [Google Scholar]
- 2.Heller DA, de Faire U, Pedersen NL, Dahlen G, McClearn GE. Genetic and environmental influences on serum lipid levels in twins. N Engl J Med. 1993;328:1150–1156. doi: 10.1056/NEJM199304223281603. [DOI] [PubMed] [Google Scholar]
- 3.Breslow JL. Genetics of lipoprotein abnormalities associated with coronary artery disease susceptibility. Annu Rev Genet. 2000;34:233–254. doi: 10.1146/annurev.genet.34.1.233. [DOI] [PubMed] [Google Scholar]
- 4.Rader DJ, Cohen J, Hobbs HH. Monogenic hypercholesterolemia: new insights in pathogenesis and treatment. J Clin Invest. 2003;111:1795–1803. doi: 10.1172/JCI18925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han Z, Heath SC, Shmulewitz D, Li W, Auerbach SB, Blundell ML, Lehner T, Ott J, Stoffel M, Friedman JM, Breslow JL. Candidate genes involved in cardiovascular risk factors by a family-based association study on the island of Kosrae, Federated States of Micronesia. Am J Med Genet. 2002;110:234–242. doi: 10.1002/ajmg.10445. [DOI] [PubMed] [Google Scholar]
- 6.Shmulewitz D, Heath SC, Blundell ML, Han Z, Sharma R, Salit J, Auerbach SB, Signorini S, Breslow JL, Stoffel M, Friedman JM. Linkage analysis of quantitative traits for obesity, diabetes, hypertension, and dyslipidemia on the island of Kosrae, Federated States of Micronesia. Proc Natl Acad Sci U S A. 2006;103:3502–3509. doi: 10.1073/pnas.0510156103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shmulewitz D, Auerbach SB, Lehner T, Blundell ML, Winick JD, Youngman LD, Skilling V, Heath SC, Ott J, Stoffel M, Breslow JL, Friedman JM. Epidemiology and factor analysis of obesity, type II diabetes, hypertension, and dyslipidemia (syndrome X) on the Island of Kosrae, Federated States of Micronesia. Hum Hered. 2001;51:8–19. doi: 10.1159/000022953. [DOI] [PubMed] [Google Scholar]
- 8.Kruglyak L. The road to genome-wide association studies. Nat Rev Genet. 2008;9:314–318. doi: 10.1038/nrg2316. [DOI] [PubMed] [Google Scholar]
- 9.Breslow JL, McPherson J, Nussbaum AL, Williams HW, Lofquist-Kahl F, Karathanasis SK, Zannis VI. Identification and DNA sequence of a human apolipoprotein E cDNA clone. J Biol Chem. 1982;257:14639–14641. [PubMed] [Google Scholar]
- 10.Wallace C, Newhouse SJ, Braund P, Zhang F, Tobin M, Falchi M, Ahmadi K, Dobson RJ, Marcano AC, Hajat C, Burton P, Deloukas P, Brown M, Connell JM, Dominiczak A, Lathrop GM, Webster J, Farrall M, Spector T, Samani NJ, Caulfield MJ, Munroe PB. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am J Hum Genet. 2008;82:139–149. doi: 10.1016/j.ajhg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandhu MS, Waterworth DM, Debenham SL, Wheeler E, Papadakis K, Zhao JH, Song K, Yuan X, Johnson T, Ashford S, Inouye M, Luben R, Sims M, Hadley D, McArdle W, Barter P, Kesaniemi YA, Mahley RW, McPherson R, Grundy SM, Bingham SA, Khaw KT, Loos RJ, Waeber G, Barroso I, Strachan DP, Deloukas P, Vollenweider P, Wareham NJ, Mooser V. LDL-cholesterol concentrations: a genome-wide association study. Lancet. 2008;371:483–491. doi: 10.1016/S0140-6736(08)60208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, Strait J, Duren WL, Maschio A, Busonero F, Mulas A, Albai G, Swift AJ, Morken MA, Narisu N, Bennett D, Parish S, Shen H, Galan P, Meneton P, Hercberg S, Zelenika D, Chen WM, Li Y, Scott LJ, Scheet PA, Sundvall J, Watanabe RM, Nagaraja R, Ebrahim S, Lawlor DA, Ben-Shlomo Y, Davey-Smith G, Shuldiner AR, Collins R, Bergman RN, Uda M, Tuomilehto J, Cao A, Collins FS, Lakatta E, Lathrop GM, Boehnke M, Schlessinger D, Mohlke KL, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, Wahlstrand B, Hedner T, Corella D, Tai ES, Ordovas JM, Berglund G, Vartiainen E, Jousilahti P, Hedblad B, Taskinen MR, Newton-Cheh C, Salomaa V, Peltonen L, Groop L, Altshuler DM, Orho-Melander M. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, Daly MJ, Hughes TE, Groop L, Altshuler D, Almgren P, Florez JC, Meyer J, Ardlie K, Bengtsson Bostrom K, Isomaa B, Lettre G, Lindblad U, Lyon HN, Melander O, Newton-Cheh C, Nilsson P, Orho-Melander M, Rastam L, Speliotes EK, Taskinen MR, Tuomi T, Guiducci C, Berglund A, Carlson J, Gianniny L, Hackett R, Hall L, Holmkvist J, Laurila E, Sjogren M, Sterner M, Surti A, Svensson M, Tewhey R, Blumenstiel B, Parkin M, Defelice M, Barry R, Brodeur W, Camarata J, Chia N, Fava M, Gibbons J, Handsaker B, Healy C, Nguyen K, Gates C, Sougnez C, Gage D, Nizzari M, Gabriel SB, Chirn GW, Ma Q, Parikh H, Richardson D, Ricke D, Purcell S. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 15.Fisher RA. Statistical Methods for Research Workers. London: Oliver and Boyd; 1932. [Google Scholar]
- 16.Mosley ST, Brown MS, Anderson RG, Goldstein JL. Mutant clone of Chinese hamster ovary cells lacking 3-hydroxy-3 -methylglutaryl coenzyme A reductase. J Biol Chem. 1983;258:13875–13881. [PubMed] [Google Scholar]
- 17.Walli AK, Seidel D. Role of lipoprotein-X in the pathogenesis of cholestatic hypercholesterolemia. Uptake of lipoprotein-X and its effect on 3-hydroxy-3-methylglutaryl coenzyme A reductase and chylomicron remnant removal in human fibroblasts, lymphocytes, and in the rat. J Clin Invest. 1984;74:867–879. doi: 10.1172/JCI111504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edwards PA, Lemongello D, Fogelman AM. Improved methods for the solubilization and assay of hepatic 3-hydroxy-3-methylglutaryl coenzyme A reductase. J Lipid Res. 1979;20:40–46. [PubMed] [Google Scholar]
- 19.Crawford DC, Carlson CS, Rieder MJ, Carrington DP, Yi Q, Smith JD, Eberle MA, Kruglyak L, Nickerson DA. Haplotype diversity across 100 candidate genes for inflammation, lipid metabolism, and blood pressure regulation in two populations. Am J Hum Genet. 2004;74:610–622. doi: 10.1086/382227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 21.Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 22.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 23.Ioannidis JP, Patsopoulos NA, Evangelou E. Heterogeneity in meta-analyses of genome-wide association investigations. PLoS ONE. 2007;2:e841. doi: 10.1371/journal.pone.0000841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chasman DI, Posada D, Subrahmanyan L, Cook NR, Stanton VP, Jr, Ridker PM. Pharmacogenetic study of statin therapy and cholesterol reduction. Jama. 2004;291:2821–2827. doi: 10.1001/jama.291.23.2821. [DOI] [PubMed] [Google Scholar]
- 25.Krauss RM, Mangravite LM, Smith JD, Medina MW, Wang D, Guo X, Rieder MJ, Simon JA, Hulley SB, Waters D, Saad M, Williams PT, Taylor KD, Yang H, Nickerson DA, Rotter JI. Variation in the 3-hydroxyl-3-methylglutaryl coenzyme a reductase gene is associated with racial differences in low-density lipoprotein cholesterol response to simvastatin treatment. Circulation. 2008;117:1537–1544. doi: 10.1161/CIRCULATIONAHA.107.708388. [DOI] [PubMed] [Google Scholar]
- 26.Kayden HJ, Hatam L, Beratis NG. Regulation of 3-hydroxy-3-methylglutaryl coenzyme A reductase activity and the esterification of cholesterol in human long term lymphoid cell lines. Biochemistry. 1976;15:521–528. doi: 10.1021/bi00648a011. [DOI] [PubMed] [Google Scholar]
- 27.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 28.Caceres JF, Kornblihtt AR. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 2002;18:186–193. doi: 10.1016/s0168-9525(01)02626-9. [DOI] [PubMed] [Google Scholar]
- 29.Istvan ES, Palnitkar M, Buchanan SK, Deisenhofer J. Crystal structure of the catalytic portion of human HMG-CoA reductase: insights into regulation of activity and catalysis. Embo J. 2000;19:819–830. doi: 10.1093/emboj/19.5.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng HH, Xu L, Kumagai H, Simoni RD. Oligomerization state influences the degradation rate of 3-hydroxy-3-methylglutaryl-CoA reductase. J Biol Chem. 1999;274:17171–17178. doi: 10.1074/jbc.274.24.17171. [DOI] [PubMed] [Google Scholar]
- 31.Khoo B, Roca X, Chew SL, Krainer AR. Antisense oligonucleotide-induced alternative splicing of the APOB mRNA generates a novel isoform of APOB. BMC Mol Biol. 2007;8:3. doi: 10.1186/1471-2199-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.