Summary
An intriguing feature of centrioles is that these highly complicated microtubule-based structures duplicate once per cell cycle and the cell has precise control over their number. Each cell contains exactly two centrioles, linked together as a pair, one of which is a mother centriole formed in a previous cell cycle and the other a daughter centriole whose assembly is templated by the mother. Neither the molecular basis nor the functional role of mother-daughter centriole linkage is understood. We have identified a mutant, asq2, with defects in centriole linkage. asq2 mutant cells have variable numbers of centrioles and defects in centriole positioning. Here, we show that ASQ2 encodes the novel conserved protein, TBCCd1, a member of a protein family that includes a tubulin folding co-chaperone and the retinitis pigmentosa protein, RP2, involved in tubulin quality control during ciliogenesis. We characterize mitosis in asq2 cells. We show that the majority of cells establish a bipolar spindle, but that cells have defects in spindle orientation. A small subset of asq2 cells have centrioles at both poles, and these cells have properly positioned spindles, indicating that centrioles at the poles may be important for spindle orientation. The defects in centriole number control, centriole positioning, and spindle orientation appear to arise from a primary defect in centriole linkage mediated by TBCCd1/ASQ2.
Results and Discussion
asq2 cells have defects in centriole linkage
Centrioles exist in pairs with one older mother centriole and one less mature daughter. This pair is held together by cohesion proteins, several of which have been identified [1-4]. Centrin localizes to the connecting fibers [5, 6] and C-NAP1 and rootletin have been functionally implicated in mother-daughter centriole cohesion [2, 4]. Each centriole will mature and serve as a mother to give rise to a new daughter each cell cycle through a template-driven synthesis process. Centrioles can form de novo [7, 8], but this pathway is apparently suppressed by the template-driven duplication pathway. We previously identified a mutant in the green alga Chlamydomonas reinhardtii [9], asq2, with defects in centriole linkage (Figure 1 B-D) that causes errors in both centriole number control and positioning. This phenotype resembles a class of mutants with variable numbers of centrioles and flagella (vfl) [10-14], but asq2 is not allelic to these mutations (data not shown). Despite defects in centriole linkage, asq2 centrioles appear to have normal molecular composition, recruiting acetylated α-tubulin, centrin, Bld10p (Figure 1B-D), and Vfl1p (Figure 1E-H), consistent with previous ultrastructural analysis [9].
Figure 1. ASQ2 encodes the conserved protein TBCCd1.
(A-D) are stained with DAPI (blue) and antibodies against centrin and acetylated α-tubulin (green) and Bld10p (red). A) Wild-type cells have two centrioles that each makes a flagellum at the apical end of the cell. Each centriole recruits centrin and acetylated α-tubulin (green) and Bld10p (red). (B-D) asq2 mutants make variable numbers of flagella due to their variable numbers of centrioles. B) asq2 cell with one centriole nucleating one flagellum. C) Biflagellate asq2 cell with two unpaired centrioles at a distance from one another. D) Triflagellate asq2 cell with a pair of centrioles at the apical surface of the cell and one lone centriole off to the side. E-H) VFL1-HA expressing cells are labeled with antibodies against HA (red), acetylated α-tubulin (green), and DAPI (blue). (E’-H’) show only HA localization. E) Wild-type cells expressing HA-tagged Vfl1p have an existing centriolar pair that localizes acetylated α-tubulin (green) and two new pro-centrioles forming nearby that localize Vfl1p (red). F) asq2 cells expressing HA-tagged Vfl1p have newly forming pro-centrioles forming near (red, arrow) and at a distance (G, arrow) from older centrioles (green and red staining). H) Newly forming centrioles can also appear in the absence of existing centrioles (arrow). I) asq2 maps to a <.3 cM region on linkage group IX (grey box defines the genetic interval that is constrained by recombination events). The hygromycin cassette (HYG) inserted on scaffold 22, while the asq2 mutation maps to scaffold 23. Both scaffolds are part of the same chromosome (Linkage Group IX). Within the genetic interval containing asq2 is the previously unannotated gene TBCCd1, a conserved protein named for its TBCC domain. J) PCR across the exon-intron boundary of exon 8 (I, arrow) demonstrates that asq2 cells have an insertion in genomic TBCCd1 and in the cDNA (K) between exon 8 and 9. L) TBCCd1 is contained within a family of proteins that is divided into three clades as previously described [18], canonical TBCC (blue box), retinitis pigmentosa 2 (RP2, green box), and TBCCd1 (yellow box). TBCC domains from mutual best-hit proteins were aligned and organized into a phylogenetic tree using Clustalw. (CR, Chlamydomonas reinhardtii, VC, Volvox carteri, HS, Homo sapiens, TB, Trypanosoma brucei, and AT, Arabidopsis thaliana).
asq2 maps to linkage group IX
The asq2 mutation was generated in an insertional screen [9] using an aphVII hygromycin resistance cassette [15]. Using a degenerate PCR strategy [16], we found this cassette inserted at position 1487009 on scaffold 22 of the Chlamydomonas genome [17] between two predicted genes (version 3.0, gene models 79905 and 191060) on linkage group IX (Supplemental Figure S1). The aphVII insertion was linked to the asq2 mutation at a distance of approximately 16.8 cM (96/572 recombinants).
asq2 cells were outcrossed to a polymorphic strain (cc-1952) and the mutation mapped to a <.3 cM region on scaffold 23 of linkage group IX between markers near gene models 148927 and 174289 (Figure 1I). Because this genomic interval contains gaps of poor sequence data, we examined a syntenic region in the closely-related green alga Volvox carteri (http://genome.jgi-psf.org/Volca1) and found a conserved gene, TBCCd1 (Figure 1I, Volvox gene model 121354). A polymorphism near the corresponding Chlamydomonas locus was tightly linked to the asq2 mutation (0/750 recombinants).
ASQ2 encodes the conserved protein TBCCd1
To determine if asq2 cells have a mutation in the previously unannotated homologue of TBCCd1, we identified a 2043 base pair (bp) open reading frame in Chlamydomonas (Figure 1I, Genbank accession number EU816954). We identified a 513 bp insertion in asq2 genomic DNA (Figure 1J), which lies in a splice junction between exons 8 and 9 of the predicted cDNA (Figure 1I, arrow). By sequencing RT-PCR products, we found that the asq2 mutant cDNA has a 168 bp insertion, but other splice products are visible (Figure 1K).
We transformed mutant cells with wild-type Chlamydomonas TBCCd1 (Supplemental Figure S2A), and found that transformed cells are morphologically normal (Supplemental Figure S2B), with wild-type centriole positioning as measured by our previously described [9] metric (Supplemental Figure S2D, mean θcentriole =21.0 ± 9.6°, n= 60, compare Supplemental Figure S2D-F), wild-type spindle orientation θspindle (Supplemental Figure S2I, mean = 94.2 ± 16.0 (n=23)), and wild-type flagellar number distribution (Supplemental Figure S2J).
ASQ2 encodes TBCCd1, a 681 amino acid protein. This protein has a predicted TBCC domain, named for the tubulin binding cofactor C protein. The insertion in asq2 cells is in the TBCC domain, presumably perturbing its function.
Three protein families with TBCC domains have been described [18]. The first is the canonical tubulin binding cofactor C (TBCC) that catalyzes tubulin folding [19]. Chlamydomonas has one mutual best hit (Figure 1L, blue box, gene model 147601). The second class of TBCC proteins contains homologues of the human gene RP2, mutated in patients with retinitis pigmentosa. The trypanosome homologue has been implicated in ciliary assembly [18]. Chlamydomonas has one mutual best-hit homologue of this gene (Figure 1L, green box, gene model 179569) and one paralogue (gene model 172780).
ASQ2 falls into the third class of TBCC proteins, named TBCCd1 for TBCC-domain containing, which have a TBCC domain, but which reportedly [18] lack a conserved catalytic arginine residue responsible for GAP activity [20, 21]. We have identified an arginine residue (Supplemental Figure S3) two positions away from the functional residue in TBCC and RP2 proteins that may suffice for GAP activity. ASQ2 encodes a mutual best hit for TBCCd1 (Figure 1L, yellow box, partial gene model 148930). TBCCd1 might help control centriole duplication through interactions with tubulin, or by regulating folding/assembly of atypical centriole-associated tubulins such as δ- and ε-tubulin [22-24].
ASQ2/TBCCd1 localizes to a region sub-proximal to the centrioles
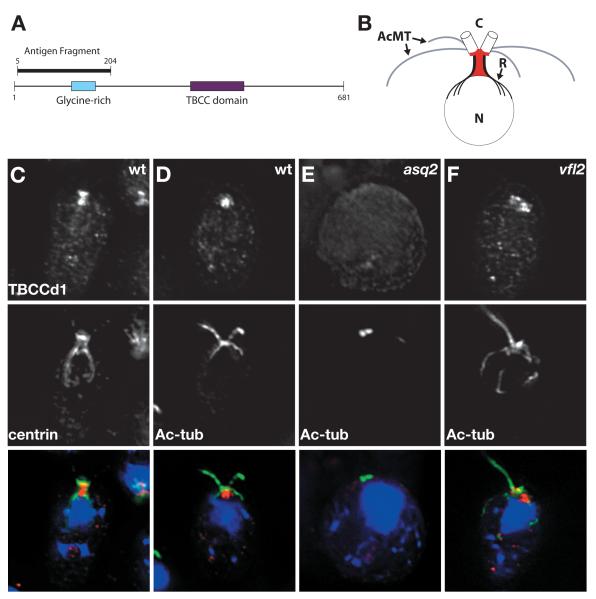
To localize ASQ2/TBCCd1, we generated an antibody to a 200 amino acid N-terminus fragment (Figure 2A). In wild-type cells, TBCCd1 localizes to a region sub-proximal to the centrioles between the centrioles and the nuclear envelope (Figure 2C and D). Some TBCCd1 was found on the centrioles themselves, but the majority localized to the region between the two nucleus-centriole connectors known as rhizoplasts (summarized in Figure 2B). To our knowledge, TBCCd1 is the first protein known to localize to this cellular region, previously described by electron microscopy as a ribosome-free region of cytoplasm [25]. TBCCd1 localization is abolished in the asq2 mutant (Figure 2E) and restored in the rescue line expressing wild-type TBCCd1 (data not shown). We also find that TBCCd1 is recruited near centrioles in vfl2 cells (Figure 2F), although this localization is sometimes mispositioned, potentially due to the lack of rhizoplasts in this mutant.
Figure 2. TBCCd1 localizes to a region sub-proximal to centrioles.
A) Cartoon of TBCCd1 protein. TBCCD1 is a 681 amino acid protein with a predicted glycine-rich domain (blue box) and a TBCC domain (purple box). An antibody was raised against an N-terminal 200 amino acid fragment of the protein. (B) Cartoon of TBCCd1 localization pattern. Acetylated microtubule bundles (AcMT) called rootlets are shown in grey. Rhizoplasts (R) that join the centrioles (C) to the nucleus (N) are shown in black. TBCCd1 (red) localizes to the region between the rhizoplasts and below the centrioles. Some of the protein can be seen at centrioles. (C-F) Cells are labeled with DAPI (blue) and antibodies against TBCCd1 (red) and centrin (C), which marks the centrioles and the rhizoplasts or acetylated α-tubulin (D-F, green), which marks the centrioles and rootlets. C and D) In wild-type cells, TBCCd1 localizes to the region between the rhizoplasts and to the centrioles. E) In asq2 cells, TBCCd1 localization at or near the centrioles is absent. F) In vfl2 cells, TBCCd1 localization is present near the centrioles but is sometimes mispositioned.
Centriole linkage is required for spindle orientation
We next examined spindle establishment in asq2 cells. In Chlamydomonas cells, each pole has a pair of centrioles, which is attached to a two-membered rootlet structure composed of acetylated α-tubulin (Figure 3A and Supplemental Figure S4) that forms a hook-like structure over the nucleus. In asq2 mutants, some centrioles are found at the poles (Supplemental Figure S4F & G), while others are unassociated with the spindle apparatus (Supplemental Figure S4F). Some asq2 cells form irregular spindles (35.5%, Supplemental Figure S4I-K) including monopolar (Supplemental Figure S4I), tripolar (Supplemental Figure S4J), or end-on-end spindles (Supplemental Figure S4K). Most asq2 cells (64.5%, Supplemental Figure S4F-H), form a bipolar spindle. Spindle irregularity does not correlate with the presence or absence of centrioles at the poles. Because most asq2 cells form bipolar spindles, but show defects in centriole position, we asked if asq2 cells would show spindle orientation defects.
Figure 3. Centriole linkage is necessary for proper spindle orientation.
A) Cartoon of a metaphase Chlamydomonas spindle. During mitosis in wild-type cells, a pair of centrioles (green) migrates to each pole to form a bipolar spindle (red). Rootlet microtubules (one two-membered pair shown in grey and one shown in black) are connected to each pair of centrioles through an interaction with one of the centrioles at each pole. B) Cartoon depiction of θspindle measurement. To measure the position of the spindle in Chlamydomonas cells, we use the pyrenoid (blue) center of mass (yellow) and the cell center of mass (purple) to define the central axis of the cell. We then pick a point at each end of the spindle (black) to define the spindle axis. θspindle is defined as the angle at which the spindle axis intersects the central cell axis. (C,F,I,L) DIC images and (D,G,J,M) immunofluorescence images labeled with antibodies against α-tubulin (red), acetylated α-tubulin (green), and DAPI (blue) of dividing Chlamydomonas cells were used to generate θspindle. E) Wild-type cells have a mean θspindle of 94.5 ± 17.2 (n=12) with a mean deviation of 14.9 ± 8.7°. H) asq2 cells have a mean θspindle of 78.9 ± 36.4 (n=36) and a mean deviation of 29.0 ± 24.2°. This distribution differs significantly from wild-type (F-test on the variance, p < 4.6e-4). K) vfl2 cells have a mean θspindle of 80.5 ± 41.3 (n=54) and a mean deviation of 33.4 ± 25.6°, which differs significantly from wild-type (F-test on the variance, p < 2.3e-4). N) vfl1 cells have a mean θspindle of 90.3 ± 38.2 (n=33) and a mean deviation of 32.7± 20.0°, which differs significantly from wild-type (F-test on the variance, p < 2.7e-3).
To measure spindle orientation, we used the pyrenoid center of mass and the cell center of mass to define a central cell axis (Figure 3B). We marked each end of the spindle the angle between the spindle axis and the central cell axis (θspindle, Figure 3B). For measurement of θspindle, we only considered bipolar spindles. In wild-type cells, the average θspindle is 94.6 ± 17.2° (Figure 3E), consistent with previous visual estimates [26, 27].
We cannot differentiate the two poles during our analysis of θspindle, hence for all statistical analysis, we measure the absolute value of the deviation from 90° of the angular measurement and will refer to this value as “deviation”. In asq2 cells, θspindle is 78.9 ± 36.4° with a mean deviation of 29.0 ± 24.2° (Figure 3H, n=36), which differs significantly from wild-type (mean deviation = 14.9 ± 8.7°, one-tailed t-test, p < 2.5e-3; F-test on the variance, p < 4.6e-4). This defect in spindle positioning is also observed in other vfl mutants with defective linkage (Figure 3I-K). vfl2 cells have a θspindle of 80.5 ± 41.3° with a mean deviation of 33.4 ± 25.6° (Figure 3K, n=54) and vfl1 cells have a mean θspindle of 90.3 ± 38.1° with a mean deviation of 32.7± 20.0° (Figure 3N, n=33). Spindle position in both mutants differs significantly from wild-type. These data suggest centriole linkage is required for proper spindle positioning.
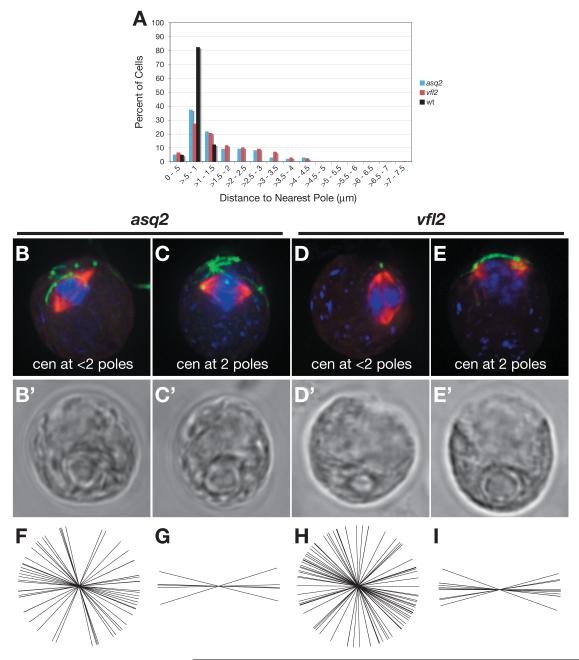
It is formally possible that the centrioles formed in asq2 mutant cells are functionally defective, perhaps as a result of formation by a de novo assembly pathway (see below). However, we noted that the majority of asq2 cells as well as vfl2 cells are missing centrioles from one or both poles (Figure 4B and D, respectively). These cells have severe defects in spindle positioning (asq2: Figure 4F, mean = 77.1 ± 33.4°, n=31, and vfl2: 4H, mean = 78.5 ± 45.3°, n=44), suggesting it could be the absence of centrioles at poles, rather than defects in the centrioles themselves, that leads to spindle mis-orientation.
Figure 4. Centrioles may be required at the poles for proper spindle orientation.
A) Distance from each centriole to the nearest pole was measured in fixed cells. Wild-type cells have a mean centriole-to-pole distance of .80 ± .19 μm (black, n= 40). asq2 cells have a mean distance of 1.51 ± .96 μm (blue, n= 96), which differs significantly from wild-type (one-tailed t-test: p < 1.9e-10). vfl2 cells have a mean distance of 1.68 ± 1.07 μm (red, n= 193), which differs significantly from wild-type (one-tailed t-test: p < 4.7e-22). (B-E) Immunofluorescence images of fixed cells labeled with DAPI (blue) and antibodies against acetylated α-tubulin (green), α-tubulin (red), and phosphorylated histone-H3 (CY5, not shown). (B’- E’) DIC images of cells shown in (B-E). The majority of asq2 cells (B) or vfl2 cells (D) do not have centrioles (green) at both poles (distance is greater than one standard deviation from wild-type mean). These cells have severe defects in spindle positioning (F, asq2 mean = 77.1 ± 33.4°, n=31, H, vfl2 mean = 78.5 ± 45.3°, n=44). A small subset of asq2 and vfl2 cells have centrioles at both poles (Figure 4C and E, respectively, distance is < 1 standard deviation from the wild-type mean distance). In these cases, the spindles are correctly positioned (asq2: Figure 4G, mean = 90.4 ± 11.2°, n=5, and vfl2: 4I, mean = 89.5 ± 9.7°, n=10).
In a small subset of asq2 and vfl2 cells, centrioles can be found at both poles (Figure 4C and E, respectively, distance is < 1 standard deviation from the wild-type mean distance). In these cases, the spindles are correctly positioned (asq2: Figure 4G, mean = 90.4 ± 11.2°, n=5, and vfl2: 4I, mean = 89.5 ± 9.7°, n=10), suggesting that centrioles are required at the pole to promote proper spindle positioning. bld2 cells which have truncated centrioles [28] that fail to attach to the cortex [9] also have a defect in spindle positioning [29] (Supplemental Figure S5), suggesting that full-length centrioles able to attach to the membrane are important for spindle orientation.
We therefore find a strong correlation between the presence of centrioles at the poles and proper spindle orientation. While we cannot determine why some centrioles are at the poles and some are not, we speculate that differences between mature and immature centrioles may play a role in their ability to interact with the spindle apparatus and/or proper cortex attachment site.
Procentriole position relative to mother centrioles in asq2 cells
We previously found that asq2 cells have defects in centriole positioning due to an inability of daughter centrioles to find the correct position on the cell surface [9]. Our previous studies were not able to tell whether the daughter centrioles lost their position during cell division, or were formed in an incorrect position in the first place. Using cells that express an HA-tagged copy of Vfl1p [11], we investigated the location of newly forming centrioles in asq2 cells. VFL1p is recruited early in centriole assembly and has been localized to nascent pro-centrioles [11]. Consistent with previous reports [11], we found that wild-type cells that have duplicated their centrioles have four Vfl1p spots (Figure 1E). Two of these spots represent the original mother-daughter pair and two represent the newly formed pro-centrioles. Only the more mature centrioles (the original mother-daughter pair) localize acetylated α-tubulin. The other two pro-centrioles only localize Vfl1p. We used this difference in staining to distinguish newly formed pro-centrioles from the older structures assembled in a previous cell cycle.
In asq2 cells, we find instances of pairs of centrioles (Figure 1F) as well as lone centrioles. In the case of paired centrioles, pro-centrioles may have formed by the template-driven assembly process (Figure 1F, arrow). Similarly, some of the lone centrioles also localize acetylated α-tubulin, suggesting that they may represent older centrioles that are not being duplicated or are late to mature. We frequently find lone centrioles that only localize Vfl1p (Figure 1G, arrow). These centrioles may represent newly born daughters that, untethered to their mothers, have wandered to uncharacteristic locations. Alternatively, these lone pro-centrioles may have assembled de novo. We also find these pro-centrioles in cells lacking mature centrioles. Since pre-existing centrioles are not known to degrade, and since pro-centriole maturation occurs within one cell cycle, the presence of pro-centrioles in cells lacking mature centrioles suggests they can arise de novo (Figure 1H, arrow), as in other vfl mutants [7].
In normal cells, de novo assembly does not occur when pre-existing centrioles are present [7, 8]. Because centrioles may assemble de novo in asq2 cells containing pre-existing centrioles (Figure 1F & G), we hypothesize TBCCd1 may be involved in suppressing the de novo synthesis pathway. This model is supported by a computational analysis (Supplemental Figure S6A) showing loss of templated duplication results in a centriole number distribution similar to that seen in asq2 and vfl mutants. Control of de novo synthesis by TBCC might be direct (Supplementary Figure S6B, 1) or indirect through the regulation of centriole linkage, which may itself suppress unlicensed de novo synthesis (Supplemental Figure S6B, 2).
Supplementary Material
Acknowledgements
The authors would like to thank L. Holt, K. Wemmer, W. Ludington, and J. Azimzadeh for critical review of the manuscript, J. Ochs for help with programming, J. Salisbury, P. Hegemann, M. Hirono, W. Mages, P. Manzanillo, J.Cox, B. Bradley, and L. Quarmby for reagents, and E. Harris and the Chlamydomonas Genetics Center for providing strains. J.L.F. was supported by an NSF Predoctoral Fellowship and UC Fletcher-Jones Fellowship. This work was supported by NIH grants R01 GM077004 and R03 HD051583, the W.M. Keck Foundation, and by the Searle Scholars Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Faragher AJ, Fry AM. Nek2A kinase stimulates centrosome disjunction and is required for formation of bipolar mitotic spindles. Mol Biol Cell. 2003;14:2876–2889. doi: 10.1091/mbc.E03-02-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mayor T, Stierhof YD, Tanaka K, Fry AM, Nigg EA. The centrosomal protein C-Nap1 is required for cell cycle-regulated centrosome cohesion. J Cell Biol. 2000;151:837–846. doi: 10.1083/jcb.151.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graser S, Stierhof YD, Nigg EA. Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. J Cell Sci. 2007;120:4321–4331. doi: 10.1242/jcs.020248. [DOI] [PubMed] [Google Scholar]
- 4.Bahe S, Stierhof YD, Wilkinson CJ, Leiss F, Nigg EA. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J Cell Biol. 2005;171:27–33. doi: 10.1083/jcb.200504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geimer S, Melkonian M. Centrin scaffold in Chlamydomonas reinhardtii revealed by immunoelectron microscopy. Eukaryot Cell. 2005;4:1253–1263. doi: 10.1128/EC.4.7.1253-1263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lechtreck KF, Grunow A. Evidence for a direct role of nascent basal bodies during spindle pole initiation in the green alga Spermatozopsis similis. Protist. 1999;150:163–181. doi: 10.1016/S1434-4610(99)70019-2. [DOI] [PubMed] [Google Scholar]
- 7.Marshall WF, Vucica Y, Rosenbaum JL. Kinetics and regulation of de novo centriole assembly. Implications for the mechanism of centriole duplication. Curr Biol. 2001;11:308–317. doi: 10.1016/s0960-9822(01)00094-x. [DOI] [PubMed] [Google Scholar]
- 8.La Terra S, English CN, Hergert P, McEwen BF, Sluder G, Khodjakov A. The de novo centriole assembly pathway in HeLa cells: cell cycle progression and centriole assembly/maturation. J Cell Biol. 2005;168:713–722. doi: 10.1083/jcb.200411126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feldman JL, Geimer S, Marshall WF. The mother centriole plays an instructive role in defining cell geometry. PLoS Biol. 2007;5:e149. doi: 10.1371/journal.pbio.0050149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adams GM, Wright RL, Jarvik JW. Defective temporal and spatial control of flagellar assembly in a mutant of Chlamydomonas reinhardtii with variable flagellar number. J Cell Biol. 1985;100:955–964. doi: 10.1083/jcb.100.3.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silflow CD, LaVoie M, Tam LW, Tousey S, Sanders M, Wu W, Borodovsky M, Lefebvre PA. The Vfl1 Protein in Chlamydomonas localizes in a rotationally asymmetric pattern at the distal ends of the basal bodies. J Cell Biol. 2001;153:63–74. doi: 10.1083/jcb.153.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taillon BE, Adler SA, Suhan JP, Jarvik JW. Mutational analysis of centrin: an EF-hand protein associated with three distinct contractile fibers in the basal body apparatus of Chlamydomonas. J Cell Biol. 1992;119:1613–1624. doi: 10.1083/jcb.119.6.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wright RL, Chojnacki B, Jarvik JW. Abnormal basal-body number, location, and orientation in a striated fiber-defective mutant of Chlamydomonas reinhardtii. J Cell Biol. 1983;96:1697–1707. doi: 10.1083/jcb.96.6.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright RL, Adler SA, Spanier JG, Jarvik JW. Nucleus-basal body connector in Chlamydomonas: evidence for a role in basal body segregation and against essential roles in mitosis or in determining cell polarity. Cell Motil Cytoskeleton. 1989;14:516–526. doi: 10.1002/cm.970140409. [DOI] [PubMed] [Google Scholar]
- 15.Berthold P, Schmitt R, Mages W. An engineered Streptomyces hygroscopicus aph 7′′ gene mediates dominant resistance against hygromycin B in Chlamydomonas reinhardtii. Protist. 2002;153:401–412. doi: 10.1078/14344610260450136. [DOI] [PubMed] [Google Scholar]
- 16.Gonzalez-Ballester D, de Montaigu A, Galvan A, Fernandez E. Restriction enzyme site-directed amplification PCR: a tool to identify regions flanking a marker DNA. Anal Biochem. 2005;340:330–335. doi: 10.1016/j.ab.2005.01.031. [DOI] [PubMed] [Google Scholar]
- 17.Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Marechal-Drouard L, Marshall WF, Qu LH, Nelson DR, Sanderfoot AA, Spalding MH, Kapitonov VV, Ren Q, Ferris P, Lindquist E, Shapiro H, Lucas SM, Grimwood J, Schmutz J, Cardol P, Cerutti H, Chanfreau G, Chen CL, Cognat V, Croft MT, Dent R, Dutcher S, Fernandez E, Fukuzawa H, Gonzalez-Ballester D, Gonzalez-Halphen D, Hallmann A, Hanikenne M, Hippler M, Inwood W, Jabbari K, Kalanon M, Kuras R, Lefebvre PA, Lemaire SD, Lobanov AV, Lohr M, Manuell A, Meier I, Mets L, Mittag M, Mittelmeier T, Moroney JV, Moseley J, Napoli C, Nedelcu AM, Niyogi K, Novoselov SV, Paulsen IT, Pazour G, Purton S, Ral JP, Riano-Pachon DM, Riekhof W, Rymarquis L, Schroda M, Stern D, Umen J, Willows R, Wilson N, Zimmer SL, Allmer J, Balk J, Bisova K, Chen CJ, Elias M, Gendler K, Hauser C, Lamb MR, Ledford H, Long JC, Minagawa J, Page MD, Pan J, Pootakham W, Roje S, Rose A, Stahlberg E, Terauchi AM, Yang P, Ball S, Bowler C, Dieckmann CL, Gladyshev VN, Green P, Jorgensen R, Mayfield S, Mueller-Roeber B, Rajamani S, Sayre RT, Brokstein P, Dubchak I, Goodstein D, Hornick L, Huang YW, Jhaveri J, Luo Y, Martinez D, Ngau WC, Otillar B, Poliakov A, Porter A, Szajkowski L, Werner G, Zhou K, Grigoriev IV, Rokhsar DS, Grossman AR. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318:245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stephan A, Vaughan S, Shaw MK, Gull K, McKean PG. An essential quality control mechanism at the eukaryotic basal body prior to intraflagellar transport. Traffic. 2007;8:1323–1330. doi: 10.1111/j.1600-0854.2007.00611.x. [DOI] [PubMed] [Google Scholar]
- 19.Tian G, Huang Y, Rommelaere H, Vandekerckhove J, Ampe C, Cowan NJ. Pathway leading to correctly folded beta-tubulin. Cell. 1996;86:287–296. doi: 10.1016/s0092-8674(00)80100-2. [DOI] [PubMed] [Google Scholar]
- 20.Scheffzek K, Ahmadian MR, Wittinghofer A. GTPase-activating proteins: helping hands to complement an active site. Trends Biochem Sci. 1998;23:257–262. doi: 10.1016/s0968-0004(98)01224-9. [DOI] [PubMed] [Google Scholar]
- 21.Bartolini F, Bhamidipati A, Thomas S, Schwahn U, Lewis SA, Cowan NJ. Functional overlap between retinitis pigmentosa 2 protein and the tubulin-specific chaperone cofactor C. J Biol Chem. 2002;277:14629–14634. doi: 10.1074/jbc.M200128200. [DOI] [PubMed] [Google Scholar]
- 22.Chang P, Stearns T. Delta-tubulin and epsilon-tubulin: two new human centrosomal tubulins reveal new aspects of centrosome structure and function. Nat Cell Biol. 2000;2:30–35. doi: 10.1038/71350. [DOI] [PubMed] [Google Scholar]
- 23.Dutcher SK, Morrissette NS, Preble AM, Rackley C, Stanga J. Epsilon-tubulin is an essential component of the centriole. Mol Biol Cell. 2002;13:3859–3869. doi: 10.1091/mbc.E02-04-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dutcher SK, Trabuco EC. The UNI3 gene is required for assembly of basal bodies of Chlamydomonas and encodes delta-tubulin, a new member of the tubulin superfamily. Mol Biol Cell. 1998;9:1293–1308. doi: 10.1091/mbc.9.6.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taillon BE. Proteins Associated with the basal body apparatus in Chlamydomonas reinhardtii: a molecular genetic and cell biological analysis. Carnegie Mellon; Pittsburgh: 1993. [Google Scholar]
- 26.Holmes JA, Dutcher SK. Cellular asymmetry in Chlamydomonas reinhardtii. J Cell Sci. 1989;94(Pt 2):273–285. doi: 10.1242/jcs.94.2.273. [DOI] [PubMed] [Google Scholar]
- 27.Kater J. Morphology and division of Chlamydomonas with reference to the phylogeny of the flagellate neuromotor system. Univ Calif Pub Zool. 1929;33:125–168. [Google Scholar]
- 28.Goodenough UW, Clair HS. BALD-2: a mutation affecting the formation of doublet and triplet sets of microtubules in Chlamydomonas reinhardtii. J Cell Biol. 1975;66:480–491. doi: 10.1083/jcb.66.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ehler LL, Holmes JA, Dutcher SK. Loss of spatial control of the mitotic spindle apparatus in a Chlamydomonas reinhardtii mutant strain lacking basal bodies. Genetics. 1995;141:945–960. doi: 10.1093/genetics/141.3.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.