Abstract
Vertebrate hematopoiesis first produces primitive (embryonic) lineages and ultimately generates the definitive (adult) blood. While definitive hematopoiesis may produce many diverse blood types via a common multipotent progenitor, primitive hematopoiesis has been thought to produce only erythrocytes or macrophages via progenitors that are unipotent for single blood lineages. Using a variety of in vivo celltracing techniques, we show that primitive blood in zebrafish derives from two different progenitor types. On the dorsal gastrula, blood progenitors are unipotential cells that divide infrequently, populate the rostral blood islands (RBI), and differentiate into macrophages. In contrast, on the ventral gastrula, blood progenitors are multipotential cells with rapid cell cycles, populate the intermediate cell mass (ICM), and differentiate into erythrocytes, neutrophils, and thrombocytes. Our results demonstrate the existence of primitive hematopoietic progenitors that are segregated very early in development and that are specified to produce either a unipotent or a multipotent blood cell lineage.
Introduction
Hematopoiesis in vertebrates occurs in two successive waves known as primitive (embryonic) and definitive (adult) (reviewed in: Godin and Cumano 2002). One of the hallmarks of vertebrate hematopoiesis is that blood cells are produced in ever changing sites throughout development (Baron, 2003; Keller et al., 1999; Lacaud et al., 2001; Orkin and Zon, 2008). Hence where a cell is produced, and not just when, often defines whether it is considered primitive or definitive. In mammals, during definitive hematopoiesis, all blood, composed of a diverse population of white blood cells (monocytes, granulocytes and lymphocytes), red blood cells (erythrocytes), and platelets (thrombocytes), is thought to derive from a common progenitor termed the hematopoietic stem cell (Graf, 2008; Orkin and Zon, 2008; Reya et al., 2001). In contrast, during primitive hematopoiesis, blood cells are not thought to originate from a multipotent blood founder, but rather from individual progenitor cells called hemangioblasts which generate only a single blood cell type along with endothelial descendents (Baron, 2003; Keller et al., 1999; Lacaud et al., 2001; Orkin and Zon, 2008).
Histological comparison of peripheral blood and the expression of homologous genes essential for hematopoiesis, show that zebrafish possess equivalent blood types to those of mammals. Two cell types, erythrocytes and macrophages (monocytes), are considered by many to be the only primitive blood of zebrafish, as in mouse (Berman et al., 2005; Davidson and Zon, 2004; Onnebo et al., 2004; Palis et al., 1999) . However, in the overlapping interval between primitive hematopoiesis and the onset of definitive hematopoiesis, initiating somewhere between 24 to 48 hours (Bertrand et al., 2007; Murayama et al., 2006; Zhang and Rodaway, 2007), three other cell types appear, these include: neutrophils (heterophilic granulocytes), thrombocytes, and lymphocytes.
Studies of gene expression and limited fate mapping show that the zebrafish blood derives from intermediate mesoderm (Davidson and Zon, 2004; Rohde et al., 2004), a narrow band of tissue extending bilaterally along either side of the head and paraxial mesoderm. The intermediate mesoderm also includes cells of the future endothelial and pronephric lineages (Liao et al., 1998; Majumdar et al., 2000). By 12 hours, the hematopoietic portion of the intermediate mesoderm segregates into the earliest blood differentiation sites roughly equivalent to the blood islands of the mammalian yolk sac (Berman et al., 2005; Davidson and Zon, 2004; Onnebo et al., 2004). One site located in the head (Fig 1A), termed the rostral blood island (RBI), produces the primitive macrophages that by 18 hours migrate away from the head and out over the yolk sac whereupon they quickly disperse throughout the mesenchyme of the embryo (Herbomel et al., 1999; Herbomel et al., 2001; Lieschke et al., 2002). The other site located in the trunk (Fig. 1A–C), called the intermediate cell mass (ICM), produces the primitive erythrocytes which migrate anteriorly towards the yolk sac where upon they enter the developing circulatory system around 26 hours (Detrich et al., 1995; Liao et al., 2002; Long et al., 1997).
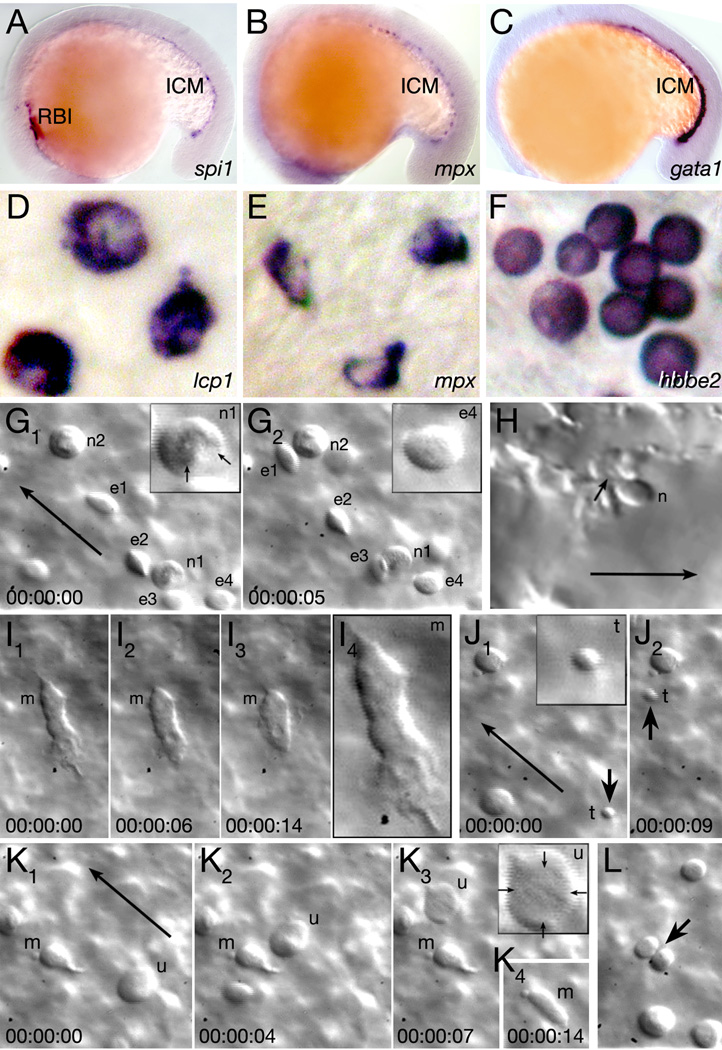
Figure 1.
Characterization of embryonic blood. (A–C) The rostral blood island (RBI) and intermediate cell mass (ICM) at 18 hours, visualized with: (A) a myeloid marker, (B) a neutrophil marker, and (C) an erythroid marker. (D–F) Individual blood cells at 26 hours, visualized by in situ hybridization: (D) macrophage cells, (E) neutrophil cells, and (F) erythrocyte cells. (G–L) Individual blood cells in the live 30 hour embryo. Each image is a single frame from a real-time recording of circulating blood. Elapsed time is indicated in lower frame, long arrows indicate flow of circulation and insets show 200x magnified view of specific cells. (G) Neutrophils (n) and circulating erythrocytes (e) in the viteline vein over the yolk sac, and (H) in the lumen of a tail blood sinus. The arrows in inset G1 indicate individual nuclear lobes of a neutrophil and small arrow in H indicates where the neutrophil anchors itself to the endothelial lumen. (I) A macrophage (m) patrolling the viteline vein. The last panel is magnified 200x. (J) A circulating thrombocyte (t and arrow). (K, L) Unidentified cells. (K) A large rare cell type (u), arrows in inset indicate its diffuse nuclear envelope, and (L) a dividing cell (arrow). A macrophage (m) is also visible in K.
Currently, there is confusion concerning the derivation of embryonic neutrophils, thrombocytes and lymphocytes. Gene expression studies indicate that neutrophils may derive from the ICM (Bennett et al., 2001; Lieschke et al., 2001; Lieschke et al., 2002; Thisse and Thisse, 2004) (Fig. 1A–B), but it has also been reported that neutrophils derive from primitive macrophage cells once they exit the RBI (Le Guyader et al., 2007) or alternatively from the earliest definitive blood progenitors located in the posterior blood island (PBI) (Bertrand et al., 2007). Regardless, cells having the characteristics of neutrophils in other species are evident throughout circulation by 26 hours (Bennett et al., 2001; Hall et al., 2007; Lieschke et al., 2001; Mathias et al., 2006; Meijer et al., 2007). Little is known of the origin of embryonic thrombocytes as they have no early markers, but they first appear in circulation around 36 hours (Gregory and Jagadeeswaran, 2002). Unlike their mammalian equivalent they do not appear to derive from megakaryocytes (Lin et al., 2005). Lymphocytes have also been reported to originate from the ICM (Willett et al., 2001), but more recent studies claim they originate from one of the initial sites of definitive hematopoiesis along the dorsal aorta, before migrating to the developing thymi (Bertrand et al., 2008).
Previous fate map studies have reported that zebrafish blood derives exclusively from the ventral margin of the gastrula-stage embryo (an area from which pronephros and endothelial cells are also derived). However, in the discussion of a report showing that primitive macrophages differentiate in the RBI and have an independent program of development from the erythrocyte portion of the blood, Lieschke et al. (2002) speculated that, based upon the known movements of zebrafish cells, the macrophage precursors could possibly be derived from a more dorsally-located region in the gastrula. Here, we re-examine the origin of zebrafish embryonic blood. By intra-cellularly labeling single marginal cells at the blastula stage, and correlating the position of these clones at varying locations along the dorsoventral axis of late blastula and early gastrula-staged embryos, we show that future blood derives from all areas of the margin, including dorsal. We further show that blood derived from the ventral margin does not produce only a single cell type, but rather many ventral blood progenitors are multipotent with lineages that include not only primitive erythrocytes, but also embryonic thrombocytes and neutrophils. These multipotent ventral blood progenitors eventually populate the site of primitive hematopoiesis in the ICM. In contrast, blood derived from the dorsal margin exclusively produces a single cell type, namely primitive macrophages. The dorsal blood progenitors eventually populate the site of primitive hematopoiesis within the RBI. This unipotent dorsal lineage divides infrequently, whereas cells from the multipotent ventral lineage divide often and continue to produce erythrocyte and neutrophil progeny after 26 hours. Both unipotent dorsal and multipotent ventral blood progenitors frequently share lineages with endothelial cells, but a common progenitor is not obligatory for blood to form. From clonal analysis calculations and observations of a mutant which undergoes a cell cycle arrest, we estimate that the number of blood progenitors to be roughly 60 out of the calculated 8000 total number of cells at the early gastrula stage.
Materials and Methods
Embryos
For fate mapping, embryos were derived from crosses of wild type strains. For in situ analysis, embryos were derived from crosses of wild type strains or of identified hrpti245 heterozygotes.
Lineage tracing and fate map construction
Lineage tracing and fate map construction were adapted from those previously described (Warga and Nüsslein-Volhard, 1999). Briefly an individual blastomere was labeled between the 1K- to 4K–cell stages with a 5% solution of lysinated rhodamine-dextran (70,000 MW; Invitrogen, formerly Molecular Probes). At stages indicated below, embryos were oriented in 3% methyl cellulose in Daniaeu’s media and analyzed using a Zeiss Axioplan microscope, VideoScope VS-2525 image intensifier and VS2000N Newvicon Camera. Images were stored directly onto a Power Macintosh 9600/350 running Cytos 3.0.1 software (Applied Scientific Instrumentation).
In the zebrafish, all embryonic fates derive from the deep cell portion of the blastoderm, not from the outer enveloping layer (EVL) (Kimmel et al., 1990). The segregation between deep cell and EVL lineages occurs a cell cycle or two after the mid-to late-blastula stages (Kimmel and Law, 1985a; Kimmel and Law, 1985b), about when our clones were established. At this time, a blastomere can divide to give rise to two deep cells or two EVL cells or a combination of one deep cell plus one EVL cell. We used this segregation to determine when a single-cell clone (of deep cells) was established. For example, if at 40 to 50% epiboly, two labeled deep cells were observed, this clone was established in the mid-blastula. If however, only one labeled deep cell was observed at 40 to 50% epiboly, as seen in Fig 2A, this clone was established in the late blastula i.e. the onset of gastrulation. For making the fate map, clones were examined at 40 to 50% epiboly, to determine distance from the margin (Fig. 2A) and again at shield stage, to determine distance from dorsal (Fig. 2B). Clones were re-examined from 24 to 48 hours of development to determine cell fate (Fig. 2F,G). Cell fate was assigned based on analysis of behavior and morphology in the live embryo; typically blood cells were analyzed for 5 to 10 minutes, a portion of these clones were also processed for in situ hybridization to confirm cell fate (Table 2). In some embryos, we also examined which blood island the clone located to (Fig. 2C) or its precise location in a blood island just before the onset of circulation (Fig. 2E).
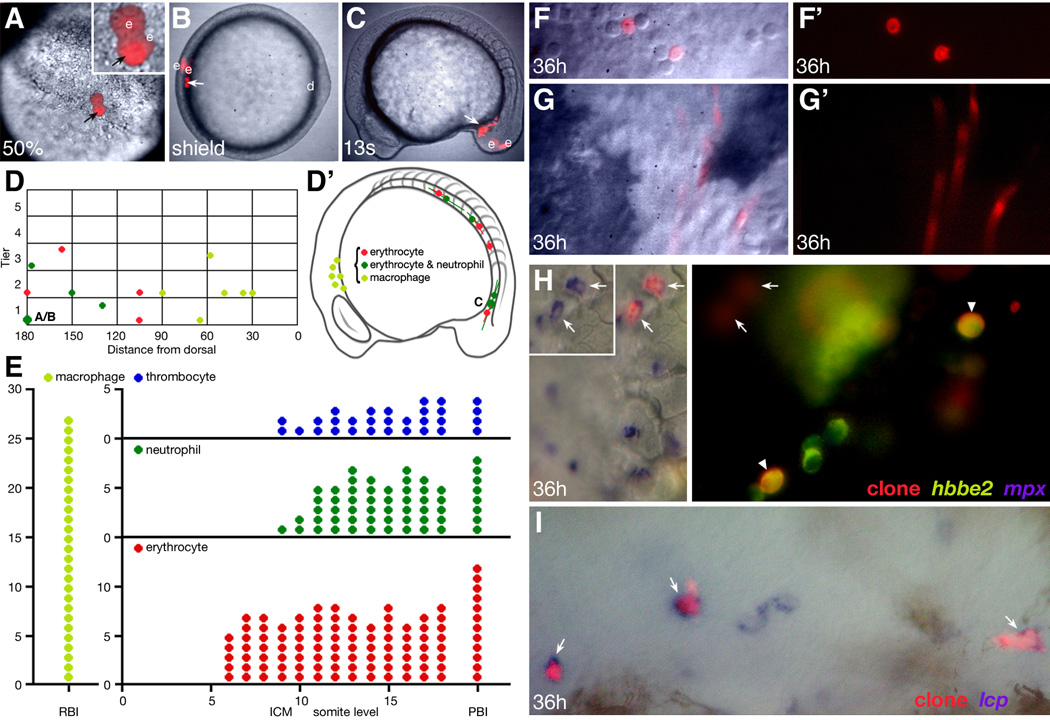
Figure 2.
Lineage tracing and fate map analysis of the embryonic blood. (A–C, F–G) Progeny of a single mid-blastula labeled cell. (A) The clone at 50% epiboly (face view), now 1 deep cell (arrow) and 2 EVLs (e) at the margin of the blastoderm; note that the single deep cell is being viewed through one of the EVL cells. (B) The clone at shield stage (animal pole view), now 2 deep cells (arrow) and 2 EVLs (e) located 180 degrees of arc from the dorsal midline (d). (C) The clone at 13-somite stage (side view), now 11 deep cells (arrow) in the posterior IMC. The EVL portion of the clone (e), now periderm, is extraembryonic. (F, G) The clone at 36 hours, now endothelial cells and circulating blood which include (F) neutrophils and (G) erythrocytes. (D, D’) Hematopoietic fate maps. Graphs depict the location of the same clones at two different stages of development. (D) 6 hour gastrula fate map, clones vs. dorsoventral location. The presentation is a side view, dorsal to the right, with clones on the right projected to the left. Each symbol is the average location of the deep cell portion of a clone relative to the margin (tier 0), in units of cell diameter, and the dorsal midline (0), in degrees of arc. The oversize symbol represents the clone shown in panels A–C and the small letters refer to the respective panel. (D’) 16 hour 14-somite stage fate map, clones vs. primitive blood island. The horizontal line through each symbol shows the anteroposterior spread of each clone. All clones except one also included endothelial cells. (E) 24 hour hematopoietic fate map, fate of clone vs. anteroposterior location (RBI, somite level within the ICM, or PBI). Each symbol represents a single clone at that anteroposterior location, however clones that spread over several somite levels are represented with multiple symbols. Fate map is based on 27 macrophage clones, 41 erythrocyte clones, 16 neutrophil clones, and 12 thrombocyte clones. These clones include all that were later verified by in situ hybridization. (H–I) Verification of in vivo blood morphology by one or two-color in situ hybridization at 36 hours. (H) A multilineage erythrocyte, neutrophil and thrombocyte clone (red fluorescence) which was visualized for co-expression of hbbe2, an erythrocyte-specific marker (green fluorescence) and mpx, a neutrophil-specific marker (NBT/BCIP purple substrate). Left panel, white light image shows 2 lineage labeled neutrophils (arrows). Inset, same cells showing mpx expression alone. Right panel, UV image shows 2 lineage labeled erythrocytes (arrowheads), and a clonally-related thrombocyte (hbbe2-negative; white circle). The neutrophil portion of clone is out of focus (arrows). (I) A unilineage macrophage clone (red fluorescence) which was visualized for co-expression of lcp, a macrophage marker (NBT/BCIP purple substrate). White light image shows 3 lineage labeled macrophages (arrows) and 2 macrophages not in the clone.
Table 2.
Verification of in vivo identified cells with in situ markers
|
in vivo identification: |
Total number | Ratio of cells in situ/in vivo | |||
|---|---|---|---|---|---|
| cells | embryos | lcp | hbbe2 | mpx | |
| macrophage | 99 | 19 | 90/91a | not done | 0/21 |
| erythrocyte | 590 | 24 | 0/160 | 571/590b | 0/344 |
| neutrophil | 39 | 12 | 0/11 | 0/34 | 26/28c |
| thrombocyte d | 55 | 10 | not done | 0/55 | 0/52 |
Clones were examined at 24 hours and again between 30 to 36 hours before being fixed and processed for in situ hybridization. In general, but not always ventral clones were examined with two different riboprobes as detailed in the methods section.
Of 91 labeled cells identified as macrophages, 90 cells expressed the marker lcp which is specific for macrophages and possibly other white blood cells.
The marker hbbe2 is specific to erthrocytes.
The marker mpx is specific for neutrophils.
Thrombocytes do not express the thrombocyte specific marker cd41 until 48 h.
For marking single cells at 26 hours, individual blastomeres in the lK-cell stage embryo were injected with a mixture of 1% phenol red and Kaede mRNA (∼100ng). As above, the resulting clone was examined at 40 to 50% epiboly and shield stage. At 26 hours, if the clone contained blood, a single blood cell was photoconverted by changing the fluorescent spectrum of the Kaede protein from green to red fluorescence (Ando et al., 2002) using 10 second pulses of 440 um light. Photoconversion was done at 630x on a Zeiss Axioplan with a specially machined field diaphragm having a pinhole aperture of less than l0µm. Progeny of this red cell and the original green clone continued to be monitored up to 48 hours of development.
in situ hybridization and confirmation of cell fate
Embryos were fixed in 4% paraformaldehyde and whole-mount RNA in situ hybridization was carried out using digoxigenin- or fluorescein-labeled riboprobes (Jowett and Yan, 1996) following the protocol by Thisse et al. (1993) and the following modifications: 1) after permeabilizing embryos with MeOH, and before the Protinase K treatment, embryos were incubated for 10 minutes in 0.6% H2O2 in PBS to quench endogenous Peroxidase activity, 2) Protinase K concentration was reduced to .25mg/ml and digestion time to no more than 5 minutes and 3) after Protinase K treatment, embryos were post-fixed in fresh paraformaldehyde for at least 1.5 hours at room temperature. Both these latter modifications decreased yolk sac loss – where most of the blood cells reside once circulation commences.
For detection of the digoxigenin-labeled riboprobe, we used the alkaline phosphatase labeled anti-digoxygenin antibody (Roche, 1:5000) and a NBT/BCIP purple substrate. For colocalization experiments involving marked clones, we additionally probed with a fluorescein-labeled riboprobe that was visualized by using a peroxidase labeled anti-fluorescein antibody (Roche, 1:1000) and teramide signal amplification using fluorescein reagents kindly provided by Vicky Prince. To see the labeled clone within in situ labeled cells it was necessary to amplify the fluorescent signal. Therefore following in situ hybridization, rhodamine-dextran labeled cells were immunodetected with an antitetramethylrhodamine antibody (Invitrogen, 1:500) and Alexa Fluor 555 or 488 goat antirabbit secondary antibodies (Invitrogen, 1:200). Embryos were cleared in 70% glycerol and photographed on a Zeiss Axioplan using a Nikon Dl or Sony F-707 digital camera
We used: spi1(Hsu et al., 2004; Lieschke et al., 2002); gata1 (Detrich et al., 1995); Icp1 (Herbomel et al., 1999); mpx (Bennett et al., 2001; Lieschke et al., 2001); and hbbe2 (Brownlie et al., 2003).
Results
Characterization of zebrafish embryonic blood
Of the five blood types observed in the embryonic zebrafish, only three populations can be unambiguously identified by gene expression by 1 day. Erythrocytes specifically express hbbe2 (Brownlie et al., 2003), macrophages express lcp1 (Herbomel et al., 1999), and neutrophils specifically express mpx (Bennett et al., 2001; Lieschke et al., 2001). Using these probes, we found cells that were morphologically distinguishable from one another by 26 hours (Fig. 1D–F). Macrophages were large; almost twice the size of other cells and had smaller nuclei relative to their size. In addition, macrophages and neutrophils had irregular cell shapes and folded nuclei. Erythrocytes were uniform in shape and had rounded nuclei. While these markers were chosen because double labeling studies indicated that each marker labels unique blood cell populations (Bennett et al., 2001), there have been a number of recent reports on whether lcp1 only marks macrophage cells (Hall et al., 2007; Le Guyader et al., 2007; Mathias et al., 2006; Meijer et al., 2007). This is because between 48 to 72 hours, some neutrophil cells seem also to express lcp1.
Because of the discrepancies in the literature concerning gene marker specificity, we were cautious in using gene expression to solely identify cell types. Thus we examined and relied upon the morphology of individual blood cells in vivo for their identification. Blood cells were easiest to observe in circulation at 30 hours over the yolk sac, before the viteline vein narrowed and displaced anteriorly or in the rostral part of the tail before there was an artery and vein. We observed five different types of in vivo cells, three of which correlated with our in situ study: (1) Macrophages: Unmistakable cells often located on the yolk sac or in the mesenchyme of the head. Their morphology and dimension (Fig. 1I) resembled their in situ stained counterparts. Macrophages used an amoeboid-type of locomotion, as has been reported (Herbomel et al., 1999), but if quiescent tended to extrude pseudopodia (Fig 1K). (2) Neutrophils: Smaller than macrophages, these cells were often anchored to the vessel wall (Fig. 1H). Like their in situ stained counterparts and Wright-Giemsa stained adult neutrophils (Bennett et al., 2001; Lieschke et al., 2001), they possessed a segmented nucleus (Fig. 1G1). (3) Erythrocytes: The predominant cell in circulation, like their in situ stained counterparts. In vivo, erythrocytes appeared concave and elliptical (Fig. 1G). (4) Thrombocytes: Tiny cells with no apparent nucleus (Fig. 1J) (Gregory and Jagadeeswaran, 2002). (5) Unidentified: Very large uniform cells (compare insets in Fig. 1K to Fig. 1G) or cells in division (Fig. 1L), these probably made up less than 5% of the total. We did not observe anything that resembled a lymphocyte.
Blood fate and blood island location are segregated in the early gastrula
To trace the origin of blood, we intra-cellularly labeled single cells with lineage tracer dye in the mid-blastula stage embryo and subsequently followed only the deep cell (embryonic) portion of the clone (Fig. 2) as outlined in the methods section. In our fate map analyses, we also noted the non-blood identities of cells contained within our clones, these included endothelial (Fig. 3D, J) and pronephric cells (Fig. 3E, K). Inherent in many models of stem cell identity is the hypothesis that fate decisions are both hierarchical and sequential. To determine at what stage individual cells became specified towards a pronephros versus a hemangioblast versus a hematopoietic fate, we catalogued the identity of all labeled cells within our clones relative to when that deep cell clone was established (for an explanation see methods section). Segregating our data into deep cell clones that were either established at the mid-blastula or the early gastrula stage, allowed us to determine 1) whether individual labeled cells gave rise to single fates or multiple fates and 2) whether there was a tendency for particular identities to group together within the same clone.
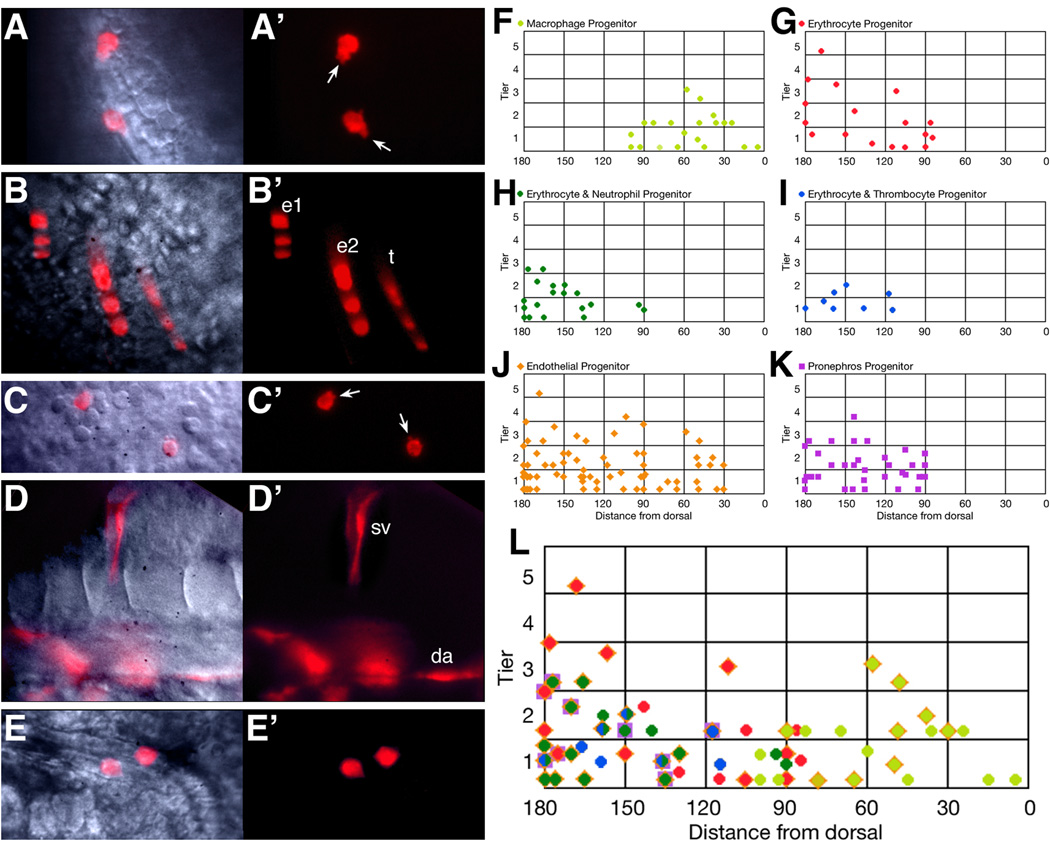
Figure 3.
Hematopoietic progenitors originate from both the dorsal and ventral gastrula. (A–E) Examples of individual derivatives. (A) macrophages, (B) erythrocytes and thrombocytes, (C) neutrophils, (D) endothelial cells, and (E) pronephric cells. Abbreviations: da, dorsal aorta; e, erythrocytes; sv, segmental vein; t, thrombocytes. Arrows indicate cellular protrusions. (F–L) 6 hour gastrula fate maps. Clones that included: (F) macrophage cells, (G) just erythrocyte cells, (H) both erythrocyte and neutrophil cells, (I) both erythrocyte and thrombocyte cells, (J) endothelial cells, and (K) pronephric cells. (L) Summary hematopoietic fate map. Superimposed symbols show individual clones that gave rise to multiple fates. The half green/half blue symbol indicates the 4 clones that included all three ventral-derived blood fates (erythrocytes, neutrophils and thrombocytes). Some of the blood clones depicted on the fate maps were also later verified by in situ hybridization.
In agreement with previous fate map analyses (Kimmel et al., 1990; Vogeli et al., 2006; Warga and Nüsslein-Volhard, 1999), we found that blood derived from the ventral portion of the gastrula stage embryo (Fig. 2B and Fig. 3G). Without exception these ventrally-derived cells and their progeny located to the ICM (Fig. 2D) before differentiating initially as primitive erythrocytes (Fig. 2G and Fig. 3B). We also found that blood derived from the dorsal portion of the gastrula. Without exception, these dorsally-derived cells located to the RBI (Fig. 2D) before differentiating as primitive macrophages (Fig. 2I and Fig. 3A). Interestingly, macrophage and erythrocyte cells never derived together from the same labeled precursor cell even though their fate map territories somewhat overlapped during gastrulation. It should be noted that often clones containing either macrophages or erythrocytes also included labeled endothelial cells (Fig. 3L) suggesting that some blood cells derived from a precursor that had the characteristics of a hemangioblast. However, passing through a hemangioblast-like state does not appear to be an absolute requirement for blood production given the high frequency of solely blood clones (Fig. 3L).
Intermediate mesoderm becomes progressively restricted to single fates
We found that mid-blastula-derived clones (Table 1a; n=67) only occasionally gave rise to blood alone (10%; rows 3, 13–14; Table 1a). In fact, the most commonly observed clones (27%; rows 2, 11–12; Table 1a) contained only blood and endothelium together, not inconsistent with a potential hemangioblast origin. Mid-blastula clones never gave rise to pronephros alone, but endothelium and pronephros often derived exclusively together (13%; row 5;Table 1a) from the same clone. A surprising number of mid-blastula clones included blood, endothelium and pronephros (12%; rows 6–7;Table 1a). However, it was rare (1%; row 9;Table 1a) for a clone to give rise to both blood and pronephros in the absence of endothelium, whereas endothelium-only clones were observed frequently (15%; rows 1 & 10;Table 1a). These results indicate that most cells in the mid-blastula are not yet restricted to a single fate in the intermediate mesoderm. We also suggest that these results indicate that there is an initial decision between hematopoietic fate (blood and endothelium) versus one leading to the production of pronephros and/or endothelium.
Table 1.
| Table 1a. Blastula Clones | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Row | Location | 36 hour Fates | # of clones | Comments | ||||||
| 1 | Dorsal | ✓ | 5 | |||||||
| 2 | ✓ | ✓ | 8 | a | ||||||
| 3 | ✓ | 3 | a | |||||||
| 4 | Ventral | ✓ | 0 | |||||||
| 5 | ✓ | ✓ | 12 | b | ||||||
| 6 | ✓ | ✓ | ✓ | 2 | c | |||||
| 7 | ✓ | ✓ | ✓ | ✓ | 6 | d | ||||
| 8 | ✓ | ✓ | 0 | |||||||
| 9 | ✓ | ✓ | ✓ | 1 | ||||||
| 10 | ✓ | 5 | ||||||||
| 11 | ✓ | ✓ | 9 | e | ||||||
| 12 | ✓ | ✓ | ✓ | 6 | a | |||||
| 13 | ✓ | 4 | e | |||||||
| 14 | ✓ | ✓ | 6 | a, c | ||||||
| Table 1b. Gastrula Clones | |||||||
|---|---|---|---|---|---|---|---|
| Row | Location | 36 hour Fates | # of clones | ||||
| 1 | Dorsal | ✓ | 4 | ||||
| 2 | ✓ | ✓ | 1 | ||||
| 3 | ✓ | 8 | |||||
| 4 | Ventral | ✓ | 9 | ||||
| 5 | ✓ | ✓ | 0 | ||||
| 6 | ✓ | ✓ | ✓ | 0 | |||
| 7 | ✓ | ✓ | ✓ | ✓ | 0 | ||
| 8 | ✓ | ✓ | 0 | ||||
| 9 | ✓ | ✓ | ✓ | 0 | |||
| 10 | ✓ | 5 | |||||
| 11 | ✓ | ✓ | 2 | ||||
| 12 | ✓ | ✓ | ✓ | 3 | |||
| 13 | ✓ | 2 | |||||
| 14 | ✓ | ✓ | 0 | ||||
two of these clones also made endoderm.
three of these clones also made muscle.
one of these clones also made muscle.
two of these clones also made muscle or heart.
one of these clones also made endoderm.
In contrast to the data obtained from mid-blastula-derived clones, gastrula-derived clones (Table 1b; n=34) tended to be restricted to a single fate, consistent with earlier fate mapping studies (Kimmel et al., 1990; Melby et al., 1996; Shih and Fraser, 1995). Thus the majority of gastrula clones gave rise to blood only (29%; rows 3 & 13;Table 1b) or endothelium only (26%; rows 1 & 10;Table 1b) or pronephros only (26%; row 4;Table 1b). However, a substantial number of clones (18%; rows 2, 11–12;Table 1b) still gave rise to blood and endothelium together, and if one excluded those containing macrophages, this amount (15%; rows 11–12;Table 1b) was remarkably consistent with the previously-reported percentage of cells regarded to be the zebrafish hemangioblast in the early gastrula (Vogeli et al., 2006).
Because gastrula clones never gave rise to blood or endothelium along with pronephros (Table 1b), our data suggests that pronephric identity is established as a single fate-producing lineage shortly before gastrulation. One other fate appears to become established by gastrulation. Clones containing only macrophages were highly prevalent (89%) when established in the gastrula, but less prevalent (11%) when established at the mid-blastula stage (compare rows 2 and 3 between Table 1a and Table 1b). Such was not the case for other blood containing clones as erythrocytes, neutrophils and thrombocytes still had a marked tendency to share a lineage with each other as well as with endothelial cells (Table 1b).
Neutrophils and thrombocytes are derived from ventral, erythrocyte-producing lineages
Our fate map data shows that a labeled ventral clone can gave rise to multiple blood cell types by 48 hours, namely erythrocytes, thrombocytes and neutrophils (rows 7, 12 and 14; Table 1a and Table 1b). However, in earlier observations at 26 hours, erythrocytes and macrophages appeared to be the only blood types present. One possibility is that neutrophils and thrombocytes derive from definitive blood cells sharing lineage with primitive erythrocytes. To test this hypothesis we reinvestigated where blood clones were located at 24 hours, before blood migrates out of the ICM and begins to circulate; we then followed these clones for the next 12 hours.
We found that clones in the anterior portion of the ICM (somite levels 6–8) produced exclusively erythrocytes. Clones in the remainder of the ICM, as well as clones in the posterior blood island (PBI, the portion of ICM behind the yolk sac extension where definitive blood is born and approximately somite levels 19–22) often produced in addition neutrophils and/or thrombocytes (Fig. 2E). Without exception, either of these two fates only derived from clones that included erythrocytes. In multilineage clones, neutrophils and erythrocytes formed, on average, a 1:9 ratio; whereas thrombocytes and erythrocytes formed, on average, a 1:4 ratio. It was also possible for all three ventral blood types to be present together in a single clone (Fig. 2H and Fig. 3L). Thus it is unlikely that neutrophils and/or thrombocytes are solely derived from the earliest definitive blood progenitors. Furthermore, clones that included two or three of these ventrally-derived blood types often also included endothelial cells (Fig. 3L) suggesting that the ventral hemangioblast progenitor identified by Vogeli et. al. (2006) is perhaps multipotent.
Further examination of these clones revealed that thrombocytes were already discernable in the ICM at 24 hours. Such labeled cells did not enter circulation immediately, but many were circulating by 30 hours (Fig. 3B). Neutrophils began to appear in the viteline vein of the yolk sac and within other blood vessels (Fig. 2F–H and Fig. 3C) in the interval between 24 and 36 hours. This period correlated with the rapid expansion of the labeled erythrocytes within our clones.
We confirmed the identity of these cells by in situ analysis (Fig. 2H–I). As shown in Table 2, we had an error rate for erythrocytes of 3%, some of which was likely due to cells resembling an erythrocyte at 36 hours, but not yet having differentiated enough to express the hbbe2 globin gene (Brownlie et al., 2003). With respect to neutrophils, the majority of cells (93%) identified as neutrophils expressed mpx, a gene specific to neutrophils (Bennett et al., 2001; Lieschke et al., 2001), and never hbbe2 or lcp which at this time is mainly expressed in only macrophage cells (Bennett et al., 2001; Herbomel et al., 1999). While the error rate for neutrophils is higher than that of erythrocytes, in situ analysis never disqualified a clone we determined in vivo to contain at least some neutrophil cells. This is because we were only occasionally in error on the precise number of neutrophils within a clone, but never on whether a clone contained neutrophils or not. Thrombocytes have no specific early markers, but are known to not express hbbe2 or mpx (Lin et al., 2005); as shown in Table 2, 100% of the cells that we identified as thrombocytes in vivo adhered to these parameters.
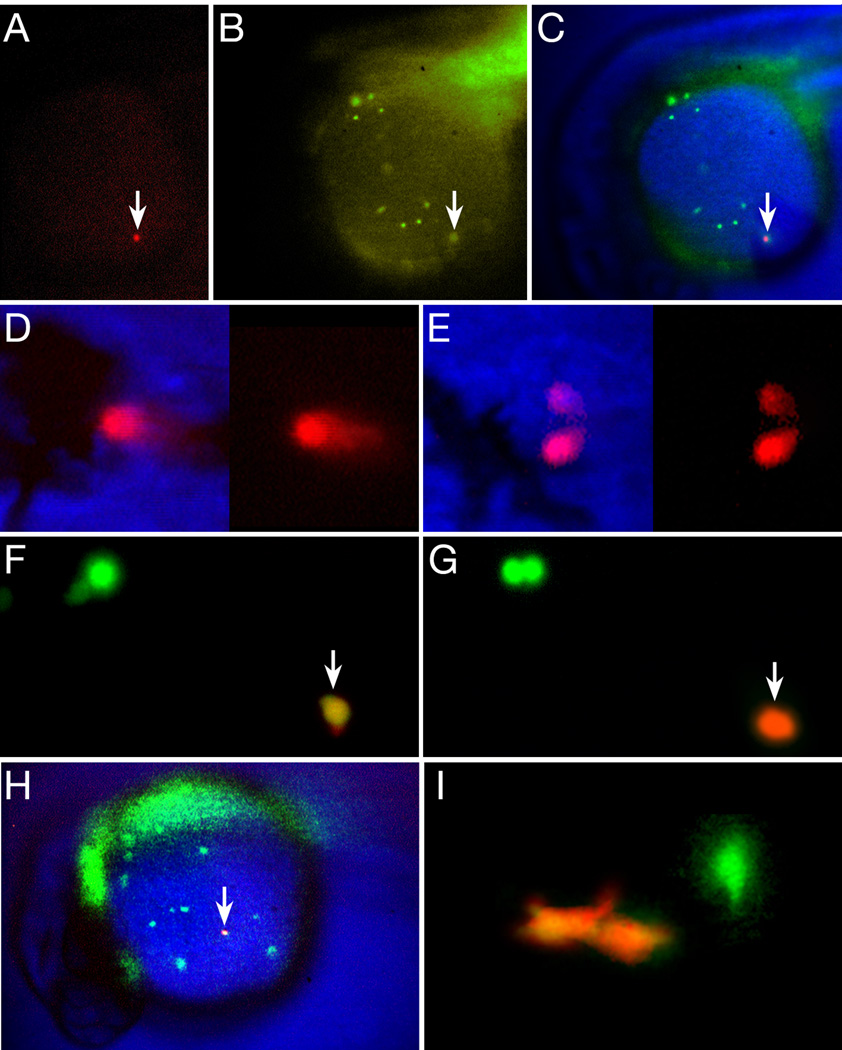
To corroborate our data obtained from single cell blastula clones, we employed a different method of marking cells using the UV-induced green-to-red photoconversion of the fluorescent Kaede protein (Ando et al., 2002). This strategy allowed us to test whether single blood cells at the onset of circulation, continue to divide and/or are multipotent when it is otherwise difficult to label blood by traditional lineage tracer techniques. In brief, we intra-cellularly injected single cells with Kaede mRNA in the mid-blastula stage embryo and at 24 to 26 hours, selected embryos having clones of Kaede expressing cells in the appropriate tissue; we then photoconverted a single cell to produce red fluorescence in that cell and all its progeny.
To avoid the possibility of labeling definitive blood believed to be lurking—not yet circulating—in the PBI (Bertrand et al., 2007), we initially photoconverted only the earliest circulating blood cells in the viteline vein of the yolk sac, i.e. cells that we would otherwise have identified as erythrocytes (Fig. 4A–C). In later experiments, we targeted noncirculating cells in the ICM. We found that photoconverted cells (n=10) usually exhibited robust division rates – in nine of these clones there were about 10 to 20 red-labeled blood cells the next day, suggesting that individual cells divide every 5 to 6 hours. This rapid cell cycle length is supported by the observation that 7 hours after marking, one clone already had 3 red-labeled blood cells indicating that at least two doublings had occurred in this interval. Not all photoconverted cells continued to divide (n=l), thus some blood has dropped out of the cell cycle by this time. Of the nine photoconverted cells that multiplied, two included neutrophils as well as erythrocytes amongst their progeny (Fig. 4D, E). In agreement with our above ICM fate map (Fig. 2E), one of these clones was descended from a cell marked in the viteline vein just as it was entering circulation (Fig. 4A–C), whereas the other was descended from a cell marked in the posterior ICM. Unlike our blastomere labeling studies reported above, we did not observe labeled thrombocytes amongst the photoconverted progeny, possibly because these cells no longer share lineages with erythrocytes at this later time. Taken together, these two different marking schemes demonstrate that neutrophils, as well as thrombocytes, originate from progenitors in the ventral gastrula that are robustly proliferative, share lineages with primitive erythrocytes and can remain multipotent rather late in development.
Figure 4.
Not all blood is committed to a unipotential lineage at 26 hours. (A–E) Single circulating blood cells give rise to neutrophil and erythrocyte progeny. (A–C) A newly photoconverted blood cell (arrow), within a ventral gastrula-derived clone. (A) Red wavelength, (B) green wavelength (note other blood cells) and (C) composite image. (D–E) The resulting red-labeled clone at 48 hours included (D) circulating erythrocytes and (E) 2 neutrophils attached to the lumen of a blood vessel. (F–I) Single macrophage cells give rise to macrophage progeny. (F–H) High magnification view of an individual macrophage (arrow) in the process of being photoconverted from green (F) to red (G). Nearby, another green-labeled macrophage (upper left panel) is preparing to divide. (H) Low magnification view of the entire Kaede clone showing the newly photoconverted macrophage (arrow) among other green-labeled macrophages and out of focus endoderm and endothelium. (I) The resulting red-labeled clone at 48 hours was solely 2 macrophages near a blood vessel. Another green-labeled macrophage is out of focus.
In notable contrast to the ventrally-derived blood, the dorsally-derived blood gave rise to only macrophages, as assayed by both morphology and in situ hybridization of labeled cells (Fig. 2I, Fig. 3A and Table 2). Hence no other primitive blood type appears to derive from the RBI (Fig. 2E). Macrophages divided infrequently relative to the erythrocyte lineage and Kaede photoconversion of individual macrophage cells at 26 hours (Fig. 4F–H) either showed that the cell did not divide further (n=2) or gave rise to 2 or 3 red-labeled macrophage progeny the next day usually located in the mesenchyme outside the circulatory system (Fig. 4I; n=7).
Blood cells derive from a small pool of gastrula progenitors
Whereas blood cells are abundant at one day of development, they appear to arise from a relatively small founder population in the blastula based on their infrequency in our entire fate map data set. An estimate of the founder population size for any cell type can be calculated by comparing the number of progeny derived from a single precursor cell labeled during fate map analysis to the total number of that cell type found in the embryo at a particular stage. For example, in our experiments, a single dorsal blood progenitor labeled at the mid-blastula stage will give rise to an average of 6±2 macrophage progeny by 26 hours. By counting the total number of cells that express the macrophage marker lcp1 in an embryo, we observed a total population of 151±37 macrophage cells present at 26 hours. These numbers allow us to estimate that the number of dorsal blood progenitors at the mid- to late-blastula stage to be in the range of 14 to 47 cells with the median being 31 cells. For the ventral blood progenitors, we counted cells expressing hbbe2 and mpx at 26 hours, arriving at estimates of 594±96 erythrocytes and 67±2 neutrophils. (This ratio, about 9 erythrocytes to 1 neutrophil, was equivalent to the 9:1 ratio found in our clonal analysis reported above.) Using the total cell counts of erythrocytes and neutrophils considered together, compared to the clonal expansion size of a single, ventral blood progenitor giving rise to 26±7 circulating cells at 26 hours, we calculated a founder population size in the range of 17 to 40 cells with a median of 29 cells for the ventral population.
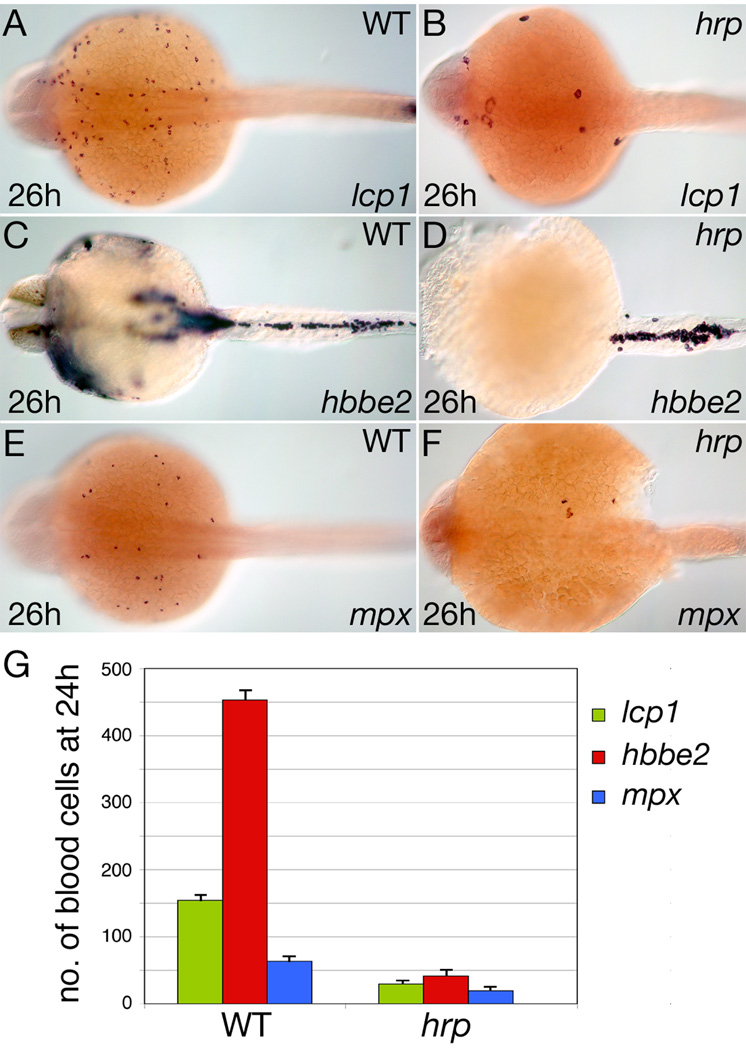
As an independent test of these calculations, we checked the numbers of blood cells produced in the cell cycle defective harpy (hrp) mutant (Kane et al., 1996), which we have confirmed by complementation and sequence analysis (our unpublished data) to harbor a mutation in the zebrafish homologue of emi1 (Zhang et al., 2008). At the mid-gastrula stage, cells in the hrp mutant begin to endocycle, replicating their DNA each Sphase, but do not undergo further cell divisions. Despite this defect, we have confirmed by lineage tracing analysis that cells in the mutant survive and eventually differentiate including within the blood lineage (R.M.W. and D.A.K. unpublished). In 26 hour hrp mutants, we found cells that expressed the macrophage marker lcp1 (Fig. 5B), the erythrocyte marker hbbe2 (Fig. 5D), and the neutrophil marker mpx (Fig. 5F). In general, the mutant cells labeled by these markers resembled larger versions of their wild type counterparts (Fig. 5A, C, E).
Figure 5.
Blood formation in the cell cycle arrested mutant harpy (hrp). (A, B) Macrophage cells, (C, D) Erythrocyte cells, and (E, F) Neutrophil cells in wild type and hrp siblings. Note the super sized cells in the hrp mutant. (G) The approximate number of blood cells found at 24 hours. Error bars show SEM. For each blood type, a total of 10 or more wild-type and mutant embryos were counted.
In Fig. 5A–F, embryos are shown at 26 hours. However, because the mutants do not start normal circulation—and sometimes begin to necrose—we fixed embryos slightly earlier for cell counts. In all cases at 24 hours (Fig. 5G), hrp mutant blood cells were fewer in number than wild type blood cells: Macrophages were reduced from about 154±4 to about 30±2 cells, a 5X reduction. Erythrocytes were reduced from 453±9 to about 41±2 cells, an 11X reduction, and neutrophils were reduced from about 64±3 to about 19±1 cells, a 3X reduction. This number for macrophages is very similar to the above calculated from counts and clonal expansion estimates, confirming our estimate of about 31 dorsal blood progenitors. The counts of erythrocytes and neutrophils considered together, is 2X higher than our estimate above from clonal analyses, of about 29 ventral gastrula founders. However, because our fate map clones were established near the onset of gastrulation and the cell cycle arrest in hrp mutants happens during mid-gastrulation, we assume that this difference is due to a round of mitotic activity normally occurring between these two developmental stages. This assumption is in line with our observation that ventral blood progenitors display more robust division rates than their dorsal counterparts.
Discussion
Primitive hematopoietic progenitors derive from hierarchical lineages
During definitive hematopoiesis in mammals, fates appear in a chronological and hierarchical order (Graf, 2008; Orkin and Zon, 2008; Reya et al., 2001). We observe a similar phenomena during development of the primitive hematopoietic lineage of zebrafish, summarized in Fig. 6A. Based upon the types and timing of restrictions observed within our blood-containing clones, the earliest hierarchical fate distinction that occurs is between a pronephros fate versus a blood/endothelial fate. This segregation of pronephros-only lineages occurs in ventrally located cells between the mid-blastula stage and the onset of gastrulation and is evidenced by the fact that many blood and endothelium-containing clones initiated at mid-blastula stages also contained pronephros cells whereas clones initiated at gastrula stages were never observed to share lineages with pronephric tissues (compare rows 4–9 in Table 1a and Table 1b). Interestingly, this observation correlates with previous studies suggesting that the hematopoietic progenitors of many vertebrate embryos derive from mesoderm that first activates a kidney program of development (Davidson and Zon, 2004). While many of the mid-blastula established clones shared lineages with other mesodermal tissues or even endodermal tissues (Table 1a), we did not observe hematopoietic-containing clones to include heart cells even though their fate map territories overlap (Keegan et al., 2004; Kimmel et al., 1990; Stainier et al., 1993; Warga and Nüsslein-Volhard, 1999).
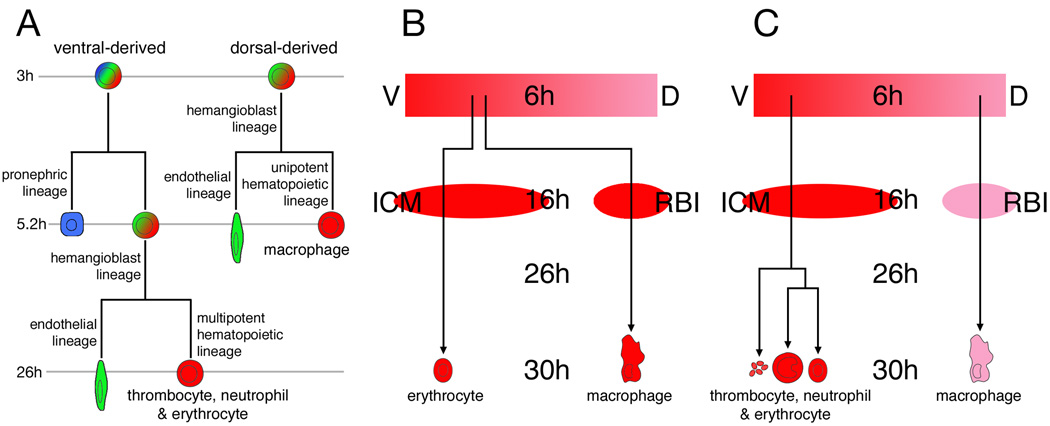
Figure 6.
Ontogeny of zebrafish embryonic blood. (A) Hierarchy of fates occurring in the dorsal and ventral hematopoietic lineages. Timeline depicts when restrictions occurred in our clonal analysis. (B) The previous view of primitive blood development. (C) A revised view based on findings described in this work. The lineage tree for the ventral blood progenitors is purely hypothetical. Abbreviations: D, dorsal; ICM, intermediate cell mass; RBI, rostral blood island; V, ventral.
A second hierarchical distinction presents itself sometime at the onset of gastrulation, separating blood and endothelial fates. Notably, these observations correlate with clonal studies showing the existence of cells called hemangioblasts in the mammalian gastrula that can generate both erythroid and vascular cells, at least in vitro (Huber et al., 2004; Kennedy et al., 2007; Tober et al., 2007), which are largely absent by the time of blood island development (Yokomizo et al., 2007). Cells fitting the description of hemangioblasts or hemangioblast precursors (i.e. those cells giving rise to endothelium, blood and one other fate) constituted the majority (62%, rows 2, 6–7, 11–12;Table 1a) of the blood producing clones (rows 2–3, 6–9, 11–14;Table 1a) at the mid-blastula stage. By the late blastula/early gastrula stage, the numbers of such cells (rows 2, 11–12;Table 1b) in our data appear to drop by more than a third of the numbers observed at mid-blastula (38% of the total blood producing clones; rows 2–3, 11–13;Table 1b) whereas the percentage of blood-only producing clones is now the majority.
A previous study by Vogeli et al. (2006), reported the inability to consistently find evidence for a hemangioblast or hemangioblast precursor before the onset of gastrulation and concluded that erythrocytes and endothelial cells rarely derived together (even with other fates) until the onset of gastrulation. One interpretation from Vogeli et al. would be that a hemangioblast represented a new entity that arises de novo at the onset of gastrulation. Our fate map data from various different stages of development, as described above and summarized in Table 1a and Table 1b, do not support this interpretation. While we do not fully understand the reason for the differences in our study and that of Vogeli et al., we interpret our observations to be consistent with the hypothesis that the embryonic blood cell lineages of zebrafish are derived in a progressively restricted manner (Fig. 6A) similar, at least in form, to the hierarchical lineage that is thought to give rise to definitive blood cell types.
Erythrocytes and macrophages are derived from distinct regions of the gastrula
In the original zebrafish fate map papers, blood was classified as being derived exclusively from the ventral side of the gastrula, however the designation of "blood" used by these authors appears to have been applied mainly, if not solely, to primitive erythrocytes (Kimmel et al., 1990; Warga and Nüsslein-Volhard, 1999). Subsequent blood fate map papers made a distinction between erythrocytes and macrophages, but also reported that zebrafish macrophages derived from the ventral side of the embryo, sometimes even from a progenitor that also gave rise to erythrocytes (Herbomel et al., 1999). Thus the current state of knowledge from the literature (summarized in Fig. 6B) was that a common progenitor cell on the ventral side of the zebrafish gastrula gives rise to progeny potent for either macrophage or erythrocyte fate depending upon which blood island it populates or which transcription factors it expresses (Galloway et al., 2005; Rhodes et al., 2005). We note that a common progenitor has also been reported to exist during primitive hematopoiesis in mammals (Kennedy et al., 2007; Palis et al., 1999).
In this present study, we now show that not all embryonic blood of the zebrafish derives from a common gastrula location. Rather like the recently revised fate map of Xenopus (Lane and Sheets, 2006), blood originates around the entire circumference of the gastrula-stage embryo, excepting the dorsal midline (Fig. 6C). In agreement with previous reports, we show that erythrocytes derive from the ventral half of the gastrula (Kimmel et al., 1990; Vogeli et al., 2006; Warga and Nüsslein-Volhard, 1999) and later the ICM (Detrich et al., 1995; Long et al., 1997). However, in contrast to previous reports (Herbomel et al., 1999), our data shows that macrophages derive exclusively from the dorsal half of the gastrula. Moreover, we never observed a clone labeled at the mid-blastula or gastrula stages that gave rise to both erythrocytes and macrophages. We suggest that the different results obtained between this and previous studies may be due to the time at which individual cells were labeled. Previous work to fate map the macrophage lineage (Herbomel et al., 1999) was based on cell labeling at the 16-cell stage and correlating cell fates in the 1 day embryo. As reported by a number of studies, there is no fixed relation to dorsoventral position at the 16-cell stage, nor correlation to particular fates due to the highly variable patterns of cell division that occur after the 5th cleavage (Kimmel and Law, 1985a; Kimmel and Warga, 1987). In addition, clones established at the 16-cell stage are extremely large and scatter widely between unlabeled cells due to the cell movements that occur during early epiboly (Kimmel and Law, 1985b; Warga and Kimmel, 1990; Wilson et al., 1995). Because we labeled cells around the 2K–cell stage, our clones were small (usually no more than 4 cells) therefore it was easier to verify the actual dorsoventral position of the clone at the gastrula stage. In summary, our data indicates that the identity of a macrophage versus an erythrocyte producing lineage can be accurately predicted solely upon its dorsoventral position within the mid-blastula/early gastrula stage embryo. Furthermore, because dorsoventral position in the gastrula correlates approximately with later anteroposterior position in the embryo (Warga and Kimmel, 1990), our finding that primitive macrophages derive from more dorsally located cells concurs with their later association in a more anterior blood island relative to the primitive erythrocyte population.
Primitive neutrophils, thrombocytes and erythrocytes share a common lineage
By intra-cellular labeling of early blastomeres, we found that neutrophils and thrombocytes derived from the same lineages as primitive erythrocytes. These results demonstrate that some hematopoietic progenitors are multipotent rather than unipotent during the initial primitive wave. The multipotent progenitors are originally located on the ventral side of the gastrula-stage embryo from which they later become positioned along the length of most of the ICM before entering circulation. In the case of zebrafish thrombocytes, our data corroborate a recent report showing that the mammalian source of this cell type in the embryonic mouse frequently derives from lineages that also include primitive erythrocytes (Tober et al., 2007). In this study, we additionally activated the Kaede molecule within individual blood cells at 26 hours to support the surprising finding that a population of zebrafish neutrophils, a white blood cell, could also be generated after the first day of development, from what appear to be primitively-derived red blood cells, namely circulating erythrocytes. Alternatively, this last result could also be interpreted as showing that a sub-population of primitive blood, which can not be distinguished superficially from an erythrocyte, must retain multipotency relatively late in development, even up to the beginning of definitive blood formation.
We note here that neutrophils have also been reported to be derived from the primitive macrophage lineage by 36 hours (Le Guyader et al., 2007). Like its mammalian counterpart, the zebrafish neutrophil possesses myeloperoxidase granules, and a number of studies agree that expression of myeloperoxidase (mpx) reliably identifies embryonic neutrophil cells (Bennett et al., 2001; Hall et al., 2007; Lieschke et al., 2001; Mathias et al., 2006; Meijer et al., 2007). As our studies were concentrated upon the primitive blood lineages and therefore did not continue far beyond the second day of development, we cannot be sure whether the cell types identified as neutrophils by Le Guyader et al. (2007) are in fact the same population of cells that we identified as embryonic neutrophils. However, when checked by in situ hybridization at 36 hours, the cells we had identified as macrophages and macrophage progeny expressed the macrophage gene lcp (99%), but never expressed the neutrophil-specific gene mpx (0%), whereas those cells that we had identified as neutrophils almost always expressed the mpx gene (93%; Table 2). Therefore, within the time frame of primitive hematopoiesis, we find no evidence for a lineal relationship between macrophages and neutrophils.
In summary, our data shows that the ventral gastrula produces some 30 or so multipotent progenitor cells that have the capacity, perhaps due in part to their rapid division rate, to generate not only all the primitive erythrocytes, but also the earliest circulating thrombocytes and neutrophils. The dorsal gastrula likewise produces about an equal number of 30 progenitor cells, however these cells divide infrequently and, in terms of a primitive blood cell fate, solely generate macrophages. Hence the ventral gastrula is not the sole provider of all blood lineages as had been previously thought, but rather there appears to be two different mechanisms that correspond to both ventral and dorsal positions within the early gastrula for producing the primitive blood of zebrafish. In the future, it will be important to determine the molecular mechanisms governing these separate unipotent and multipotent programs as well as the precise relationships between different blood types in both the primitive and definitive blood populations.
Acknowledgements
We thank Dr. Rachel Müeller for comments on earlier versions of this manuscript and useful discussions, as well as members of the Western Michigan University and University of Chicago biology communities for helpful input. We thank Josh Chetta and Robert Risley for expert fish care. This work was supported by funds from Western Michigan University and NIH grants R01-DK68286 and R01-GM067714 to R.K.H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci. 2002;99:12651–12656. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baron MH. Embryonic origins of mammalian hematopoiesis. Exp Hematol. 2003;31:1160–1169. doi: 10.1016/j.exphem.2003.08.019. [DOI] [PubMed] [Google Scholar]
- Bennett CM, Kanki JP, Rhodes J, Liu TX, Paw BH, Kieran MW, Langenau DM, Delahaye-Brown A, Zon LI, Fleming MD, Look AT. Myelopoiesis in the zebrafish, Danio rerio. Blood. 2001;98:643–651. doi: 10.1182/blood.v98.3.643. [DOI] [PubMed] [Google Scholar]
- Berman JN, Kanki JP, Look AT. Zebrafish as a model for myelopoiesis during embryogenesis. Exp Hematol. 2005;33:997–1006. doi: 10.1016/j.exphem.2005.06.010. [DOI] [PubMed] [Google Scholar]
- Bertrand JY, Kim AD, Teng S, Traver D. CD41+ cmyb+ precursors colonize the zebrafish pronephros by a novel migration route to initiate adult hematopoiesis. Development. 2008;135:1853–1862. doi: 10.1242/dev.015297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand JY, Kim AD, Violette EP, Stachura DL, Cisson JL, Traver D. Definitive hematopoiesis initiates through a committed erythromyeloid progenitor in the zebrafish embryo. Development. 2007;134:4147–4156. doi: 10.1242/dev.012385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlie A, Hersey C, Oates AC, Paw BH, Falick AM, Witkowska HE, Flint J, Higgs D, Jessen J, Bahary N, et al. Characterization of embryonic globin genes of the zebrafish. Dev Biol. 2003;255:48–61. doi: 10.1016/s0012-1606(02)00041-6. [DOI] [PubMed] [Google Scholar]
- Davidson AJ, Zon LI. The ‘definitive’ (and ‘primitive’) guide to zebrafish hematopoiesis. Oncogene. 2004;23:7233–7246. doi: 10.1038/sj.onc.1207943. [DOI] [PubMed] [Google Scholar]
- Detrich HWr, Kieran MW, Chan FY, Barone LM, Yee K, Rundstadler JA, Pratt SJ, Ransom D, Zon LI. Intraembryonic hematopoietic cell migration during vertebrate development. Proc Natl Acad Sci. 1995;92:10713–10717. doi: 10.1073/pnas.92.23.10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway JL, Wingert RA, Thisse C, Thisse B, Zon LI. Loss of gata1 but not gata2 converts erythropoiesis to myelopoiesis in zebrafish embryos. Dev Cell. 2005;8:109–116. doi: 10.1016/j.devcel.2004.12.001. [DOI] [PubMed] [Google Scholar]
- Graf T. Immunology: Blood lines redrawn. Nature. 2008;452:702–703. doi: 10.1038/452702a. [DOI] [PubMed] [Google Scholar]
- Gregory M, Jagadeeswaran P. Selective labeling of zebrafish thrombocytes: quantitation of thrombocyte function and detection during development. Blood Cells Mol Dis. 2002;28:418–427. doi: 10.1006/bcmd.2002.0527. [DOI] [PubMed] [Google Scholar]
- Hall C, Flores MV, Storm T, Crosier K, Crosier P. The zebrafish lysozyme C promoter drives myeloid-specific expression in transgenic fish. BMC Dev Biol. 2007;7:42. doi: 10.1186/1471-213X-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbomel P, Thisse B, Thisse C. Ontogeny and behavior of early macrophages in the zebrafish embryo. Development. 1999;126:3735–3745. doi: 10.1242/dev.126.17.3735. [DOI] [PubMed] [Google Scholar]
- Herbomel P, Thisse B, Thisse C. Zebrafish early macrophages colonize cephalic mesenchyme and developing brain, retina, and epidermis through a M-CSF receptor-dependent invasive process. Dev Biol. 2001;238:274–288. doi: 10.1006/dbio.2001.0393. [DOI] [PubMed] [Google Scholar]
- Hsu K, Traver D, Kutok JL, Hagen A, Liu TX, Paw BH, Rhodes J, Berman JN, Zon LI, Kanki JP, Look AT. The pu.1 promoter drives myeloid gene expression in zebrafish. Blood. 2004;104:1291–1297. doi: 10.1182/blood-2003-09-3105. [DOI] [PubMed] [Google Scholar]
- Huber TL, Kouskoff V, Fehling HJ, Palis J, Keller G. Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature. 2004;432:625–630. doi: 10.1038/nature03122. [DOI] [PubMed] [Google Scholar]
- Jowett T, Yan YL. Double fluorescent in situ hybridization to zebrafish embryos. Trends Genet. 1996;12:387–389. doi: 10.1016/s0168-9525(96)90091-8. [DOI] [PubMed] [Google Scholar]
- Kane DA, Maischein H-M, Brand M, Eeden FJMv, Furutani-Seiki M, Granato M, Haffter P, Hammerschmidt M, Heisenberg C-P, Jiang Y-J, et al. The zebrafish early arrest mutants. Development. 1996;123:57–66. doi: 10.1242/dev.123.1.57. [DOI] [PubMed] [Google Scholar]
- Keegan BR, Meyer D, Yelon D. Organization of cardiac chamber progenitors in the zebrafish blastula. Development. 2004;131:3081–3091. doi: 10.1242/dev.01185. [DOI] [PubMed] [Google Scholar]
- Keller G, Lacaud G, Robertson S. Development of the hematopoietic system in the mouse. Exp Hematol. 1999;27:777–787. doi: 10.1016/s0301-472x(99)00024-7. [DOI] [PubMed] [Google Scholar]
- Kennedy M, D'Souza SL, Lynch-Kattman M, Schwantz S, Keller G. Development of the hemangioblast defines the onset of hematopoiesis in human ES cell differentiation cultures. Blood. 2007;109:2679–2687. doi: 10.1182/blood-2006-09-047704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Law RD. Cell lineage of zebrafish blastomeres: I. cleavage pattern and cytoplasmic bridges between cells. Dev Biol. 1985a;108:78–85. doi: 10.1016/0012-1606(85)90010-7. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Law RD. Cell lineage of zebrafish blastomeres: III. clonal analyses of the blastula and gastrula stages. Dev Biol. 1985b;108:94–101. doi: 10.1016/0012-1606(85)90012-0. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Warga RM. Indeterminate cell lineage of the zebrafish embryo. Dev Biol. 1987;124:269–280. doi: 10.1016/0012-1606(87)90478-7. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Warga RM, Schilling TF. Origin and organization of the zebrafish fate map. Development. 1990;108:581–594. doi: 10.1242/dev.108.4.581. [DOI] [PubMed] [Google Scholar]
- Lacaud G, Robertson S, Palis J, Kennedy M, Keller G. Regulation of hemangioblast development. Ann N Y Acad Sci. 2001;938:96–108. doi: 10.1111/j.1749-6632.2001.tb03578.x. [DOI] [PubMed] [Google Scholar]
- Lane MC, Sheets MD. Heading in a new direction: implications of the revised fate map for understanding Xenopus laevis development. Dev Biol. 2006;296:12–28. doi: 10.1016/j.ydbio.2006.04.447. [DOI] [PubMed] [Google Scholar]
- Le Guyader D, Redd MJ, Colucci-Guyon E, Murayama E, Kissa K, Briolat V, Mordelet E, Zapata A, Shinomiya H, Herbomel P. Origins and unconventional behavior of neutrophils in developing zebrafish. Blood. 2007;111:132–141. doi: 10.1182/blood-2007-06-095398. [DOI] [PubMed] [Google Scholar]
- Liao EC, Paw BH, Oates AC, Pratt SJ, Postlethwait JH, Zon LI. SCL/Tal-1 transcription factor acts downstream of cloche to specify hematopoietic and vascular progenitors in zebrafish. Genes Dev. 1998;12:621–626. doi: 10.1101/gad.12.5.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao EC, Trede NS, Ransom D, Zapata A, Kieran M, Zon LI. Non-cell autonomous requirement for the bloodless gene in primitive hematopoiesis of zebrafish. Development. 2002;129:649–659. doi: 10.1242/dev.129.3.649. [DOI] [PubMed] [Google Scholar]
- Lieschke GJ, Oates AC, Crowhurst MO, Ward AC, Layton JE. Morphologic and functional characterization of granulocytes and macrophages in embryonic and adult zebrafish. Blood. 2001;98:3087–3096. doi: 10.1182/blood.v98.10.3087. [DOI] [PubMed] [Google Scholar]
- Lieschke GJ, Oates AC, Paw BH, Thompson MA, Hall NE, Ward AC, Ho RK, Zon LI, Layton JE. Zebrafish spi-1 (pu.1) marks a site of myeloid development independent of primitive erythropoiesis: implications for axial patterning. Dev Biol. 2002;246:274–295. doi: 10.1006/dbio.2002.0657. [DOI] [PubMed] [Google Scholar]
- Lin HF, Traver D, Zhu H, Dooley K, Paw BH, Zon LI, Handin RI. Analysis of thrombocyte development in CD41-GFP transgenic zebrafish. Blood. 2005;106:3803–3810. doi: 10.1182/blood-2005-01-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q, Meng A, Wang H, Jessen JR, Farrell MJ, Lin S. GATA-1 expression pattern can be recapitulated in living transgenic zebrafish using GFP reporter gene. Development. 1997;124:4105–4111. doi: 10.1242/dev.124.20.4105. [DOI] [PubMed] [Google Scholar]
- Majumdar A, Lun K, Brand M, Drummond IA. Zebrafish no isthmus reveals a role for pax2.1 in tubule differentiation and patterning events in the pronephric primordia. Development. 2000;127:2089–2098. doi: 10.1242/dev.127.10.2089. [DOI] [PubMed] [Google Scholar]
- Mathias JR, Perrin BJ, Liu TX, Kanki J, Look AT, Huttenlocher A. Resolution of inflammation by retrograde chemotaxis of neutrophils in transgenic zebrafish. J Leukoc Biol. 2006;80:1281–1288. doi: 10.1189/jlb.0506346. [DOI] [PubMed] [Google Scholar]
- Meijer AH, van der Sar AM, Cunha C, Lamers GEM, Laplante MA, Kikuta H, Bitter W, Becker TS, Spaink HP. Identification and real-time imaging of a myc-expressing neutrophil population involved in inflammation and mycobacterial granuloma formation in zebrafish. Dev Comp Immunol. 2007;32:36–49. doi: 10.1016/j.dci.2007.04.003. [DOI] [PubMed] [Google Scholar]
- Melby AE, Warga RM, Kimmel CB. Specification of cell fates at the dorsal margin of the zebrafish gastrula. Development. 1996;122:2225–2237. doi: 10.1242/dev.122.7.2225. [DOI] [PubMed] [Google Scholar]
- Murayama E, Kissa K, Zapata A, Mordelet E, Briolat V, Lin HF, Handin RI, Herbomel P. Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity. 2006;25:963–975. doi: 10.1016/j.immuni.2006.10.015. [DOI] [PubMed] [Google Scholar]
- Onnebo SMN, Yoong SHS, Ward AC. Harnessing zebrafish for the study of white blood cell development and its perturbation. Exp Hematology. 2004;32:789–796. doi: 10.1016/j.exphem.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palis J, Robertson S, Kennedy M, Wall C, Keller G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development. 1999;126:5073–5084. doi: 10.1242/dev.126.22.5073. [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Rhodes J, Hagen A, Hsu K, Deng M, Liu TX, Look AT, Kanki JP. Interplay of pu.l and gatal determines myelo-erythroid progenitor cell fate in zebrafish. Dev Cell. 2005;8:97–108. doi: 10.1016/j.devcel.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Rohde LA, Oates AC, Ho RK. A crucial interaction between embryonic red blood cell progenitors and paraxial mesoderm revealed in spadetail embryos. Dev Cell. 2004;7:1–20. doi: 10.1016/j.devcel.2004.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih J, Fraser SE. Distribution of tissue progenitors within the shield region of the zebrafish. Development. 1995;121:2755–2765. doi: 10.1242/dev.121.9.2755. [DOI] [PubMed] [Google Scholar]
- Stainier DYR, Lee RK, Fishman MC. Cardiovascular development in the zebrafish: I. Myocardial fate map and heart tube formation. Development. 1993;119:31–40. doi: 10.1242/dev.119.1.31. [DOI] [PubMed] [Google Scholar]
- Thisse B, Thisse C. Fast Release Clones: A High Throughput Expression Analysis. 2004 ZFIN Direct Data Submission ZDB-PUB-040907-1. [Google Scholar]
- Tober J, Koniski A, McGrath KE, Vemishetti R, Emerson R, de Mesy-Bentley KK, Waugh R, Palis J. The megakaryocyte lineage originates from hemangioblast precursors and is an integral component both of primitive and of definitive hematopoiesis. Blood. 2007;109:1433–1441. doi: 10.1182/blood-2006-06-031898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogeli KM, Jin SW, Martin GR, Stainier DY. A common progenitor for haematopoietic and endothelial lineages in the zebrafish gastrula. Nature. 2006;443:337–339. doi: 10.1038/nature05045. [DOI] [PubMed] [Google Scholar]
- Warga RM, Kimmel CB. Cell movements during epiboly and gastrulation in zebrafish. Development. 1990;108:569–580. doi: 10.1242/dev.108.4.569. [DOI] [PubMed] [Google Scholar]
- Warga RM, Nüsslein-Volhard C. Origin and development of the zebrafish endoderm. Development. 1999;126:827–838. doi: 10.1242/dev.126.4.827. [DOI] [PubMed] [Google Scholar]
- Willett CE, Kawasaki H, Amemiya CT, Lin S, Steiner LA. Ikaros expression as a marker for lymphoid progenitors during zebrafish development. Dev Dyn. 2001;222:694–698. doi: 10.1002/dvdy.1223. [DOI] [PubMed] [Google Scholar]
- Wilson ET, Cretekos CJ, Helde KA. Cell mixing during early epiboly in the zebrafish embryo. Dev Genetics. 1995;17:6–15. doi: 10.1002/dvg.1020170103. [DOI] [PubMed] [Google Scholar]
- Yokomizo T, Takahashi S, Mochizuki N, Kuroha T, Ema M, Wakamatsu A, Shimizu R, Ohneda O, Osato M, Okada H, et al. Characterization of GATA-1(+) hemangioblastic cells in the mouse embryo. EMBO J. 2007;26:184–196. doi: 10.1038/sj.emboj.7601480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Kendrick C, Jülich D, Holley SA. Cell cycle progression is required for zebrafish somite morphogenesis but not segmentation clock function. Development. 2008;135:2065–2070. doi: 10.1242/dev.022673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XY, Rodaway AR. SCL-GFP transgenic zebrafish: in vivo imaging of blood and endothelial development and identification of the initial site of definitive hematopoiesis. Dev Biol. 2007;307:179–194. doi: 10.1016/j.ydbio.2007.04.002. [DOI] [PubMed] [Google Scholar]