Abstract
We report for the first time abnormalities in cardiac ventricular electrophysiology in a genetically modified murine model lacking the Scn3b gene (Scn3b−/−). Scn3b−/− mice were created by homologous recombination in embryonic stem (ES) cells. RT-PCR analysis confirmed that Scn3b mRNA was expressed in the ventricles of wild-type (WT) hearts but was absent in the Scn3b−/− hearts. These hearts also showed increased expression levels of Scn1b mRNA in both ventricles and Scn5a mRNA in the right ventricles compared to findings in WT hearts. Scn1b and Scn5a mRNA was expressed at higher levels in the left than in the right ventricles of both Scn3b−/− and WT hearts. Bipolar electrogram and monophasic action potential recordings from the ventricles of Langendorff-perfused Scn3b−/− hearts demonstrated significantly shorter ventricular effective refractory periods (VERPs), larger ratios of electrogram duration obtained at the shortest and longest S1–S2 intervals, and ventricular tachycardias (VTs) induced by programmed electrical stimulation. Such arrhythmogenesis took the form of either monomorphic or polymorphic VT. Despite shorter action potential durations (APDs) in both the endocardium and epicardium, Scn3b−/− hearts showed ΔAPD90 values that remained similar to those shown in WT hearts. The whole-cell patch-clamp technique applied to ventricular myocytes isolated from Scn3b−/− hearts demonstrated reduced peak Na+ current densities and inactivation curves that were shifted in the negative direction, relative to those shown in WT myocytes. Together, these findings associate the lack of the Scn3b gene with arrhythmic tendencies in intact perfused hearts and electrophysiological features similar to those in Scn5a+/− hearts.
Keywords: Sodium channel, Scn3b, Ventricular tachycardia, Brugada syndrome
1. Introduction
Voltage-gated sodium (Na+) channels, critical for the excitation of cardiac muscle (Keating and Sanguinetti, 2001), are formed of pore-forming α-subunits and one or more β-subunits (Isom, 2001). Experiments using the two-electrode voltage clamp in Xenopus oocytes demonstrated that the α-subunits alone can form functional Na+ channels (Noda et al., 1986). Thus far nine α-subunits, Nav1.1–Nav1.9 are each encoded by different genes and have been identified and cloned in mammals. Mutations in SCN5A which encodes for the cardiac α-subunit, Nav1.5, can result in arrhythmic conditions, including the Brugada (BrS) and Long QT syndrome (LQTS) (Tan et al., 2003).
However, the electrophysiological roles of β-subunits in the function of cardiac Na+ channels are less well understood. Six β-subunits have been identified and are encoded by 4 different genes; SCN1B, SCN2B, SCN3B and SCN4B (Isom et al., 1992, 1995a; Kazen-Gillespie et al., 2000; Morgan et al., 2000; Qin et al., 2003; Yu et al., 2003). These genes, which encode for β1, β2, β3 and β4 subunits respectively, are expressed in human tissues including the brain, skeletal muscle and the heart (Candenas et al., 2006; Stevens et al., 2001). Of these, the β1 and β3 subunits have been demonstrated in the transverse tubules, and β2 and β4 subunits in the intercalated disks of murine ventricular myocytes (Maier et al., 2004).
It is possible that the presence of β-subunits can affect both the function and expression of the Na+ channel. Firstly, studies in heterologous expression systems provided by Xenopus oocytes, Chinese hamster ovary (CHO) cells and human embryonic kidney (HEK) cells demonstrated that β-subunits modify gating kinetics, including activation, inactivation and the recovery from inactivation of the Na+ channel (Chen et al., 2002; Meadows et al., 2002; Morgan et al., 2000; Patton et al., 1994). However, these alterations vary with the particular cell types used to express the Na+ channel subunits (Isom et al., 1995b; Morgan et al., 2000; Qu et al., 2001). Secondly, these subunits can also alter the Na+ current amplitude (Fahmi et al., 2001; Lopez-Santiago et al., 2007; Watanabe et al., 2008). Thirdly, β-subunits contain domains that are structurally homologous to the V-set of the Ig superfamily, which includes cell adhesion molecules (CAMs) (Isom et al., 1995a). This permits cell–cell interaction and adhesion in cells expressing these subunits (Malhotra et al., 2000). Finally, the β-subunits have been shown to regulate the expression density of the Na+ channel in the plasma membrane. Experiments using CHO and 1610 cell lines have demonstrated a 2- to 4-fold greater Na+ channel expression when α-subunits were co-expressed with β1 subunits, than when these were expressed alone (Isom et al., 1995b).
The presence or absence of β-subunits has recently been associated with electrophysiological abnormalities in cardiac tissue. Surface electrocardiogram experiments carried out on genetically modified mice, lacking the Scn1b gene, have demonstrated a LQT-like phenotype, characterised by a prolonged QT interval (Lopez-Santiago et al., 2007). However, only two clinical reports thus far have associated mutations in the gene SCN4B with LQTS (Medeiros-Domingo et al., 2007) and SCN1B, encoding both β1 and β1B subunits with BrS (Watanabe et al., 2008). In the present study, we assessed, for the first time, electrophysiological properties in the ventricles of Scn3b−/− hearts compared to those shown in WT hearts. Firstly, Scn3b−/− mice were created using homologous recombination in embryonic stem (ES) cells. The phenotype of the resulting Scn3b−/− mice, including body weight and temperature, appeared similar to that of WT mice. Secondly, ventricles of such mice showed altered expression levels of Scn3b, Scn1b and Scn5a mRNA. Thirdly, studies in Langendorff-perfused Scn3b−/− hearts demonstrated arrhythmogenic features. Finally, these corresponded to changes in the biophysical properties of the Na+ channel, with features similar to those previously reported in Scn5a+/− hearts (Papadatos et al., 2002; Stokoe et al., 2007a).
2. Methods
2.1. Generation of Scn3b-deficient (−/−) mice
PCR amplification was used to generate homologous arms from the 129SvEv mouse genome, using primers designed to amplify a 5′ arm spanning 2.0 kb of the Scn3b locus terminating downstream of the third coding exon splice acceptor (5′armF; tttgcggccgCAGCCTTGTATGAACCCAGGGTCTTTC and 5′armR; tttactagtCACCTCCTGGTGGCCATTTCGATACTC) and a 3′ arm spanning 4.1 kb of the Scn3b locus commencing just upstream of the third coding exon splice donor (3′armF; tttggcgcgccGGTAAGCCTGAGGCCTGTAGTCTCTTC and 3′armR; tttggccggccGTGGACTTTAGTCCCATGTCCTCATTG).
The arms were cloned into the plasmid pTK5IBLMNL (Paradigm Therapeutics Ltd, Cambridge, U.K.) using the restriction sites incorporated in the arm primers, such that the 5′ and 3′ arms flank an IRES-fronted LacZ reporter gene followed by a loxP-flanked PGKNeopA selectable marker. After homologous recombination, 191 bp of the 226 bp third coding exon is replaced by the IRESLacZ/PGKneo cassette. Putative targeted embryonic stem (ES) cells were identified by PCR using an external 5′ screening primer (5′scr: CTGACATCTTCTCAGCAGATAACTGAC) in combination with a vector specific primer (DR2; ATCATGGCCCTACCATGCGCTAAACAC). Successfully targeted ES cells were further confirmed by Southern blot of AflII and BclI digested genomic DNA, using an external probe amplified from a genomic region downstream of the end of the 3′ arm (using the primers 3′prF; AGATGTCCAGCGATACTGTTGAGGCAG and 3′prR; TGTGAGTCTTATTGGAGGTACAGTGTG), indicating that the integration had occurred legitimately.
Correctly targeted 129 SvEv ES cells, containing the Scn3b knockout (KO) allele were injected into host blastocysts as previously described (Bradley et al., 1984), generating male chimeras which were subsequently mated with 129 SvEv females. Pups from these crosses were screened with the original target screening PCR (using 5′scr and vector specific primer, DR2), to identify heterozygote animals and in further generations by a multiplex PCR designed to amplify a 204 bp region specific to the WT allele and a 334 bp region specific to the Scn3b KO allele. This allowed differentiation of each of the three possible genotypes (using primers hetF, GTCGTCTGCAGTGGAATGGGAGCAAAG; hetR, TGAAGAGACTACAGGCCTCAGGCTTAC; and Asc306, AATGGCCGCTTTTCTGGATTCATCGAC). All genotypes were observed at the expected Mendelian ratios. Routinely homozygous matings were established to produce the experimental cohorts and 129SvEv stock used as the WT controls. Male and female offspring of WT and Scn3b−/− mice were randomly selected for use in all experiments. Mice obtained were kept in cages at a room temperature of 21 ± 1 °C in an animal facility with 12-hour light/dark cycles. The mice had free access to sterile rodent chow and water.
A series of tests were used to assess baseline metabolic and behavioural parameters and were performed on cohorts of WT and Scn3b−/− mice. Each cohort typically consisted of 5–8 mice of either sex at the age of 3–4 months. The metabolic tests included measuring the body weight and temperature. Each mouse was weighed monthly for up to 3 months and body temperature was measured using a rectal probe (World Precision Instruments, Florida, USA). The behavioural tests measured visual tracking, locomotor activity, touch sensitivity and anxiety. Firstly, visual tracking was assessed using an optokinetic drum apparatus (Paradigm Therapeutics, Cambridge, U.K.). Secondly, locomotor activity was measured as the distance travelled, in one hour, using the LABORAS system (Metris, Hoofddorp, The Netherlands). Finally, the percentage time spent in the open and closed arms of an elevated plus-maze apparatus was used to assess aversion to open spaces in the mice (Lister, 1987). All procedures conformed to the UK Animals (Scientific Procedures) Act 1986.
Randomly selected male and female mice (age 5–8 months) were killed by cervical dislocation in all RT-PCR and ex vivo experiments (Schedule 1: UK Animals (Scientific Procedures) Act 1986).
2.2. Analysis of sodium channel α- and β-subunit transcripts
To quantify changes in the mRNA expression levels of sodium α- and β-subunits in hearts obtained from WT and Scn3b−/− mice, RT-PCR experiments were performed on an ABI 7500 Fast cycler (Applied Biosystems, Warrington, U.K.). Total RNA was isolated from left atria (LA), right atria (RA), left ventricle (LV) and right ventricle (RV) of WT and Scn3b−/− mice (n = 3 each) using a Qiagen RNAeasy kit. Excised tissues were stored in RNAlater® (Ambion, Warrington, U.K.) to maintain the integrity of the RNA before isolation. The total RNA was reverse transcribed into cDNA using random hexamer primers and a SuperScript III kit (Invitrogen, Paisley, U.K.). Oligos for Scn3b, Scn5a, and Scn1b were FAM/TAMRA labelled (Applied Biosystems). All experiments were performed in triplicate.
The number of the copies of mRNA was calculated from its respective threshold cycle (CT) using a standard curve. Each value was normalized for the expression value of the housekeeper gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and expressed as a percentage of GAPDH expression (i.e. 2ΔCT × 100).
2.3. Preparation of Langendorff-perfused hearts for electrophysiological recordings
The experiments used Langendorff-perfused murine hearts as previously described (Balasubramaniam et al., 2003). Hearts were rapidly excised whilst minimizing contact with the atria and ventricles, then submerged in ice-cold bicarbonate-buffered Krebs–Henseleit solution containing (mM): 119 NaCl, 25 NaHCO3, 4.0 KCl, 1.2 KH2PO4, 1.0 MgCl2, 1.8 CaCl2, 10 glucose and 2.0 sodium-pyruvate (pH 7.4) and bubbled with 95% O2/5% CO2 gas mixture (British Oxygen Company, Manchester, U.K.). Under the ice-cold buffer, excess tissues surrounding the heart were removed, leaving a 2–3 mm section of the aorta. This was cannulated and sealed, using a micro-aneurysm clip (Harvard Apparatus, Edenbridge, U.K.) to a 21-gauge tailor-made cannula. The latter was pre-filled with ice-cold buffer solution using a 1 ml syringe. The preparation was transferred and attached to a Langendorff system, and then retrogradely perfused, using the bicarbonate-buffered Krebs–Henseleit solution described above, warmed to 37 °C via a water jacket and circulator (Techne model C-85 A, Cambridge, U.K.). The warmed perfusate was initially passed through a 200 μm and 5 μm filter membrane (Millipore, Watford, U.K.), before being introduced into the aorta at a constant flow of 2–2.5 ml min−1 using a peristaltic pump (Watson–Marlow Bredel model 505S, Falmouth, Cornwall, U.K.). The cannulated hearts were perfused for 5 min before further testing. Viable, healthy hearts then regained a homogenous pink colouration and spontaneous rhythmic contraction. Hearts that did not demonstrate these features upon perfusion were instantly discarded.
2.4. Bipolar electrogram recording
In all experiments analysing the ventricles of the isolated, perfused murine heart, paired platinum stimulating electrodes (1 mm interpole spacing) were positioned over the epicardial surface of the right ventricle. Ventricular activity was examined by recording from the epicardial surface of the left ventricle using a silver chloride (2 mm tip diameter) recording electrode (Linton Instruments, Harvard Apparatus, U.K.), which was manually positioned. The electrical signals recorded from these hearts resulted in bipolar electrogram (BEG) recordings. The paired platinum stimulating electrodes paced the epicardial surface of the right ventricle and the stimulation used a 2 ms square-wave stimuli at three times excitation threshold (Grass-Telefactor, U.K., Slough, U.K.). These signals were amplified and high-pass filtered for recordings of murine heart (30–1 kHz) using a Gould 2400S amplifier (Gould-Nicolet Technologies, Ilford, Essex, U.K.) and then digitised using an analogue to digital converter (CED 1401plus MKII, Cambridge Electronic Design, Cambridge, U.K.) at a sampling frequency of 5 kHz. All digitised data was then captured and analysed using Spike 2 software (Cambridge Electronic Design).
2.5. Monophasic action potential recording and regular pacing
In addition to ventricular BEGs, monophasic action potentials (MAPs) of the ventricles were also recorded using an established contact-electrode technique (Franz, 1999; Killeen et al., 2007; Knollmann et al., 2001). The stimulating electrodes were positioned on the basal surface of the right ventricular epicardium and used to deliver electrical stimulation, as described above. Each isolated perfused heart underwent a period of regular pacing using 2 ms square-wave stimuli, delivered at three times the excitation threshold, at a basic cycle length (BCL) of 125 ms, for up to 20 min to record both epicardial and endocardial MAPs. Epicardial MAPs were recorded from the basal surface of the left ventricular epicardium using the silver chloride recording electrode. Endocardial MAPs were also obtained, from the left ventricular endocardium using a custom-made endocardial MAP electrode, constructed from two Teflon-coated (0.25 mm diameter) silver wires (99.99%) (Advent Research Materials Ltd. Oxford, U.K.), that were twisted together and galvanically chlorided. This electrode was inserted through a small access window carefully created in the interventricular septum (Casimiro et al., 2001). The tip of the electrode was rotated and positioned against the left ventricular free wall, maintaining stable contact pressure. Signals obtained from the recording electrode were amplified and low-pass filtered between 0.1 Hz and 1.0 kHz using the Gould 2400S amplifier (Gould-Nicolet Technologies) and digitised using the CED 1401plus MKII analogue to digital converter (Cambridge Electronic Design) as described above. Both epicardial and endocardial MAP waveforms were analysed using Spike2 software (Cambridge Electronic Design).
MAPs that were used for analysis, satisfied previously established waveform characteristics including a stable baseline, a rapid upstroke phase with consistent amplitude, a smooth contoured repolarization, a stable duration and 100% repolarization achieved by a full return to the baseline (Stokoe et al., 2007a). These MAP waveforms were used to obtain action potential durations (APDs) at 30% (APD30), 50% (APD50), 70% (APD70) and 90% (APD90) repolarization. Transmural gradients of repolarization (ΔAPD90) were calculated as endocardial APD90 value minus the epicardial APD90 value (Killeen et al., 2007; Thomas et al., 2007).
2.6. Programmed electrical stimulation
Programmed electrical stimulation (PES) was used to assess ventricular arrhythmogenesis in each heart preparation and recorded as BEGs and MAPs (Balasubramaniam et al., 2003; Saumarez and Grace, 2000). The PES protocol first paced the heart for 25 s at the BCL. This was followed by cycles each consisting of an eight stimuli (S1) drive train at a CL of 125 ms followed by a ninth extra-stimulus (S2). The first S1–S2 interval (between the eighth S1 and S2 stimuli) equalled the pacing interval. Successive cycles progressively reduced the S1–S2 interval by 1 ms until the S2 stimulus could no longer evoke a ventricular deflection, at which point, the whole heart preparation reached the ventricular effective refractory period (VERP). VERP was hence defined as the longest S1–S2 interval that could not elicit a ventricular deflection.
From the ventricular BEGs obtained, conduction curves were constructed, using paced electrogram fractionation analysis (PEFA), a procedure applied in clinical practice (Head et al., 2005; Saumarez and Grace, 2000). These curves plotted the response latencies of each S2 extra-stimulus, defined as the time difference between the extra-stimulus and the peak and troughs of the resulting BEG, against the corresponding S1–S2 interval. The electrogram duration (EGD) is characterised as the time difference between the first and last BEG peak and trough. The EGD ratios from corresponding conduction curves were calculated by normalizing the EGD of the BEG obtained from the shortest S1–S2 interval with the EGD of the longest S1–S2 interval (Stokoe et al., 2007a). The initial conduction latency was defined as the time difference from the initial S2 stimulus applied at the S1–S2 interval of 125 ms and the initial deflection of the resulting electrogram response.
2.7. Ventricular myocyte isolation
A solution consisting of (in mM): 120 NaCl, 5.4 KCl, 5 MgSO4, 5.5 sodium-pyruvate, 10 glucose, 20 taurine and 10 HEPES was used as the basic isolation buffer. This was used to make the following solutions. Firstly, an NTA buffer consisted of the isolation buffer and 5 mM nitrilotriacetic acid (NTA). The pH of this buffer was adjusted to 6.95. Secondly, a digestion buffer, which contained the isolation buffer to which 0.25 mM CaCl2, 1 mg ml−1 collagenase type 2 (Worthington, NJ, U.S.A.) and 1 mg ml−1 hyaluronidase (Sigma–Aldrich, Poole, U.K.) was added. Thirdly, a stop buffer was created, which contained the isolation buffer and 2 mg ml−1 bovine serum albumin (Sigma–Aldrich). Fourthly, the isolation buffer to which 0.6 mM CaCl2 was added, was created as the wash buffer. Finally, a storage buffer, similar to the wash buffer but with 1.2 mM CaCl2, was made. All the buffers created, except for the NTA buffer, had their pH adjusted to 7.4.
Hearts of adult mice were rapidly excised and submerged in ice-cold isolation buffer, prior to whole-heart isolation. The hearts were then perfused for 2 min with NTA buffer in a retrograde fashion using a variable speed peristaltic pump at a flow rate of 3.5 ml ms−1 (Autoclude EV045, Essex, U.K.). Before perfusion, the buffer was warmed to 37 °C with a water jacket and circulator (Techne model RB-5 A, Cambridge, U.K.). Following this, the heart was then perfused in the same fashion with the digestion buffer for 12–15 min. Using watchmaker's forceps, ventricular tissue was taken from the whole heart and placed in the stop buffer. After transferring into a 15 ml test tube, gentle trituration was used to isolate single myocytes from the harvested cardiac tissue. After 5 min for the cells to settle, the supernatant was then gently removed. The isolated cells were then washed using wash buffer. Following a further 5 min, the isolated myocytes were resuspended and stored in the storage buffer. After each step after harvesting the cells, isolated myocytes were checked under a light microscope to confirm cell integrity. Live myocytes appeared as smooth and rod-shaped cells.
2.8. INa measurements
The biophysical experiments studied the activation, inactivation and recovery from inactivation in the Na+ channels of WT and Scn3b−/− myocytes. These experiments were carried out at a physiological temperature of 37 °C, using the whole-cell configuration, with an Axopatch 700B amplifier (Molecular Devices, Berkshire, U.K.). Borosilicate glass pipettes (0.86 mm outer diameter; Harvard Apparatus, Kent, U.K) with a resistance of 1.5–2.5 MΩ were used. These pipettes were filled with (in mM): 70.26 CsCl, 24.74 Cs-aspartate, 10 HEPES, 1 Na2ATP, 4 MgATP, 1.37 MgCl2, and 10 Cs-BAPTA, adjusted to pH 7.2 using CsOH. Cells were loaded into a chamber which was mounted on the stage of a Nikon TE2000 Eclipse microscope (Nikon, Kingston, U.K.). These cells were initially perfused with an extracellular bath solution, which contained (in mM): 145 NaCl, 5 KCl, 10 glucose, 10 HEPES, 1 MgCl2 and 1 CaCl2, adjusted to pH 7.4 using NaOH. To achieve optimum voltage control, Na+ currents were recorded in a low [Na+] solution. This solution contained (in mM): 10 NaCl, 140 CsCl, 10 glucose, 10 HEPES, 1 MgCl2 and 1 CaCl2, adjusted to pH 7.4 using CsOH. The currents obtained were filtered at a 10 kHz high frequency cut off and digitised at 100 kHz using pClamp 10 software (Molecular Devices). In each experiment, the series resistance was compensated by at least 70% and each protocol used a holding potential of −100 mV.
Firstly, the voltage dependence of activation was studied by applying depolarizing pulses of 20 ms duration. These pulses were applied over series of voltage steps from −100 mV, in 10 mV increments, to 70 mV. The resulting currents were normalized against the cell capacitance (Cm) and plotted against time. The peak current densities (pA/pF) were calculated by dividing the peak currents amplitude by Cm. The current–voltage relationship was obtained by plotting the values of the peak current density against each corresponding membrane voltage (Vm). The values of peak Na+ conductance (gNa) values were determined from the equation
where INa was the peak current density and the ENa was the [Na+] reversal potential. The ENa was assumed to be 42.6 mV. This was based on the Nernst equation;
where R is the universal gas constant, T is the absolute temperature at 310 K, z is the valence of the ionic species at 1, F is Faraday's constant, [Na+]extracellular = 10 mM and [Na+]intracellular = 2 mM. The gNa values calculated from each myocyte were normalized against the corresponding peak gNa value. When plotted against the test-pulse voltage, the data assumed a sigmoid function, increasing with membrane potential.
Secondly, the voltage dependence of inactivation was examined using pre-pulses of 500 ms duration. These pre-pulses were applied over a voltage range from −120 mV to −20 mV in 10 mV increments. Each pre-pulse was followed by a test pulse to −20 mV at a duration of 10 ms. The peak currents obtained were normalized against the peak current recorded at the pre-pulse of −120 mV. The normalized currents were then plotted against the pre-pulse voltage. The data assumed a sigmoid function, decreasing with membrane potential. Both inactivation and activation data were fitted with a Boltzmann function:
yielding the half-maximal voltage (V1/2) and slope factor (k) values. The y values corresponded to the normalized current values.
Finally, to determine the time course of recovery from inactivation, a protocol consisting of 18 cycles was employed. Each cycle consisted of a 1 s pre-pulse to −20 mV, used to achieve inactivation of the Na+ channels, followed by a return to the voltage of −100 mV to allow recovery of the channels. With each cycle, the duration of recovery increased from 1 to 250 ms. A 50 ms test pulse to −20 mV determined the current from the recovered channels. The currents recorded were normalized against the current obtained from each pre-pulse applied. A bi-exponential function was fitted to the recovery from inactivation data,
where Af and As are the amplitude of the fast and slow inactivating components, and τf and τs are the time constants of the fast and slow components, respectively. The y values correspond to the normalized current values. Data was analysed using Signal 2.16 (Cambridge Electronic Design) and all curve fits were performed using OriginPro 8 (OriginLab Corporation, MA, USA).
2.9. Data analysis and statistical procedures
For experiments in mRNA expression levels, the single-factor analysis of variance for independent samples (ANOVA) test was used to compare the difference between WT and Scn3b−/− tissue samples (Microsoft EXCEL Analysis ToolPak) to show significant changes in expression of the mRNA.
Whole-heart BEG and MAP data were initially imported into Microsoft EXCEL. The ANOVA test was used to compare WT and Scn3b−/− data sets (Microsoft EXCEL Analysis ToolPak). Calculated values of P < 0.05 were considered significant. Results are shown as means ± S.E.M values.
An F-test was used firstly to confirm the normal distribution of the data acquired from the Na+ current recordings (Microsoft EXCEL Analysis ToolPak) made in WT and Scn3b−/− myocytes. Depending on the type of variance, the appropriate form of the Student t test was applied to analyze the differences between two groups of data.
3. Results
3.1. Gene targeting of Scn3b and generation of knockout mice
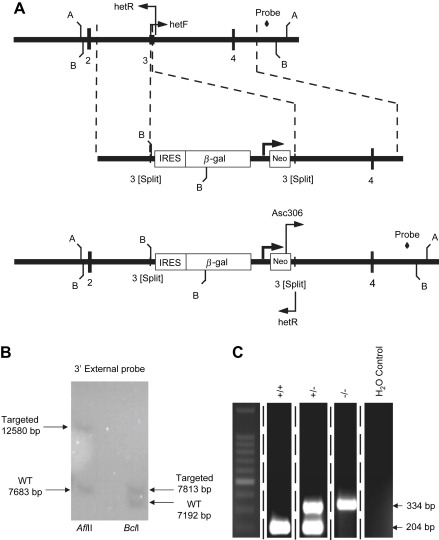
To disrupt the β3 coding region, 191 bp of the 226 bp third exon of the β3 sequence was replaced by the IRESLacZ/pGKneo cassette (Fig. 1A). The 5′ arm of the plasmid used to generate the mutant contained a 2.0 kb fragment of the Scn3b locus terminating downstream of the third coding exon splice acceptor, and a 3′ arm which spanned 4.1 kb of the Scn3b locus commencing just upstream of the third coding exon splice donor. Successfully targeted ES cells were screened by Southern blot using an external 3′ probe (amplified from mouse genomic DNA using 3′prF; AGATGTCCAGCGATACTGTTGAGGCAG and 3′prR; TGTGAGTCTTATTGGAGGTACAGTGTG). Digestion with AflII yielded a 7.7 kb endogenous band and a 12.6 kb bp targeted band as predicted. Similar experiments using BclI resulted in a 7.2 kb endogenous band and the predicted 7.8 kb bp targeted band (Fig. 1B). Homozygous mutant (Scn3b−/−) offspring were identified using PCR screening (Fig. 1C). The primers used (hetF, GTCGTCTGCAGTGGAATGGGAGCAAAG; hetR, TGAAGAGACTACAGGCCTCAGGCTTAC; and Asc306, AATGGCCGCTTTTCTGGATTCATCGAC) resulted in the generation of a 204 bp band from the wild-type (WT) (+/+) allele, and a 334 bp band from the Scn3b−/− (−/−) allele. Both bands were present in the heterozygote (+/−) allele. A water control was used to confirm the specificity of the PCR primers.
Fig. 1.
Targeted deletion of the Scn3b gene. (A) Schematic representation of the Scn3b allele that was targeted for deletion. The top line shows a partial restriction map of the Scn3b locus, indicating the restriction sites for AflII (A) and BclI (B). The centre line represents the targeted vector and the predicted targeted allele after homologous recombination shown in the bottom line. The location of the PCR primers and the probe used for screening the correctly targeted embryonic stem (ES) cells and the mice are shown. IRES; internal ribosome entry site, β-gal; beta-galactosidase gene and Neo; neomycin resistance gene. (B) Southern blot analysis of a successfully targeted ES cell. Digestion with AflII yielded a 7.7 kb endogenous band and a 12.6 kb bp targeted band as predicted. Digestion with BclI resulted in a 7.2 kb endogenous band and the predicted 7.8 kb bp targeted band. (C) PCR analysis identified band sizes of 204 bp in the wild-type (+/+) allele and 334 bp in homozygote knockout (−/−) allele. Both bands were observed in the heterozygote (+/−) allele. A water control was used to assess the specificity of the primers used. All 5 lanes shown were obtained from the same gel.
To assess the phenotype of the Scn3b−/− mice, both metabolic and behavioural tests were used. The body weight and temperature of these mice were similar to those obtained from WT mice (Table 1). Additionally, vision, locomotor activity and anxiety tests showed similar results in both WT and Scn3b−/− mice (Table 1).
Table 1.
Metabolic and behavioural tests applied to WT and Scn3b−/− mice.
| Phenotypic test | WT mice | Scn3b−/− mice | Result |
|---|---|---|---|
| Body temperature (°C) | 37.7 ± 0.1 (n = 35) | 38.09 ± 0.1 (n = 9) | P > 0.05 |
| Body weight at 3 months (g) | |||
| Males | 26.6 ± 0.7 (n = 20) | 26.9 ± 1.2 (n = 5) | P > 0.05 |
| Females | 21.4 ± 0.5 (n = 23) | 21.9 ± 0.8 (n = 4) | P > 0.05 |
| Visual tracking | Present | Present | |
| Distance travelled (m) | 17.69 ± 1.8 (n = 10) | 14.28 ± 2.3 (n = 9) | P > 0.05 |
| Plus Maze | |||
| % time in closed arm | 56.1 ± 4.6 (n = 10) | 52.0 ± 7.1 (n = 10) | P > 0.05 |
| % time in open arm | 2.2 ± 1.0 (n = 10) | 2.2 ± 0.9 (n = 10) | P > 0.05 |
3.2. mRNA expression levels of Na+ channel subunits in murine ventricles
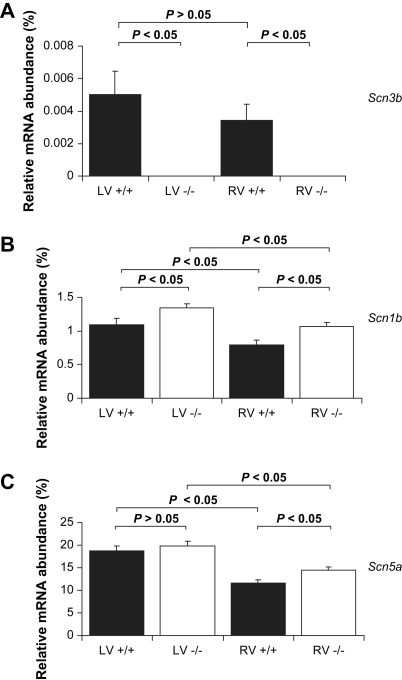
The mRNA expression studies demonstrated that Scn3b mRNA was not present and an increase in Scn1b mRNA in both the left and right ventricles of Scn3b−/− hearts. An increase in the Scn5a mRNA was observed only in the right ventricles of Scn3b−/− hearts. In all cases, the mRNA expression in the left ventricles was consistently higher. Reverse-transcriptase PCR (RT-PCR) was used to investigate mRNA expression levels of sodium (Na+) channel β-subunits Scn3b, Scn1b, and the cardiac Na+ channel α-subunit, Scn5a, in tissue obtained from the right and left ventricles of mouse wild-type (WT) (RV+/+ and LV+/+; n = 3) and Scn3b−/− hearts (RV−/− and LV−/−; n = 3). The number of the copies of mRNA was calculated from its respective threshold cycle (CT) using a standard curve. Each value was normalized to the corresponding expression of the housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
This made comparison between samples possible by correcting for any variation between the quantity of mRNA isolated from the WT and transgenic mouse hearts. Transcript expression was expressed as relative abundances of mRNA expression, calculated as a % of the GAPDH expression (i.e. 2ΔCT × 100). Firstly, the mRNA encoding the β3 subunits, Scn3b, was expressed at statistically similar levels in both the left and right ventricles of WT hearts (Fig. 2A, black column, n = 3, P < 0.05). In contrast, both the ventricles of Scn3b−/− hearts (n = 3), as expected, did not show any detectable expression of Scn3b mRNA.
Fig. 2.
RT-PCR analysis of mRNA expression. The relative abundance of mRNA (normalized and calculated as % of GAPDH expression) of encoding Na+ channel subunit transcripts. The histograms show data from the left and right ventricles (LV and RV respectively) of wild-type (black) and Scn3b−/− (white) mice. (A) mRNA transcripts for Scn3b was present in ventricles of WT (left; LV +/+, right; RV +/+, n = 3) and not detectable in ventricles of Scn3b−/− mice (left; LV −/−, right; LV −/−, n = 3). (B) The expression levels of Scn1b mRNA were significantly higher in both the right and left ventricles of Scn3b−/− hearts than shown in WT hearts (P < 0.05). (C) The Scn5a transcripts were significantly higher in the right but not in the left ventricles of Scn3b−/− hearts. mRNA expression levels of Scn3b, Scn1b and Scn5a were consistently higher in the left than in the right ventricles of both WT and Scn3b−/− hearts (n = 3 each, P < 0.05) with the exception of Scn3b mRNA expression levels in Scn3b−/− hearts.
Secondly, both the left and right ventricles of the Scn3b−/− hearts (Fig. 2B, white column) showed an increase in the expression levels of Scn1b mRNA, relative to corresponding values in WT hearts (P < 0.05). Finally, Fig. 2C confirms the expression of the mRNA encoding for the α-subunit of the Na+ channel, Scn5a, in both ventricles of WT (black columns) and Scn3b−/− (white columns) hearts. The left ventricles of WT and Scn3b−/− hearts demonstrated similar levels of Scn5a mRNA expression (P > 0.05). However, the right ventricles of the Scn3b−/− hearts demonstrated a small but significant increase in the expression of Scn5a mRNA, than that shown in WT hearts (P < 0.05). Thus, mRNA expression levels of Scn1b and Scn5a were always higher in the left than in the right ventricles of WT and Scn3b−/− hearts (n = 3 each, P < 0.05).
3.3. Analysis of BEG recordings during PES
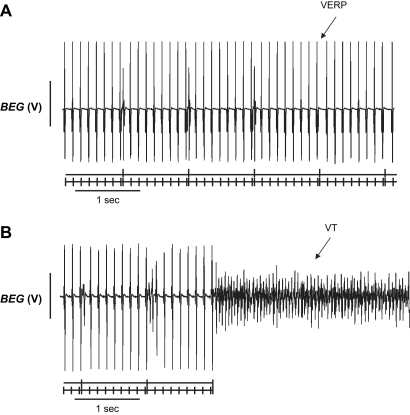
The mRNA expression changes were correlated with a ventricular arrhythmogenic phenotype in Scn3b−/− hearts. This was demonstrated in electrophysiological experiments that assessed ventricular arrhythmogenicity in isolated, Langendorff-perfused WT and Scn3b−/− murine hearts. This was achieved by obtaining bipolar electrogram (BEG) (first series; 6 WT hearts and 7 Scn3b−/− hearts) and monophasic action potential (MAP) (second series; 14 WT hearts and 17 Scn3b−/− hearts, third series; 4 WT hearts and 4 Scn3b−/− hearts) recordings from the left ventricles. BEGs and MAPs were obtained once the preparations had regained a steady state after 10 min following mounting. BEG recordings were obtained during programmed electrical stimulation (PES). This procedure has been used in clinical situations to assess arrhythmogenicity and applied to murine hearts as described on previous occasions (Balasubramaniam et al., 2003; Saumarez and Grace, 2000). Hearts were initially paced at a basic cycle length (BCL) of 125 ms for 25 s. Subsequent cycles used a drive train each consisting of eight stimuli (S1) at the same BCL, followed by a premature extra-stimulus (S2). The initial S2 stimulus was imposed at an S1–S2 interval of 125 ms. With each cycle, this interval was reduced by 1 ms, until either the ventricular effective refractory period (VERP) was reached, at which point the premature S2 did not trigger a response or resulted in ventricular tachycardia (VT).
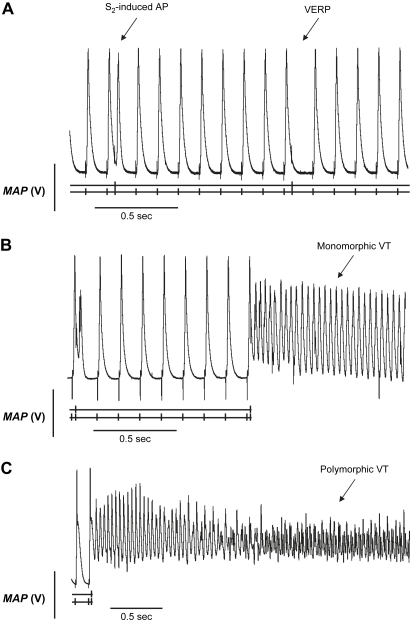
Fig. 3 shows examples of BEG traces obtained during PES. Each trace demonstrates results from the final cycles that corresponded to the shortest S1–S2 intervals. The single vertical marks below each recording indicate the successive S1 stimuli and the double vertical marks, the S2 extra-stimuli. Furthermore, each trace contains stimulus artefacts that occurred at regular CL, followed by their resulting electrogram. In the case of WT hearts (Fig. 3A), shortening of the S1–S2 interval led to an S2 extra-stimulus that did not elicit an electrogram. This S1–S2 interval corresponded to the VERP. In contrast, under such circumstances, a significant proportion of Scn3b−/− hearts showed ventricular arrhythmias (Fig. 3B).
Fig. 3.
BEG recordings of programmed electrical stimulation (PES) from WT and Scn3b−/− hearts. (A) PES of an isolated, WT Langendorff-perfused heart. Ventricular effective refractory period (VERP) was obtained from all WT hearts studied (n = 6). (B) BEG recording showing PES of an isolated, Scn3b−/− Langendorff-perfused heart. PES induced ventricular tachycardia (VT) in 5 out of 7 Scn3b−/− heart preparations.
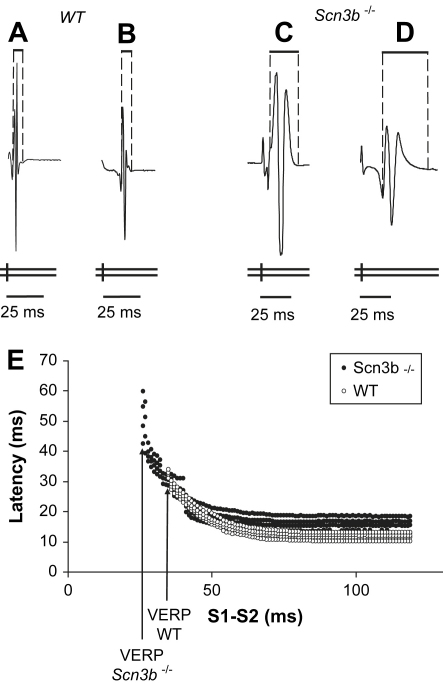
Conduction curves were then derived from the BEG recordings obtained during PES, using paced electrogram fractionation analysis (PEFA), a procedure that has been used to assess clinical risk of ventricular arrhythmogenicity on earlier occasions (Saumarez and Grace, 2000). It has also been previously applied to assess arrhythmogenicity in Kcne1−/− (Balasubramaniam et al., 2003), Scn5a+/ΔKPQ (Head et al., 2005; Stokoe et al., 2007b) and Scn5a+/− (Stokoe et al., 2007a) murine hearts. Fig. 4A–D illustrates S2-induced BEG waveforms at a faster time base than shown in Fig. 3 in order to demonstrate differences in electrogram duration (EGD). This was defined as the time interval between the first and last deflection of each electrogram waveform. These waveforms were obtained during PES of WT (Fig. 4A, B) and Scn3b−/− (Fig. 4C, D) hearts. The waveforms in Fig. 4A, C were induced by the S2 stimuli at the longest S1–S2 intervals that were identical to the BCL, employed at the beginning of the procedure. The duration of these waveforms were defined as the initial EGD. The waveforms in Fig. 4B, D were induced by the S2 stimuli at the shortest S1–S2 intervals that could elicit an electrogram response, at the end of the procedure. The duration of these waveforms were defined as the final EGD. The EGD ratio was calculated as the final EGD divided by the initial EGD. The initial conduction latency was calculated as the time difference from the initial S2 extra-stimulus applied at the S1–S2 interval of 125 ms and the resulting BEG.
Fig. 4.
Electrogram waveforms and conduction curves from programmed electrical stimulation (PES) and bipolar electrogram (BEG) recordings. The electrograms recorded at the longest S1–S2 interval (basic cycle length of 125 ms) in WT hearts (A) were significantly shorter in duration than in Scn3b−/− hearts (C). Similarly, electrograms obtained prior to the heart becoming refractory or arrhythmic were consistently shorter in duration in WT hearts (B) than those recorded in Scn3b−/− hearts (D). (E) Conduction curves were created by the application of paced electrogram fractionation analysis (PEFA) to PES data obtained from WT (white circles) and Scn3b−/− hearts (black circles). The vertical axis represents latency (ms) and the horizontal axis, the S1–S2 interval (ms). The representative conduction curve demonstrates a greater increase in electrogram duration at the shortest S1–S2 interval and shorter VERP in Scn3b−/− hearts, than recorded in WT hearts.
Fig. 4E shows examples of the conduction curves in WT and Scn3b−/− hearts. Each of the curves plots the latencies of each peak and trough of each deflection contained in the electrogram from the S2 stimulus against the corresponding S1–S2 interval. Such curves demonstrate increases in the degree of spread of each electrogram and conduction latency as S1–S2 interval shortens. However, these changes are more marked in Scn3b−/− hearts. A greater increase in the EGD was observed in Scn3b−/− hearts with shortening of the S1–S2 interval, (D, filled circles) compared to the EGD shown in WT hearts (B, clear circles). Fig. 4E also demonstrates the significantly shorter VERP observed in Scn3b−/− hearts than in WT hearts. Tables 2 and 3 summarise the VERP, initial and final EGD values, EGD ratios and conduction latencies obtained from the PEFA analysis of WT and Scn3b−/− hearts. It was possible to determine a VERP in only 2 out 7 Scn3b−/− hearts, with the remaining 5 hearts demonstrating PES-induced VT (Table 3). Nevertheless, the mean S1–S2 interval at which VERP and arrhythmogenicity occurred in Scn3b−/− hearts was 24.7 ± 1.6 ms (n = 7 hearts), which was significantly shorter than the VERP of the WT hearts studied (31.2 ± 1.5 ms, n = 6, P < 0.05). Scn3b−/− hearts showed significantly larger initial and final EGD values (17.1 ± 3.8 ms and 34.6 ± 6.6 ms respectively, n = 7; Table 3) compared to those of WT hearts (2.3 ± 0.4 ms and 2.8 ± 0.4 ms, n = 6, P < 0.05; Table 2). Furthermore, Scn3b−/− hearts demonstrated a larger mean EGD ratio than that shown in WT hearts (2.1 ± 0.2 ms, n = 7 vs. 1.3 ± 0.1 ms n = 6, P < 0.05). WT and Scn3b−/− hearts both showed similar conduction latencies (8.0 ± 0.4 ms vs. 8.3 ± 0.2 ms P > 0.05; Tables 2 and 3).
Table 2.
Ventricular effective refractory period, EGDs and latencies measured in BEG recordings of (“−” = absent; “+” = present).
| WT | ||||||
|---|---|---|---|---|---|---|
| Heart | Arrhythmogenic? | VERP (ms) | Initial EGD (ms) | Final EGD (ms) | EGD ratio | Latency (ms) |
| 1 | − | 27 | 1.87 | 3 | 1.6 | 7.9 |
| 2 | − | 35 | 2.2 | 2.2 | 1.0 | 9.6 |
| 3 | − | 28 | 4.06 | 4.6 | 1.1 | 8.3 |
| 4 | − | 36 | 1.4 | 1.8 | 1.3 | 8.2 |
| 5 | − | 32 | 1.3 | 1.9 | 1.5 | 6.7 |
| 6 | − | 29 | 2.84 | 3.4 | 1.2 | 7.4 |
| Mean ± S.E.M. | 31.2 ± 1.5 | 2.3 ± 0.4 | 2.8 ± 0.4 | 1.3 ± 0.1 | 8.0 ± 0.4 | |
Table 3.
Ventricular effective refractory period, S1–S2 interval of PES-induced VT, EGDs and latencies measured in BEG recordings of Scn3b−/− hearts.
|
Scn3b−/− | ||||||
|---|---|---|---|---|---|---|
| Heart | Arrhythmogenic? | VERP (ms) | Initial EGD (ms) | Final EGD (ms) | EGD ratio | Latency (ms) |
| 1 | + | <21 | 30.5 | 62.2 | 2.0 | 8.3 |
| 2 | + | <23 | 16.0 | 36.2 | 2.3 | 8.2 |
| 3 | + | <33 | 31.4 | 55.0 | 1.8 | 8.6 |
| 4 | − | 24 | 12.8 | 26.4 | 2.1 | 7.5 |
| 5 | − | 26 | 5.6 | 17.3 | 3.1 | 9.6 |
| 6 | + | <26 | 11.9 | 21.8 | 1.8 | 8.0 |
| 7 | + | >20 | 11.6 | 23.1 | 2.0 | 8.3 |
| Mean ± S.E.M. | 24.7 ± 1.6a | 17.1 ± 3.8a | 34.6 ± 6.6a | 2.1 ± 0.2a | 8.3 ± 0.2b | |
3.4. Analysis of MAP recordings during PES
Analysis of MAP recordings permitted a closer differentiation of hearts with arrhythmogenic and non-arrhythmogenic phenotypes as well as of monomorphic and polymorphic VT. MAP waveforms were compared during PES in a second series of isolated, perfused WT and Scn3b−/− hearts. Each heart was subjected to an application of a single procedure of PES. This made it possible to determine when VT occurred, whether it was monomorphic or polymorphic. Fig. 5 illustrates MAP recordings obtained from WT (A) and Scn3b−/− (B, C) hearts during the final cycles of a PES procedure that corresponded to the shortest S1–S2 intervals. In each panel, the single vertical mark below each recording indicates the S1 stimuli and the double vertical mark the S2 extra-stimulus. The traces also show the corresponding stimulus artefacts that occurred at a regular CL, closely followed by their elicited MAP. MAPs in response to S1 stimuli were consistent in waveform in both WT and Scn3b−/− hearts, with each AP demonstrating a stable baseline, rapid upstroke and smooth repolarisation phase (Fabritz et al., 2003; Guo et al., 1999; Killeen et al., 2007). Neither demonstrated spontaneous events such as early or delayed afterdepolarizations that were reported in hypokalaemic and Scn5a+/ΔKPQ murine hearts (Killeen et al., 2007; Thomas et al., 2008).
Fig. 5.
Monophasic action potential (MAP) recordings of PES from WT and Scn3b−/− hearts. PES of isolated Langendorff-perfused WT (A) and Scn3b−/− (B, C) hearts. S2-induced action potential (AP) and VERP was observed in all WT hearts studied (n = 14). (B) PES-induced episodes of VT occurred in 10 out of 17 Scn3b−/− hearts, with five such preparations demonstrating a monomorphic VT. (C) An episode of PES-induced VT in a Scn3b−/− heart preparation began as a monomorphic VT and deteriorated into polymorphic VT.
Fig. 5A shows a typical MAP recording obtained during the final cycles of the PES procedure in a WT heart. The first S2 extra-stimulus that is displayed, gave rise to an action potential. However, the heart was refractory in response to the second S2 extra-stimulus, thereby permitting the determination of the VERP. In contrast, Scn3b−/− hearts showed episodes of either monomorphic (Fig. 5B) or polymorphic VT (Fig. 5C) following an S2 extra-stimulus. Monomorphic VT showed successive ventricular deflections that were of similar waveform and amplitude. Polymorphic VT demonstrated varying intervals between each successive deflection with irregular amplitudes and waveforms. Table 4 summarises the properties of the MAP waveforms during PES in WT (n = 14) and Scn3b−/− hearts (n = 17). It lists the VERP shown in non-arrhythmogenic hearts. Where the heart was arrhythmogenic, it lists the S1–S2 interval value, which would be less than the VERP. The WT hearts had a mean VERP of 39.3 ± 1.6 ms (n = 14). PES did not induce VT in any of the WT hearts. In contrast to WT hearts, only 7 out of 17 Scn3b−/− hearts were non-arrhythmogenic. These hearts had a mean VERP of 43.3 ± 4.4 ms (n = 7). This was not significantly different from the corresponding mean VERP, recorded in WT hearts (n = 14, P > 0.05). The remaining Scn3b−/− hearts were arrhythmogenic in response to a mean S1–S2 interval of <29.3 ± 2.4 ms (n = 10). This was significantly shorter than the VERP of the WT (n = 14) and non-arrhythmogenic Scn3b−/− hearts (n = 7; P < 0.05). The mean duration of the VT was 98.85 ± 39.8 s (Fig. 5B, C). In 5 out of the 10 arrhythmogenic hearts, the hearts showed monomorphic VT (Fig. 5B). The mean duration of episodes of monomorphic VT was 100.6 ± 46.4 s (Table 5). In 4 hearts out of 10 arrhythmogenic hearts, the hearts demonstrated polymorphic VT. The mean duration of the polymorphic VT was 27.3 ± 13.8 s. Finally, PES-induced VT in the remaining heart began as a monomorphic VT and deteriorated into polymorphic VT (Fig. 5C). This persisted until spontaneous termination of the episode.
Table 4.
Ventricular effective refractory periods and S1–S2 (+) intervals of PES-induced VT in MAP recordings of WT and Scn3b−/− hearts.
| Heart | WT |
Heart |
Scn3b−/− |
||||
|---|---|---|---|---|---|---|---|
| Arrhythmogenic? | VERP (ms) | Arrhythmogenic? | VERP (ms) | Duration of VT (sec) | Type of VT | ||
| 1 | − | 46 | 1 | − | 40 | – | – |
| 2 | − | 42 | 2 | − | <41 | 20.715 | Poly |
| 3 | − | 35 | 3 | − | 50 | – | – |
| 4 | − | 38 | 4 | − | 31 | – | – |
| 5 | − | 32 | 5 | − | <36 | 21.54 | Mono |
| 6 | − | 36 | 6 | − | <31 | 376.176 | Mono → Poly |
| 7 | − | 46 | 7 | − | 30 | – | – |
| 8 | − | 49 | 8 | + | 45 | – | – |
| 9 | − | 36 | 9 | + | <22 | 150.386 | Mono |
| 10 | − | 30 | 10 | + | <31 | 1.11 | Poly |
| 11 | − | 35 | 11 | + | 64 | – | – |
| 12 | − | 40 | 12 | + | <31 | 21.24 | Poly |
| 13 | − | 47 | 13 | + | <22 | 72.5 | Mono |
| 14 | − | 38 | 14 | + | <37 | 3.1 | Mono |
| 15 | + | 43 | – | – | |||
| 16 | + | <24 | 255.62 | Mono | |||
| 17 | + | <18 | 66.16 | Poly | |||
| Mean ± S.E.M | 39.3 ± 1.6 | Arrhythmogenic (+) | <29.3 ± 2.4a | 98.9 ± 39.8 (n = 10) | – | ||
| Non-arrhythmogenic (−) | 43.3 ± 4.4b | ||||||
Poly: Polymorphic VT.
Mono: Monomorphic VT.
P < 0.05 vs. WT.
P > 0.05 vs. WT.
Table 5.
Number of arrhythmogenic Scn3b−/− hearts, in MAP recordings, showing PES-induced monomorphic ventricular tachycardia (mVT), polymorphic ventricular tachycardia (pVT) or both (mVT + pVT).
| Type of VT | No. of hearts | Mean duration ± S.E.M. (sec) |
|---|---|---|
| mVT | 5 | 100.6 ± 46.4 |
| pVT | 4 | 27.3 ± 13.8 |
| mVT + pVT | 1 | 376.2 |
| Total VT | 10 | 98.9 ± 39.8 |
3.5. Endocardial and epicardial MAP recordings in WT and Scn3b−/− hearts
Comparison of endocardial and epicardial recordings suggested that Scn3b−/− hearts showed a shorter endocardial and epicardial APD90 which nevertheless demonstrated a normal transmural gradient of repolarization, ΔAPD90. Previous studies have demonstrated that arrhythmogenic properties were associated with abnormal differences in epicardial and endocardial APDs (Killeen et al., 2007; Stokoe et al., 2007b). The present investigation made similar experiments, which recorded MAPs at the BCL from the left ventricular epicardium and endocardium of a third series of hearts. Measurements were obtained from five separate 20 s periods during regular pacing. For each period taken, the recording electrode was repositioned to obtain independent readings from different regions of the ventricle. This was to ensure the values obtained were comparable. Action potential duration (APD) at 30% (APD30), 50% (APD50), 70% (APD70) and 90% (APD90) repolarization values were obtained from the MAPs.
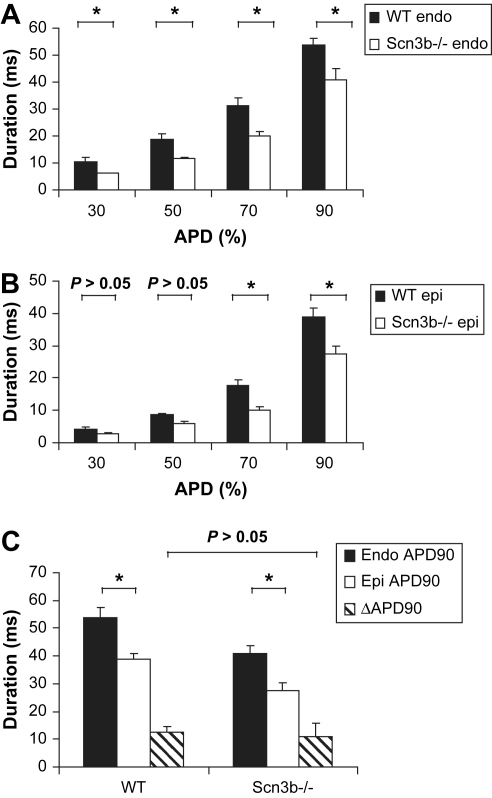
Tables 6 and 7 summarise the APD values obtained from the epicardium and endocardium. Endocardial APD values in Scn3b−/− hearts were significantly shorter than their corresponding WT endocardial values. Thus, the endocardial APD30, APD50, APD70 and APD90 recorded in WT hearts (n = 4) were 10.5 ± 1.4, 18.8 ± 2.1, 31.2 ± 2.9 and 53.6 ± 2.7 ms respectively (Fig. 6A). The same measurements made in Scn3b−/− hearts demonstrated APD values of 6.2 ± 0.1 ms, 12.3 ± 1.2 ms, 20.2 ± 1.6 ms and 41.0 ± 3.8 ms (n = 4, P < 0.05). Furthermore, Scn3b−/− epicardial APD70 and APD90 values were also significantly shorter than their respective WT APD70 and APD90 values. Thus, the epicardial APD70 and APD90 values obtained from WT hearts were 17.7 ± 1.6 ms and 38.7 ± 2.8 ms respectively (Fig. 6B). Equivalent measurements obtained from Scn3b−/− hearts were 10.2 ± 0.8 ms and 27.5 ± 2.3 ms respectively (n = 4, P < 0.05). Epicardial APD30 and APD50 values were both similar in WT and Scn3b−/− hearts (P > 0.05). The transmural gradient of repolarization (ΔAPD90) was quantified as the difference between the values of the endocardial APD90 and epicardial APD90 (Killeen et al., 2007). The Scn3b−/− and WT hearts showed similar positive ΔAPD90 (14.0 ± 3.4 ms vs. 15.4 ± 4.4 ms, P > 0.05) (Fig. 6C). This is despite a decrease in epicardial and endocardial values observed in Scn3b−/− hearts.
Table 6.
Endocardial monophasic APDs of WT and Scn3b−/− hearts.
| Parameters (ms) | Endocardial |
|
|---|---|---|
| Wild-type (n = 4) | Scn3b−/− (n = 4) | |
| APD30 | 10.5 ± 1.4 | 6.2 ± 0.1a |
| APD50 | 18.8 ± 2.1 | 12.3 ± 1.3a |
| APD70 | 31.2 ± 2.9 | 22.0 ± 1.4a |
| APD90 | 53.6 ± 2.7 | 41.0 ± 3.8a |
P < 0.05 vs. WT.
Table 7.
Epicardial monophasic APDs of WT and Scn3b−/− hearts.
| Parameters (ms) | Epicardial |
|
|---|---|---|
| Wild-type (n = 4) | Scn3b−/− (n = 4) | |
| APD30 | 4.1 ± 0.8b | 2.8 ± 0.4b |
| APD50 | 8.5 ± 0.6b | 5.8 ± 1.0b |
| APD70 | 17.7 ± 1.6b | 10.2 ± 0.8a,b |
| APD90 | 38.7 ± 2.8b | 27.5 ± 2.3a,b |
P < 0.05 vs. WT.
P < 0.05 vs. Endocardial value (in Table 6).
Fig. 6.
Action potential durations (APDs) in WT and Scn3b−/− hearts. Comparison of endocardial (endo) (A) and epicardial (epi) (B) APD values, measured at 30%, 50%, 70% and 90% repolarization for WT (n = 4) and Scn3b−/− hearts (n = 4) (black and white columns respectively). All endocardial APD recordings, epicardial APD70 and APD90 were significantly shorter. (C) Endocardial and epicardial APD measured at 90% repolarization (APD90) values (black and white columns respectively), alongside ΔAPD90 values for WT and Scn3b−/− hearts, which showed no significant difference (striped column; P > 0.05). *P < 0.05.
3.6. Na+ current recordings in isolated ventricular myocytes
The electrophysiological changes in the Scn3b−/− whole heart was correlated with a reduction in the peak Na+ current and a shift in the inactivation curve in the negative direction. The activation properties and recovery from inactivation remained similar in WT and Scn3b−/− myocytes. The effects of the expression of β3 subunits on Na+ currents were assessed using the whole-cell patch-clamp technique on cardiac myocytes isolated from WT and Scn3b−/− hearts. In all experiments, the series resistance was compensated by at least 70% and each protocol used a holding voltage of −100 mV.
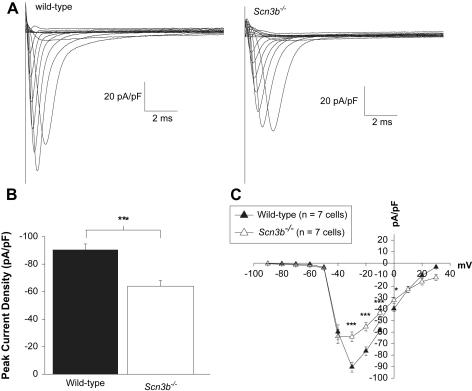
Firstly, Na+ current densities and voltage dependence of activation were assessed using a pulse protocol which applied depolarizing pulses of 20 ms duration, from a holding potential of −100 mV, to test potentials between −90 and 70 mV in 10 mV increments. Fig. 7A compares families of representative current traces obtained from WT (left panel) and Scn3b−/− (right panel) myocytes in response to voltage steps, between the test potentials of −100 to 30 mV. In all the WT and Scn3b−/− myocytes examined, the greatest magnitude of peak Na+ current density was observed at voltage steps to either −30 or −40 mV. However, the maximum value of the peak Na+ current density was significantly smaller in the Scn3b−/− myocytes, compared to that shown in WT myocytes (Fig. 7A). Thus, Fig. 7B demonstrates that the maximum value of the peak current density at −30 mV was significantly reduced by 30% in Scn3b−/− myocytes when compared to that shown in WT myocytes (90.4 ± 4.3 pA/pF, n = 7 vs. 63.3 ± 4.8 pA/pF, n = 7 each, P < 0.001; Table 8). Furthermore, Fig. 7C plots the peak Na+ current densities obtained from WT and Scn3b−/− myocytes against the corresponding test potentials. The peak Na+ current densities in Scn3b−/− myocytes (Fig. 7C, white triangles) at −30 (P < 0.001), −20 (P < 0.001), −10 (P < 0.001) and 0 mV (P < 0.05) were significantly smaller than the corresponding densities recorded in WT myocytes (black triangles, n = 7 each).
Fig. 7.
Peak Na+ current densities in WT and Scn3b−/− myocytes. Representative Na+ current traces recorded from WT (A, left panel) and Scn3b−/− (A, right panel) myocytes. (B) The peak Na+ current densities were significantly smaller in Scn3b−/− myocytes (white column, n = 7) compared to those shown in WT myocytes (black column, n = 7). (C) The current–voltage relationship demonstrated a smaller Na+ current in Scn3b−/− myocytes (white triangles, n = 7) between the voltage steps of −30 and 0 mV compared to those shown in WT myocytes (black triangles, n = 7). *P < 0.05, ***P < 0.001. All recordings were obtained at 37 °C.
Table 8.
INa properties in myocytes.
| Wild-type (n = 7) | Scn3b−/−(n = 7) | ||
|---|---|---|---|
| Peak INa density (pA/pF) | −90.4 ± 4.3 | −63.3 ± 4.8*** | |
| Voltage dependence of activation | V1/2 (mV) | −41.0 ± 0.2 | −43.6 ± 0.4 |
| k (mV) | 2.3 ± 0.3 | 2.1 ± 0.2 | |
| Voltage dependence of inactivation | V1/2 (mV) | −59.5 ± 0.4 | −67.0 ± 0.4** |
| k (mV) | 5.7 ± 0.3 | 6.4 ± 0.4 | |
| Recovery from inactivation | τ-Fast (ms) | 2.3 ± 0.5 | 2.1 ± 0.2 |
| Af | 0.83 ± 0.07 | 0.97 ± 0.04 | |
| τ-Slow (ms) | 27.1 ± 7.5 | 39.5 ± 5.3 | |
| As | 0.3 ± 0.05 | 0.27 ± 0.02 | |
** P < 0.01, *** P < 0.001.
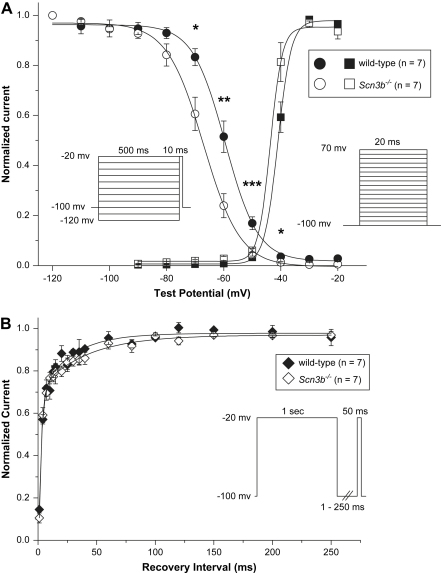
Secondly, to examine changes in the biophysics of the Na+ channel activation and inactivation properties were assessed in WT and Scn3b−/− myocytes. The activation function of Na+ channels (Fig. 8A, squares) in WT (black) and Scn3b−/− (white) myocytes was obtained by plotting the peak Na+ conductance (gNa) (peak gNa) values against Vm. Peak gNa values were calculated by the following formula;
where INa was the peak current density, Vm was membrane voltage and the ENa was the reversal potential of Na+ ions. This was calculated from the intracellular and extracellular Na+ concentration as an assumed value of 42.6 mV. The peak gNa values obtained were normalized to the corresponding maximum peak gNa values.
Fig. 8.
Steady-state activation, inactivation and recovery from inactivation properties in WT and Scn3b−/− hearts. (A) Steady-state voltage dependence of activation (square symbols) and inactivation (circle symbols) in WT (black symbols) and Scn3b−/− (white symbols) myocytes. The solid lines represent the Boltzmann fits. The normalized inactivation currents were significantly smaller in Scn3b−/− myocytes (n = 7) between the voltage steps of −70 and −40 mV compared to those shown in WT myocytes (n = 7). The pulse protocols used to study the voltage dependence of activation and inactivation are shown in the inset. (B) Recovery from inactivation in WT (black diamonds) and Scn3b−/− (white diamonds) myocytes. The solid lines represent double-exponential fits. The derived biophysical properties are summarised in Table 8. *P < 0.05, **P < 0.01, ***P < 0.001. All recordings were obtained at 37 °C.
Thirdly, the voltage dependence of inactivation was assessed using pre-pulses of 500 ms duration. These pre-pulses were applied over a voltage range from −120 to −20 mV in 10 mV increments. A test pulse to −20 mV of duration of 10 ms immediately followed this. The resulting currents were normalized against the Na+ current recorded at the pre-pulse at −120 mV. The inactivation function (Fig. 8A, circles) in WT (black) and Scn3b−/− (white) myocytes was obtained by plotting the normalized currents against Vm. The continuous lines display the Boltzmann functions fitted to the activation and inactivation data. The data was used to obtain the half-maximal voltages (V1/2) and slope factor (k) values of both activation and inactivation in WT and Scn3b−/− myocytes (Table 8). The activation function of the Na+ channel in WT and Scn3b−/− myocytes remained similar, despite a smaller Na+ current recorded in Scn3b−/− myocytes. Thus, the V1/2 and k values of WT (−41.0 ± 0.2 mV and 2.4 ± 0.3 mV) were similar to those in Scn3b−/− hearts (−43.6 ± 0.4 mV and 2.1 ± 0.2 mV, n = 7 each, P > 0.05). However, the inactivation curve of the Na+ channels in Scn3b−/− myocytes was shifted in the negative direction relative to the corresponding curve in WT myocytes. Thus, the V1/2 in the voltage dependence of inactivation of WT and Scn3b−/− myocytes was −59.5 ± 0.4 mV and −67.0 ± 0.4 mV respectively (n = 7 each, P < 0.01).
However, the slope factors of the inactivation curves were similar in WT and Scn3b−/− myocytes (WT; 5.7 ± 0.3 mV vs. Scn3b−/−; 6.4 ± 0.4 mV, n = 7 each, P > 0.05). At the holding potential of −100 mV, the normalized inactivation current was 0.95 in both WT and Scn3b−/− myocytes (Fig. 8A). However, at a depolarized potential of −80 mV, the normalized values were 0.93 and 0.84 in WT and Scn3b−/− myocytes respectively. This suggests that at −80 mV, an additional 10% of the channels in Scn3b−/− myocytes were already inactivated, compared to those in WT myocytes. Furthermore, the percentage of inactivated channels in Scn3b−/− myocytes, compared to WT myocytes, increases to 37% at a potential of −70 mV. This was calculated from the normalized current values of 0.83 and 0.61 in WT and Scn3b−/− myocytes respectively.
Finally, the time course of recovery from inactivation was examined using a 1 s pre-pulse to −20 mV. This was followed by a return to the holding voltage of −100 mV to allow recovery, at varying durations between 1 and 250 ms. A test pulse at a duration of 50 ms to −20 mV determined the current observed in fractions of recovered channels. The currents recorded were normalized against the current obtained from each of the 1 s pre-pulses applied. To determine the relative proportion of channels that were in the fast (τ-fast) and slow (τ-slow) phase, a 2-exponential function was fitted to the data (Fig. 8B, Table 8). No significant differences were observed in the time constants and fractions of the τ-fast (WT; 2.3 ± 0.5 ms and 0.83 ± 0.07, n = 7 vs. Scn3b−/−; 2.1 ± 0.2 ms and 0.97 ± 0.04, n = 7, P > 0.05) and τ-slow (WT; 27.1 ± 7.5 ms and 0.3 ± 0.05, n = 7 vs. Scn3b−/−; 39.5 ± 5.3 ms and 0.27 ± 0.02, n = 7, P > 0.05) values obtained (Fig. 8B).
4. Discussion
Sodium (Na+) channel function is critical for normal initiation, conduction and propagation of cardiac electrical activity (Grant, 2001). Thus, mutations in the SCN5A gene, which encodes for the pore-forming α-subunit of the cardiac Na+ channel Nav1.5, result in clinical arrhythmic conditions that include the Brugada syndrome (BrS) (Brugada and Brugada, 1992; Chen et al., 1998) and long QT syndrome 3 (LQT3) (Wang et al., 1995). These conditions have been reproduced in genetically modified, Scn5a+/− and Scn5a+/ΔKPQ, murine models, respectively (Head et al., 2005; Papadatos et al., 2002). Murine models have also been useful in studying the consequences of alterations in the potassium (K+) channel, such as mutations in the KCNE1 subunit resulting in LQT5 (Balasubramaniam et al., 2003; Vetter et al., 1996) and of acute conditions such as hypokalaemia (Killeen et al., 2007).
There have been fewer studies of β- than α-subunits of ion channels in murine systems, particularly those associated with the Na+ channel. One or more of such β-subunits associate with an α-subunit to form the complete channel (Catterall, 2000). So far, six types of Na+ channel β-subunits have been identified and cloned. Of these, the β1 (Isom et al., 1992), β2 (Isom et al., 1995a), β3 (Morgan et al., 2000) and β4 subunits (Yu et al., 2003) are encoded by SCN1B, SCN2B, SCN3B and SCN4B respectively. The remaining β1A (Kazen-Gillespie et al., 2000) and β1B (Qin et al., 2003) subunits are generated by splice variants of SCN1B. mRNA transcripts encoding for these β1, β2, β3 and β4 subunits are expressed in a wide range of human tissues that include brain, skeletal (Candenas et al., 2006) and cardiac muscle (Isom et al., 1995b; Makita et al., 1994; Morgan et al., 2000; Qin et al., 2003; Yu et al., 2003). The expression of these subunits is thought to be important in normal surface expression (Isom et al., 1995b) and determining the biophysical characteristics of the Na+ channel (Chen et al., 2002; Meadows et al., 2002; Morgan et al., 2000; Patton et al., 1994). It has also been suggested that cleaved forms of the β-subunits may act as transcription factors for mRNA transcripts encoding for Na+ channel subunits. Thus, the cleaved intracellular domain of the β2 subunit has been shown to increase the expression of Nav1.1 α-subunits (Kim et al., 2007). Furthermore, the ventricles of Scn1b−/− hearts have greater expression levels of Scn5a mRNA than do WT hearts (Lopez-Santiago et al., 2007). Modifications in β-subunits thus have potential effects on cardiac electrophysiology.
We report, for the first time, that deficiencies in the Scn3b gene in a murine system lead to altered biochemical and biophysical properties of the Na+ channel, resulting in abnormal ventricular electrophysiology. Scn3b−/− mice were first created using homologous recombination in embryonic stem (ES) cells. Such mice showed no significant changes in behavioural and metabolic tests compared to those assessed in WT mice. These findings are in contrast to those shown in Scn1b−/− mice, which demonstrate ataxic gait, generalized seizures and growth retardation (Chen et al., 2004). Reverse-transcriptase polymerase chain reaction (RT-PCR) analysis not only confirmed the presence or absence of Scn3b mRNA in WT and Scn3b−/− hearts respectively, but also that the Scn3b mRNA or protein may exert a regulatory role on the expression of Scn1b and Scn5a mRNA in murine ventricles. Firstly, it demonstrated that Scn3b mRNA was expressed in wild-type (WT) with similar expression in both the left and right ventricles. This was in agreement with previous studies that had demonstrated that β3 subunits were expressed in the ventricular myocytes obtained from WT and Scn1b−/− mouse hearts, specifically in the transverse tubules (Lopez-Santiago et al., 2007; Maier et al., 2004). As expected, Scn3b mRNA was not expressed in Scn3b−/− hearts. Secondly, the Scn1b and Scn5a mRNA in both WT and Scn3b−/− ventricles were expressed at higher levels compared to the expression of Scn3b mRNA in WT hearts, confirming a previous report concerning such experiments in WT hearts (Marionneau et al., 2005). Thirdly, Scn1b mRNA was expressed at higher levels in Scn3b−/− hearts than in WT hearts. These levels of expression were greater in the left than in the right ventricles in both WT and Scn3b−/− hearts. Finally, Scn5a mRNA was expressed at similar levels in the left ventricles of WT and Scn3b−/− hearts. It was expressed at higher levels in the right ventricles of Scn3b−/− hearts, compared to levels in the right ventricles of WT hearts.
Electrophysiological studies in the ventricles of the whole intact Scn3b−/− hearts also showed arrhythmogenesis and conduction abnormalities. This arrhythmogenesis took the form of either monomorphic or polymorphic ventricular tachycardias (VTs). These electrophysiological findings showed both similarities and contrasts to earlier results obtained from experiments on arrhythmogenic murine Scn5a+/−, Scn5a+/ΔKPQ, Kcne1−/− and hypokalaemic hearts, as well as human BrS (Table 9(A)). The murine systems had been used to model human BrS, LQT3, LQT5 and hypokalaemia, respectively (Balasubramaniam et al., 2003; Head et al., 2005; Killeen et al., 2007; Papadatos et al., 2002). Firstly, the Scn3b−/− hearts showed VT during programmed electrical stimulation (PES) and paced electrogram fractionation analysis (PEFA) of these results demonstrated abnormal conduction curves, in common with the remaining examples in Table 9(A). These curves showed that Scn3b−/− hearts had electrogram duration (EGD) ratios that were significantly longer but conduction latencies that were similar at the S1–S2 interval of 125 ms as those shown in WT hearts. Secondly, the Scn3b−/− hearts showed shorter ventricular effective refractory periods (VERPs) than in WT hearts, in common with findings from hypokalaemic hearts and clinical cases of BrS but in contrast to features shown by the remaining murine models. Finally, Scn3b−/− hearts had shorter endocardial and epicardial APD90 values than did WT hearts. Nevertheless, the difference between these values in Scn3b−/− hearts, which offers a measure of the transmural gradient of repolarization in the form of the ΔAPD90, was similar to that shown by WT hearts. This is in common with findings in Scn5a+/− hearts but in contrast to the remaining examples.
Table 9.
Alterations in electrophysiological features in the hearts of Scn3b−/− mice relative to WT hearts, compared with corresponding changes in arrhythmogenic murine models and clinical Brugada Syndrome.
| Features | Scn3b−/− | Arrhythmogenic Scn5a+/− | Arrhythmogenic Scn5a+/ΔKPQ | Kcne1−/− | Hypokalaemic | Clinical BrS | References |
|---|---|---|---|---|---|---|---|
| (A) Whole heart properties | |||||||
| PES VT | + | + | + | + | + | + | 1–5 |
| Increased EGD | + | + | + | + | Not assessed | + | 2, 3, 5, 6 |
| Initial conduction latency | No change | No change | Increased3 | No change | Not assessed | Not assessed | 2, 3, 5, 7 |
| No change7 | |||||||
| VERP | Shorter | No change2 | No change | No change | Shorter | Short | 2, 3, 5, 7–10, 23 |
| Longer9 | Longer3 | ||||||
| ΔAPD90 | No change | No Change | Negative | Negative | Negative | Varied | 2, 4, 7, 11–13 |
| (B) Single cell properties | |||||||
| Peak Na+ current density | Reduced | Reduced | Greater | N/A | N/A | Reduced | 3, 9, 14 |
| Na+ channel activation | No change | No change | No Change | N/A | N/A | Positive shift | 15–19 |
| Negative shift | 20 | ||||||
| No Change | 21 | ||||||
| Na+ channel inactivation | Negative shift | No change | No change | N/A | N/A | Positive shift | 17, 21, 22 |
| Negative shift | 15, 16, 18, 20 | ||||||
| No Change | 18, 20 | ||||||
| Na+ channel recovery from inactivation | No change | Not assessed | No Change | N/A | N/A | Faster | 17–20, 22 |
| Slower | 21 | ||||||
| No change | 15, 16, 20 | ||||||
Key: 1 = Morita et al., 2003, 2 = Stokoe et al., 2007a, 3 = Head et al., 2005, 4 = Killeen et al., 2007, 5 = Balasubramaniam et al., 2003, 6 = Saumarez et al., 2003, 7 = Stokoe et al., 2007b, 8 = Watanabe et al., 2005, 9 = Papadatos et al., 2002, 10 = Sabir et al., 2007, 11 = Antzelevitch, 2001, 12 = Kurita et al., 2002, 13 = Thomas et al., 2007, 14 = Chen et al., 1998, 15 = Baroudi and Chahine, 2000, 16 = Wan et al., 2001, 17 = Zhang et al., 2007, 18 = Akai et al., 2000, 19 = Dumaine et al., 1999, 20 = Rook et al., 1999, 21 = Chen et al., 1998, 22 = Wang et al., 2000, 23 = Wilde et al., 2002. (“−” = absent; “+” = present).
The use of the whole-cell patch-clamp technique demonstrated the loss of function of the cardiac Na+ channel in ventricular myocytes isolated from Scn3b−/− hearts. These experiments were carried out at the same temperature of 37 °C as the studies used on intact hearts in contrast to previous investigations that had carried out similar single cell experiments at room temperature (Lopez-Santiago et al., 2007; Papadatos et al., 2002). Table 9(B) compares the resulting Na+ channel properties in Scn3b−/− myocytes with those shown in Scn5a+/− and Scn5a+/ΔKPQ myocytes and points to a number of important comparisons. Firstly, Scn3b−/− myocytes showed significantly smaller peak Na+ current densities, in response to depolarizing voltage steps from a −100 mV holding potential, than did WT myocytes. This was in common with previous findings in Scn5a+/− myocytes but in contrast to those shown in Scn5a+/ΔKPQ myocytes (Table 9(B)). This suggests a reduced expression or proportion of functional Na+ channels in the plasma membrane of Scn3b−/− ventricles compared to that shown in WT ventricles. The present experiments were performed under conditions of higher ratios of extracellular and intracellular [Na+]. This may account for the larger Na+ current densities observed in the present study, compared to those observed in previous investigations (Nuyens et al., 2001; Papadatos et al., 2002). Both latter studies used lower extracellular and intracellular [Na+] ratios of 2:1 and 2:2 respectively and carried out single cell experiments at room temperature (22–23 °C). Such conditions had resulted in peak Na+ current densities of ∼15 pA/pF and 21 ± 5 pA/pF respectively. Secondly, the Na+ channel activation properties in Scn3b−/− myocytes were similar to those of WT myocytes in common with findings in Scn5a+/− and Scn5a+/ΔKPQ myocytes. However, the present findings were obtained at a higher temperature of 37 °C than used in the previous studies (Lopez-Santiago et al., 2007; Papadatos et al., 2002). This may account for the similarly highly rapid time courses of activation and small slope factors of steady-state activation observed in both the WT and Scn3b−/− myocytes. Thirdly, the position of the inactivation curves in Scn3b−/− myocytes was nevertheless shifted in the negative direction relative to that shown in WT myocytes, whereas both Scn5a+/− and Scn5a+/ΔKPQ myocytes showed normal inactivation curves. At a typical resting membrane potential between −80 and −70 mV (Guo et al., 1999; Remme et al., 2006; Yao et al., 1998), such a shift would result in the inactivation of a further 10 to 37% of the Na+ channels in Scn3b−/− myocytes, compared to that shown in WT myocytes. Finally, the time course in the recovery from inactivation in Scn3b−/− myocytes were similar to that shown in WT myocytes in common with findings in Scn5a+/ΔKPQ myocytes.
Together, the altered peak Na+ current density and inactivation features shown in Scn3b−/− myocytes are compatible with a reduction of the total in vivo Na+ current. This is compatible with findings in BrS. Thus, although the clinical cases of BrS have shown a wide range of alterations in the activation, inactivation and recovery from inactivation properties, all show reduced Na+ currents. The present findings also add a further example of BrS that involves mutations in the Na+ channel β-subunit. A previous investigation had identified a single mutation in the SCN1B gene and two mutations in the splice variant of the same gene in patients with BrS (Watanabe et al., 2008). It used a heterologous expression system provided by Chinese hamster ovary (CHO) cells, to study the consequent changes in the biophysical properties of the Na+ channel resulting from the mutations. In agreement with the present example, cells expressing both the WT Nav1.5 and such mutant β-subunits showed significantly reduced peak Na+ currents. In contrast, activation and inactivation properties of the Na+ channel shifted in the positive direction in relation to findings in cells expressing WT Nav1.5 and WT β-subunits. The present findings also complement other investigations in which alterations in the β-subunit expression can produce other electrophysiological changes that include a LQT-like phenotype. For example, Lopez-Santiago et al. (2007) associated a deletion of the Scn1b gene in a murine system with an increase in peak Na+ currents and QT prolongation.
The present findings complement reports of altered Na+ channel biophysical properties resulting from β3 expression in heterologous expression systems by describing the consequences of similar expression in vivo. However, the properties of such in vitro expression systems may not correspond to features observed in vivo and have demonstrated conflicting results despite the transfection of identical sets of genes. Such systems lack functionally important proteins such as ankyrin proteins that form complexes with the Na+ channel in vivo (Abriel and Kass, 2005). Thus, on the one hand, Xenopus oocytes injected with cRNA for Nav1.5 and β3 subunits demonstrated significantly larger peak Na+ currents and positive shifts in the inactivation curves than in cells expressing Nav1.5 only (Fahmi et al., 2001). In contrast, CHO cells expressing Nav1.5 and β3 subunits resulted in similar peak Na+ currents with negative shifts in the activation and inactivation curves compared to cells expressing Nav1.5 only (Ko et al., 2005).
In conclusion, we have for the first time associated the lack of the Scn3b gene with a ventricular arrhythmogenesis in intact perfused hearts whose features are similar to those shown in Scn5a+/− hearts.
Acknowledgements
Supported by grants from the British Heart Foundation, the Medical Research Council, the Wellcome Trust, the Helen Kirkland Fund for Cardiac Research and the Papworth Hospital Trust.
References
- Abriel H., Kass R.S. Regulation of the voltage-gated cardiac sodium channel Nav1.5 by interacting proteins. Trends Cardiovasc. Med. 2005;15:35–40. doi: 10.1016/j.tcm.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Akai J., Makita N., Sakurada H., Shirai N., Ueda K., Kitabatake A., Nakazawa K., Kimura A., Hiraoka M. A novel SCN5A mutation associated with idiopathic ventricular fibrillation without typical ECG findings of Brugada syndrome. FEBS Lett. 2000;479:29–34. doi: 10.1016/s0014-5793(00)01875-5. [DOI] [PubMed] [Google Scholar]
- Antzelevitch C. The Brugada syndrome: diagnostic criteria and cellular mechanisms. Eur Heart J. 2001;22:356–363. doi: 10.1053/euhj.2000.2461. [DOI] [PubMed] [Google Scholar]
- Balasubramaniam R., Grace A.A., Saumarez R.C., Vandenberg J.I., Huang C.L.-H. Electrogram prolongation and nifedipine-suppressible ventricular arrhythmias in mice following targeted disruption of KCNE1. J. Physiol. 2003;552:535–546. doi: 10.1113/jphysiol.2003.048249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroudi G., Chahine M. Biophysical phenotypes of SCN5A mutations causing long QT and Brugada syndromes. FEBS Lett. 2000;487:224–248. doi: 10.1016/s0014-5793(00)02360-7. [DOI] [PubMed] [Google Scholar]
- Bradley A., Evans M., Kaufman M.H., Robertson E. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature. 1984;309:255–256. doi: 10.1038/309255a0. [DOI] [PubMed] [Google Scholar]
- Brugada P., Brugada J. Right bundle branch block, persistent ST segment elevation and sudden cardiac death: a distinct clinical and electrocardiographic syndrome. A multicenter report. J. Am. Coll. Cardiol. 1992;20:1391–1396. doi: 10.1016/0735-1097(92)90253-j. [DOI] [PubMed] [Google Scholar]
- Candenas L., Seda M., Noheda P., Buschmann H., Cintado C.G., Martin J.D., Pinto F.M. Molecular diversity of voltage-gated sodium channel alpha and beta subunit mRNAs in human tissues. Eur. J. Pharmacol. 2006;541:9–16. doi: 10.1016/j.ejphar.2006.04.025. [DOI] [PubMed] [Google Scholar]
- Casimiro M.C., Knollmann B.C., Ebert S.N., Vary J.C., Jr., Greene A.E., Franz M.R., Grinberg A., Huang S.P., Pfeifer K. Targeted disruption of the Kcnq1 gene produces a mouse model of Jervell and Lange–Nielsen Syndrome. Proc. Natl. Acad. Sci. U.S.A. 2001;98:2526–2531. doi: 10.1073/pnas.041398998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catterall W.A. From ionic currents to molecular mechanisms: the structure and function of voltage-gated sodium channels. Neuron. 2000;26:13–25. doi: 10.1016/s0896-6273(00)81133-2. [DOI] [PubMed] [Google Scholar]
- Chen C., Bharucha V., Chen Y., Westenbroek R.E., Brown A., Malhotra J.D., Jones D., Avery C., Gillespie P.J., 3rd, Kazen-Gillespie K.A., Kazarinova-Noyes K., Shrager P., Saunders T.L., Macdonald R.L., Ransom B.R., Scheuer T., Catterall W.A., Isom L.L. Reduced sodium channel density, altered voltage dependence of inactivation, and increased susceptibility to seizures in mice lacking sodium channel beta 2-subunits. Proc. Natl. Acad. Sci. U.S.A. 2002;99:17072–17077. doi: 10.1073/pnas.212638099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Westenbroek R.E., Xu X., Edwards C.A., Sorenson D.R., Chen Y., McEwen D.P., O'Malley H.A., Bharucha V., Meadows L.S., Knudsen G.A., Vilaythong A., Noebels J.L., Saunders T.L., Scheuer T., Shrager P., Catterall W.A., Isom L.L. Mice lacking sodium channel beta1 subunits display defects in neuronal excitability, sodium channel expression, and nodal architecture. J. Neurosci. 2004;24:4030–4042. doi: 10.1523/JNEUROSCI.4139-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Kirsch G.E., Zhang D., Brugada R., Brugada J., Brugada P., Potenza D., Moya A., Borggrefe M., Breithardt G., Ortiz-Lopez R., Wang Z., Antzelevitch C., O'Brien R.E., Schulze-Bahr E., Keating M.T., Towbin J.A., Wang Q. Genetic basis and molecular mechanism for idiopathic ventricular fibrillation. Nature. 1998;392:293–296. doi: 10.1038/32675. [DOI] [PubMed] [Google Scholar]
- Dumaine R., Towbin J.A., Brugada P., Vatta M., Nesterenko D.V., Nesterenko V.V., Brugada J., Brugada R., Antzelevitch C. Ionic mechanisms responsible for the electrocardiographic phenotype of the Brugada syndrome are temperature dependent. Circ. Res. 1999;85:803–809. doi: 10.1161/01.res.85.9.803. [DOI] [PubMed] [Google Scholar]
- Fabritz L., Kirchhof P., Franz M.R., Eckardt L., Monnig G., Milberg P., Breithardt G., Haverkamp W. Prolonged action potential durations, increased dispersion of repolarization, and polymorphic ventricular tachycardia in a mouse model of proarrhythmia. Basic Res. Cardiol. 2003;98:25–32. doi: 10.1007/s00395-003-0386-y. [DOI] [PubMed] [Google Scholar]
- Fahmi A.I., Patel M., Stevens E.B., Fowden A.L., John J.E., 3rd, Lee K., Pinnock R., Morgan K., Jackson A.P., Vandenberg J.I. The sodium channel beta-subunit SCN3b modulates the kinetics of SCN5a and is expressed heterogeneously in sheep heart. J. Physiol. 2001;537:693–700. doi: 10.1111/j.1469-7793.2001.00693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz M.R. Current status of monophasic action potential recording: theories, measurements and interpretations. Cardiovasc. Res. 1999;41:25–40. doi: 10.1016/s0008-6363(98)00268-5. [DOI] [PubMed] [Google Scholar]
- Grant A.O. Molecular biology of sodium channels and their role in cardiac arrhythmias. Am. J. Med. 2001;110:296–305. doi: 10.1016/s0002-9343(00)00714-2. [DOI] [PubMed] [Google Scholar]
- Guo W., Xu H., London B., Nerbonne J.M. Molecular basis of transient outward K+ current diversity in mouse ventricular myocytes. J. Physiol. 1999;521:587–599. doi: 10.1111/j.1469-7793.1999.00587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head C.E., Balasubramaniam R., Thomas G., Goddard C.A., Lei M., Colledge W.H., Grace A.A., Huang C.L.-H. Paced electrogram fractionation analysis of arrhythmogenic tendency in DeltaKPQ Scn5a mice. J. Cardiovasc. Electrophysiol. 2005;16:1329–1340. doi: 10.1111/j.1540-8167.2005.00200.x. [DOI] [PubMed] [Google Scholar]
- Isom L.L. Sodium channel beta subunits: anything but auxiliary. Neuroscientist. 2001;7:42–54. doi: 10.1177/107385840100700108. [DOI] [PubMed] [Google Scholar]
- Isom L.L., De Jongh K.S., Patton D.E., Reber B.F., Offord J., Charbonneau H., Walsh K., Goldin A.L., Catterall W.A. Primary structure and functional expression of the beta 1 subunit of the rat brain sodium channel. Science. 1992;256:839–842. doi: 10.1126/science.1375395. [DOI] [PubMed] [Google Scholar]
- Isom L.L., Ragsdale D.S., De Jongh K.S., Westenbroek R.E., Reber B.F., Scheuer T., Catterall W.A. Structure and function of the beta 2 subunit of brain sodium channels, a transmembrane glycoprotein with a CAM motif. Cell. 1995;83:433–442. doi: 10.1016/0092-8674(95)90121-3. [DOI] [PubMed] [Google Scholar]
- Isom L.L., Scheuer T., Brownstein A.B., Ragsdale D.S., Murphy B.J., Catterall W.A. Functional co-expression of the beta 1 and type IIA alpha subunits of sodium channels in a mammalian cell line. J. Biol. Chem. 1995;270:3306–3312. doi: 10.1074/jbc.270.7.3306. [DOI] [PubMed] [Google Scholar]
- Kazen-Gillespie K.A., Ragsdale D.S., D'Andrea M.R., Mattei L.N., Rogers K.E., Isom L.L. Cloning, localization, and functional expression of sodium channel beta1A subunits. J. Biol. Chem. 2000;275:1079–1088. doi: 10.1074/jbc.275.2.1079. [DOI] [PubMed] [Google Scholar]
- Keating M.T., Sanguinetti M.C. Molecular and cellular mechanisms of cardiac arrhythmias. Cell. 2001;104:569–580. doi: 10.1016/s0092-8674(01)00243-4. [DOI] [PubMed] [Google Scholar]
- Killeen M.J., Thomas G., Gurung I.S., Goddard C.A., Fraser J.A., Mahaut-Smith M.P., Colledge W.H., Grace A.A., Huang C.L.-H. Arrhythmogenic mechanisms in the isolated perfused hypokalaemic murine heart. Acta Physiol. (Oxf.) 2007;189:33–46. doi: 10.1111/j.1748-1716.2006.01643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.Y., Carey B.W., Wang H., Ingano L.A., Binshtok A.M., Wertz M.H., Pettingell W.H., He P., Lee V.M., Woolf C.J., Kovacs D.M. BACE1 regulates voltage-gated sodium channels and neuronal activity. Nat. Cell Biol. 2007;9:755–764. doi: 10.1038/ncb1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knollmann B.C., Katchman A.N., Franz M.R. Monophasic action potential recordings from intact mouse heart: validation, regional heterogeneity, and relation to refractoriness. J. Cardiovasc. Electrophysiol. 2001;12:1286–1294. doi: 10.1046/j.1540-8167.2001.01286.x. [DOI] [PubMed] [Google Scholar]
- Ko S.H., Lenkowski P.W., Lee H.C., Mounsey J.P., Patel M.K. Modulation of Na(v)1.5 by beta1- and beta3-subunit co-expression in mammalian cells. Pflugers Arch. 2005;449:403–412. doi: 10.1007/s00424-004-1348-4. [DOI] [PubMed] [Google Scholar]
- Kurita T., Shimizu W., Inagaki M., Suyama K., Taguchi A., Satomi K., Aihara N., Kamakura S., Kobayashi J., Kosakai Y. The electrophysiologic mechanism of ST-segment elevation in Brugada syndrome. J. Am. Coll. Cardiol. 2002;40:330–334. doi: 10.1016/s0735-1097(02)01964-2. [DOI] [PubMed] [Google Scholar]
- Lister R.G. The use of a plus-maze to measure anxiety in the mouse. Psychopharmacology (Berl.) 1987;92:180–185. doi: 10.1007/BF00177912. [DOI] [PubMed] [Google Scholar]
- Lopez-Santiago L.F., Meadows L.S., Ernst S.J., Chen C., Malhotra J.D., McEwen D.P., Speelman A., Noebels J.L., Maier S.K., Lopatin A.N., Isom L.L. Sodium channel Scn1b null mice exhibit prolonged QT and RR intervals. J. Mol. Cell Cardiol. 2007;43:636–647. doi: 10.1016/j.yjmcc.2007.07.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier S.K., Westenbroek R.E., McCormick K.A., Curtis R., Scheuer T., Catterall W.A. Distinct subcellular localization of different sodium channel alpha and beta subunits in single ventricular myocytes from mouse heart. Circulation. 2004;109:1421–1427. doi: 10.1161/01.CIR.0000121421.61896.24. [DOI] [PubMed] [Google Scholar]
- Makita N., Sloan-Brown K., Weghuis D.O., Ropers H.H., George A.L., Jr. Genomic organization and chromosomal assignment of the human voltage-gated Na+ channel beta 1 subunit gene (SCN1B) Genomics. 1994;23:628–634. doi: 10.1006/geno.1994.1551. [DOI] [PubMed] [Google Scholar]
- Malhotra J.D., Kazen-Gillespie K., Hortsch M., Isom L.L. Sodium channel beta subunits mediate homophilic cell adhesion and recruit ankyrin to points of cell–cell contact. J. Biol. Chem. 2000;275:11383–11388. doi: 10.1074/jbc.275.15.11383. [DOI] [PubMed] [Google Scholar]
- Marionneau C., Couette B., Liu J., Li H., Mangoni M.E., Nargeot J., Lei M., Escande D., Demolombe S. Specific pattern of ionic channel gene expression associated with pacemaker activity in the mouse heart. J. Physiol. 2005;562:223–234. doi: 10.1113/jphysiol.2004.074047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows L.S., Chen Y.H., Powell A.J., Clare J.J., Ragsdale D.S. Functional modulation of human brain Nav1.3 sodium channels, expressed in mammalian cells, by auxiliary beta 1, beta 2 and beta 3 subunits. Neuroscience. 2002;114:745–753. doi: 10.1016/s0306-4522(02)00242-7. [DOI] [PubMed] [Google Scholar]
- Medeiros-Domingo A., Kaku T., Tester D.J., Iturralde-Torres P., Itty A., Ye B., Valdivia C., Ueda K., Canizales-Quinteros S., Tusie-Luna M.T., Makielski J.C., Ackerman M.J. SCN4B-encoded sodium channel beta4 subunit in congenital long-QT syndrome. Circulation. 2007;116:134–142. doi: 10.1161/CIRCULATIONAHA.106.659086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan K., Stevens E.B., Shah B., Cox P.J., Dixon A.K., Lee K., Pinnock R.D., Hughes J., Richardson P.J., Mizuguchi K., Jackson A.P. Beta 3: an additional auxiliary subunit of the voltage-sensitive sodium channel that modulates channel gating with distinct kinetics. Proc. Natl. Acad. Sci. U.S.A. 2000;97:2308–2313. doi: 10.1073/pnas.030362197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita H., Morita S.T., Nagase S., Banba K., Nishii N., Tani Y., Watanabe A., Nakamura K., Kusano K.F., Emori T., Matsubara H., Hina K., Kita T., Ohe T. Ventricular arrhythmia induced by sodium channel blocker in patients with Brugada syndrome. J. Am. Coll. Cardiol. 2003;42:1624–1631. doi: 10.1016/j.jacc.2003.06.004. [DOI] [PubMed] [Google Scholar]
- Noda M., Ikeda T., Suzuki H., Takeshima H., Takahashi T., Kuno M., Numa S. Expression of functional sodium channels from cloned cDNA. Nature. 1986;322:826–828. doi: 10.1038/322826a0. [DOI] [PubMed] [Google Scholar]
- Nuyens D., Stengl M., Dugarmaa S., Rossenbacker T., Compernolle V., Rudy Y., Smits J.F., Flameng W., Clancy C.E., Moons L., Vos M.A., Dewerchin M., Benndorf K., Collen D., Carmeliet E., Carmeliet P. Abrupt rate accelerations or premature beats cause life-threatening arrhythmias in mice with long-QT3 syndrome. Nat. Med. 2001;7:1021–1027. doi: 10.1038/nm0901-1021. [DOI] [PubMed] [Google Scholar]
- Papadatos G.A., Wallerstein P.M., Head C.E., Ratcliff R., Brady P.A., Benndorf K., Saumarez R.C., Trezise A.E., Huang C.L.-H., Vandenberg J.I., Colledge W.H., Grace A.A. Slowed conduction and ventricular tachycardia after targeted disruption of the cardiac sodium channel gene Scn5a. Proc. Natl. Acad. Sci. U.S.A. 2002;99:6210–6215. doi: 10.1073/pnas.082121299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton D.E., Isom L.L., Catterall W.A., Goldin A.L. The adult rat brain beta 1 subunit modifies activation and inactivation gating of multiple sodium channel alpha subunits. J. Biol. Chem. 1994;269:17649–17655. [PubMed] [Google Scholar]
- Qin N., D'Andrea M.R., Lubin M.L., Shafaee N., Codd E.E., Correa A.M. Molecular cloning and functional expression of the human sodium channel beta1B subunit, a novel splicing variant of the beta1 subunit. Eur. J. Biochem. 2003;270:4762–4770. doi: 10.1046/j.1432-1033.2003.03878.x. [DOI] [PubMed] [Google Scholar]
- Qu Y., Curtis R., Lawson D., Gilbride K., Ge P., DiStefano P.S., Silos-Santiago I., Catterall W.A., Scheuer T. Differential modulation of sodium channel gating and persistent sodium currents by the beta1, beta2, and beta3 subunits. Mol. Cell Neurosci. 2001;18:570–580. doi: 10.1006/mcne.2001.1039. [DOI] [PubMed] [Google Scholar]
- Remme C.A., Verkerk A.O., Nuyens D., van Ginneken A.C., van Brunschot S., Belterman C.N., Wilders R., van Roon M.A., Tan H.L., Wilde A.A., Carmeliet P., de Bakker J.M., Veldkamp M.W., Bezzina C.R. Overlap syndrome of cardiac sodium channel disease in mice carrying the equivalent mutation of human SCN5A-1795insD. Circulation. 2006;114:2584–2594. doi: 10.1161/CIRCULATIONAHA.106.653949. [DOI] [PubMed] [Google Scholar]
- Rook M.B., Bezzina Alshinawi C., Groenewegen W.A., van Gelder I.C., van Ginneken A.C., Jongsma H.J., Mannens M.M., Wilde A.A. Human SCN5A gene mutations alter cardiac sodium channel kinetics and are associated with the Brugada syndrome. Cardiovasc. Res. 1999;44:507–517. doi: 10.1016/s0008-6363(99)00350-8. [DOI] [PubMed] [Google Scholar]
- Sabir I.N., Fraser J.A., Cass T.R., Grace A.A., Huang C.L.-H. A quantitative analysis of the effect of cycle length on arrhythmogenicity in hypokalaemic Langendorff-perfused murine hearts. Pflugers Arch. 2007;454:925–936. doi: 10.1007/s00424-007-0255-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saumarez R.C., Chojnowska L., Derksen R., Pytkowski M., Sterlinski M., Huang C.L., Sadoul N., Hauer R.N., Ruzyllo W., Grace A.A. Sudden death in noncoronary heart disease is associated with delayed paced ventricular activation. Circulation. 2003;107:2595–2600. doi: 10.1161/01.CIR.0000068342.96569.A1. [DOI] [PubMed] [Google Scholar]
- Saumarez R.C., Grace A.A. Paced ventricular electrogram fractionation and sudden death in hypertrophic cardiomyopathy and other non-coronary heart diseases. Cardiovasc. Res. 2000;47:11–22. doi: 10.1016/s0008-6363(00)00096-1. [DOI] [PubMed] [Google Scholar]
- Stevens E.B., Cox P.J., Shah B.S., Dixon A.K., Richardson P.J., Pinnock R.D., Lee K. Tissue distribution and functional expression of the human voltage-gated sodium channel beta3 subunit. Pflugers Arch. 2001;441:481–488. doi: 10.1007/s004240000449. [DOI] [PubMed] [Google Scholar]
- Stokoe K.S., Balasubramaniam R., Goddard C.A., Colledge W.H., Grace A.A., Huang C.L.-H. Effects of flecainide and quinidine on arrhythmogenic properties of Scn5a+/− murine hearts modelling the Brugada syndrome. J. Physiol. 2007;581:255–275. doi: 10.1113/jphysiol.2007.128785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stokoe K.S., Thomas G., Goddard C.A., Colledge W.H., Grace A.A., Huang C.L.-H. Effects of flecainide and quinidine on arrhythmogenic properties of Scn5a+/Delta murine hearts modelling long QT syndrome 3. J. Physiol. 2007;578:69–84. doi: 10.1113/jphysiol.2006.117945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H.L., Bezzina C.R., Smits J.P., Verkerk A.O., Wilde A.A. Genetic control of sodium channel function. Cardiovasc. Res. 2003;57:961–973. doi: 10.1016/s0008-6363(02)00714-9. [DOI] [PubMed] [Google Scholar]
- Thomas G., Killeen M.J., Grace A.A., Huang C.L.-H. Pharmacological separation of early afterdepolarizations from arrhythmogenic substrate in DeltaKPQ Scn5a murine hearts modelling human long QT 3 syndrome. Acta Physiol. (Oxf.) 2008;192:505–517. doi: 10.1111/j.1748-1716.2007.01770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G., Killeen M.J., Gurung I.S., Hakim P., Balasubramaniam R., Goddard C.A., Grace A.A., Huang C.L.-H. Mechanisms of ventricular arrhythmogenesis in mice following targeted disruption of KCNE1 modelling long QT syndrome 5. J. Physiol. 2007;578:99–114. doi: 10.1113/jphysiol.2006.118133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter D.E., Mann J.R., Wangemann P., Liu J., McLaughlin K.J., Lesage F., Marcus D.C., Lazdunski M., Heinemann S.F., Barhanin J. Inner ear defects induced by null mutation of the isk gene. Neuron. 1996;17:1251–1264. doi: 10.1016/s0896-6273(00)80255-x. [DOI] [PubMed] [Google Scholar]
- Wan X., Chen S., Sadeghpour A., Wang Q., Kirsch G.E. Accelerated inactivation in a mutant Na(+) channel associated with idiopathic ventricular fibrillation. Am. J. Physiol. Heart Circ. Physiol. 2001;280:H354–H360. doi: 10.1152/ajpheart.2001.280.1.H354. [DOI] [PubMed] [Google Scholar]
- Wang D.W., Makita N., Kitabatake A., Balser J.R., George A.L., Jr. Enhanced Na(+) channel intermediate inactivation in Brugada syndrome. Circ. Res. 2000;87:E37–E43. doi: 10.1161/01.res.87.8.e37. [DOI] [PubMed] [Google Scholar]
- Wang Q., Shen J., Splawski I., Atkinson D., Li Z., Robinson J.L., Moss A.J., Towbin J.A., Keating M.T. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell. 1995;80:805–811. doi: 10.1016/0092-8674(95)90359-3. [DOI] [PubMed] [Google Scholar]
- Watanabe H., Chinushi M., Sugiura H., Washizuka T., Komura S., Hosaka Y., Furushima H., Watanabe H., Hayashi J., Aizawa Y. Unsuccessful internal defibrillation in Brugada syndrome: focus on refractoriness and ventricular fibrillation cycle length. J. Cardiovasc. Electrophysiol. 2005;16:262–266. doi: 10.1046/j.1540-8167.2005.40579.x. [DOI] [PubMed] [Google Scholar]
- Watanabe H., Koopmann T.T., Le Scouarnec S., Yang T., Ingram C.R., Schott J.J., Demolombe S., Probst V., Anselme F., Escande D., Wiesfeld A.C., Pfeufer A., Kaab S., Wichmann H.E., Hasdemir C., Aizawa Y., Wilde A.A., Roden D.M., Bezzina C.R. Sodium channel beta1 subunit mutations associated with Brugada syndrome and cardiac conduction disease in humans. J. Clin. Invest. 2008;118:2260–2268. doi: 10.1172/JCI33891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilde A.A., Antzelevitch C., Borggrefe M., Brugada J., Brugada R., Brugada P., Corrado D., Hauer R.N., Kass R.S., Nademanee K., Priori S.G., Towbin J.A. Proposed diagnostic criteria for the Brugada syndrome: consensus report. Circulation. 2002;106:2514–2519. doi: 10.1161/01.cir.0000034169.45752.4a. [DOI] [PubMed] [Google Scholar]
- Yao A., Su Z., Nonaka A., Zubair I., Lu L., Philipson K.D., Bridge J.H., Barry W.H. Effects of overexpression of the [Ca2+]i exchanger on Na+-Ca2+ transients in murine ventricular myocytes. Circ. Res. 1998;82:657–665. doi: 10.1161/01.res.82.6.657. [DOI] [PubMed] [Google Scholar]
- Yu F.H., Westenbroek R.E., Silos-Santiago I., McCormick K.A., Lawson D., Ge P., Ferriera H., Lilly J., DiStefano P.S., Catterall W.A., Scheuer T., Curtis R. Sodium channel beta4, a new disulfide-linked auxiliary subunit with similarity to beta2. J. Neurosci. 2003;23:7577–7585. doi: 10.1523/JNEUROSCI.23-20-07577.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z.S., Tranquillo J., Neplioueva V., Bursac N., Grant A.O. Sodium channel kinetic changes that produce Brugada syndrome or progressive cardiac conduction system disease. Am. J. Physiol. Heart Circ. Physiol. 2007;292:H399–H407. doi: 10.1152/ajpheart.01025.2005. [DOI] [PubMed] [Google Scholar]