Abstract
The human lens proteins β-crystallins are subdivided into acidic (βA1-βA4) and basic (βB1-βB3) subunit groups. These structural proteins exist at extremely high concentrations and associate into oligomers under physiological conditions. Crystallin acidic-basic pairs tend to form strong heteromolecular associations. The long N-terminal extensions of β-crystallins may influence both homo- and heteromolecular interactions. However, identification of the critical regions of the extensions mediating protein associations have not been previously addressed. This was studied by comparing the self association and heteromolecular associations of wild-type recombinant βA3 and βB1 crystallins and their N-terminal truncated counterparts (βA3ΔN30 and βB1ΔN56) using several biophysical techniques including analytical ultracentrifugation and fluorescence spectroscopy. Removal of the N-terminal extension of βA3 had no effect on dimerization or heteromolecular tetramer formation with βB1. In contrast, the self association of βB1ΔN56 increased resulting in homotetramer formation and heteromolecular association with βA3 was blocked. Limited proteolysis of βB1 produced βB1ΔN47, which similar to intact protein formed dimers but in contrast showed enhanced heteromolecular tetramer formation with βA3. The tryptic digestion was of physiological significance, corresponding to protease processing sites observed in-vivo. Molecular modeling of the N-terminal βB1 extension indicates structural features which position a mobile loop in the vicinity of these processing sites. The loop is derived from residues 48-56 which appear critical for mediating protein interactions with βA3-crystallin.
Keywords: β-crystallins, N-terminal extension, protein association
Crystallins are the major structural proteins of the lens, where at very high concentrations they are responsible for the transparency and high refractive index (1). In the mammalian lenses, crystallins can be grouped in two families, small heat shock-related α-crystallins and βγ-crystallins (1;2). All βγ-crystallins are composed of one, two or multiple βγ-crystallin domains, where each domain is made up of two β-stranded Greek-key motifs. The β-crystallins are distinguished from γ crystallins by having either N- and C-termini extensions, for basic β-crystallins (βB1, βB2, βB3), or only an N-terminal extension, for acidic forms (βA1/A3, βA2, βA4) (3;4). Whereas γ crystallins exist as monomers, β-crystallins are known to associate into dimers, tetramers, and higher-order complexes under physiological conditions (5;6). Based on the size-exclusion chromatography of lens extract, β-crystallins form three size classes of aggregates: βH (octamers of 160-200 kDa), βL1 (tetramers of 70-100 kDa), and βL2 (dimers of 46-50 kDa) (7;8). Intact βB1- crystallin is present only in the largest aggregate, βH, and absent in both the dimeric βL2, and the intermediate βL1 (9-11). Lower molecular weight aggregates contained only truncated forms of βB1 (11).
Because lens crystallins do not turnover with age, they are susceptible over time to a wide variety of post-translational modifications such as acetylation, deamidation, methylation, oxidation, phosphorylation, and truncation of terminal extensions by thiol proteases (12-16). These modifications may perturb protein stability and structure, enhancing further association and aggregation and leading to cataract development (12;17;18). Age-related proteolytic processing of human lens β-crystallins occurs mainly at the N-terminal extensions (19-21) and the first crystallins modified are βB1 and βA1/A3 (17;19-21). Processing of βB1 (also βA1/A3) was noticeable in human lenses less then one year old, and although the proportion of these truncated proteins increased with age most had occurred by the age of 20 (22). Water-soluble high molecular weight protein fractions of cataractous lenses also showed truncated βB1 and βA3 (22).
Earlier studies suggested that the N-terminal extension of β-crystallin played an important role in oligomerization (23). The role of these extensions in β-crystallin association has been studied by comparing the biophysical properties of N-terminally deleted variants with intact protein. That these studies have not given a clearer picture is due among other things to the varying extent of the N-terminal deletions made. It was shown, for example, that residues 1-22 of the N-terminal extension of βA3 are not required for self-association (24); however, loss of residues 1-30 increases its tendency to self-associate (25) and significantly increases the enthalpy and entropy of binding relative to full length protein (26). Gupta et al. (17) showed that the loss of 21, 22, and 30 N-terminal residues resulted in oligomerization without changes in secondary structure. Also, truncated forms of βB1 with N-terminal deletions of 6 and 41 residues show increased self-association as determined using light scattering (27). Removal of N-terminal 41 residues of βB1, which are extended outside the globular domain of the protein has also been shown to suppress oligomerization and prevent protein crystallization (28). It has also been reported that deletion of 15 residues from the N-terminus of βB2 causes no apparent changes in physical properties compared to intact protein (26;29). However, deletion of 41 residues from N-terminal arm of βB1 did not result in major structural alterations (28).
All acidic-basic pairs of β-crystallins, except βA4:βB's, show strong hetero-molecular interactions (30) and in vivo, most β-crystallins are present as hetero-dimers (31). At low concentrations, βA3:βB2 form hetero-dimers (6;32) and at higher concentration undergo further oligomerization to form hetero-tetramers (6;9). In the βA3:βB2 complex, both the flexible N- and C- terminal extensions of βB2 are solvent shielded whereas the N-terminal extension of βA3 is solvent accessible (6). This suggests terminal extensions of βB2 play a role in the heteromolecular complex formation More recently it was shown that deamidation of residues at the interface in the βA3 dimer decreased formation of the hetero-tetramer with βB1 and heterodimer with βB2 (33).
Previously we demonstrated that βB1 and βA3 spontaneously form a reversible heteromolecular tetramer complex (34). In our present work we used N-truncated forms of βA3 (βA3ΔN30) and βB1 (βB1ΔN56) to study the effects of the deletions on this association. To gain additional insight into the role of the N-terminal extension of βB1, we used limited proteolytic digestion to produce the βB1ΔN47. Structural models and sequences of the crystallins described in the study are shown in Figure 1. Our results indicate that the N-terminal extensions stabilize the crystallins as their removal increases the tendency of both βA3 and βB1 to self-association. Whereas βB1ΔN56 does not form heteromolecular complexes with the βA3 (or βA3ΔN30) the tryptic digest βB1ΔN47 forms tight heteromolecular complexes with βA3. These results are discussed in relationship to the structure of the N-terminal extensions and the in-vivo processing of the crystallins.
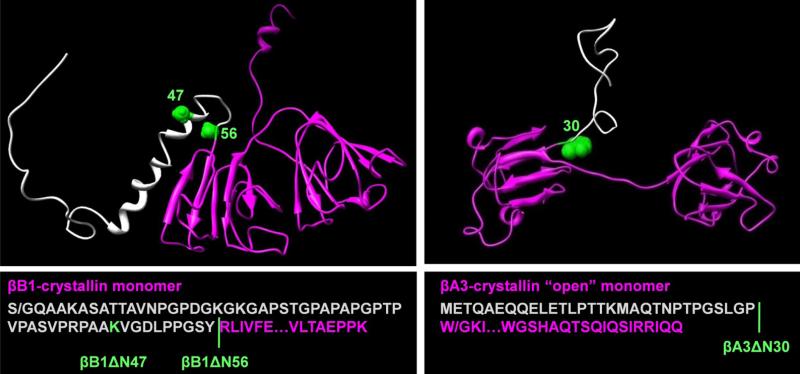
Figure 1. Three-dimensional models and sequences of βB1 and βA3 and N-terminal deletion variants.
The models of murine βB1 and human βA3 were generated by homology modeling as previously described (25). In βB1, the N-terminal extension consists of four regions where residues 1-28 are disordered, 29-41 and 43-47 form two short helical stretches and 48-57 a loop region connecting helix-2 and the main structural domain. The N-terminal extensions are colored grey, the sites of truncations, green, and the core structures, magenta. In βB1, S/G indicates mutation of Ser to Gly; in βA3, W/G indicates the mutation of Trp to Gly.
Materials and Methods
Expression and purification of βB1 and βB1ΔN56
Murine βB1 was expressed and purified as described previously (34). Truncated mutant (βB1ΔN56), where N-terminal residues 1-56 were deleted, was expressed using the plasmid pET/βB1ΔN56 in BL21(DE3) pLysS E .coli strain (Invitrogen, CA). While βB1 is expressed in E.coli cells as both soluble and aggregated (inclusion body) protein, βB1ΔN56 formed only inclusion bodies (Fig. 2A, lanes P and S1). As we were unable to fold aggregated βB1ΔN56 extracted with urea or guanidine hydrochloride we attempted to express βB1ΔN56 as a soluble protein. The effects of IPTG concentration (0, 0.1, 0.5, 1.0 mM), temperature (16, 21, 28, 37 °C), and induction times (0.5, 1, 2, 6, 12, 16, and 24 h) were studied and optimum conditions for soluble protein expression were: 0mM IPTG, 21°C, and 16 h (Fig. 2A, line S2); however, the yield of soluble protein was still low compared to the full length βB1. Cells were harvested by centrifugation at 5000 × g, 4°C for 15 min and the pellets were resuspended in 50 mM Tris-HCl, 1 mM EDTA, 0.15 M NaCl, 1 mM DTT, 50 μM TCEP, at pH 7.5 (Buffer A). The suspended cells were disrupted by sonication, and centrifuged at 14000 rpm for 30 min at 4°C. Briefly, purification of βB1ΔN56 was performed by DEAE anion exchange, size-exclusion and monoQ anion exchange chromatographies. The purity and identity of βB1 and βB1ΔN56 were confirmed by SDS-PAGE and liquid chromatography mass spectrometry, respectively.
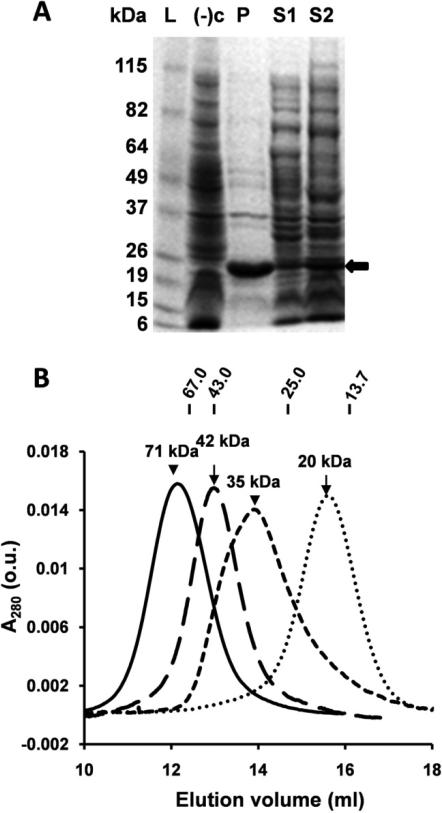
Figure 2. E. coli expression of β-crystallins and size fractionation of purified proteins.
(A) SDS-PAGE of βB1ΔN56; L, molecular weight marker; (−)c, BL21(DE3) competent cells only; P, pellet (insoluble fraction); S1, supernatant (soluble fraction) 1 mM IPTG, 37°C, 2 h; S2, supernatant, no IPTG, 21°C, 16 h (βB1ΔN56 protein indicated by black arrow); (B) Chromatography using Superdex 75 of βB1(short dash line), βB1ΔN56 (solid line), βA3 (dash line), and βA3ΔN30 (dotted line). Elution positions and molecular weights of protein standards are shown uppermost in figure.
Expression and purification of βA3 and βA3ΔN30
Human βA3 and βA3ΔN30 were expressed as a soluble proteins with high yields in E. coli and purified by anion-exchange and size-exclusion chromatography as described previously (34). In truncated βA3 (βA3ΔN30), N-terminal residues 1-30 were deleted and residue 31 was replaced by glycine (W/G). cDNA encoding βA3ΔN30 was cloned into pET-20b(+) then used to transform competent BL21(DE3) cells (Invitrogen, CA). Bacterial cultures were grown to an OD600nm of 0.6-0.9, and induced for 2 h with 0.5 mM IPTG at 37°C. The harvested cell pellet resuspended in Buffer A with Complete Protease Inhibitor cocktail (Roche, IN), sonicated on ice and centrifuged. Supernatant was dialyzed overnight against 2 l of Buffer B (50 mM Tris-HCl, 1 mM EDTA, 1 mM DTT, 50 μM TCEP, pH 7.8), then loaded on a 5 ml HiTrap DEAE FF anion exchange chromatography column (GE Healthcare, NJ), and eluted with a gradient of 0 to 1 M NaCl in Buffer B. 2.5 ml fractions containing βA3ΔN30 were pooled, concentrated and applied to a 120 ml Superdex 75 HR16/60 SEC column previously equilibrated with Buffer A at a flow rate 0.5 ml/min and 0.5 ml fractions were collected.
Tryptic digestion of βB1
Murine βB1 (0.5mg/ml) in 50mM Tris-HCl, 150 mM NaCl, 1mM EDTA, and 1 mM TCEP, pH 7.5 (Buffer C) was incubated with 2.5% w/w trypsin (Trypsin Gold, Promega) for 8 min at 37°C. The reaction was quenched with 2 mM AEBSF (Pefabloc SC, Roche, IN), 1.5 M urea added and the sample applied to a Superdex 200 column (1.6 cm × 50 cm) equilibrated with Buffer C. The peak fractions were collected and analyzed by SDS-PAGE and mass spectrometry.
Liquid Chromatography - Mass Spectrometry
A HP1100 LC-MS electrospray mass spectrometer (Agilent) coupled to a Zorbac C-3, 2.1 mm × 15 cm column was used. Protein (~ 0.1 – 0.5mg/ml) was diluted with H2O or 10% formic acid 1:20 to 1:50 and sample (5 μl) applied to the column which was washed for 15 minutes with 0.1% formic acid, 5% acetonitrile then a 35 min gradient 5% to 100% acetonitrile applied. The flow rate was 0.2 ml/min and gradient eluate was analyzed by MS.
Circular dichroism
Circular dichroic spectra were recorded in the far-UV region (185 – 260 nm) using a Jasco 500A spectropolarimeter and 0.02 cm path-length cells. Samples were in 10 mM sodium phosphate buffer with 50 μM TCEP (pH 7.0) All spectra were averaged over 10 scans and were corrected by subtraction of a buffer blank. Mean residue ellipticities were expressed for all wavelengths as deg × cm2 × d.mol−1 and were calculated from the equation [θ] = θobs × M /10 × d × c, where θobs is the measured ellipticity in degrees, M, the mean residue molecular weights (113, 116, and 117 for βB1, βB1ΔN47, and βB1ΔN56, respectively), d the optical path in cm and c the protein concentration in grams per milliliter.
Fluorescence spectroscopy
Fluorescence emission spectra of individual and mixed crystallins (10 μM) were recorded after 0 and 24 h of incubation at room temperature in 10 mM sodium phosphate buffer, 50 μM TCEP, pH 7.0 plus or minus 8 M urea. Spectra were measured using a Cary Eclipse Fluorescence Spectrophotometer (Varian, CA). Proteins were excited at 285 nm and the emission recorded 300 - 400 nm. The tryptophan analogue N-acetyl-L-tryptophanamide was used as a model for the intrinsic tryptophan emission.
Analytical Ultracentrifugation
Sedimentation equilibrium experiments were performed using a Beckman Optima XL-I analytical ultracentrifuge, absorption optics, an An-60 Ti rotor, and standard double-sector centerpiece cells. All analyses were performed using duplicate protein samples. Prior to a centrifugation proteins, >95% purity on SDS-PAGE, were incubated for 1h at room temperature with 10 mM DTT and 1.5 M urea as previously described (25;26;34), dialyzed for 24 h at 4 °C against Buffer A, then adjusted to ~ 0.4 mg/ml. Data were collected after 16 hours of centrifugation at 14,500 - 17,500 rpm at 20°C. The baselines were established by overspeeding at 45,000 rpm for another 3 hours. Equilibria profiles were analyzed by standard Optima XL-I Origin-based data analysis software. Solvent density was estimated as previously described (35). Monomeric molecular weights and molar extinction coefficients were used for calculation of dissociation constants (Kd). Mr and Kd values were averaged from at least two experimental runs using duplicate samples.
Association studies of β-crystallins
Purified proteins were dialyzed overnight against Buffer A in Slide-a-Lyser casettes (Pierce, IL) at 4°C. Protein concentrations were estimated from A280nm using absorbencies calculated from cDNA sequences (NCBI sequence βB1, NP_076184 and βA3, NP_005199) and adjusted to 0.5 mg/ml. Equimolar mixtures were incubated at room temperature. Aliquots for SEC (250 μL and native gel (10 μL) were sampled at various time points up to 24h, immediately frozen in ethanol-dry ice and stored at −80°C.
Size Exclusion Chromatography
Aliquots (250 μL) of individual β-crystallins or crystallin mixtures were applied to an analytical grade 24 ml Superdex 75 HR10/30 column, precalibrated with low-molecular weight protein standards (bovine serum albumin, 67 kDa, ovalbumin, 43 kDa, chymotrypsinogen, 25 kDa, and ribonuclease A, 13.7 kDa; Sigma, MO). The column was equilibrated in Buffer A, eluted at a flow rate of 0.5 ml/min and 0.5 ml fractions collected. Native gel electrophoresis on 4–15% gradient Tris-glycine gels (Bio-Rad, CA) was performed using 10 μl aliquots of protein controls or β-crystallin mixtures.
Results
Expression and physical properties of β-crystallins
In previous work we showed that βB1 exhibited mixed expression in E. coli as both soluble and aggregated (inclusion body) protein was produced (34). Removal of N-terminal extension resulted in expression of only inclusion body protein (Fig. 2A, lanes P and S1). Many proteins can be readily folded from denaturant solubilized aggregates but βB1ΔN56 was refractory to this approach. By modifying the expression conditions by, for example, using lower induction temperatures (36;37), we were able to generate soluble protein (Fig. 2 lane S2). Only the soluble fraction was used for protein purification and the biophysical studies described herein. The complete aggregation of expressed βB1ΔN56 was a first indication of the reduced solubility or instability compared to wild type protein. This has not been an issue before as only partially deleted N-terminal extensions were previously studied and these proteins were soluble when produced by in vitro proteolytic digestion (5;38), recombinant expression (27;28;39) or directly isolated from the lens (11;19). Because of the proteins history, we carefully checked βB1ΔN56 for protein modification and conformational perturbation (described below). Mass spectrometry gave a mass of 22,776.29 Da consistent with the cDNA sequence which indicated the protein was intact and that no residues had been modified, factors that may lead to selective solubility during expression.
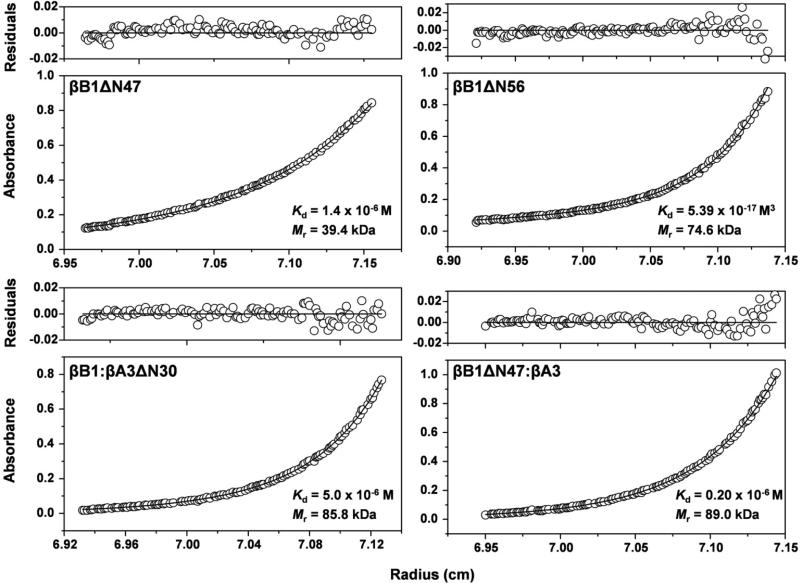
Gel filtration of βB1ΔN56 indicated an apparent molecular weight of ~ 75 kDa (Fig. 2B) which is about twice that of homodimeric βB1 (35 kDa). Removal of the N-terminal extension clearly promotes higher order protein association compared to dimeric wild type protein. A more detailed picture was obtained using sedimentation equilibrium analysis. Equilibrium profiles of βB1ΔN56 were best fitted to a monomer – tetramer system with a Kd = 5.39 ± 0.06 × 10−17 M3 and average molecular weight of 74.6 kDa (Fig. 3).
Figure 3. Sedimentation equilibrium of single and complexed β–crystallins.
The protein concentration profiles, absorbance (280 nm) vs radial distance, are indicated. The solid lines indicate the predicted monomer-dimer (βB1ΔN47) monomer-tetramer (βB1ΔN56) and heterodimer-heterotetramer (βB1ΔN47: βA3 and βB1: βA3ΔN30) systems. Open circles represent the experiment values. The upper panels of each graph show the residuals of the fitted curves to the data points.
Removal of N-terminal extension from βA3 had no effect on E. coli expression and unlike βB1ΔN56, was expressed as a soluble protein. Sedimentation analyses of βA3ΔN30 indicated a monomer – dimer system similar to wild type protein although the truncation resulted in tighter dimer formation (Table 1).
Table 1.
Physical properties of homo- and hetero-oligomers of recombinant β-crystallins
| β-crystallin | Calculated Mr1 | SEC | AUC | association model (reference) | ||
|---|---|---|---|---|---|---|
| monomer | dimer | Mr2 | Mr3 | Kd | ||
| self-association | ||||||
| mouse B1 | 28.0 | 56.0 | 35 | 47.3 | 1.5 × 10−6 M | monomer-dimer (34) |
| mouse B1ΔN56 | 22.9 | 45.8 | 71 | 74.6 | 5.4 × 10−17M3 | monomer-tetramer |
| mouse B1ΔN47 | 23.6 | 47.2 | - | 39.4 | 1.4 × 10−6 M | monomer-dimer |
| human A3 | 25.1 | 50.2 | 42 | 42.6 | 4.5 × 10−6 M | monomer-dimer |
| human A3ΔN30 | 21.9 | 43.8 | 20* | 44.3 | 0.1 × 10−6 M | dimer |
| mouse A3 | 25.2 | 50.4 | 40 | 42.6 | 5.1 × 10−6 M | monomer-dimer (26) |
| mouse A3ΔN30 | 21.8 | 43.6 | 23* | 42.7 | 0.7 × 10−6 M | monomer-dimer (26) |
| mouse B2 | 23.4 | 46.8 | 37 | 37.6 | 4.6 × 10−6 M | monomer-dimer (26) |
| mouse B2ΔN17 | 21.7 | 43.4 | 36 | 34.5 | 9.7 × 10−6 M | monomer-dimer (26) |
| hetero-association | ||||||
| B1:A3 | 28.0:25.1 | 53.1 | 71 | 96.0 | 1.1 × 10−6 M | dimer-tetramera (34) |
| B1:A3ΔN30 | 28.0:21.8 | 49.8 | 70 | 85.8 | 5.0 × 10−6 M | dimer-tetramer |
| B1ΔN56:A3 | 22.9:25.1 | 48.1 | 47 | - | - | no association |
| B1ΔN47:A3 | 23.6:25.1 | 48.7 | - | 89.0 | 0.2 × 10−6 M | dimer-tetramer |
| B1ΔN56:A3ΔN30 | 22.9:21.8 | 44.8 | - | - | - | no association |
| B2:A3 | 23.4:25.2 | 48.6 | 49 | 50.0 | - | hetero-dimerb (6) |
| B2:A3 | 23.4:25.2 | 48.6 | - | 90.0 | - | hetero-tetramerc (6) |
Polypeptide molecular weights in kDa were calculated from the protein sequences (consistent with NCBI sequence of βB1-NP_076184, βA3 - NP_005199, and βB2 - NP_031799). Murine βA3 and βB2 show 95%, and 97% identity with their human species, respectively with ~50% sequence identity shared among the pairs of β-crystallins independently on species; the globular domains of human and mice βB1-crystallin are about 80% identical and ~92% similar by their amino acid properties which provide a reasonable basis for comparison a role of their globular domains and terminal extensions on β-crystallin association (34)
Apparent molecular weights in kDa analyzed by size exclusion chromatography
Weight-average molecular weights in kDa determined by AUC; Kd (M) dissociation constants from analytical ultracentrifugation. Samples were analyzed in the concentration range of 0.5-2 mg/mla, <1 mg/mlb, >1 mg/mlc; 4The wild-type and truncated βB1 and βA3 were expressed in E.coli (34) and βB2 was expressed in baculovirus system (26) or isolated from bovine lenses (6)
Proteins which elutes on SEC as apparent monomers due to the interaction with the column matrix).
Identification of a flexible and solvent accessible region of the βB1 N-terminal extension
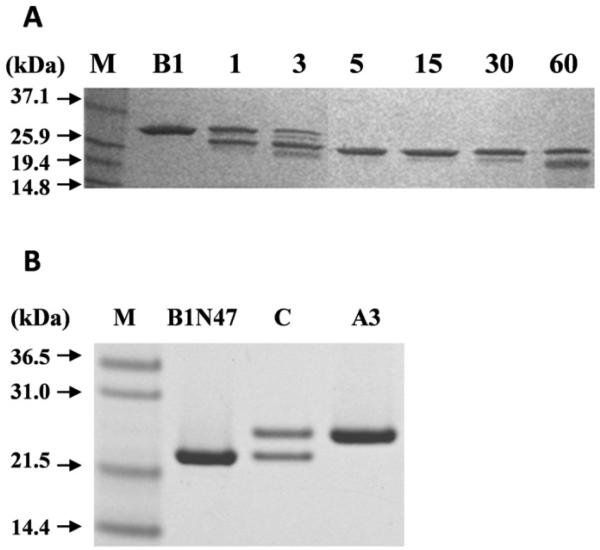
As removal of the complete N-terminal extension of βB1 had such a profound effect on the physical properties of βB1 we asked if there was a minimal length or a cut-off point in this region which switches the behavior of the protein. As a simple first approach the structure of βB1 was probed using limited proteolytic digestion. Trypsin was used because of its very reliable specificity and fact that there is good distribution of basic residues throughout the protein. For example, in the N-terminal extension of the murine protein there are basic residues at positions 5, 19, 21, 43, 47 and 57 (all these sites are matched in the human sequence except at position 43). The time course for digestion (Fig. 4A) shows a single highly specific cleavage occurring in less than 5 min. Mass spectrometry (LC-MS) determined that the digestion product had a mass of 23,601.8 Da. This corresponded to residues 48 – 240 and that tryptic cleavage occurred C-terminal to lysine 47. This cleavage, because of both the rate and selectivity must occur at an accessible and highly mobile region, is significant as it corresponds to the in-vivo sites processed by gelatinase B (40) and m-calpain (41). Although the C-terminal region has been identified as accessible to protease activity (5) under the condition used, we did not observe any major processing in this region with trypsin even after extended digestion times.
Figure 4. Tryptic digestion of βB1: analytical scale time course and preparative scale.
(A) SDS-PAGE of tryptic digestion of βB1 (B1) with 2.5% (w/w) trypsin, times indicated in min. Black arrows indicate molecular mass of the protein marker (M). (B) SDS-PAGE of large scale βB1 tryptic digestion followed by gel filtration purification: βB1ΔN47 (B1N47); equimolar mixture of βB1ΔN47: βA3 incubated for 24h at RT (C) and used for analytical ultracentrifugation; βA3 used for complex formation studies (A3).
To study βB1ΔN47 in detail, the tryptic digestion was scaled up and the processed protein isolated by gel filtration (Fig. 4A). Analysis by sedimentation equilibrium indicated a monomer – dimer system with a Kd = 1.4 × 10−6M and average molecular weight of 39.4 kDa (Fig. 3) values similar to those of the wild type protein (Table 1). This is in marked contrast to that of βB1ΔN56 which, as described above, undergoes further association to form tetramers (Fig. 3).
Conformational analyses of β-crystallins
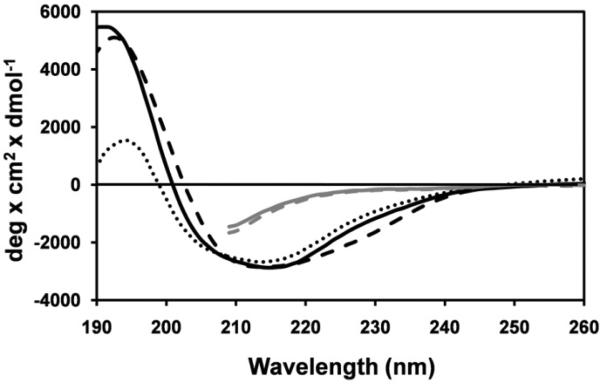
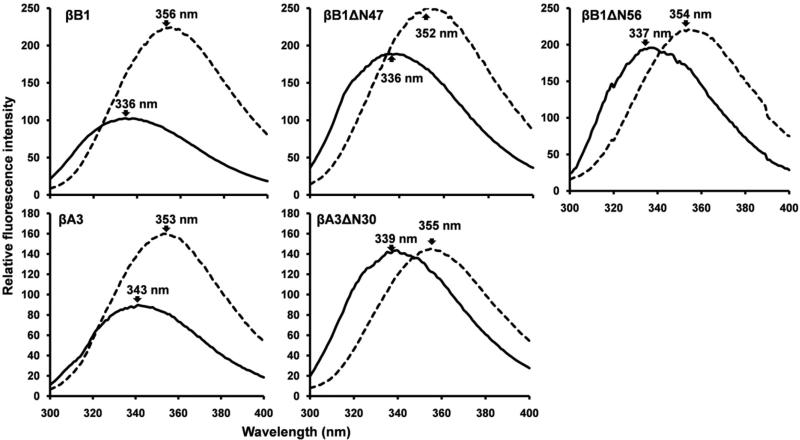
The Far-UV circular dichroic spectra of βB1 and the two truncated variants βB1ΔN56 and βB1ΔN47 all indicated predominately β-sheet secondary signatures characterized by low molar ellipticities and broad negative peaks (troughs) at ~ 214 – 215 nm (Fig. 5). The small spectral difference between these proteins and the wild type standard were eliminated by unfolding the proteins with 8M urea (Fig. 5). Thus, despite major differences in the physical properties of proteins with complete (Δ56) or partial (Δ47) deletions, no significant difference in the secondary structure was observed. There is also little structural perturbation between βA3 and βA3ΔN30 (42). Conformation was also probed using fluorescence emission, very sensitive to tertiary folding, using proteins (~10 μM) at higher concentration than the Kd values of the associating systems (Table 1). Emission spectra of both βB1 and βA3 are red-shifted upon denaturation with concomitant ~ 2-fold increases in intensity (Fig. 6). As exposure of buried tryptophan on unfolding normally decreases emission intensity due to solvent exposure (higher polarity), in the native proteins there must have been quenching by neighboring residues. Unfolding the truncated variants (βB1ΔN47, βB1ΔN56 and βA3ΔN30) all resulted in red-shifts in emission maxima, similar to their wild type counterparts, with small, or no, increases in emission intensity (Fig. 6). In both βB1 and βA3 there are no aromatic residues in the N-terminal extensions, so changes in Trp emission resulting from their removal must either be due to the removal of quenching residue (s) located in N-terminal extensions or due to local conformational shifts (Table 1).
Figure 5. Far-UV circular dichroic spectra of βB1-crystallins.
Samples were in 10 mM sodium phosphate, pH 7.0. βB1, βB1ΔN56, and βB1ΔN47 are indicated by black solid, dashed, and dotted lines respectively. βB1 and βB1ΔN56 unfolded with 8M urea are indicted by gray solid and, dashed lines, respectively. Full far-UV spectra of unfolded proteins are not shown because the absorption by 8M urea precludes measurements < 210 nm (54).
Figure 6. Tryptophan fluorescence emission spectra of βB1 and βA3 and N-terminal deletion variants.
Native and unfolded proteins are indicated by solid and dotted lines, respectively. Proteins were excited at 285 nm and emission spectra recorded from 300 - 400 nm. Proteins (10μM) were in 10 mM phosphate buffer, pH 7.0, 50 μM TCEP with or without 8 M urea.
The overall results of this section indicate that the global folding of the individual crystallin N-terminal variants is native-like with no major structural perturbations but small localized conformational shifts cannot be ruled out, especially for βB1ΔN56.
Heteromolecular association of β-crystallins
βB1 associates with βA3 or βA3ΔN30 to form heterotetramer complexes
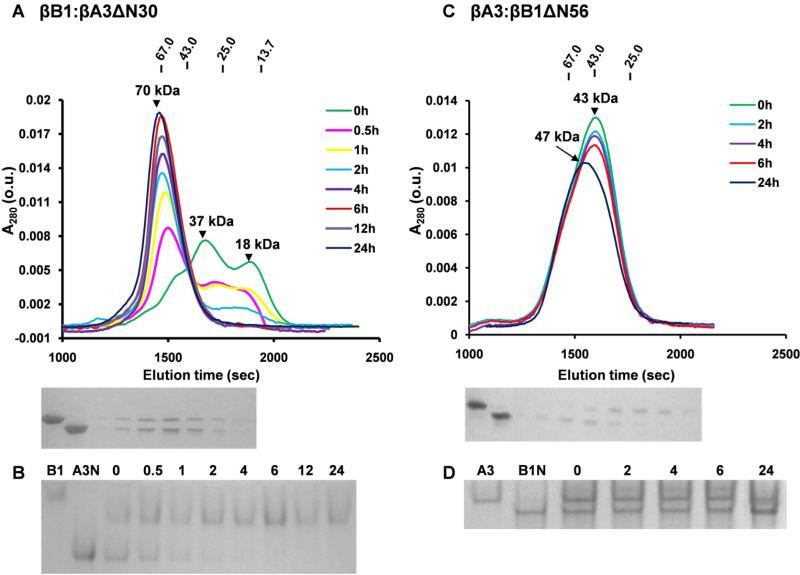
Heteromolecular complex formation between βB1 and βA3ΔN30 was monitored by gel filtration and native gel electrophoresis. After 2 h of incubation a single peak ~70 kDa formed (Fig. 7A) which corresponded to single band of intermediate mobility on electrophoresis (Fig. 7B). We previously demonstrated that βB1 and βA3 form a tetrameric complex by (hetero-dimer) – (hetero-dimer) association and using this model, the association of βB1 and βA3ΔN30 was best fitted with a Kd = 5.0 μM (Fig. 3 and Table 1). So although gel filtration showed the rate of complex formation was faster than that with wild type βA3, the overall affinity appears weaker (34).
Figure 7. Size-exclusion chromatography and native gel electrophoresis of crystalline complexes.
(A) Chromatography of 10μM equimolar mixture of βB1 and βA3ΔN30 preincubated for indicated times (h). Molecular weights of major peaks were estimated as 18 kDa and 37 kDa at 0h and 70 kDa at > 4h. Molecular weights of standards are shown uppermost of Figure. SDS-PAGE of the fractions from the 24 h chromatogram is shown below the chromatograms where Lane 1 and 2 are βB1 and βA3ΔN30 standards. (B) Native gel electrophoresis of βB1 and βA3ΔN30 mixture incubated for indicated times (h) and where lanes 1 and 2 indicate βB1 and βA3ΔN30 (C) Chromatography of a 10μM equimolar mixture of βA3 and βB1ΔN56 with matching SDS-PAGE of the fractions from 24 h chromatogram, Lanes 1 and 2 are βA3 and βB1ΔN56. (D) Native gel electrophoresis of the βA3 and βB1ΔN56 mixture incubated for indicated times (h) where βA3 and βB1ΔN56 standards are indicated.
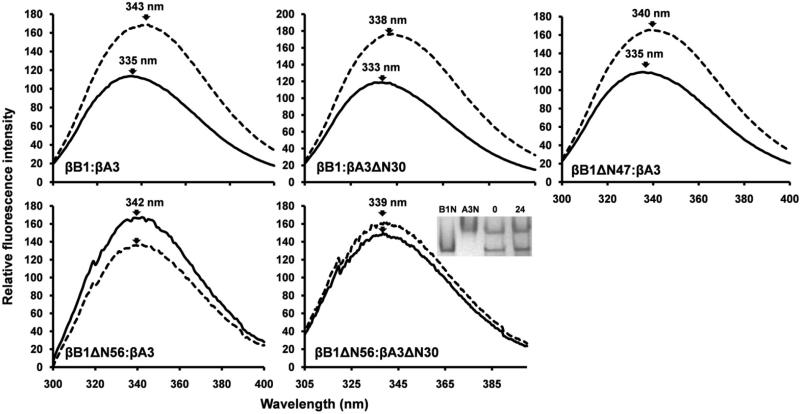
To examine potential conformational changes on complex formation, fluorescence emission spectra of mixed proteins were compared with those of individual summated (non-mixed) samples. Both βB1: βA3 and βB1: βA3ΔN30 pairs exhibited blue shifted spectra (5 – 8 nm) with reduced intensities (Fig. 8). These changes are reminiscent of protein folding and indicate surface tryptophan shielding and quenching upon protein association and are analogous to those described for βB1 with βA1/βA3 and βA4 (43). From accessibility calculations of βB1(44), four Trp are exposed or partially exposed and in the homology model of βA3 (26), five Trp are exposed or partially exposed. Some of these surface tryptophan residues in the crystallin core structures must constitute part of the hetero-dimer interaction surfaces and provide a useful monitor of protein association.
Figure 8. Tryptophan fluorescence emission spectra of β-crystallins complexes.
Spectra of equimolar mixtures following 24 h incubation are indicated by solid lines and the corresponding summated spectra of the individual (non-mixed) β-crystallin pairs by dotted lines. The insert shows native gel electrophoresis of βB1ΔN56 and βA3ΔN30 incubated for 0 and 24h, B1N and A3N refer to βB1ΔN56 and βA3ΔN30.
Tetramer formation of βB1ΔN56 is suppressed in the presence of βA3
βB1ΔN56 did not interact with either βA3 or βA3ΔN30 as only the individual dimeric proteins were observed during gel filtration (Fig. 7C). This is consistent with analytical ultracentrifugation which indicated non-interacting dimeric proteins (Table 1) and non blue-shifted emission spectra (Fig. 8). As there appears to be no stable interaction between these proteins is was of interest that tetrameric βB1ΔN56, present when protein is analyzed alone, was not observed. One possibility is that heterodimers are formed which are stable and do not undergo further association. Native gel electrophoresis; however, shows that complex formation is either very slow or does not occur (Fig. 7B). Alternatively, transitory heteromolecular interactions may occur, for example, at the surfaces (presumably hydrophobic) which mediate tetramerization of βB1ΔN56. Suppressed association may weaken over time as gel filtration peaks of the unresolved homodimers shift to higher masses with extended preincubation times, where the leading edge of the peak is more populated with βB1ΔN56 than βA3 despite its lower monomer mass (Fig. 7C).
βB1ΔN47 associates with βA3 to form a stable heterotetramer complex
We have seen that truncation of the N-terminal extension of βB1 abrogates its ability to undergo heteromolecular interaction with βA3 crystallin while at the same time increasing its tendency for higher order self-association. The removal of over 80% the βB1 extension by limited tryptic digestion; however, does not impair heteromolecular association but results in tighter complex formation. The association of βB1ΔN47 with βA3 was measured by sedimentation equilibrium and corresponded to a heterodimer- heterodimer (tetramer) association (Fig. 3). The Kd (0.2 × 10−6 M) of the system indicated a five-fold stronger affinity than previously reported for unmodified βB1 (34). Heteromolecular complex formation was also demonstrated by fluorescence spectroscopy which shows a characteristic blue shifted emission spectrum, almost identical to that of the βB1: βA3 complex (Fig. 8).
Summary of molecular interactions
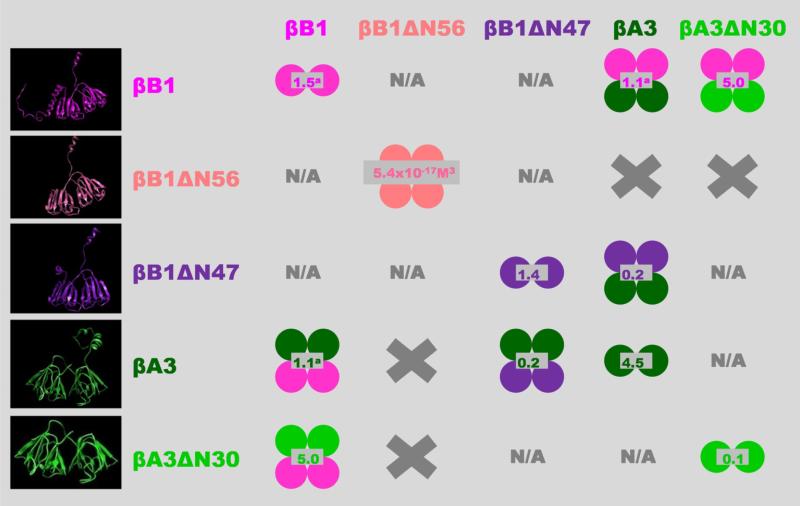
A schematic overview of the main findings of this study are summarized in Figure 10 and the physical properties of various individual and complexed recombinant crystallin, both from this study and those previously published, are given in Table 1.
Figure 10. Overview of β-crystallin homo- and hetero-association.
The indicated shifts in the oligomeric equilibria as a result of N-terminal truncations may also occur in the lens but this is difficult to prove directly due to the high in-vivo protein concentrations. Some support for this, however, is that the proteolytic processing of crystallins causes cataract formation in mice (40). Single circles represent monomeric units; homo-oligomers are shown in single colors; hetero-oligomers bicolored; values in gray indicate dissociation constants (Kd) in μM (except for βB1ΔN56); left panel show β-crystallins models from top to bottom: βB1 full length and βB1 with N-terminal truncation of 56 (βB1ΔN56) and 47 (βB1ΔN47) residues; βA3 full length and with N-terminal truncation of 30 residues (βA3ΔN30). X, no association between indicated β-crystallins and N/A. interaction not characterized.
a Values previously determined (34).
Discussion
The available crystallin structures βB1 and βB2 (PDB: 1blb, 1oki) do not include the N-terminal extensions although based on their susceptibility to both in-vivo and in -vitro protease processing are likely to be solvent accessible, flexible and probably unstructured (38;44;45). Although we cannot directly assign structure to these regions, based on homology modeling they may include microdomains with helical structure ((16) and Fig. 10). Related to the lack of structural detail is the fact that functional roles of N-terminal extensions are also unclear despite much study. That they stabilize crystallin structure is suggested by their partial removal often leading to increased protein association and aggregation (25;27;38;46). This is complicated by the fact that crystallins exist as complexes and N-terminal modifications may also stabilize or destabilize heteromolecular interactions (6;9;31). To clarify the role of the N-terminal extensions of the two major eye lens crystallins βB1 and βA3 we prepared recombinant deletion mutants corresponding to the boundaries of the core structures of these proteins. The physical and conformational properties of these mutants βA3ΔN30 and βB1ΔN57, lacking 30 and 57 N-terminal residues were studied. Specifically, we used analytical ultracentrifugation for the direct determination of molecular masses and equilibrium binding constants (summarized in Table 1). Fluorescence emission measurements were useful for assessing both the conformational integrity of the individual N-terminal deletion mutants (Fig. 6) and for monitoring their heteromolecular protein interactions (Fig. 8).
Crystallin N-terminal extensions mediate protein interaction
From the results of this study it is clear that removal of the N-terminal extension of βB1 has major effects on the physical properties of the protein by increasing the self-association potential and by blocking heteromolecular associations with βA3 ((34), Figs. 3 and 7). On the other hand, removal of the N-terminal extension of βA3 has little effect on either its dimerization potential or its ability to complex with βB1. Complex formation between βA3 and βB2 also indicates lack of involvement of the N-terminal extension of the former which remains solvent accessible (6). The long 56 residue N-terminal extension of βB1 may directly interact with βA3 at the dimer interface allowing the formation of heterodimers which then further associate into tetramers. This type of ‘holdase‘ role of terminal extensions has been discussed earlier for other proteins (47) and although we do not have direct structural evidence for such a role in βB1 we have shown that its removal abrogates heteromolecular interaction with βA3.
Apart from potential direct interactions, the presence or absence of the N-terminal extensions may have other consequences, namely, shifting the balance between so-called ‘closed’ and ‘open’ conformational isomers (25). The ‘open’ conformation was determined experimentally for βB2 (PDB: 1blb) (48). Later it was experimentally confirmed that β-crystallins might adopt the ‘closed’ conformation as shown in the structure of βB1 (PDB file: 1oki) (44). Hence in βB1, the large N-terminal arm could affect the kinetics and equilibrium position of these two conformations. This would be important if one conformation presented a favored interface for interaction with βA3.
Although N-terminal arms of crystallins may directly affect the first step in association pathways, namely, homodimer and/or heterodimer formations, other interfaces in the main core domains are probably more importance for mediating higher order associations. This is indicated by the blue shifted fluorescence emission accompanying complex formation between, for example, βB1 and βA3, and is due to solvent shielding of surface Trp(s) on core domains (Fig. 8). While removal of the N-terminal extension from βB1 results in higher order self association to form homotetramer (Fig. 8), heteromolecular interactions with βA3 are blocked. If we assume homotetramer is formed by dimer – dimer association, the interface between dimers is likely to be similar to that which stabilizes heterotetramers. Future determination of the structure of heteromolecular complexed crystallins will provide the detailed information presently lacking.
The structural basis of N-terminal extension processing
It is well documented that the N-terminal extensions of crystallins are very susceptible to in-vivo proteolytic processing (40;49-51); however, it is less clear which proteases are physiologically significant and what, if any, are the triggering mechanisms. Based on the dramatic effects of completely removing the N-terminal of βB1 we were concerned that although the deleted sequence conformed to the structural domain determined by protein crystallography (44) it had no physiological relevance as no in-vivo processing at this site has been reported. Other groups, however, have also considered fully N-terminal truncated βB1 relevant and indeed whilst we were preparing this manuscript a report appeared showing that truncated βB1 had degraded physical properties including inability to complex with βA3 (52). As different techniques and His-tagged proteins were used in this study it is difficult to compare in detail these results with our data.
As complete removal of the N-terminal domain of βB1 has such a large impact on its physical properties we were interested in defining more precisely the structural boundaries of the N-terminal region. This cannot be approached directly because as mentioned, the N-terminal domains are not included in any X-ray models, so limited proteolytic digestion is an obvious first choice given the sensitivity of this region to in-vivo processing (40;44;45;51). We have described the remarkable selectivity with which tryptic processing of βB1 occurs resulting in deletion of over 80% of the N-terminal extension (Figs. 1 and 4). The trypsin digested product βB1ΔN47 retained the physical properties of the intact protein exhibiting stable monomer-dimer association and also the ability to form stable hetero-molecular tetramer complexes with βA3 (Fig. 3). We note that the deletion proteins, βB1ΔN47 and βB1ΔN56, do not have a stable oligomeric structure at low protein concentrations as they are in dynamic equilibria where monomers/dimers are associating and dissociating. Therefore, there is no difference in the effect of the N-terminal extension on association for mutant proteins prepared by either site-directed mutagenesis or proteolytic digestion. It is interesting to note that the circular dichroic spectrum of βB1ΔN47 is very similar to that of βB1ΔN57 (Fig. 5) and deconvolution of either spectrum indicates apparent change in secondary structure compared to wild type protein (data not shown). Yet this difference cannot account for the altered physical properties, as βB1ΔN47 behaves similar to wild type protein. More likely they point to the difficulties in estimating secondary structure of proteins with high β-sheet content (53) and the fact that the N-terminal domain is partially structured.
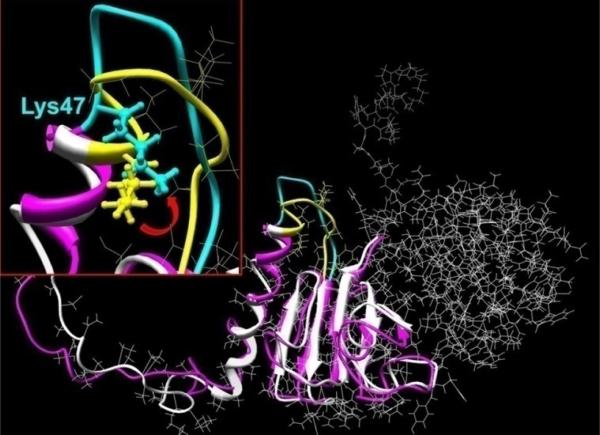
The selectivity of the tryptic digestion may be explained on a structural basis (Fig. 1). Molecular modeling suggests that the βB1 N-terminal extension comprises an unstructured region (residues 1 to 28), two helical segments (29- 41 and 43- 47) and a loop-like domain (resides 48-57). In contrast to typical α-helices, which are saturated with hydrogen bonds, the stability of the N-terminal helices is derived from proline residues. Helix-1 is formed by 6 repeats of the dipeptide Pro-X and the shorter Helix-2 contains two such repeats. In both helices, only a few hydrogen bonds were found. Several changes in the N-extension were observed during molecular dynamics simulations (Fig. 9). First after 20 ps of structure equilibration, a partial ‘melting’ of the proline-based helices occurred with conformational change in a highly mobile loop region in which lysine 47 has a high solvent accessibility (Fig. 9 insert). These structural features provide basis for the susceptibility of this region for tryptic processing and for the in-vivo processing by gelatinase B (40) and m-calpain (41). Furthermore, they indicate that the post-translational processing of the βB1 N-terminal extension is defined by its structure and conformation and not the specificity of any particular protease.
Figure 9. Molecular model the βB1 N-terminal extension.
The murine βB1 crystallin structure was determined using homology modeling and human truncated βB1-crystallin (PDB file: 1oki) as the structural template. Hydrogen atoms were added to the structure which was regularized using OPLS_2005 potentials with 12 Å non-bonded cut-offs. Molecular dynamics in water were performed with the Impact module of the Maestro program software (version 8.0.308: Schrödinger Inc., New York). The initial structure of βB1 monomer is shown in white ribbons and after 20 ps MD equilibration as pink ribbons (details shown for residues 1 to 142). The insert shows structural details of residues 47-57 where the initial and equilibrated structures are indicated by yellow and cyan, respectively. The location of the tryptic cleavage site (between K47V48) is indicated by the ball and stick rendition and the red arrow shows approximate direction of Lys 47 and the peptide movement.
Conclusions
The crystallin N-terminal extensions are susceptible to temporal in-vivo proteolytic processing and this appears to impact protein interaction potentials. As the in-vivo processing endpoints are variable (40;49-51) we used structural boundaries to define the lengths of the extensions. In βB1, removal of the extension strengthens the propensity for self-association thus weakening or blocking ability to participate in heteromolecular associations with βA3. Removal of the corresponding extension from βA3 has little effect on dimerization and did not block heteromolecular association with βB1. We have suggested that the N-terminal arms mainly influence initial homo- and heteromolecular dimerization and that interfaces in the main core domains mediate the higher order associations. In-vitro proteolytic processing of βB1 simulates in-vivo processing and provided structure – function insight by showing that removal of over 80% of the βB1 extension rather than inhibiting heteromolecular associations actually enhances them. Molecular modeling suggests that the βB1 N-terminal although largely disordered and flexible contain regions with helical propensity which position a flexible mobile loop region creating a protease “hot spot”. Cleavage specificity is, thus, structure driven and the physiological significance of any one particular proteases reported to process this region remains to be more clearly established. The structural significance of the small stretch of βB1 sequence 48-57 also requires further investigation as its presence or absence has a profound effect on protein interactions.
Acknowledgment
The authors wish to thank to Dr. Oleg Voloshin (NIDDK, NIH) for assistance with circular dichroism and to Dr. Fielding Hejtmancik (NEI, NIH) for useful discussions and criticism.
Abbreviations
- βA3
human βA3-crystallin
- βA3ΔN30
residues 1-30 deleted
- βB1
murine βB1-crystallin
- βB1ΔN56 and βB1ΔN47
deletion of 1-56 and 1-47 residues, respectively
- IPTG
isopropyl-β-D-thiogalactopyranoside
- SEC
size-exclusion chromatography
- AUC
analytical ultracentrifugation
- Tris
tris-hydroxymethylaminomethane
- EDTA
ehtylendiaminetetraacetic acid
- DTT
1,4-Dithio-DL-threitol
- TCEP
Tris[2-carboxyethyl]phosphine hydrochloride
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- AEBSF
(Pefabloc SC), 4-(2-Aminoethyl)-benzenesulfonyl fluoreide hydrochloride
- LC-MS
Liquid Chromatography Mass Spectrometry
- PTMs
post-translational modifications
References
- 1.Delaye M, Tardieu A. Short-range order of crystallin proteins accounts for eye lens transparency. Nature. 1983;302:415–417. doi: 10.1038/302415a0. [DOI] [PubMed] [Google Scholar]
- 2.Wistow GJ, Piatigorsky J. Lens crystallins: the evolution and expression of proteins for a highly specialized tissue. Annu. Rev. Biochem. 1988;57:479–504. doi: 10.1146/annurev.bi.57.070188.002403. [DOI] [PubMed] [Google Scholar]
- 3.Bloemendal H, de JW, Jaenicke R, Lubsen NH, Slingsby C, Tardieu A. Ageing and vision: structure, stability and function of lens crystallins. Prog. Biophys. Mol. Biol. 2004;86:407–485. doi: 10.1016/j.pbiomolbio.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 4.Lubsen NH, Aarts HJ, Schoenmakers JG. The evolution of lenticular proteins: the beta- and gamma-crystallin super gene family. Prog. Biophys. Mol. Biol. 1988;51:47–76. doi: 10.1016/0079-6107(88)90010-7. [DOI] [PubMed] [Google Scholar]
- 5.Lampi KJ, Oxford JT, Bachinger HP, Shearer TR, David LL, Kapfer DM. Deamidation of human beta B1 alters the elongated structure of the dimer. Exp. Eye Res. 2001;72:279–288. doi: 10.1006/exer.2000.0950. [DOI] [PubMed] [Google Scholar]
- 6.Werten PJ, Lindner RA, Carver JA, de Jong WW. Formation of betaA3/betaB2-crystallin mixed complexes: involvement of N- and C-terminal extensions. Biochim. Biophys. Acta. 1999;1432:286–292. doi: 10.1016/s0167-4838(99)00123-5. [DOI] [PubMed] [Google Scholar]
- 7.Zigler JS, Jr., Horwitz J, Kinoshita JH. Human beta-crystallin. I. Comparative studies on the beta 1, beta 2 and beta 3-crystallins. Exp. Eye Res. 1980;31:41–55. doi: 10.1016/0014-4835(80)90089-5. [DOI] [PubMed] [Google Scholar]
- 8.Bindels JG, Koppers A, Hoenders HJ. Structural aspects of bovine beta-crystallins: physical characterization including dissociation-association behavior. Exp. Eye Res. 1981;33:333–343. doi: 10.1016/s0014-4835(81)80056-5. [DOI] [PubMed] [Google Scholar]
- 9.Slingsby C, Bateman OA. Qarternary interactions in eye lens beta-crystallins: basic and acidic subunits of beta-crystallins favor heterologous association. Biochemistry. 1990;29:6592–6599. doi: 10.1021/bi00480a007. [DOI] [PubMed] [Google Scholar]
- 10.Berbers GAM, Boerman OC, Bloemendal H, de Jong WW. Primary gene products of bovine beta-crystallin and reassociation behavior of its aggregates. Eur. J. Biochem. 1982;128:495–502. doi: 10.1111/j.1432-1033.1982.tb06992.x. [DOI] [PubMed] [Google Scholar]
- 11.Ajaz MS, Ma Z, Smith DL, Smith JB. Size of human lens beta-crystallin aggregates are distinguished by N-terminal truncation of betaB1. J. Biol. Chem. 1997;272:11250–11255. doi: 10.1074/jbc.272.17.11250. [DOI] [PubMed] [Google Scholar]
- 12.Hanson SR, Hasan A, Smith DL, Smith JB. The major in vivo modifications of the human water-insoluble lens crystallins are disulfide bonds, deamidation, methionine oxidation and backbone cleavage. Exp. Eye Res. 2000;71:195–207. doi: 10.1006/exer.2000.0868. [DOI] [PubMed] [Google Scholar]
- 13.Lampi KJ, Ma Z, Hanson SR, Azuma M, Shih M, Shearer TR, Smith DL, Smith JB, David LL. Age-related changes in human lens crystallins identified by two-dimensional electrophoresis and mass spectrometry. Exp. Eye Res. 1998;67:31–43. doi: 10.1006/exer.1998.0481. [DOI] [PubMed] [Google Scholar]
- 14.Han J, Schey KL. MALDI tissue imaging of ocular lens alpha-crystallin. Invest Ophthalmol. Vis. Sci. 2006;47:2990–2996. doi: 10.1167/iovs.05-1529. [DOI] [PubMed] [Google Scholar]
- 15.Robinson NE, Lampi KJ, Speir JP, Kruppa G, Easterling M, Robinson AB. Quantitative measurement of young human eye lens crystallins by direct injection Fourier transform ion cyclotron resonance mass spectrometry. Mol. Vis. 2006;12:704–711. [PubMed] [Google Scholar]
- 16.Sergeev YV, David LL, Chen H-C, Hope JN, Hejtmancik JF. Local microdomain structure of terminal extensions in betaA3- and betaB2-crystallins. Molecular Vision 4. 1998 [PubMed] [Google Scholar]
- 17.Gupta R, Srivastava K, Srivastava OP. Truncation of motifs III and IV in human lens betaA3-crystallin destabilizes the structure. Biochemistry. 2006;45:9964–9978. doi: 10.1021/bi060499v. [DOI] [PubMed] [Google Scholar]
- 18.Harding JJ, Crabbe MJC. The lens. Development, proteins, metabolism and cataract. In: Davson H, editor. The Eye vol.IB. 3 ed. Academic Press; Orlando: 1984. pp. 207–492. [Google Scholar]
- 19.David LL, Lampi KJ, Lund AL, Smith JB. The sequence of human betaB1-crystallin cDNA allows mass spectrometric detection of betaB1 protein missing portions of its N-terminal extension. J. Biol. Chem. 1996;271:4273–4279. doi: 10.1074/jbc.271.8.4273. [DOI] [PubMed] [Google Scholar]
- 20.Lampi KJ, Ma Z, Shih M, Shearer TR, Smith JB, Smith DL, David LL. Sequence analysis of betaA3, betaB3, and betaA4 crystallins completes the identification of the major proteins in young human lens. J. Biol. Chem. 1997;272:2268–2275. doi: 10.1074/jbc.272.4.2268. [DOI] [PubMed] [Google Scholar]
- 21.Takemoto L, Takemoto D, Brown G, Takehana M, Smith J, Horwitz J. Cleavage from the N-terminal region of beta Bp crystallin during aging of the human lens. Exp. Eye Res. 1987;45:385–392. doi: 10.1016/s0014-4835(87)80125-2. [DOI] [PubMed] [Google Scholar]
- 22.Harrington V, McCall S, Huynh S, Srivastava K, Srivastava OP. Crystallins in water soluble-high molecular weight protein fractions and water insoluble protein fractions in aging and cataractous human lenses. Mol. Vis. 10. 2004:476–489. [PubMed] [Google Scholar]
- 23.Berbers GAM, Hoekman WA, Bloemendal H, de Jong WW, Kleinschmidt T, Braunitzer G. Proline- and alanine-rich N-terminal extension of the basic bovine beta-crystallin B1 chains. FEBS Lett. 1983;161:225–229. doi: 10.1016/0014-5793(83)81013-8. [DOI] [PubMed] [Google Scholar]
- 24.Werten PJL, Carver JA, Jaenicke R, de Jong WW. The elusive role of the N-terminal extension of betaA3- and betaA1-crystallin. Protein. Eng. 1996;9:1021–1028. doi: 10.1093/protein/9.11.1021. [DOI] [PubMed] [Google Scholar]
- 25.Sergeev YV, Wingfield PT, Hejtmancik JF. Monomer-dimer equilibrium of normal and modified beta A3-crystallins: experimental determination and molecular modeling. Biochemistry. 2000;39:15799–15806. doi: 10.1021/bi001882h. [DOI] [PubMed] [Google Scholar]
- 26.Sergeev YV, Hejtmancik JF, Wingfield PT. Energetics of Domain-Domain Interactions and Entropy Driven Association of beta-Crystallins. Biochemistry. 2004;43:415–424. doi: 10.1021/bi034617f. [DOI] [PubMed] [Google Scholar]
- 27.Bateman OA, Lubsen NH, Slingsby C. Association behaviour of human betaB1-crystallin and its truncated forms. Exp. Eye Res. 2001;73:321–331. doi: 10.1006/exer.2001.1038. [DOI] [PubMed] [Google Scholar]
- 28.Annunziata O, Pande A, Pande J, Ogun O, Lubsen NH, Benedek GB. Oligomerization and phase transitions in aqueous solutions of native and truncated human beta B1-crystallin. Biochemistry. 2005;44:1316–1328. doi: 10.1021/bi048419f. [DOI] [PubMed] [Google Scholar]
- 29.Kroone RC, Elliott GS, Ferszt A, Slingsby C, Lubsen NH, Schoenmakers JG. The role of the sequence extensions in beta-crystallin assembly. Protein Eng. 1994;7:1395–1399. doi: 10.1093/protein/7.11.1395. [DOI] [PubMed] [Google Scholar]
- 30.Liu BF, Liang JJ. Protein-protein interactions among human lens acidic and basic beta-crystallins. FEBS Lett. 2007;581:3936–3942. doi: 10.1016/j.febslet.2007.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jaenicke R, Slingsby C. Lens crystallins and their microbial homologs: structure, stability, and function. Crit Rev. Biochem. Mol. Biol. 2001;36:435–499. doi: 10.1080/20014091074237. [DOI] [PubMed] [Google Scholar]
- 32.Hejtmancik JF, Wingfield P, Chambers C, Russell P, Chen H-C, Sergeev YV, Hope JN. Association properties of beta-B2- and betaA3-crystallin: ability to form dimers. Protein. Eng. 1997;10:1347–1352. doi: 10.1093/protein/10.11.1347. [DOI] [PubMed] [Google Scholar]
- 33.Takata T, Woodbury LG, Lampi KJ. Deamidation alters interactions of beta-crystallins in hetero-oligomers. Mol. Vis. 2009;15:241–249. [PMC free article] [PubMed] [Google Scholar]
- 34.Chan MP, Dolinska M, Sergeev YV, Wingfield PT, Hejtmancik JF. Association properties of betaB1- and betaA3-crystallins: ability to form heterotetramers. Biochemistry. 2008;47:11062–11069. doi: 10.1021/bi8012438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding SE, Rowe AJ, Horton JC, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Royal Society for Chemistry; Cambridge, United Kingdom: 1992. pp. 90–125. [Google Scholar]
- 36.Ventura S. Sequence determinants of protein aggregation: tools to increase protein solubility. Microb. Cell Fact. 2005;4:11. doi: 10.1186/1475-2859-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scharnagl C, Reif M, Friedrich J. Stability of proteins: temperature, pressure and the role of the solvent. Biochim. Biophys. Acta. 2005;1749:187–213. doi: 10.1016/j.bbapap.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 38.Lampi KJ, Kim YH, Bachinger HP, Boswell BA, Lindner RA, Carver JA, Shearer TR, David LL, Kapfer DM. Decreased heat stability and increased chaperone requirement of modified human betaB1-crystallins. Mol. Vis. 2002;8:359–366. [PubMed] [Google Scholar]
- 39.Coop A, Goode D, Sumner I, Crabbe MJ. Effects of controlled mutations on the N- and C-terminal extensions of chick lens beta B1 crystallin. Graefes Arch. Clin. Exp. Ophthalmol. 1998;236:146–150. doi: 10.1007/s004170050055. [DOI] [PubMed] [Google Scholar]
- 40.Descamps FJ, Martens E, Proost P, Starckx S, Van den Steen PE, Van DJ, Opdenakker G. Gelatinase B/matrix metalloproteinase-9 provokes cataract by cleaving lens betaB1 crystallin. FASEB J. 2005;19:29–35. doi: 10.1096/fj.04-1837com. [DOI] [PubMed] [Google Scholar]
- 41.Shih M, David LL, Lampi KJ, Ma H, Fukiage C, Azuma M, Shearer TR. Proteolysis by m-calpain enhances in vitro light scattering by crystallins from human and bovine lenses. Curr. Eye Res. 2001;22:458–469. doi: 10.1076/ceyr.22.6.458.5483. [DOI] [PubMed] [Google Scholar]
- 42.Hope JN, Chen HC, Hejtmancik JF. BetaA3/A1-crystallin association: role of the amino terminal arm. Protein. Eng. 1994;7:445–451. doi: 10.1093/protein/7.3.445. [DOI] [PubMed] [Google Scholar]
- 43.Bateman OA, Sarra R, Van Genesen ST, Kappe G, Lubsen NH, Slingsby C. The stability of human acidic beta-crystallin oligomers and hetero-oligomers. Exp. Eye Res. 2003;77:409–422. doi: 10.1016/s0014-4835(03)00173-8. [DOI] [PubMed] [Google Scholar]
- 44.van Montfort RL, Bateman OA, Lubsen NH, Slingsby C. Crystal structure of truncated human betaB1-crystallin. Protein Sci. 2003;12:2606–2612. doi: 10.1110/ps.03265903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Norledge BV, Trinkl S, Jaenicke R, Slingsby C. The X-ray structure of a mutant eye lens beta B2-crystallin with truncated sequence extensions. Protein Sci. 1997;6:1612–1620. doi: 10.1002/pro.5560060802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trinkl S, Glockshuber R, Jaenicke R. Dimerization of beta B2-crystallin: the role of the linker peptide and the N- and C-terminal extensions. Protein Sci. 1994;3:1392–1400. doi: 10.1002/pro.5560030905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bergdoll M, Remy MH, Cagnon C, Masson JM, Dumas P. Proline-dependent oligomerization with arm exchange. Structure. 1997;5:391–401. doi: 10.1016/s0969-2126(97)00196-2. [DOI] [PubMed] [Google Scholar]
- 48.Nalini V, Bax B, Driessen H, Moss DS, Lindley PF, Slingsby C. Close packing of an oligomeric eye lens beta-crystallin induces loss of symmetry and ordering of sequence extensions. J. Mol. Biol. 1994;236:1250–1258. doi: 10.1016/0022-2836(94)90025-6. [DOI] [PubMed] [Google Scholar]
- 49.David LL, Shearer TR, Shih M. Sequence analysis of lens beta-crystallins suggests involvement of calpain in cataract formation. J. Biol. Chem. 1993;268:1937–1940. [PubMed] [Google Scholar]
- 50.David LL, Shearer TR. Beta-crystallins insolubilized by calpain II in vitro contain cleavage sites similar to beta-crystallins insolubilized during cataract. FEBS Lett. 1993;324:265–270. doi: 10.1016/0014-5793(93)80131-d. [DOI] [PubMed] [Google Scholar]
- 51.Shih M, Lampi KJ, Shearer TR, David LL. Cleavage of beta-crystallins dring maturation of bovine lens. Mol. Vis. 4. 1998 [PubMed] [Google Scholar]
- 52.Srivastava K, Gupta R, Chaves JM, Srivastava OP. Truncated Human B1-Crystallin Shows Altered Structural Properties and Interaction with Human A3-Crystallin. Biochemistry. 2009 doi: 10.1021/bi900313c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Evans P, Bateman OA, Slingsby C, Wallace BA. A reference dataset for circular dichroism spectroscopy tailored for the betagamma-crystallin lens proteins. Exp. Eye Res. 2007;84:1001–1008. doi: 10.1016/j.exer.2007.01.016. [DOI] [PubMed] [Google Scholar]
- 54.Srivastava OP, Kirk MC, Srivastava K. Characterization of covalent multimers of crystallins in aging human lenses. J. Biol. Chem. 2004;279:10901–10909. doi: 10.1074/jbc.M308884200. [DOI] [PubMed] [Google Scholar]
- 55.Ma Z, Hanson SR, Lampi KJ, David LL, Smith DL, Smith JB. Age-related changes in human lens crystallins identified by HPLC and mass spectrometry. Exp. Eye Res. 1998;67:21–30. doi: 10.1006/exer.1998.0482. [DOI] [PubMed] [Google Scholar]
- 56.Griko Y, Sreerama N, Osumi-Davis P, Woody RW, Woody AY. Thermal and urea-induced unfolding in T7 RNA polymerase: calorimetry, circular dichroism and fluorescence study. Protein Sci. 2001;10:845–853. doi: 10.1110/ps.39701. [DOI] [PMC free article] [PubMed] [Google Scholar]