Abstract
Residue conservation is an important, established method for inferring protein function, modularity and specificity. It is important to recognize that it is the 3D spatial orientation of residues that drives sequence conservation. Considering this, we have built a new computational tool, VENN that allows researchers to interactively and graphically titrate sequence homology onto surface representations of protein structures. Our proposed titration strategies reveal critical details that are not readily identified using other existing tools. Analyses of a bZIP transcription factor and receptor recognition of Fibroblast Growth Factor using VENN revealed key specificity determinants. Weblink: http://sbtools.uchc.edu/venn/.
INTRODUCTION
In order to gain insight into protein function, scientists often compare orthologous protein sequences (from different species) to identify important residues that are conserved throughout evolution. However, sequences are only a 1D representation of 3D proteins. In this context, it is the spatial configuration of amino acids, not the protein sequence itself, which is under evolutionary pressure. The 3D aspects of the conserved structural motif are not readily decoded from a protein sequence. For example, a binding surface or enzyme active site may have several conserved residues spread over its entire sequence, but in 3D space the residues are consolidated into a localized binding surface.
Many tools such as BLAST have been developed for generating sequence alignments (1). While computational tools such as ConSurf (2) and the Evolutionary Trace Server (3) are very useful to visualize sequence similarity embedded on protein structure, fixed non-interactive selection of similar sequences limits their usefulness. This constraint can obscure details that are critical for understanding proteins and protein families. Here we report VENN, a new program that addresses this limitation. Because it maps the intersect of sequence and structure to evaluate function, it is named after John Venn for his work on Venn diagrams (4).
RESULTS
VENN is a Java application interfaced to a local MySQL database. Users begin by selecting a protein structure, which is retrieved from the Protein Data Bank and displayed using the Jmol molecular viewer (http://www.jmol.org). A BLAST alignment identifies up to 500 putative homologs. Users interactively select among these homologs, and the calculated amino acid conservation at each position is mapped onto the protein structure as a heat map. The application and help videos are at http://sbtools.uchc.edu/venn/.
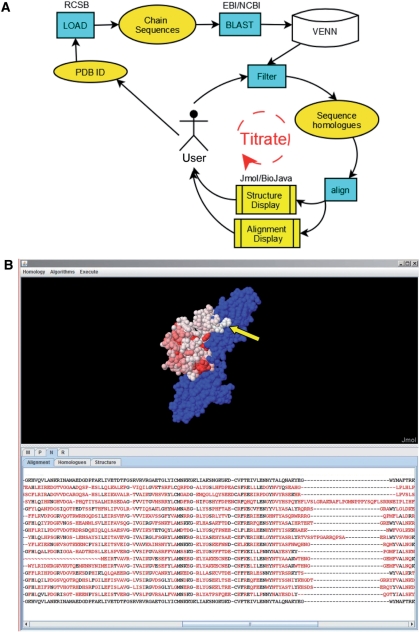
The VENN workflow is shown in Figure 1A. The user loads the protein structure and sequence into VENN via the Protein Data Bank (PDB) accession number (5). Similar matches to the individual chain sequences (which are putative orthologs or paralogs) in the structure are remotely retrieved from EBI (6) or locally via NCBI using BLAST and stored in the local VENN database. The user selects a set of sequences and initiates an alignment of these filtered sequences, shown in the alignment display. Sequence conservation at each position is calculated from the sequence alignment and used to generate a heat map that is used to color the protein structure in the Jmol structural display window. The user can repeat the filtration process selecting more, fewer, or different groups of sequences to titrate the sequence homology and map it onto the surface of the protein structure. A screen shot of the structural display and alignment windows is shown in Figure 1B.
Figure 1.
(A) Data processing model for VENN. Processes are shown as boxes (cyan); products are ellipses (orange); displays are yellow. (B) Screen shot of VENN analyzed with a complex of Fibroblast Growth Factor 8 (FGF8) bound to a FGF receptor 2c homodimer (blue) (2FDB). Arrow indicates non-conserved specificity residues Thr-Phe in human FGF1/3/4/5/6/7/8/10/11/16/19/20/21/22/23. Residues R1052/G1094/E1131/E1135 in FGF are nearly completely conserved among 15 different FGF family members and contact the FGF receptor.
We have identified four principal strategies for using homolog titration in VENN; users are encouraged to create their own, novel titration protocols: (i) Select all orthologs or paralogs; choosing proteins with the same name can be used for this analysis. This allows a user to determine which regions of the protein are evolutionarily conserved (e.g. Figure 2A); (ii) Select sequences with similar BLAST scores that include different proteins from different species. This reveals important functional sites that are conserved in protein families (e.g. Figure 2B); (iii) Select sparsely distributed sequences with a wide range of BLAST scores. In addition to identifying conserved functional sites in gene families, non-conserved residues can provide clues to the specificity of family members (e.g. Figures 1B and 2C); and (iv) Select sequences that have low BLAST scores to reveal the modularity of functional sites in proteins (e.g. Figure 2D).
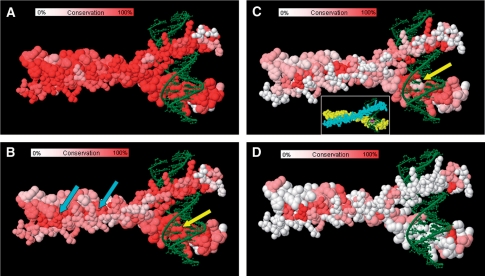
Figure 2.
Homology titration of C/EBPβ using VENN. (A–D) Images from VENN analysis of C/EBPβ homodimer (1GU4); chain A is shown using larger spheres. DNA (green) and a heatmap coloring code of residue conservation are shown. Residue conservation maps for putative C/EBPβ homologs are shown: (A) orthologs from four species, (B) homologs with 50 highest BLAST scores, (C) every 20th sequence from the top 160 BLAST scores; inset shows chain A (yellow) with Val 285 (magenta) and chain B (cyan), (D) comparison to coil–coil regions of human myosins and centrosomal protein (290 kDa). Arrows indicate the dimerization interface (cyan) and Val 285 in the DNA-binding site (yellow).
To demonstrate the utility of VENN we explored these four strategies by examining CCAAT/enhancer-binding protein β (C/EBPβ; PDB: 1GU4), a transcription factor of the bZIP family. The automated BLAST analysis identified 500 C/EBPβ homologs for homology titration. Comparing four orthologs from human, frog, flounder and pufferfish shows high conservation of almost all residues (Figure 2A). As the user titrates in the 50 sequences with the highest BLAST scores representing C/EBP family α, β, δ and γ members from many species, functional sites for coil–coil homodimerization and DNA binding emerge (Figure 2B, cyan and yellow arrows, respectively). At the dimerization interface, residues L306, N310, L313, L320, E323, L324 and L327 are completely conserved among distant homologs and form contacts at the dimerization interface. Residues R278, N281, N282, A284, K287, S288, R289 and R295 comprise a DNA-binding site.
To identify differences among closely related members of the bZIP protein family, we selected every 20th sequence in the top 160 BLAST scores (Figure 2C). Within the highly conserved DNA-binding site, V285 (yellow arrow) was poorly conserved. Closer examination reveals that this residue is juxtaposed to a guanine base in the DNA. A literature search revealed that this residue is known to be important for base selectivity in bZIP transcription factors (7). In a similar type of analysis, VENN was used to identify a similar recognition determinant among 15 different FGF family members for binding their receptors (Figure 1B). From this analysis we hypothesize that the critical Thr–Phe residues are specificity determinants for FGF receptor recognition of FGF8 ligands; this was previously recognized for FGF8 isoforms (8).
The BLAST results also revealed several myosins and centrosomal proteins that are not thought to bind DNA, which is supported by a VENN analysis. When the conservation between these proteins is plotted onto the transcription factor, it is clear that the coil–coil dimerization interface remains conserved while the DNA-binding region is not (Figure 2D).
VENN has other unique capabilities. VENN accommodates all protein chains in structures of protein complexes in a single analysis which facilitates analysis of multiprotein complexes. VENN also provides different sequence alignment strategies. A neutral sequence alignment places no weight on any individual amino acid, whereas a BLOSUM alignment weights residues based on the BLOSUM62 matrix (9). VENN also offers a parametric sequence alignment where weights of alignment can be based on the existence of chemical and physical properties of amino acids (for instance, aliphatic, aromatic, acidic, basic, polar). From the visualization perspective, VENN can be used to interactively identify and color regions of protein by searching for a regular expression. Thus, a user could search with ‘P.P’ to identify any motif that has two prolines separated by one residue. Alternatively, by entering a single amino acid ‘M’ all methionines can be colored. These features can be used to examine the 3D location of conservation motifs or residues.
DISCUSSION
VENN is an interactive software application that allows users to titrate and map sequence conservation onto protein structures. VENN performs a type of conservation analysis that is distinct from the many programs for pairwise and multiple sequence alignments and from programs such as DALI which is used to identify proteins with similar structural folds (10). Other programs have been published that integrate sequence similarity and protein structure to identify functional sites. VENN is most similar to ConSurf (2), Evolutionary Trace (3) and HomolMapper (11), however VENN has a number of important distinctions that enable new types of discovery. For this section it is helpful to compare an analysis of C/EBPβ with VENN (Figure 2) to that with ConSurf and Evolutionary Trace (Figure 3). HomolMapper has much more limited capabilities.
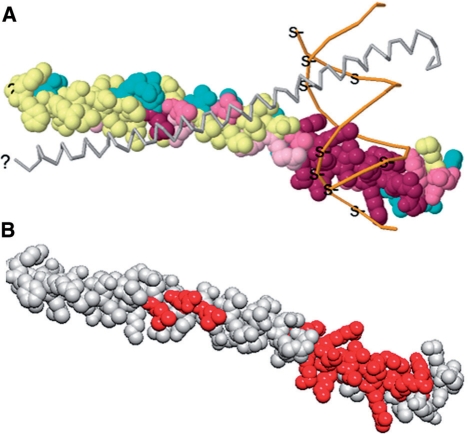
Figure 3.
Comparison of ConSurf and Evolutionary Trace analysis of C/EBPβ. (A) Image from ConSurf analysis of C/EBPβ homodimer (1GU4); chain A is shown using larger spheres and chain B backbone is shown; DNA (orange). Color progression from teal to maroon indicates increased conservation; yellow spheres indicate insufficient data. (B) Image from analysis of C/EBPβ homodimer (1GU4) with Evolutionary Trace. Red residues indicate conservation when plotted with the highest Z-score (7.146). Orientations are similar to those for the VENN analysis of the same protein in Figure 2.
Advantages of VENN
VENN has a number of unique features that distinguish it from ConSurf and Evolutionary Trace; however, VENN can be used synergistically with these programs. Most notably, VENN is interactive database-driven program which enables filtration, and iterative selection of different sets of sequences. This is important because it streamlines a number of different strategies for protein sequence selection. Several protein sequence groups can be automatically selected based on species, protein family, motifs, mass, pI, protein length and presence of a user-defined motif. Homologs can also be sorted by BLAST score (default), name, or taxonomy. In order to select different sets of sequences in ConSurf or Evolutionary Trace a user must first perform a multiple sequence alignment and upload a sequence alignment file. This is a limitation: C/EBPβ specificity determinants are only revealed through an ordered interactive titration of homologous sequences (Figure 2C) and in this case not by analyses with ConSurf or Evolutionary Trace (Figure 3). We expect that VENN's flexibility in protein selection and manipulation will enable new types of strategies that we have not yet explored.
VENN also automatically identifies and searches all chains present in a PDB accession number. Therefore, no prior knowledge of the number or identity of chains is required. This user-friendly aspect in VENN is important for exploring multiprotein complexes or complexes of proteins with other molecules. Large structures of complexes, such as nucleosomes, clathrin coats and ribosomes can be analyzed in a single analysis. Often interpretation of a conserved functional site is much easier when visualized in the context of its association with another molecule as exemplified by the conserved residues juxtaposed to a DNA molecule in the structure of the C/EBPβ : DNA complex (Figures 2 and 3). Each chain must be analyzed individually with Evolutionary Trace. While ConSurf can display multiple chains, analysis of multiprotein complexes is slowed by the fact that only one chain at a time can be analyzed.
A number of other features of VENN allow users to readily identify important functional regions in proteins. VENN enables users to select specific residues in the alignment tab; these can be selected and colored on the structure. Motifs can also be selected and colored in the Structure tab; likewise entire domains or protein chains can be colored using either of these functions. Specific residues that are conserved can be identified by examining a multiple sequence alignment in the Alignment tab. Alternatively, holding a mouse over a residue or atom in the structure reveals a popup balloon with its identity. In addition to standard neutral and BLOSUM sequence alignment matrices, VENN also allows flexibility in alignment strategies based on emphasis of different attributes with the aforementioned parametric alignment; e.g. users can heavily weight hydrophobic residues, hydrophilic, etc. ConSurf offers Bayesian or Maximum Likelihood methods for calculating amino acid similarity. By using the Execute Custom Command from the menu a user can enter any Jmol command to modify the display of structure. This flexibility allows users to generate images for publication. While VENN does not have an output function for structure images and alignments, these can be readily captured using a screenshot program (e.g. Snipping tool in VISTA) and the alignments can be cut and pasted into any text editor. VENN can also be used to identify conserved structural features in proteins or proteins families. For example, we used VENN to identify a novel asparagine finger in dynein light chain (1M9L; data not shown) (12).
Synergistic functions in similar software tools
Other tools can be used to complement or precede an analysis with VENN. The ConSurf server, for example, is web based and can be utilized for a quick, automated viewing of highly conserved residues for a single chain in structures of close family members. The Evolutionary trace (ET) program uses a ranking and clustering strategy to map functional sites. Both ConSurf and ET enable more customizable features as well. Other tools, such as SwissPDBViewer (13) and Chimera (14) enable structural modeling and comparison, including alignment of multiple PDB sequences to generate a structural model that relates an entire family of proteins. Such models can serve as novel inputs to VENN for subsequent sequence titration. SwissPDBViewer and Chimera can also be used to manually generate individual figures which resemble those made by VENN by menu and command driven operations. VENN differs from these tools in that it is entirely interactive, integrated with proteome data sources, requires no intermediary file formats for any of its analysis features, and embeds a database and data model of protein sequences/meta data which can directly automate the aforementioned sequence selection, filtration and titration strategies.
Limitation of mapping homology onto protein structures
VENN, ConSurf and Evolutionary Trace have the major limitation that a protein structure is needed to perform an analysis and there are only ∼55 000 structures in the latest release of the PDB. One possible solution is to use the ModBase (http://modbase.compbio.ucsf.edu/modbase-cgi/index.cgi) (15) or the Swiss-Model Repository (http://swissmodel.expasy.org/repository/) (16), databases that have millions of structural models that can be downloaded as PDB files. All three programs can read user-defined PDB files. Alternatively, if the query protein is homologous to a protein of known structure, then Swiss-Model can be first used to generate a model structure in PDB file format (17).
CONCLUSIONS
VENN is a novel cross-platform software tool which provides biologists with a highly integrated methodology for visualizing conservation of various functional groups and taxonomical families on the 3D structure of a protein of interest. The ability to readily combine the vast proteomic sequence space with structural information in an automatic fashion can reveal functional attributes which have not been reported using similar tools.
FUNDING
National Institutes of Health (grant numbers GM079689 and AI078708 to M.R.S. and EB001496 to M.R.G.). Funding for open access charge: National Institute of General Medical Sciences, grant number GM079689.
Conflict of interest statement. None declared.
Footnotes
Present address: Martin R. Schiller, School of Life Sciences, University of Nevada, Las Vegas, 4505 Maryland Parkway, Las Vegas, NV 89154-4004, USA
REFERENCES
- 1.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morgan DH, Kristensen DM, Mittelman D, Lichtarge O. ET viewer: an application for predicting and visualizing functional sites in protein structures. Bioinformatics. 2006;22:2049–2050. doi: 10.1093/bioinformatics/btl285. [DOI] [PubMed] [Google Scholar]
- 4.Venn J. On the diagrammatic and mechanical representation of propositions and reasonings. J. Science. 1880;9:1–18. [Google Scholar]
- 5.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Labarga A, Valentin F, Anderson M, Lopez R. Web services at the European bioinformatics institute. Nucleic Acids Res. 2007;35:W6–W11. doi: 10.1093/nar/gkm291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujii Y, Shimizu T, Toda T, Yanagida M, Hakoshima T. Structural basis for the diversity of DNA recognition by bZIP transcription factors. Nat. Struct. Biol. 2000;7:889–893. doi: 10.1038/82822. [DOI] [PubMed] [Google Scholar]
- 8.Olsen SK, Li JYH, Bromleigh C, Eliseenkova AV, Ibrahimi OA, Lao ZM, Zhang FM, Linhardt RJ, Joyner AL, Mohammadi M. Structural basis by which alternative splicing modulates the organizer activity of FGF8 in the brain. Genes Dev. 2006;20:185–198. doi: 10.1101/gad.1365406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henikoff S, Henikoff JG. Amino acid substitution matrices from protein blocks. Proc. Natl Acad. Sci. USA. 1992;89:10915–10919. doi: 10.1073/pnas.89.22.10915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holm LF, Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 11.Rockwell NC, Lagarias JC. Flexible mapping of homology onto structure with homolmapper. Bmc Bioinformatics. 2007;8:1–13. doi: 10.1186/1471-2105-8-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu HW, Maciejewski MW, Takebe S, King SM. Solution structure of the Tctex1 dimer reveals a mechanism for dynein-cargo interactions. Structure. 2005;13:213–223. doi: 10.1016/j.str.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 13.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 14.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera – a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 15.Pieper U, Eswar N, Braberg H, Madhusudhan MS, Davis FP, Stuart AC, Mirkovic N, Rossi A, Marti-Renom MA, Fiser A, et al. MODBASE, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 2004;32:D217–D222. doi: 10.1093/nar/gkh095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kopp J, Schwede T. The SWISS-MODEL Repository of annotated three-dimensional protein structure homology models. Nucleic Acids Res. 2004;32:D230–D234. doi: 10.1093/nar/gkh008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwede T, Kopp J, Guex N, Peitsch MC. SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res. 2003;31:3381–3385. doi: 10.1093/nar/gkg520. [DOI] [PMC free article] [PubMed] [Google Scholar]