Abstract
H-NS is a bacterial DNA-binding protein that regulates gene expression and DNA transposition. In the case of Tn10, H-NS binds directly to the transposition machinery (i.e. the transpososome) to influence the outcome of the reaction. In the current work we evaluated the binding affinity of H-NS for two forms of the Tn10 transpososome, including the initial folded form and a pre-unfolded form. These two forms differ in that IHF is bound to the former but not the latter. IHF binding induces a bend (or fold) in the transposon end that facilitates transpososome formation. However, the continued presence of IHF in the transpososome inhibits intermolecular transposition events. We show that H-NS binds particularly strongly to the pre-unfolded transpososome with an apparent Kd of ∼0.3 nM. This represents the highest affinity interaction between H-NS and a binding partner documented to date. We also show that binding of H-NS to the transpososome stabilizes this structure and propose that both high-affinity binding and stabilization result from the combined interaction between H-NS and DNA and H-NS and transposase within the transpososome. Mechanistic implications for tight binding of H-NS to the transpososome and transpososome stabilization are considered.
INTRODUCTION
Lateral gene transfer (LGT) is an important force in the evolution of bacteria and is largely responsible for the prevalence of antibiotic resistance genes in clinically important bacteria (1). The nucleoid-binding protein H-NS (histone-like nucleoid-structuring protein) contributes to LGT transfer in gram-negative bacteria in two ways. First, it acts as a potent transcriptional repressor to selectively silence genes carried on newly acquired DNA segments (2–5). This is beneficial to the host because it allows gradual integration of newly acquired gene regulators into the host cells gene expression circuitry. Second, H-NS upregulates selective DNA transposition systems (6–8) and transposons are important agents in LGT. This article focuses on the mechanism by which H-NS promotes Tn10 transposition.
Tn10 (Figure 1A transposes by a nonreplicative mechanism wherein the transposon is first cleanly excised from a donor site and then the excised transposon integrates into another site (9,10). Prior to the chemical steps in transposition transposase binds to DNA sequences close to the transposon termini or ‘ends’ and organizes the transposon into a higher-order complex called the transpososome (11,12). Within the transpososome two transposon ends [two outside ends (OEs) for Tn10 and an OE and an inside end (IE) for IS10] are held together by a combination of protein–protein and protein–DNA interactions. This organization permits the two transposon ends to integrate into a single target site. )
Figure 1.
Structure of Tn10 and transpososome dynamics. (A) Tn10 is 9147 bp in length. The outside end (OE) of Tn10 contains terminal (T) (bp 1–29) and subterminal (ST) (bp 43–75) transposase-binding regions plus an IHF-binding site (bp 30–42). The inside end (IE) does not contain an IHF-binding site. IS10-Right encodes a functional transposase (T’ase) and other genes encoded by Tn10 are shown. Flanking donor DNA (FD) is depicted by an orange rectangle. Half arrows indicate the terminal inverted repeat (TIR). The origin of the two short, linear OE substrates used in this work is depicted below IS10-Right. (B) Model for H-NS–transpososome interactions and transpososome dynamics. Tn10 is initially assembled into a folded transpososome (i) containing a dimer of transposase (blue ovals), in which one or both arms of the transpososome is/are folded by IHF (yellow sphere) the model shows both arms folded. H-NS (magenta sphere) is shown binding the folded transpososome at three sites (H-NSFLANK, H-NSTIR-PROX and H-NSTIR-DIST) (ii) and inducing transpososome unfolding (iii). After excision the unfolded excised transpososome (iv) may go on to interact with target DNA (purple) via a random collision pathway (v), which increases the probability of intermolecular transposition events occurring. Alternatively, the unfolded excised transpososome may be refolded by IHF (vi) in which case intramolecular target interactions are favored (vii). H-NS binding to the folded, excised transpososome (vi) converts this species to an unfolded transpososome (iv) that is poised for random collision target interactions. An unfolded transpososome, free of H-NS, may exist in the pathway if transpososome unfolding occurs in an H-NS-independent manner or if H-NS dissociates from the transpososome at any time after inducing unfolding (not shown).
Formation of the transpososome is facilitated by integration host factor (IHF) (Figure 1B). IHF binds to the OE and introduces a U-bend into the DNA (12). This promotes transpososome formation by allowing a subterminal (ST) portion of the OE to contact transposase, presumably stabilizing the transposase–end interaction (12,13). The presence of the U-bend in one or both OE’s causes the transpososome to adopt a compact or folded conformation. Subsequent to initial transpososome formation, IHF is released from the transpososome resulting in the loss of subterminal contacts and thus the unfolding of the transpososome. There are at least two important functional consequences of IHF release and subsequent transpososome unfolding. First, transpososome unfolding is coupled to one or more of the chemical steps in the transposon excision reaction (14,15). Second, transpososome unfolding influences target DNA interactions. If IHF rebinds the transpososome (postexcision), the folded state of the transposon ends favors integration events into the transposon itself. In contrast, if IHF is not bound to the postexcision transpososome and thus the transpososome is unfolded, intermolecular transposition events are favored (16) (Figure 1B).
H-NS was first implicated as positive regulator of Tn10 transposition when it was found that disruption of the hns gene caused a large decrease in the frequency of Tn10 transposition (7). It was subsequently shown in vitro that H-NS binds with high specificity to both folded and unfolded forms of the Tn10 transpososome (17). Furthermore, the results from DNA footprinting experiments are consistent with there being as many as three distinct H-NS-binding sites in the transpososome (Figure 1B) as three new regions of protection were observed when H-NS was included in transpososome assembly reactions (17,18). Determinants for H-NS binding to one or more of these sites include the presence of distorted DNA structures within the transpososome and the transposase protein itself (18).
Addition of H-NS to in vitro Tn10 transposition reactions increases the frequency of intermolecular strand transfer events and inhibits a specific class of intramolecular events that are characteristic of a folded transpososome (19). A component of this stimulation appears to be that H-NS binding to the transpososome facilitates IHF release. Evidence for this includes the observations that: (i) H-NS binding to a folded transpososome induces a pattern of chemical nuclease hypersensitivity at the transposon–donor junction that is characteristic of an unfolded transpososome and (ii) IHF binding to the IHF site, as measured by chemical nuclease footprinting, is reduced in the H-NS-bound transpososome (18). At this point it is unclear if H-NS is the major factor in the cell that induces transpososome unfolding, as there is also evidence that Mg2+ induces transpososome unfolding (14).
Addition of the polyanion heparin provides a means of generating an unfolded transpososome in vitro. In this case heparin acts as a surrogate for DNA and traps IHF molecules that dissociate from the transpososome, allowing the transpososome to be captured in an unfolded state. Notably, it is important to study the interaction between H-NS and the transpososome when the transpososome is in the unfolded state because H-NS may function in vivo to induce transpososome unfolding and/or to maintain the transpososome in the unfolded state at critical times in the transposition pathway.
In the current work, we set out to further characterize the H-NS interaction with the Tn10 transpososome. Towards this end we have looked at the relative binding affinity of H-NS for folded and pre-unfolded (heparin treated) forms of the transpososome. We show that H-NS binds particularly tightly to the pre-unfolded form of the transpososome. Moreover, tight binding of H-NS to the transpososome has a significant stabilizing effect on this complex, which in the absence of H-NS is prone to dissociation. We also show that the stabilization of the transpososome by H-NS correlates with the ability of H-NS to both bind the transpososome in a DNA structure-specific manner and to form protein–protein contacts with transposase. Taken together, these results provide a more comprehensive view of the mechanism by which H-NS promotes Tn10 transposition.
MATERIALS AND METHODS
Plasmids and OE substrate and proU preparation
pDH406 was constructed by cloning the 149 bp BglII-SalI OE fragment from pNK1935 (12) into BglII-SalI cut pTZ18U. pDH497 is pDH406 with bp 44 converted from a G-C to a C-G by Quick-Change site-directed mutagenesis to create a unique HpaI site. OE substrate 1 (65 bp of the OE of IS10-Right + 40 bp of flanking donor DNA from the HisG1 Tn10 insertion hotspot) was generated by cutting pDH497 with BamHI and NdeI. OE substrate 2 is described in (17). OE fragments were gel purified and were indicated 5′ end labeled with T4 polynucleotide kinase and γ32P ATP using standard procedures. proU fragment DNA was generated by PCR using primers proU-FPR2 (5′ GGTGGGTTCAATCAGGCG 3′) and proU-FPF (5′ GCATCAATATTCATGCC 3′) and the plasmid pKKproU.
Protein purification
Tn10 transposase, IHF, H-NS and StpA were purified as described in references (20–22), respectively. All H-NS purifications (i.e. WT, P116S and 1-64) were from Escherichia coli BL21(DE3)hns::KanR expressing the respective hns genes from pET3a derivatives. Similarly, StpA was purified from the above hns deletion derivative expressing StpA from pET3a-StpA. Protein concentrations were determined by the BCA assay (Pierce).
Preparation of transpososomes
Transpososomes were formed in the following way: (i) 32P-labeled OE fragment was mixed with IHF for 30 min; (ii) Transposase was added and incubation was continued for an additional hour; (iii) Where indicated H-NS was added and incubation was continued for 30 min. Buffer conditions were as described in (12). Molar ratios of OE DNA, IHF, transposase and H-NS in a standard assembly reaction were 1 : 2 : 1.1 : 3, respectively, with a 1× reaction containing these components at final concentrations of 9, 18, 10 and 54 nM, in a 20 µl volume. The total reaction volume was increased according to the number of individual reactions desired per experiment. Transpososome unfolding was induced by addition of heparin sulfate to transpososome assembly reactions and incubating for 30 min. Where proU was added (Figure 3), incubation was for 30 min before addition of H-NS and then incubation was continued for an additional 30 min before samples were analyzed on a 5% native polyacrylamide gel as described in (12). In the heparin challenge experiments (Figures 4 and 5) heparin challenge, following the 30-min H-NS treatment, lasted for 30 min before samples were analyzed by gel electrophoresis as above. All incubations were at 25°C. We calculated the approximate concentration of transpososome formed in each reaction from knowledge of the input amount of OE fragment and from the proportion of total signal in the transpososome band as determined by phosphorimaging.
Figure 3.
Competitive binding assay. (A) A transpososome assembly reaction prepared with OE substrate 2 was divided in two and heparin (30 nM) was added to one of the two aliquots to unfold the transpososome. After further subdividing the samples, varying amounts of unlabeled proU fragment were added and then where indicated H-NS (20 nM) was added (except in lane 1). Samples were analyzed as in Figure 2B, except that the voltage was reduced from 200 to 150V over the 2-h period of electrophoresis. The concentration of the initial input transpososome was ∼1.0 nM. The amount of proU DNA added is given below each gel (proU to T’some ratio). In (B) pre-unfolded transpososome formed in (A) was diluted and a fixed amount of proU was added as competitor. Controls (lanes 1–4) include ‘undiluted’ transpososome subjected to the indicated treatments. A darker exposure of a portion of the gel (right hand panel) shows that at the highest proU to transpososome ratios there was significant inhibition of H-NS–transpososome formed. (C) Quantification of the experiment in (A).
Figure 4.
Transpososome stabilization assay. (A) A transpososome assembly reaction carried out with OE substrate 2 was divided in two and H-NS (30 nM) was added to one of the two aliquots. Heparin was then added (where indicated) to aliquots of each of the two reactions. The heparin concentrations used were as follows: 0.008 µM (lanes 2 and 9); 0.030 µM (lanes 3 and 10); 0.125 µM (lanes 4 and 11); 0.5 µM (lanes 5 and 12); 2 µM (lanes 6 and 13); 8 µM (lanes 7 and 14). Samples were then analyzed as in Figure 3. (B) The relative amounts of DNA populating the different species in (A) are plotted against the heparin concentration.
Quantitative H-NS-binding analysis
Transpososomes were formed by mixing 32P-labeled OE DNA, IHF and transposase in the same proportions as in the previous section but the amount of input OE DNA was reduced from 9 to 0.2 nM. The efficiency of transpososome assembly was typically 10–15% so that the transpososome concentration was ∼0.02 nM. Transpososome unfolding was induced by adding heparin to a final concentration of 20 nM to an aliquot of folded transpososome. Varying amounts of H-NS were then added and incubation was carried out at 25°C for 1 h before subjecting samples to electrophoresis on a native 5% polyacrylamide gel as previously described with the exception that the electrophoresis time was increased from 1 to 2 h in Figure 2B in order to separate H-NS transpososome from unfolded transpososome. For H-NS titrations with folded and pre-unfolded transpososome the range of H-NS concentrations used was from 6 to 400 nM and from 0.25 to 11.5 nM, respectively. Gel images were analyzed using ImageQuant software. The equilibrium dissociation constant (Kd) was determined using the equation:
where Θ is the fraction of bound transpososome and Pt equals total protein concentration. Plots were generated using GraphPad Prism software (v5.0). Data over three independent experiments were fit to a curve by nonlinear regression. Error bars represent the standard error on the mean.
Figure 2.
Electrophoretic mobility shifts and quantification of H-NS binding to the pre-unfolded transpososome. Transpososomes were assembled with OE substrate 1 shown in Figure 1A as described in ‘Materials and Methods’ section. Varying amounts of H-NS were then incubated with the transpososome preparations and samples were applied to a 5% native polyacrylamide gel. In (A) H-NS was added directly to the folded transpososome preparation whereas in (B and C) the transpososome preparation was treated with heparin to induce transpososome unfolding prior to H-NS addition. Note that the time of electrophoresis was doubled in (B) relative to (C) in order to clearly separate unfolded transpososome from H-NS–transpososome and consequently free DNA and IHF complex was run off the gel. In (C) we show that at the H-NS concentrations used in (B) no non-specific binding of H-NS to OE DNA is detected. Note also that under these electrophoresis conditions H-NS–transpososome is not clearly separated from unfolded transpososome. In (A) H-NS was present at 6 nM (lane 2), 12 nM (lane 3), 24 nM (lane 4), 48 nM (lane 5), 96 nM (lane 6) and 192 nM (lane 7). In (B) H-NS was present at 0.25 nM (lane 3), 0.5 nM (lane 4), 1 nM (lane 5), 2 nM (lane 6), 4 nM (lane 7), 8 nM (lane 8) and 16 nM (lane 9). In (C) H-NS was present at 1 nM (lane 3), 2 nM (lane 4), 4 nM (lane 5), 8 nM (lane 6) and 16 nM (lane 7). (H-NS-T’some) H-NS-bound transpososome; (f-T’some) folded transpososome; (uf-T’some) unfolded transpososome; (IHFc) IHF complex. (D) is a binding isotherm of fractional saturation as a function of H-NS concentration for two identical (open squares) and one nonidentical (filled circles) repeats of the experiment in (B). That is, a different range of H-NS concentration was used in one of the three experiments.
Protein–protein crosslinking
50× (volume) transpososome assembly reactions were prepared with unlabeled OE DNA and were, where indicated, treated with heparin (30 nM) to induce transpososome unfolding. WT H-NS (200 nM) or P116S H-NS (800 nM) was added to give full conversion of unfolded transpososome to H-NS transpososome and then assembly reactions were concentrated by microfiltration from the initial volume of 1–0.05 ml. During this process the assembly buffer was changed so that Hepes was used in place of Tris. Samples were then treated with 20 µl of the chemical crosslinker EDC (50 mM) plus NHS (12.5 mM) (both prepared in water) for 30 min or with 4 µl of BS3 (final concentration, 2 mM) prepared in water, for 30 min. Crosslinking reactions were immediately applied to a 5% native polyacrylamide gel. After staining the gel with ethidium bromide an H-NS–transpososome-containing gel slice was obtained. Proteins were eluted out of the gel slice, concentrated and subjected to immunoblot analysis as previously described (18). Purified transposase and H-NS were subjected to EDC/NHS crosslinking as previously described (18).
RESULTS
Binding of H-NS to folded and pre-unfolded transpososomes
Details of the interaction between H-NS and a variety of preferred H-NS DNA-binding sites in bacterial genomes have previously been reported (23). The strongest H-NS-binding region identified to date is the negative regulatory element (NRE) of the proU promoter in E. coli. The apparent equilibrium binding constant (Kd) (global) measured at 20°C for this region, which contains multiple H-NS-binding sites, is 15 nM (24). To further characterize the interaction between H-NS and the Tn10 transpososome, and to provide a point of reference of binding strength relative to the H-NS-proU interaction, we used an electrophoretic mobility shift assay (EMSA) to measure the binding affinity of H-NS for both folded and pre-unfolded forms of the transpososome. In the experimental set up used, the initial folded transpososome was formed by mixing a short (105 bp) 32P-labeled DNA fragment that includes the terminal 65 bp of the OE from IS10-Right (substrate 1 in Figure 1A) with IHF and then transposase. The pre-unfolded transpososome was subsequently generated by treating the initial transpososome with heparin, which titrates IHF off of the transpososome. An important distinguishing feature between the two forms is that in the folded transpososome IHF is freely available to compete with H-NS for transpososome binding, while in the pre-unfolded transpososome IHF is not available to compete with H-NS.
Kd calculations for H-NS binding to different forms of the transpososome are complicated by the fact that the transpososome is a minority component (typically 15–25%) of transpososome assembly reactions and the other reaction components (particularly unbound OE DNA) can also bind H-NS. Under our standard transpososome assembly conditions we typically see full conversion of folded transpososome to H-NS-bound transpososome (i.e. H-NS–transpososome) at ratios of H-NS to total DNA that do not yield detectable levels of H-NS–OE complex or H-NS-IHF-OE complex (17,18). This raised the possibility that we could accurately measure a Kd value for H-NS–transpososome interactions using transpososome assembly reactions as the source of transpososomes. However, when we increased the H-NS concentration, relative to our standard transpososome assembly conditions, in order to ensure that the amount of H-NS in the H-NS–transpososome complex was only a small fraction of the total H-NS concentration, we found that both transpososome and unbound OE DNA underwent an H-NS mobility shift (lanes 4–7 in Figure 2A). This partitioning of H-NS between two different binding partners prevented us from accurately measuring a Kd value.
In contrast, we found that we could get close to complete conversion (>95%) of pre-unfolded transpososome to an H-NS-bound form at H-NS concentrations where no significant binding of H-NS to free OE DNA was detected [Figure 2B (lanes 7–9) and C (lanes 5–7)] and the amount of H-NS in complex with transpososome was in the order of 200-fold less than the total amount of H-NS in the reaction. Note that in the experiment shown, we analyzed reactions under two different conditions of electrophoresis. We used a 2-h electrophoresis time to get clear separation of unfolded transpososome from H-NS–transpososome, which also resulted in free OE DNA running off the gel (Figure 2B). A 1-h electrophoresis time was used to assess H-NS binding to free OE DNA (Figure 2C). Taken together, the experiments in Figure 2A–C show that H-NS binds to the pre-unfolded transpososome at considerably lower concentrations than we observed for the folded transpososome and at these lower H-NS concentrations there was no detectable binding of H-NS to the free OE DNA. For example, ∼90% of the pre-unfolded transpososome was shifted to the H-NS form at 4 nM H-NS (lane 7 in Figure 2B and lane 5 in Figure 2C), while at 6 nM H-NS only ∼5% of the folded transpososome was converted to an H-NS form (lane 2, Figure 2A). One similarity between the two experiments was that in both cases H-NS binding to the transpososome showed considerable cooperativity. This is apparent from the burst of H-NS binding to both forms of the transpososome observed over a very narrow H-NS concentration range (compare lanes 3 and 4 in Figure 2A and lanes 5 and 6 in Figure 2B). Highly cooperative binding of H-NS to the proU DNA fragment has previously been reported (24).
From the data in Figure 2B (as well as two additional experiments) we generated a binding curve for the H-NS-pre-unfolded transpososome interaction (Figure 2D) and obtained a Kd of 0.3 ± 0.1 nM. Note that we also measured the global Kd for the H-NS–proU interaction under our standard transpososome assembly conditions (see Figure S1) and obtained Kd of 13 nM. Taken together this analysis shows that H-NS binds the pre-unfolded transpososome about 40-fold more tightly than it binds proU.
We directly compared the binding of H-NS to Tn10 transpososomes and proU by performing competition experiments. A fixed concentration of 32P-labeled transpososome (folded or pre-unfolded) was mixed with varying amounts of unlabeled proU DNA and then a fixed amount of H-NS was added. Reactions were then analyzed by EMSA. Our expectation was that pre-unfolded transpososome would strongly out-compete proU for H-NS binding. Note that in this experiment we used a slightly smaller OE substrate (substrate 2 in Figure 1A), as it was easier with this substrate to separate unfolded transpososome from H-NS–transpososome.
We show in Figure 3A that when the molar ratio of proU DNA to folded transpososome was 1 there was ∼50% inhibition of conversion of folded transpososome to H-NS–transpososome (lane 5 of Figure 3A and see also Figure 3C). As the proU to folded transpososome ratio decreased (lanes 3–4) there was less inhibition of H-NS–transpososome formation. While there was no obvious increase in inhibition as the proU to transpsosome ratio increased (i.e. >1) (lanes 6–7), this may be linked to an apparent destabilization of transpososome in these reactions as there was an increase in unbound OE DNA so the total transpososome level would be artificially low. In contrast, when the ‘competition experiment’ was carried out with pre-unfolded transpososome (lanes 9–13), there was essentially no inhibition of H-NS–transpososome formation by proU at any of the proU to pre-unfolded transpososome ratios initially tested (i.e. ∼4-fold excess; lane 13). In fact, as shown in Figure 3B, it was not until proU was in 16-fold excess relative to pre-unfolded transpososome (lane 6) that we started to see competition (manifested as the presence of unfolded transpososome). When proU was in 160-fold excess relative to pre-unfolded transpososome (lane 9) there was just over 50% inhibition of H-NS–transpososome formation. Note that the slight increase in H-NS–transpososome mobility seen in Figure 3 at increasing proU concentrations is likely due to titration of nonspecifically bound H-NS from the transpososome.
The results in Figure 3 are therefore consistent with H-NS binding the pre-unfolded transpososome substantially more tightly than either proU or the folded transpososome. In addition, the finding that conversion of folded transpososome to H-NS–transpososome was ∼50% inhibited when folded transpososome and proU were present at an equimolar ratio is consistent with H-NS binding the folded transpososome and proU with very similar affinities.
One concern in the competition experiments was that heparin, which is present only in the pre-unfolded transpososome reactions, might interfere with the ability of H-NS to bind proU DNA. We tested this and found that at the concentration of heparin used to unfold the transpososome (30 nM), there was no affect on H-NS binding to proU (data not shown).
H-NS stabilizes the unfolded form of the transpososome
One aspect of Tn10 transpososome dynamics that has not received much attention is the relative stabilities of the different transpososomes formed during transposition. The loss of subterminal contacts associated with IHF release and transpososome unfolding should in principle destabilize the unfolded transpososome. In fact, titration of IHF off of the transpososome with heparin typically does result in a decrease in the level of transpososome recovered (see below). Thus, while IHF release is important in promoting intermolecular transposition in the Tn10 system, it also has the potential to decrease the amount of transpososome present in the cell. As H-NS binds tightly and selectively to the Tn10 transpososome, we asked if it might also increase the stability of the transpososome.
We tested this idea by first allowing H-NS to bind the transpososome and then challenging the H-NS–transpososome complex with heparin. We show in a control experiment, where the folded transpososome was treated with heparin in the absence of H-NS (Figure 4A), that at the higher heparin concentrations there was a decrease in the amount of unfolded transpososome detected (compare lane 2 to lanes 6–7). Importantly, the decrease in transpososome signal coincided with an increase in the amount of unbound OE DNA (Figure 4B). This indicates that the loss of transpososome signal is due to transpososome dissociation as opposed to the transpososome adopting forms that are not easily detected in the gel. A comparison of transpososome abundance in lanes 2 and 7 of Figure 4A shows that at the highest heparin concentration used, there was as much as a 10-fold decrease in transpososome signal. In contrast, when H-NS was assembled onto the transpososome before heparin treatment, very little destabilization of the transpososome was observed (compare lane 8 with lanes 9–14 of Figure 4A and see Figure 4B). The slight increase in H-NS–transpososome mobility observed over the heparin titration in Figure 4 (also see Figure 5) is likely due to removal of non-specifically bound H-NS dimers from the transpososome.
Figure 5.
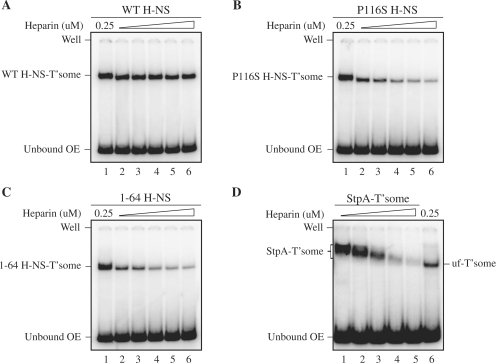
Determinants for H-NS-mediated transpososome stabilization. Heparin challenge was performed as in Figure 4 except that 0.25 µM heparin was initially used to unfold the transpososome. Subsequent to addition of either 75 nM WT-H-NS (A), 200 nM P116S H-NS (B), 250 nM 1–64 H-NS (C) or 250 nM StpA (D), heparin challenge was carried out by adding additional heparin to reactions in lanes 2–6 such that the final heparin concentrations in these reactions was 4 µM (lane 2), 8 µM (lane 3), 16 µM (lane 4), 32 µM (lane 5) and 64 µM (lane 6).
In the control experiment in Figure 4A transpososome destabilization is linked to the transition from the folded to the unfolded form because the ‘challenge’ agent (heparin) both induces transpososome unfolding and destabilization. Thus, in principle, a treatment that prevents transpososome unfolding could also prevent destabilization. However, the available evidence is consistent with H-NS inducing, as opposed to inhibiting transpososome unfolding (17) and therefore, H-NS-directed transpososome stabilization through inhibition of transpososome unfolding is extremely unlikely. To further strengthen this argument, we show in Figure S2 that when H-NS was added to a folded transpososome IHF occupancy decreased to <2% the level of IHF found in folded transpososomes (Figure S2).
Requirements for H-NS-directed transpososome stabilization
We used mutant forms of H-NS to gain insight into the mechanism through which H-NS stabilizes the unfolded transpososome. P116S H-NS is defective in structure-specific DNA-binding activity but retains nonspecific DNA-binding activity (25,26), whereas 1–64 H-NS has no appreciable DNA-binding activity (26,27). Both of these mutants retain dimerization activity (26,28) and can bind to the unfolded (but not the folded) transpososome (18). Retention of transpososome binding by 1–64 H-NS implies that this protein depends on interactions with transposase for recruitment into the unfolded transpososome. Based on these properties we reasoned that the P116S mutant could be used to ask if structure-specific DNA binding is required for H-NS-directed stabilization of the unfolded transpososome, while the 1–64 mutants could be used to ask if H-NS–transposase interactions are sufficient for H-NS-directed stabilization of the unfolded transpososome. We tested these ideas by binding the respective forms of H-NS to pre-unfolded transpososomes and then challenging these species with heparin treatment. In this regimen transpososome unfolding was induced using a relatively low concentration of heparin to allow binding of the mutant forms of H-NS to the transpososome and then higher concentrations of heparin were used in the challenge.
We show in Figure 5A that pre-unfolded transpososome loaded with WT H-NS remained stable over the full range of heparin concentrations used in the challenge. In contrast, there was a significant reduction in transpososome levels when the heparin challenge was performed with pre-unfolded transpososomes loaded with either P116S H-NS or 1–64 H-NS. For example, in the P116S ‘heparin challenge’ experiment the level of transpososome decreased as much as 20-fold. A similar result was obtained in the 1–64 ‘heparin challenge’. Thus, the results suggest that neither nonspecific DNA binding by H-NS to the pre-unfolded transpososome nor H-NS-binding mediated exclusively through interactions with transposase are sufficient to stabilize the pre-unfolded transpososome against heparin challenge.
We also asked if the transpososome-stabilizing effect observed above is specific to H-NS. StpA is an H-NS paralog that shares 58% amino acid identity and has the capacity to act as a molecular back up to H-NS in the control of gene expression (22). We show in Figure 5D that StpA is capable of binding pre-unfolded transpososome, as there was a clear reduction in complex mobility when StpA was added to assembly reactions (compare lanes 1–4 with lane 6). However, at a heparin concentration where the H-NS–transpososome remained intact (Figure 5A, lane 6), <5% of the StpA transpososome remained intact (Figure 5D, lane 5). Thus, while StpA can clearly bind the pre-unfolded transpososome, it fails to provide the same stabilizing function as H-NS.
H-NS-mediated transpososome stabilization correlates with the formation of structure-specific H-NS contacts with DNA and transposase–H-NS contacts
One way in which H-NS might stabilize the unfolded transpososome is to directly interact with both transpososome DNA and transposase. We tested this possibility by asking if P116S H-NS, which binds but does not stabilize the pre-unfolded transpososome (Figure 5B), also fails to form H-NS–transposase crosslinks. Pre-unfolded transpososomes were mixed with H-NS (WT or P116S) and treated with the protein–protein crosslinker EDC/NHS, a zero-length crosslinker that in proteins links carboxyl and amino groups. After gel isolation of transpososomes, we probed for transposase–H-NS crosslinks by immunoblotting as previously described (18). Briefly, this involved first probing the blot with an antibody to an epitope tag on transposase (T7 gene 10 peptide on the N-terminus of transposase) (Figure 6A), stripping the blot and then reprobing with an H-NS antibody (part B). We infer that species ‘a’ and ‘b’ were generated by crosslinks between transposase and H-NS because they comigrated on the two blots and were detected by both antibodies. Also, both species were only detected when EDC/NHS treatment was carried out. Notably, EDC/NHS treatment yielded very little crosslinked transposase dimer. Thus, it is not surprising that transposase–H-NS crosslinked species migrated faster on the gel than transposase dimers. Accordingly, we expect that ‘b’ is transposase crosslinked to an H-NS monomer and ‘a’ is transposase crosslinked to a crosslinked H-NS dimer.
Figure 6.
H-NS–transposase crosslinking. Transpososomes assembled with H-NS (either WT or P116S) were subjected to crosslinker treatment, either EDC/NHS (A) or BS3 (C). Reaction components were then separated on a native 5% polyacrylamide gel, and after imaging, gel slices containing H-NS–transpososome were obtained. Proteins were eluted from the gel slices and then applied to a 10% SDS gel whereupon immunoblot analysis was carried out using an antibody to an epitope tag on transposase. The blot in (A) was stripped after probing with transposase antibody and reprobed with H-NS antibody (B). Only lanes 6 and 7 are shown in (B) because these were the only lanes that gave a signal other than the molecular weight marker lane (not shown). Signal from marker lane was used to align the blots in (A) and (B). Species ‘a’ and ‘b’ are crosslinked products containing transposase and H-NS and ‘c’ is a putative crosslinked transposase–IHF product. Note that in (B) H-NS monomers are not detected because they were run off the gel.
Comparison of WT and P116S H-NS crosslinking reactions in Figure 6A and B revealed that WT H-NS efficiently formed crosslinks with transposase but P116S H-NS did not. The transposase (monomer) signal in Figure 6A provides an indication of the amount of H-NS–transpososome recovered, as only the H-NS-shifted transpososome was isolated after crosslinking. The total transposase signal was roughly 2-fold higher in the WT H-NS reaction relative to the P116S H-NS reaction, whereas the signal for transposase–H-NS crosslinked products (‘a’ + ‘b’) was roughly 20-fold higher in both transposase and H-NS blots. Thus, differences in transpososome recovery cannot account for the differences in the amounts of ‘a’ and ‘b’ in this experiment and we are left to conclude that H-NS must retain structure-specific DNA-binding activity in order to efficiently form crosslinks with transposase in the context of the transpososome.
Note that we have not performed the analogous experiment with StpA–transpososomes, thus we do not know if the inability of StpA to promote transpososome stability can be explained by the failure of StpA to interact with transposase.
We were intrigued by the loss of the relatively small amount of crosslinked transposase dimer in the EDC/NHS crosslinking experiment in Figure 6A upon incorporation of H-NS into the transpososome (compare lanes 5 and 6). To get a better idea of the magnitude of this effect, we repeated the above experiment using the crosslinker BS3, as we have found that yields of crosslinked transposase dimer are significantly higher with this reagent compared to EDC/NHS (data not shown). BS3 is a homobifunctional crosslinker that links amino groups in proteins through a 11.4 Å spacer arm. A comparison of Figure 6A and C confirms this as crosslinked transposase dimer makes up roughly 3-fold more of the total ‘transposase’ signal when pre-unfolded transpososome was crosslinked with BS3 versus EDC/NHS. When BS3 crosslinking was performed on the WT H-NS assembly reaction there was a complete loss of signal for crosslinked transposase dimer and a relatively large signal for species ‘b’ (Figure 6C, lanes 2 and 3). P116S H-NS was less effective than WT H-NS in preventing the formation of crosslinked transposase dimers and, consistent with the result in Figure 6A, failed to support the formation of a significant amount of transposase–H-NS crosslinked products (lane 4). We speculate that increased crosslinking efficiency of transposase dimers with BS3 versus EDC/NHS may be due to the relatively long spacer arm in BS3, as this would permit a greater number of residues in the dimer interface to participate in crosslink formation.
Finally, the identity of species ‘c’ in Figure 6A may be of interest. ‘c’ was only observed in the crosslinking reaction with the folded transpososome (lane 4 in Figure 6A) and therefore could be transposase crosslinked to IHF. Notably, IHF and transposase footprints overlap in the context of the folded transpososome, so there is good reason to think that these proteins are in close enough proximity to interact (13).
Overall, the results in this section show that WT but not P116S H-NS interacts directly with transposase. From this we infer that the transposase–H-NS interaction is dependent on H-NS binding the transpososome in a DNA structure-specific manner. The interaction of H-NS with transposase could play an important role in stabilizing the unfolded transpososome. In addition, incorporation of WT H-NS into the transpososome influences the amount of crosslinked transposase dimer detected in both EDC/NHS and BS3 crosslinking experiments. This could be an indication that H-NS incorporation into the transpososome has a significant influence on the structure of transposase.
DISCUSSION
The nucleoid-binding protein H-NS interacts with the initial IHF-folded Tn10 transpososome in vitro and induces transpososome unfolding. Transpososome unfolding is important for transposon excision. In addition, maintaining the transpososome in the unfolded form (post-excision) by preventing rebinding of IHF is critical for inhibiting self-destructive intramolecular transposition events and promoting intermolecular transposition events. In the current work we have looked at the interaction between H-NS and the initial-folded transpososome and a pre-unfolded transpososome. We show that H-NS binds the pre-unfolded transpososome much more tightly (∼40-fold) than either the folded transpososome or proU DNA, the latter being a well-characterized H-NS-binding partner. To our knowledge the binding of H-NS to the pre-unfolded transpososome represents the highest-affinity interaction defined to date for H-NS and a binding partner. We also evaluated the impact of H-NS binding on the relative stability of the transpososome. H-NS was shown to stabilize the transpososome against heparin challenge. This previously unrecognized property could help ensure that the transpososome stays intact throughout the course of the transposition reaction. Support for a model in which both transpososome stabilization and high-affinity H-NS binding to the transpososome is explained by H-NS binding simultaneously to transposon end DNA and transposase is provided.
H-NS has an extremely high affinity for the pre-unfolded transpososome
Based on Kd calculations we conclude that H-NS binds the pre-unfolded transpososome in the order of 40-fold more tightly than proU. Competitive binding assays confirmed that the pre-unfolded transpososome binds H-NS considerably more strongly than proU, as it was found that proU needed to be present at a 120-fold molar excess relative to pre-unfolded transpososome in order to reduce H-NS–transpososome formation by just over 50%. In contrast, when proU and folded transpososome were present at roughly equal concentrations, H-NS–transpososome formation was inhibited by about 50%. This suggests that H-NS binds proU and folded transpososome with similar strength.
One important question that arises from our comparative binding analyses is that if H-NS induces transpososome unfolding, why do we see a difference in the relative affinities of H-NS for the different transpososome forms? The most likely answer is that the Kd value determined for the pre-unfolded transpososome describes the intrinsic affinity between these two species. The H-NS binding assays with the pre-unfolded, but not the folded transpososome, were carried out in the presence of heparin. This prevents IHF from rebinding the unfolded transpososome so that there is effectively no competition between H-NS and IHF for transpososome binding (14,29,30). IHF is expected to bind the transpososome with a Kd in the low nM range (∼1 nM) (31). Thus, when H-NS and IHF are present at comparable concentrations and IHF is not titrated off the transpososome by heparin treatment, cycles of IHF and H-NS displacement from the transpososome are expected and this would increase the Kd of H-NS for the transpososome relative to the pre-unfolded transpososome.
Evidence that IHF and H-NS compete for transpososome binding comes in several forms. In this work we have shown by immunoblot analysis that H-NS addition to the folded transpososome greatly reduced the amount of IHF that copurified with the transpososome (Figure S2). In earlier work we showed that H-NS addition to the folded transpososome reduced the extent of the IHF footprint in the transpososome (18). In addition, when the concentration of IHF in an in vitro transposition reaction was increased so that IHF was in excess over H-NS (opposite to our standard reaction conditions), the impact of H-NS on strand transfer product (STP) distributions was essentially blocked. Normally, H-NS addition promotes the formation of STPs arising from random collision between the transpososome and target DNA and inhibits the formation of STPs arising from a highly constrained transpososome (19). Note that STP distributions provide an indirect readout of transpososome conformation immediately prior to integration.
Ultimately, the predominant form (unfolded versus folded) of the fully cleaved transpososome will be dictated by the concentrations of IHF and H-NS available to bind this species. H-NS is present at about twice the concentration of IHF in exponentially growing cells (32). Given the comparable Kd values for the binding of these proteins to the transpososome, it is likely that subtle changes in the expression of these genes could significantly alter the proportion of transpososome that populate the folded and unfolded forms. In this regard it may be significant that IHF expression tends to increase as cells enter stationary phase (33) as this could result in a decrease in intermolecular transposition events during this growth stage. In addition, cooperative binding of H-NS to DNA is strongly affected by temperature and the level of DNA supercoiling in the cell (24). It follows that changes in either parameter would be expected to affect the size of the H-NS pool that is able to redistribute along the DNA and compete with IHF for transpososome binding.
A mechanism for high-affinity H-NS binding to Tn10 transpososomes
H-NS is known to bind different DNA sequences with appreciably different affinities. Recent work has shown that high-affinity binding of H-NS to proU is governed by H-NS interactions with a specific DNA sequence that is present in two copies. Cloning of one copy of this sequence into another DNA fragment was sufficient to recreate a high-affinity H-NS-binding site at the inserted sequence as well as increase the affinity of two binding sites situated ∼50 and 60 bp downstream of the high-affinity site. Importantly, the high-affinity H-NS-binding site within proU has been shown to adopt a distorted DNA structure in the absence of H-NS leading to the idea that sequence-specific H-NS binding is governed by an indirect rather than direct sequence read-out mechanism. Overall these observations are strongly supportive of a cooperative binding model wherein H-NS binding to a high-affinity site via DNA structure-specific interactions promotes additional binding events with neighboring sequences that otherwise have intrinsically low affinities for H-NS. Presumably oligomerization of H-NS dimers bound at the ‘nucleation’ and secondary sites is a key component of the cooperativity (24). In addition, the propensity of the DNA sequence between nucleation and secondary binding sites to bend is expected to be an important factor in cooperative binding by H-NS. Notably, H-NS binds A-T-rich planar curved DNA more tightly than non-curved DNA (34), leading to the expectation that the presence of a nucleation site within an A-T-rich DNA sequence would lead to the highest-affinity H-NS interactions in a bacterial genome. This is supported by results from genome-wide H-NS ChIP-on-chip experiments and bioinformatics analysis (35).
The above discussion provides a context for considering a mechanism for the tight binding of H-NS to both the folded and pre-unfolded Tn10 transpososomes. Key factors to consider include (i) results from DNA footprinting studies showing protected regions within both folded and pre-unfolded transpososomes containing H-NS, two of which are immediately adjacent to transposase (see Figure 1B); (ii) results from protein–protein crosslinking studies showing H-NS forms contacts with transposase in the context of the transpososome; (iii) results from EMSA studies showing H-NS binding to the transpososome is highly cooperative (current work); (iv) results from EMSA studies with P116S suggesting that H-NS binding to the folded transpososome requires that H-NS retains structure-specific DNA recognition capabilities. Considering points (i) and (ii), it seems likely that a component of the observed tight binding would be the occurrence of transposase interactions with H-NS bound at H-NSTIR-PROX and/or H-NSTIR-DIST. Considering point (iv) H-NSTIR-PROX overlaps a region in the transpososome (folded and unfolded) that is hypersensitive to cleavage with the chemical nuclease OP-Cu and thus expected to adopt a distorted DNA structure (13,36). H-NS–transposase contacts might then form subsequent to H-NS forming initial contacts with a distorted DNA structure within H-NSTIR-PROX. Finally, the cooperative nature of H-NS binding to both folded and unfolded transpososomes (point iii) could be explained by H-NS–H-NS interactions from dimers bound to separate ends. For example, if the two ends are aligned in anti-parallel fashion, as in the Tn5 transpososome (37), then H-NS bound to H-NSTIR-PROX at one end might be able to interact with H-NS bound to H-NSTIR-DIST at the other end (see Figure 1B). If H-NSTIR-PROX were the higher-affinity binding site, then this interaction would strengthen the H-NS interaction with H-NSTIR-DIST. Notably, none of the putative H-NS-binding sites in the transpososome is a close fit to the consensus binding site for H-NS (5′ tCGATAAATT 3′; 80% A-T). However, like the consensus binding site, all three sites are A-T rich (H-NSTIR-PROX 5′ TGATGAA 3′ [66% A-T]; H-NSTIR-DIST 5′ AATGATTTT 3′ [88% A-T] and H-NSFLANK 5′ AATTAAT 3′ [100% A-T]).
An important feature of the general model described above is that the extremely tight binding of H-NS to the unfolded transpososome can be explained by H-NS forming contacts with transposase, which is tightly bound to the DNA. The CAP–RNA polymerase interaction provides a classic example of one DNA-binding protein increasing the affinity of another DNA-binding protein for a specific DNA site through protein–protein interactions (38).
A corollary of the above model is that H-NS incorporation into the transpososome would also increase the stability of the transpososome and this is exactly what we have observed.
Biological significance of H-NS-mediated transpososome stabilization
Transpososome unfolding has been linked to transposon excision and maintaining the transpososome in the unfolded form is important for promoting intermolecular target capture and strand transfer. We have shown that treatment of what formally would be a pre-excision transpososome (because flanking donor DNA is still present in our OE substrate) with heparin had a strong destabilizing effect on the transpososome when heparin was present in relatively large excess relative to the transpososome. However, if the transpososome was allowed to associate with WT H-NS prior to heparin treatment, transpososome destabilization was not observed. From this we suggest that in addition to its role in promoting transpososome unfolding, H-NS may be important in helping to stabilize the transpososome when it undergoes the unfolding transition. This would increase the probability that a transpososome is able to complete all of the chemical steps in the Tn10 transposition reaction.
It is difficult to directly relate the in vitro heparin challenge data to the likelihood of transpososome instability being a significant factor in determining the transposition frequency of Tn10 in vivo. However, if one considers heparin (in vitro) to be a surrogate for DNA (in vivo), then if transposase-Tn10 end interactions are even slightly compromised by the unfolding transition, it is reasonable to surmise that the high concentration of DNA within the cell could titrate transposase off of the transposon end DNA during this transition, leading to transpososome dissociation.
In addition to affecting transpososome dynamics in the Tn10 system, we have previously shown that H-NS has a positive effect on transpososome assembly in the Tn5 in vitro system. Here, inclusion of H-NS under suboptimal conditions for transpososome assembly increased the efficiency of transpososome formation by up to 17-fold. As in the Tn10 system, H-NS binds in very close proximity to transposase in the Tn5 transpososome and thus there is a good possibility that the above effect of H-NS is linked to H-NS helping to tether the transposase to the transposon end DNA (8). Alternatively, or in addition, the H-NS-stabilizing effect in both Tn10 and Tn5 systems might be related to the capacity of H-NS to bridge DNA molecules (39). That is, the interaction between H-NS dimers bound to different transposon end sequences could also help to hold the two ends together. The observation that neither P116S H-NS nor 1-64 H-NS can stabilize the unfolded transpososome could in part be explained by the inability of these mutant forms to participate in H-NS tetramer formation (28).
We also found that the H-NS paralog StpA failed to promote stabilization of the Tn10 transpososome. While StpA was shown to bind the unfolded transpososome, the binding interaction is likely nonspecific in nature as heparin effectively titrated StpA off of the transpososome. This mimics the behavior of P116S H-NS and 1–64 H-NS, leading us to believe that StpA lacks either the ability to recognize specific DNA structures within the transpososome and/or the ability to interact with transposase. StpA also failed to positively affect transpososome assembly in the Tn5 system (8). Thus, while StpA can serve as a molecular replacement for H-NS in the regulation of gene expression, it is clear that with respect to affecting aspects of Tn10 and Tn5 transpososome dynamics, H-NS and StpA behave very differently. This observation is suggestive that only a select group of H-NS homologues, of which there are many in gamma-proteobacteria (40) and at least one in mycobacteria (41), may be able to directly influence transposition reactions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant to D.B.H. from the Canadian Institutes of Health Research (MOP11281). Funding for open access charge: CIHR Research grant.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank D. Edgell for comments on the manuscript. They also thank D. Jin for providing H-NS antibody, S. Goodman for providing IHF antibody, M. Belfort for providing H-NS and StpA overexpression strains and S. Rimsky for providing the H-NS 1-64 overexpression vector and template plasmid for amplification of proU.
REFERENCES
- 1.Gal-Mor O, Finlay BB. Pathogenicity islands: a molecular toolbox for bacterial virulence. Cell Microbiol. 2006;8:1707–1719. doi: 10.1111/j.1462-5822.2006.00794.x. [DOI] [PubMed] [Google Scholar]
- 2.Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JC. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006;2:e81. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 4.Doyle M, Fookes M, Ivens A, Mangan MW, Wain J, Dorman CJ. An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. Science. 2007;315:251–252. doi: 10.1126/science.1137550. [DOI] [PubMed] [Google Scholar]
- 5.Oshima T, Ishikawa S, Kurokawa K, Aiba H, Ogasawara N. Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res. 2006;13:141–153. doi: 10.1093/dnares/dsl009. [DOI] [PubMed] [Google Scholar]
- 6.Rouquette C, Serre MC, Lane D. Protective role for H-NS protein in IS1 transposition. J. Bacteriol. 2004;186:2091–2098. doi: 10.1128/JB.186.7.2091-2098.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Swingle B, O'C;arroll M, Haniford D, Derbyshire KM. The effect of host-encoded nucleoid proteins on transposition: H-NS influences targeting of both IS903 and Tn10. Mol. Microbiol. 2004;52:1055–1067. doi: 10.1111/j.1365-2958.2004.04051.x. [DOI] [PubMed] [Google Scholar]
- 8.Whitfield CR, Wardle SJ, Haniford DB. The global bacterial regulator H-NS promotes transpososome formation and transposition in the Tn5 system. Nucleic Acids Res. 2009;37:309–321. doi: 10.1093/nar/gkn935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haniford DB, Chelouche AR, Kleckner N. A specific class of IS10 transposase mutants are blocked for target site interactions and promote formation of an excised transposon fragment. Cell. 1989;59:385–394. doi: 10.1016/0092-8674(89)90299-7. [DOI] [PubMed] [Google Scholar]
- 10.Bender J, Kleckner N. Genetic evidence that Tn10 transposes by a nonreplicative mechanism. Cell. 1986;45:801–815. doi: 10.1016/0092-8674(86)90555-6. [DOI] [PubMed] [Google Scholar]
- 11.Haniford DB, Benjamin HW, Kleckner N. Kinetic and structural analysis of a cleaved donor intermediate and a strand transfer intermediate in Tn10 transposition. Cell. 1991;64:171–179. doi: 10.1016/0092-8674(91)90218-n. [DOI] [PubMed] [Google Scholar]
- 12.Sakai J, Chalmers RM, Kleckner N. Identification and characterization of a pre-cleavage synaptic complex that is an early intermediate in Tn10 transposition. EMBO J. 1995;14:4374–4383. doi: 10.1002/j.1460-2075.1995.tb00112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crellin P, Chalmers R. Protein-DNA contacts and conformational changes in the Tn10 transpososome during assembly and activation for cleavage. EMBO J. 2001;20:3882–3891. doi: 10.1093/emboj/20.14.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crellin P, Sewitz S, Chalmers R. DNA looping and catalysis; the IHF-folded arm of Tn10 promotes conformational changes and hairpin resolution. Mol. Cell. 2004;13:537–547. doi: 10.1016/s1097-2765(04)00052-8. [DOI] [PubMed] [Google Scholar]
- 15.Humayun S, Wardle SJ, Shilton BH, Pribil PA, Liburd J, Haniford DB. Tn10 transposase mutants with altered transpososome unfolding properties are defective in hairpin formation. J. Mol. Biol. 2005;346:703–716. doi: 10.1016/j.jmb.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 16.Chalmers R, Guhathakurta A, Benjamin H, Kleckner N. IHF modulation of Tn10 transposition: sensory transduction of supercoiling status via a proposed protein/DNA molecular spring. Cell. 1998;93:897–908. doi: 10.1016/s0092-8674(00)81449-x. [DOI] [PubMed] [Google Scholar]
- 17.Wardle SJ, O'C;arroll M, Derbyshire KM, Haniford DB. The global regulator H-NS acts directly on the transpososome to promote Tn10 transposition. Genes Dev. 2005;19:2224–2235. doi: 10.1101/gad.1338905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ward CM, Wardle SJ, Singh RK, Haniford DB. The global regulator H-NS binds to two distinct classes of sites within the Tn10 transpososome to promote transposition. Mol. Microbiol. 2007;64:1000–1013. doi: 10.1111/j.1365-2958.2007.05708.x. [DOI] [PubMed] [Google Scholar]
- 19.Singh RK, Liburd J, Wardle SJ, Haniford DB. The nucleoid binding protein H-NS acts as an anti-channeling factor to favor intermolecular Tn10 transposition and dissemination. J. Mol. Biol. 2008;376:950–962. doi: 10.1016/j.jmb.2007.12.035. [DOI] [PubMed] [Google Scholar]
- 20.Chalmers RM, Kleckner N. Tn10/IS10 transposase purification, activation, and in vitro reaction. J. Biol. Chem. 1994;269:8029–8035. [PubMed] [Google Scholar]
- 21.Surette MG, Chaconas G. A protein factor which reduces the negative supercoiling requirement in the Mu DNA strand transfer reaction is Escherichia coli integration host factor. J. Biol. Chem. 1989;264:3028–3034. [PubMed] [Google Scholar]
- 22.Zhang A, Rimsky S, Reaban ME, Buc H, Belfort M. Escherichia coli protein analogs StpA and H-NS: regulatory loops, similar and disparate effects on nucleic acid dynamics. EMBO J. 1996;15:1340–1349. [PMC free article] [PubMed] [Google Scholar]
- 23.Navarre WW, McClelland M, Libby SJ, Fang FC. Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 2007;21:1456–1471. doi: 10.1101/gad.1543107. [DOI] [PubMed] [Google Scholar]
- 24.Bouffartigues E, Buckle M, Badaut C, Travers A, Rimsky S. H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat. Struct. Mol. Biol. 2007;14:441–448. doi: 10.1038/nsmb1233. [DOI] [PubMed] [Google Scholar]
- 25.Spurio R, Falconi M, Brandi A, Pon CL, Gualerzi CO. The oligomeric structure of nucleoid protein H-NS is necessary for recognition of intrinsically curved DNA and for DNA bending. EMBO J. 1997;16:1795–1805. doi: 10.1093/emboj/16.7.1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Badaut C, Williams R, Arluison V, Bouffartigues E, Robert B, Buc H, Rimsky S. The degree of oligomerization of the H-NS nucleoid structuring protein is related to specific binding to DNA. J. Biol. Chem. 2002;277:41657–41666. doi: 10.1074/jbc.M206037200. [DOI] [PubMed] [Google Scholar]
- 27.Williams RM, Rimsky S, Buc H. Probing the structure, function, and interactions of the Escherichia coli H-NS and StpA proteins by using dominant negative derivatives. J. Bacteriol. 1996;178:4335–4343. doi: 10.1128/jb.178.15.4335-4343.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stella S, Spurio R, Falconi M, Pon CL, Gualerzi CO. Nature and mechanism of the in vivo oligomerization of nucleoid protein H-NS. EMBO J. 2005;24:2896–2905. doi: 10.1038/sj.emboj.7600754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Junop MS, Haniford DB. Factors responsible for target site selection in Tn10 transposition: a role for the DDE motif in target DNA capture. EMBO J. 1997;16:2646–2655. doi: 10.1093/emboj/16.10.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakai JS, Kleckner N, Yang X, Guhathakurta A. Tn10 transpososome assembly involves a folded intermediate that must be unfolded for target capture and strand transfer. EMBO J. 2000;19:776–785. doi: 10.1093/emboj/19.4.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lynch TW, Read EK, Mattis AN, Gardner JF, Rice PA. Integration host factor: putting a twist on protein-DNA recognition. J. Mol. Biol. 2003;330:493–502. doi: 10.1016/s0022-2836(03)00529-1. [DOI] [PubMed] [Google Scholar]
- 32.Ali Azam T, Iwata A, Nishimura A, Ueda S, Ishihama A. Growth phase-dependent variation in protein composition of the Escherichia coli nucleoid. J. Bacteriol. 1999;181:6361–6370. doi: 10.1128/jb.181.20.6361-6370.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ditto MD, Roberts D, Weisberg RA. Growth phase variation of integration host factor level in Escherichia coli. J. Bacteriol. 1994;176:3738–3748. doi: 10.1128/jb.176.12.3738-3748.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jordi BJ, Fielder AE, Burns CM, Hinton JC, Dover N, Ussery DW, Higgins CF. DNA binding is not sufficient for H-NS-mediated repression of proU expression. J. Biol. Chem. 1997;272:12083–12090. doi: 10.1074/jbc.272.18.12083. [DOI] [PubMed] [Google Scholar]
- 35.Lang B, Blot N, Bouffartigues E, Buckle M, Geertz M, Gualerzi CO, Mavathur R, Muskhelishvili G, Pon CL, Rimsky S, et al. High-affinity DNA binding sites for H-NS provide a molecular basis for selective silencing within proteobacterial genomes. Nucleic Acids Res. 2007;35:6330–6337. doi: 10.1093/nar/gkm712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allingham JS, Haniford DB. Mechanisms of metal ion action in Tn10 transposition. J. Mol. Biol. 2002;319:53–65. doi: 10.1016/S0022-2836(02)00297-8. [DOI] [PubMed] [Google Scholar]
- 37.Davies DR, Goryshin IY, Reznikoff WS, Rayment I. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science. 2000;289:77–85. doi: 10.1126/science.289.5476.77. [DOI] [PubMed] [Google Scholar]
- 38.Straney DC, Straney SB, Crothers DM. Synergy between Escherichia coli CAP protein and RNA polymerase in the lac promoter open complex. J. Mol. Biol. 1989;206:41–57. doi: 10.1016/0022-2836(89)90522-6. [DOI] [PubMed] [Google Scholar]
- 39.Noom MC, Navarre WW, Oshima T, Wuite GJ, Dame RT. H-NS promotes looped domain formation in the bacterial chromosome. Curr. Biol. 2007;17:R913–914. doi: 10.1016/j.cub.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 40.Tendeng C, Bertin PN. H-NS in Gram-negative bacteria: a family of multifaceted proteins. Trends Microbiol. 2003;11:511–518. doi: 10.1016/j.tim.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 41.Gordon BR, Imperial R, Wang L, Navarre WW, Liu J. Lsr2 of Mycobacterium represents a novel class of H-NS-like proteins. J. Bacteriol. 2008;190:7052–7059. doi: 10.1128/JB.00733-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.