Abstract
Unnoticed cell culture contamination by viruses, Mycoplasma, or other cell lines is not uncommon and a threat to laboratory safety and the quality of scientific results. We developed and validated a novel high-throughput Multiplex cell Contamination Test (McCT), which is currently able to detect 37 contamination markers in a single reaction. The assay is based on multiplex PCR with target-specific primers and subsequent hybridization of amplimers to specific oligonucleotide probes. McCT proved to be highly specific, sensitive and robust, and allows to analyze more than 1000 cell lysates per week. In conclusion, the novel McCT assay is a powerful high-throughput tool in assessing cell line purity.
INTRODUCTION
Contaminations of eukaryotic cell cultures with microbial organisms as well as with viruses or other eukaryotic cell lines are a major problem in cell culture-related research. Although contamination of cell cultures is known since >50 years, it is still a widespread cause for erroneous research results, for reduced reproducibility and even for unusable therapeutic products (1,2). It is an essential prerequisite for cell culture-based research to work with contamination-free cell lines as otherwise all publications arising from the use of such cell lines will be invalid. Moreover, contaminations with microbial organism or cross-contaminations with other eukaryotic lines may lead to diminished cell growth and could result in loss of the original culture.
About 15–35% of all cell lines in several laboratories were found to be infected with Mycoplasma species, mostly of human, pig or bovine origin (2–4). A frequent source for Mycoplasma contaminations is contaminated cell culture medium ingredients or the experimenter. Mycoplasma can cause impairment of many cellular functions such as inhibition of cell proliferation, protein biosynthesis, alteration of immunological reactions, microarray gene expression profiles and virus replication (5–7). Due to their small cell size, Mycoplasmas are difficult to detect with a conventional microscope. Because Mycoplasmas lack a cell wall; some antibiotics (e.g. ampicillin) are not effective to suppress Mycoplasma growth. Therefore, specific methods are required to detect Mycoplasma in a cell culture (8).
Recently, squirrel monkey retrovirus (SMRV) was found to be a widespread cause for cell contaminations. For example, cells used for commercial production of interferon were found to be contaminated with SMRV (1). This type D retrovirus is classified in biological risk group 2 (9,10). Infection of other species is not described, but since contaminations of numerous vertebrate cell lines have been reported (11,12), the potential for in vivo infection cannot be excluded. Therefore, the German Central Commission for Biological Safety (ZKBS) recommended the testing of all used cell lines for SMRV. Contaminated cell lines must either be eliminated or used under conditions of biological safety level 2.
Other viruses including human herpes viruses (HHV), hepatitis B (HBV) and C (HBC) and HIV can be present in donor tissues and, thus, pose a risk to personnel working with cell lines cultured from primary explants or subcultures but also cultures of human lymphocytes, fetal cell mixtures or hepatocytes.
The most difficult type of contamination to detect may well be contamination of one cell type by another. Cell lines may be contaminated with other cell lines belonging to either the same (intra-) or another species (inter-species contamination). If the cells lines are similar in appearance, this cross-contamination or a final replacement of the slower growing cell line may remain unnoticed. In the past, frequently used HeLa cells were found to be one of the most prominent cross-contaminations (13). But also nowadays, cell lines have been often reported to be of different origin or species from that being originally described (14–17). Cross-contaminations may be due to insufficient care during cell culture handling, or mislabeling of culture flask and stocks.
A large number of different methods for detecting contaminants in cell cultures have already been described and include, e.g. polymerase chain reaction (PCR), RNA hybridization, isozyme typing, short tandem repeat profiling, microscopic analysis and microbiological colony assay (18). All of them have been shown in numerous studies to exhibit their specific advantages and disadvantages. However, the main disadvantage certainly remains that these assays usually detect only a single type or a small group of cell contamination, such as Mycoplasma or inter-species contaminations, respectively. Thus, while being able to detect specific contaminations, they fail to assess numerous other possible contaminants simultaneously.
Here, we describe the Multiplex cell Contamination Test (McCT), a rapid, high-throughput assay for reliably determining the cell line purity by analyzing 37 microbial and inter-species contaminations in mammalian cell cultures simultaneously.
MATERIALS AND METHODS
Cell lysate preparation
Cultured cells (∼106) were pelleted by centrifugation for 5 min (600 g), subsequently resuspended in 100 μl of phosphate buffered saline (PBS) and lysed at 95°C for 15 min. After a second centrifugation step, the supernatant was stored at −20°C.
Multiplex primer and probe design
Nucleotide sequences were obtained from the National Center for Biotechnology Information (NCBI) nucleotide sequence database (GenBank). The 16S RNA region of Mycoplasma orale, M. hyorhinis, M. arginini, M. fermentans, M. salivarium, M. hominis, M. pneumoniae, M. synoviae, M. pirum, M. gallisepticum and Acholeplasma laidlawii; the E1A region of Adenovirus types 1, 2, 5 and 6; and the species-specific β-globin genes, respectively, were aligned with ClustalW (19), in order to identify conserved sequences flanking inner polymorphic regions for primer design. All primer sequences were assessed for primer–dimer formation and for unspecific annealing of biotinylated primers to oligonucleotide probes by using Fast-PCR (20). One primer per pair was 5′-biotinylated for subsequent staining of the amplified product. All primers were tested in single- and multiplex PCR and subsequent gel electrophoresis and/or Luminex hybridization. For each target gene and organism, oligonucleotide probes were chosen in polymorphic sequences amplified by primers mentioned above. Oligonucleotide probes were selected to be highly specific for the gene or organism of interest. Probe sequences were tested for unspecific hybridization to biotinylated oligonucleotide primers by Fast-PCR and in Luminex hybridization experiments.
Multiplex PCR
The 50 µl reactions comprised 1× Multiplex PCR Kit buffer (Qiagen, Hilden, Germany), containing 3 mM MgCl2, dNTP mix, 0.5× Q solution and HotStartTaq DNA polymerase, 0.1–0.2 µM of each primer and 1 µl of cell lysate (equivalent to 104 –105 cells). A 15 min enzyme activation step at 95°C was followed by 40 cycles of amplification in a Mastercycler (Eppendorf, Hamburg, Germany). Each cycle included a denaturation step at 94°C for 30 s, an annealing step at 61°C for 90 s and an extension step at 72°C for 60 s. The final extension was prolonged for further 10 min and reactions were stored at 4°C.
Coupling of oligonucleotide probes
Oligonucleotide probes with 5′-amino-modified C12-linkers (Eurofins MWG Operon, Ebersberg, Germany) were coupled to carboxylated beads as described recently (21).
Multiplex hybridization
Following multiplex PCR amplification, 10 µl of each reaction mixture was transferred to 96-well plates containing in each well 33 µl of tetramethylammonium chloride (TMAC) hybridization solution (0.15 M TMAC, 75 mM Tris–HCl, 6 mM ethylen diamin tetraacetate (EDTA), 1.5 g/l Sarkosyl, pH 8), 7 µl of 1× TE and a mixture of 2000 probe-coupled beads of each set as recently described (21,22). The mixture was heated to 95°C for 10 min, immediately placed on ice for 1 min and then moved to a thermomixer for hybridization at 41°C for 30 min under agitation. The samples were transferred to a 96-well wash plate (Millipore, Bedford, MA, USA) and pre-equilibrated with washing buffer (PBS, 0.02% Tween). Subsequently, the beads were washed once with 100 µl of washing buffer on a vacuum wash station (Millipore). On a horizontal shaker at room temperature, beads were resuspended for 20 min in 50 µl of streptavidin-R-phycoerythrin (Molecular Probes, Eugene, OR, USA) diluted in the ratio 1:1600 in 2 M TMAC, 75 mM Tris–HCl, 6 mM EDTA, 1.5 g/l Sarkosyl, pH 8. Beads were then washed three times with 100 µl washing buffer and finally resuspended in 100 µl washing buffer for 5 min on a shaker. Beads were analyzed for internal bead color and R-phycoerythrin reporter fluorescence on a Luminex 100 analyzer. The median reporter fluorescence intensity (MFI) of at least 100 beads was computed for each bead set in the sample.
Cut-off definition and statistics
For each probe, MFI values in reactions with no PCR product added to the hybridization mixture were considered as background values. Net MFI values were computed by subtraction of 1.2 times the median background value. Net MFI values >5 MFI were defined as positive reactions. For all probes, this cut-off value was above the mean background plus three times the SD.
Venor®GeM Mycoplasma kit
The Venor®GeM kit (Minerva Biolabs, Berlin, Germany) was performed according to the manufacturer's instructions. Briefly, 2 µl of DNA extracts were amplified in 25 µl containing 1× AmpliTaq Gold buffer, 2.5 µl of the primer/dNTP mix, 2.5 mM MgCl2, 2.5 µl of the internal control DNA and 1 U of DNA AmpliTaq Gold polymerase (Applied Biosystems, Branchburg, NJ, USA). A 15-min denaturation step at 94°C was followed by 39 cycles of amplification with a Mastercycler (Eppendorf). Each cycle included a denaturation step at 94°C for 30 s, an annealing step at 55°C for 30 s and an elongation step at 72°C for 30 s. Positive controls with defined templates and negative controls without template were included in each experiment. For final analysis, 5 µl of PCR products were visualized on a 1.5% agarose gel.
RESULTS
Assay design
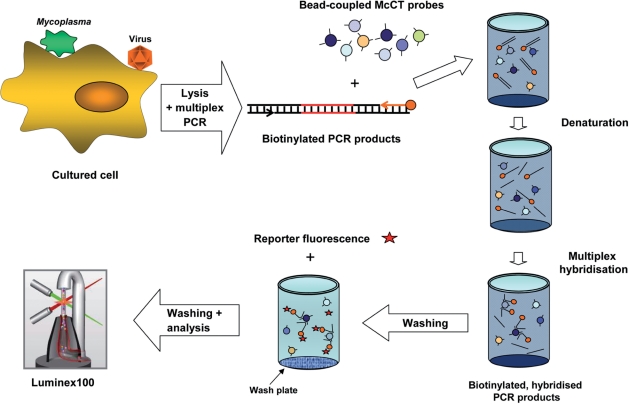
After cell lysis, the multiplex PCR amplified the contaminations with 37 specific primer pairs targeting viral, bacterial and cellular genome regions. For some targets, two gene regions were included to increase specificity; e.g. the detection of mouse and rat contaminations was based on β-globin and mitochondrial 16S ribosomal RNA gene amplification (Table 1). Multiplex PCR products were denatured and hybridized to 45 target-specific oligonucleotide probes coupled to spectrally distinct beads, allowing PCR products from 96 individual cell cultures to be processed in parallel (Figure 1). After transfer into wash plates with filter bottoms, unbound DNA was removed. Subsequently, biotinylated PCR products were stained by streptavidin-R-phycoerythrin conjugate as reporter. After further washing steps, beads were analyzed in the Luminex100 instrument, which contains two lasers to identify the bead set by the internal bead color and to quantify the reporter fluorescence on the bead surface.
Table 1.
Targets of McCT
| Contamination/marker | Gene region |
|---|---|
| Mycoplasma spec. | 16S ribosomal RNA |
| M. orale | |
| M. hyorhinis | |
| M. arginini | |
| M. fermentans | |
| M. salivarium | |
| M. hominis | |
| M. pneumonia | |
| M. synoviae | |
| M. pirum, | |
| Acholeplasma laidlawii | |
| HPV18 | E7 |
| Hepatitis B virus | Protein S |
| Adenovirus 5 | E1A |
| SMRV | Gag, Env |
| SV40 | T-antigen, VP1, VP3 |
| HHVs | UL42 family |
| HSV1 | |
| HSV2 | |
| VZV | |
| EBV | |
| CMV | |
| HHV6 | |
| HHV7 | |
| Kaposi's sarcoma associated herpesvirus (HHV8) | |
| Mammalian DNA | PolA |
| Human | β-Globin, TSPYL1 |
| Rat | β-Globin, mitochondrial 16S ribosomal RNA |
| Mouse | |
| Green monkey | |
| Chinese hamster | Mitochondrial 16S ribosomal RNA |
| Dog | |
| Cat | |
| Rabbit | |
| Guinea pig | |
| Pig | |
| Y chromosome | Sex-determining region of chromosome Y |
| Human/monkey | |
| Mouse | |
Figure 1.
Schematic overview of the McCT (picture of the Luminex analyser taken from Luminex Corp. webpage).
The multiplex assay was able to specifically detect and type the 10 different Mycoplasma species (M. orale, M. hyorhinis, M. arginini, M. fermentans, M. salivarium, M. hominis, M. pneumoniae, M. synoviae, M. pirum and A. laidlawii). An even broader spectrum of Mycoplasma was covered by two universal probes (Table 2). The assay was able to specifically detect the presence of 12 viruses [SMRV, HBV, human herpes simplex virus (HSV1 and HSV2), varicella zoster virus (VZV), Epstein–Barr virus (EBV), cytomegalovirus virus (CMV), HHV-6, -7 and -8, human papillomavirus 18 (HPV18) and monkey polyomavirus SV40]. Since some of these viruses are present in established human cell lines, they can be used as nonexclusive marker for specific human cell lines, e.g. HEK293T cells are positive for Adenovirus 5, and SV40 VP1 and T-antigen, while HeLa is positive for HPV18 (Tables 2 and 3). Inter-species contaminations were assessed by detecting specific human, green monkey, mouse, rat, Chinese hamster, cat, pig, dog, rabbit and guinea pig sequences. Analysis of male-specific sequences of human/primate or mouse allowed detection of contaminating male cells in female cell lines. Internal DNA and PCR quality was assessed by detecting the mammalian PolA sequence.
Table 2.
Hybridization results for McCT using different cell lysates
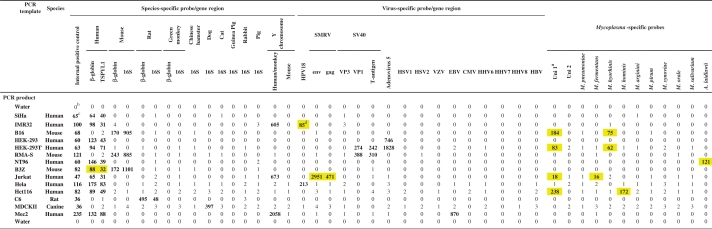 |
aTwo universal probes designed to detect Mycoplasma species not covered by species-specific probes.
bNet MFI values of PCR products hybridized to a mixture of 45 distinct bead sets, negative values set to 0.
cSignals above cut-off (value given in bold).
dIdentified cell culture contaminations (yellow boxes).
Table 3.
Nonexclusive markers for widely used cell lines
| Cell line | Species | Sex | Viral sequences |
|---|---|---|---|
| HeLa | Human | Female | HPV18 |
| HEK293T | Human | Female | Adenovirus 5, SV40 VP1 and T-antigen |
| HEK293 | Human | Female | Adenovirus 5 |
| Namalwa | Human | Female | EBV |
| SW480 | Human | Male | |
| Alexander | Human | Female | HBV |
| Cos7 | Monkey | Female | SV40 VP1, VP3 and T-antigen |
| CHO | Chinese Hamster | Female | |
| MDCK II | Dog | unknown | |
| NIH3T3 | Mouse | Female |
Specificity of McCT
Multiplex PCR was performed on crude lysates prepared from cell cultures from different species containing nonexclusive markers, or highly purified plasmid DNA (SMRV, SV40) in cellular carrier DNA. Target-specific probes were coupled to defined bead sets and hybridized to 10 µl (100–300 ng DNA) of PCR products (Tables 4 and 5). Detection of all targets was highly specific even for highly related viruses or target genes, such as the 16S ribosomal RNA gene that was used for the detection of different species. Moreover, endogenous retroviruses or HIV DNA present in biological samples did not lead to unspecific signals with the SMRV Gag- and Env-specific probes (data not shown).
Table 4.
Specificity of McCT PCR product detection: viral templates
| PCR template | Virus-specific probe |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SMRV |
HPV18 | HSV1 | HSV2 | VZV | EBV | CMV | HHV6 | HHV7 | HHV8 | HBV | Adeno virus 5 | VP1 | SV40 VP3 | T-antigen | ||
| env | gag | |||||||||||||||
| SMRV | 1687a | 488 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 |
| HPV18 | 0 | 0 | 299 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| HSV1 | 0 | 0 | 1 | 1116 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| HSV2 | 0 | 0 | 1 | 1 | 350 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| VZV | 1 | 0 | 0 | 0 | 0 | 259 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| EBV | 0 | 1 | 0 | 0 | 0 | 0 | 122 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| CMV | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 429 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 0 |
| HHV6 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 254 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| HHV7 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1732 | 0 | 1 | 0 | 1 | 0 | 1 |
| HHV8 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 355 | 0 | 0 | 0 | 1 | 0 |
| HBV | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 2660 | 0 | 0 | 0 | 0 |
| Adeno virus 5 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 1585 | 1 | 0 | 1 |
| SV40 VP3 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 210 | 1 | 0 |
| SV40 VP1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 450 | 0 |
| SV40 T-antigen | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 1068 |
aEach line represents hybridization results of the multiplex PCR product of one template hybridized to a mixture of 45 distinct probes; results of only 16 probes are shown, no positive signals were observed with any of the 29 other probes; numbers are net MFI values computed as described in ‘Materials and Methods’ section, values >5 are considered positive.
Table 5.
Specificity of McCT PCR product detection: mammalian species-specific probes
| PCR template | Species-specific probe/gene region |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Human |
Mouse |
Rat |
Monkey |
Chinese hamster | Dog | Cat | Guinea pig | Rabbit | Quality control | |||||
| β-globin | TSPYL1 | β-globin | 16S | β-globin | 16S | β-globin | 16S | 16S | 16S | 16S | 16S | 16S | PolA | |
| Human | 701a | 189 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 324 |
| Mouse | 0 | 3 | 68 | 739 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 229 |
| Rat | 0 | 0 | 0 | 0 | 496 | 62 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 231 |
| Green monkey | 2 | 14b | 0 | 1 | 0 | 0 | 310 | 374 | 0 | 0 | 0 | 0 | 0 | 147 |
| Chinese hamster | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 672 | 0 | 0 | 0 | 0 | 207 |
| Dog | 0 | 0 | 0 | 0 | 1 | 2 | 1 | 0 | 1 | 1166 | 2 | 0 | 2 | 182 |
| Cat | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 866 | 1 | 0 | 174 |
| Guinea pig | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1088 | 0 | 220 |
| Rabbit | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 155 | 173 |
| H2O-PCR | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
aEach line represents hybridization results of the multiplex PCR product of one template hybridized to a mixture of 45 distinct probes; results of only 14 probes are shown, no positive signals were observed with any of the 31 other probes; numbers are net MFI values computed as described in ‘Materials and Methods’ section, values >5 are considered positive.
bWeak cross-reactivity.
Analytic sensitivity of McCT
Analytic sensitivity was first determined for SMRV detection using SMRV plasmids (ATCC Nr. 45034). Ten-fold endpoint dilution series was prepared in 100 mM human placenta (HP-) DNA in a total volume of 30 µl. Despite the co-amplification and co-detection of human β-globin and PolA, Gag- and Env-specific SMRV oligonucleotide probes were detected as little as 100 copies of SMRV per PCR reaction in five of five replicates assayed on different days.
Next, we determined the sensitivity for the detection of HPV18, β-globin and PolA sequences in HeLa cells. DNA lysates of 5 × 106 HeLa cells were prepared, followed by a 10-fold dilution series in water. Less than five HeLa cells were sufficient to detect HPV18 and human β-globin. PolA positivity was reached using less than 50 HeLa cells indicating a high sensitivity of McCT.
Analytical sensitivity under highly competing conditions was determined for the simultaneous detection of various contaminations in a 3-fold dilution series of a cell mixture comprising 10 different cell lines (Table 6; ID 1–10) in a background of either human BJA-B Burkitt's lymphoma cells or mouse Sp2/0–Ag8 myeloma cells. Prior to cell lysis, the mixture of 10 cell lines was titrated to obtain mixes with 300 to 0.1 cells of each cell type in 6000 background cells per PCR reaction. This number of background cells was in accordance with the expected amount of cell equivalents per PCR reaction when following the lysate preparation protocol. Despite the presence of 74 primers in a single PCR reaction and simultaneous co-amplification of up to 16 amplimers, the assay proved to be very sensitive. For example, the detection limit for contaminating HeLa cells, as determined by HPV18 detection, was below three cells in 6000 background cells (<0.06%). Cross-species contaminating cells were detected with even higher sensitivity (<0.02%) (Table 6).
Table 6.
Detection limits for contaminating cell lines based on the detection of nonexclusive markers
| Cell line | ID | Nonexclusive marker | Detection limit for contaminating cells in 6000 background cells/PCR |
|
|---|---|---|---|---|
| Cell no. | (%) | |||
| CEM | 1 | Human | <1 | <0.02 |
| NIH3T3 | 2 | Mouse | <1 | <0.02 |
| a-tag R4 | 3 | Rat | <1 | <0.02 |
| CHO | 4 | Chinese hamster | <1 | <0.02 |
| MDCKII | 5 | Dog | <1 | <0.02 |
| SW480 | 6 | Human Y chromosome | <100 | <2.0 |
| HeLa | 7 | HPV18 | <3 | <0.05 |
| Namalwa | 8 | EBV | <100 | <2.0 |
| Alexander | 9 | HBV | <1 | <0.02 |
| HEK293T | 10 | SV40 Tag | <10 | <0.2 |
| HEK293T | 10 | SV40 VP1 | <10 | <0.2 |
| HEK293T | 10 | Adenovirus 5 | <3 | <0.05 |
Reproducibility of McCT
Over a period of 4 weeks, McCT was repeated 12 times using an artificial cell line mixture. The mixture comprised a total of 16 McCT PCR targets whose products were detected by 20 distinct probes; a quadruple Mycoplasma infection was detected by one universal and four specific probes. Of the total 240 detections (12 × 20 probes), only the M. hominis detection was negative on a single day resulting in a very high reproducibility of 99.6% despite the simultaneous co-amplification and co-detection of 16 amplimers (data not shown). As the missed M. hominis reaction was only borderline positive in the remaining 11 cases, this emphasizes the robustness of the assay even around the detection limit.
Comparison of Myoplasma detection by McCt and a commercial Mycoplasma test
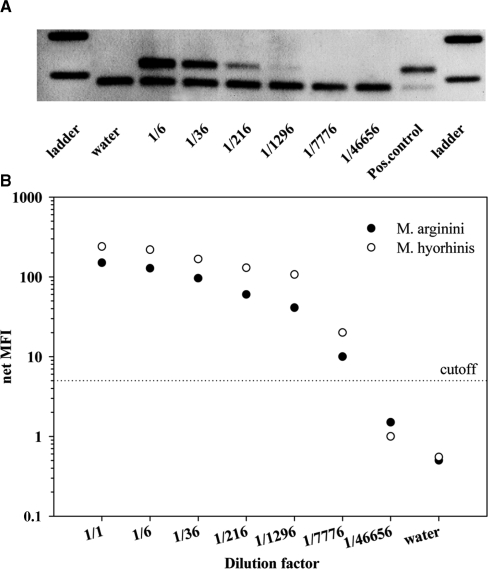
In order to compare the McCT assay to a commercial Mycoplasma test (Venor®GeM), a M. hyorhinis and M. arginini positive cell lysate was 6-fold serially diluted in a Mycoplasma negative lysate (each containing 106 human genome equivalents per 100 µl of lysate). After gel electrophoresis of the Venor®GeM amplimers, 1296-fold diluted DNA extracts could be detected, while the internal PCR control was positive for all dilution steps (Figure 2A). McCT was more sensitive in detecting and simultaneously typing Mycoplasma species in 7800-fold diluted DNA extracts (Figure 2B) despite the co-detection of PolA, human β-globin, human Y-chromosome, adenoviral and EBV DNA that was present in the DNA extract (data not shown).
Figure 2.
Detection of Mycoplasma (M.) dilutions by Venor®GeM (A) and McCT (B). Both assays were performed using 6-fold dilution series of one cell lysate positive for M. arginini and M. hyorhinis. (A) PCR products from the commercial Mycoplasma kit were loaded on a 2% agarose gel showing the internal PCR control (191 bp) and a Mycoplasma-specific band (267 bp). (B) Net MFI values obtained after hybridization of the McCT PCR products to Mycoplasma species-specific probes are shown. The cut-off is indicated by the dotted line.
McCT analysis of 713 cell lysates from different cell lines
We tested a broad spectrum of mammalian cell line lysates (n = 713, including ∼5% repeats) from different laboratories in Germany between March and July 2008. Two cell lysates were excluded from data analysis due to insufficient PolA detection. SMRV was found positive with both Gag and Env probes in nine different cell lines, some of which were received repeatedly from different labs (altogether 18 cases). Affected cell lines comprised HeLa, T-cell lines CEM, JE6.1 and Jurkat, Panc1, ASPC-1, Akata and the monkey cell lines B95.8 and BI60. Mycoplasma was the most frequent contamination (22.6%) with M. fermentans (9.4%), M. hyorhinis (8.2%) M. arginini (5.6%) and M. orale (1.8%) being most prevalent. Double infections with Mycoplasma were found in 27 cases (3.8%) and a triple infection in one case (0.1%). Contaminating HeLa cells were found in seven cases. Among inter-species contaminations (n = 49), mouse cells in human cells and vice versa were the most frequent contamination pairs.
Inter-laboratory variation of contaminations was analyzed from 21 laboratories that each had provided at least 10 cell lysates (range 10–85, median 21). The rate of Mycoplasma infections had a median of 20%, but ranged widely from 0% to 63%. Within Mycoplasma-positive samples from the same laboratory, usually one Mycoplasma species was predominant. Inter-species contaminations were detected with a median frequency of 5% but ranged widely from 0% to 55%. In general, laboratories with high rates of Mycoplasma infections tended to have increased also the rates of inter-species contaminations.
DISCUSSION
Cell culture contamination is a serious problem in biological research that is often neglected as testing for all sorts of possible contaminants is difficult, expensive and time-consuming. For some of the most frequent contaminants, the novel McCT assay is solving these problems by multiplexing more than 37 primer pairs in a single PCR reaction, and subsequent hybridization to 45 specific oligonucleotide probes coupled with Luminex beads. The spectrum of detectable contaminants reaches from 10 Mycoplasma species and 12 viruses to cross-species contaminations. Nonexclusive cell line markers enable the assessment of intra-species contaminations for frequently used cell lines, such as HeLa, HEK293(T), Namalwa and Cos7. Given the robustness of the multiplex PCR and the size of the Luminex array comprising 100 sets of beads, primers and probes for further contaminants could easily be integrated into the assay.
McCT facilitated the simultaneous and specific detection of more than 16 targets in the same sample without skewing of the results. Analytical sensitivity reached the level of 10 copies per contaminant and thus was in agreement with conventional singleplex PCR assays. Moreover, even small amounts of contaminants, <2% for any of the targets, could be detected in background cells. Investigation of the reproducibility revealed a high degree of robustness even for weak signals. No cross-reactivity was observed for all Luminex probes with unrelated amplimers. In addition to specific probes, we also designed and included two universal Mycoplasma probes in the assay that recognized all species analyzed. Cell lines positive with the universal probes but negative with the Mycoplasma-specific probes could indicate the presence of known Mycoplasma species for which no specific probe has been included, potentially also an as yet unknown new species, or a variant missed by the specific probes. The 96-well format allows fast, simple and highly reproducible analyzes of up to 1000 lysates from cell lines in <1 week without automation.
In a first validation, the McCT assay was used for an unrepresentative analysis of cell lysates from different laboratories in Germany. We discovered a Mycoplasma contamination rate that was comparable with those described in the literature (2–4), with large inter-laboratory variation. SMRV was detected in several cell lines, all of human or monkey origin. We have tested only a limited number of nonexclusive makers that do not cover all human cell lines. Thus, intra-species contaminations in cell lines could only be analyzed to a limited degree and were found less frequently than inter-species contaminations. However, our findings with the rather limited set of intra-species contamination markers indicate that intra-species contaminations are not necessarily a very rare event. We are currently developing a high-throughput human cell identification test to address this aspect.
To date, no gold standard exists for the monitoring of most cell culture contaminations. Instead, researchers are relying on single assays. One of the most frequently used commercial tests in Germany for the detection of Mycoplasma is the Venor®GeM kit. While this assay has been advertized to be very sensitive, it cannot genotype the respective species. We obtained evidence for a higher sensitivity of McCT for detecting and typing Mycoplasma species. Two factors that probably contribute to the higher sensitivity are improved amplification by novel Mycoplasma primers and improved detection by Luminex probes. Most remarkably, this was also true when multiple targets were simultaneously amplified. Current per sample costs for the commercial Mycoplasma kit (containing PCR primers and positive controls) range between 5 and 10 €, whereas the McCT reagents (primers, PCR components, beads, probes and detection reagents) amount to ∼10 €.
In conclusion, McCT appears to be a powerful tool in assessing cell line purity and would be valuable for specialized laboratories, such as core facilities at scientific institutions for routine testing of cell lines.
FUNDING
DKFZ and the Studienstiftung des Deutschen Volkes (PhD Grant to M.S.). Funding for open access charge: DKFZ.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank M. Löchelt (DKFZ) for SMRV; C. Knopf (DKFZ) for HSV1 and 2; K. Korn (University Erlangen) for VZV and HHV7; HJ. Delecluse (DKFZ) for HHV8; and H. zur Hausen (DKFZ) for CMV and HHV6. We thank numerous colleagues from various institutes for supplying cell lines for analysis. We thank L. Gissmann and O. Wiestler (DKFZ) for discussion, and Eva Tritsch, Daniela Höfler and Saskia Ziegler for experimental help.
REFERENCES
- 1.Pienkowska M, Seth A. Detection of squirrel monkey retroviral sequences in interferon samples. J. Hepatol. 1998;28:396–403. doi: 10.1016/s0168-8278(98)80312-7. [DOI] [PubMed] [Google Scholar]
- 2.Hay RJ, Macy ML, Chen TR. Mycoplasma infection of cultured cells. Nature. 1989;339:487–488. doi: 10.1038/339487a0. [DOI] [PubMed] [Google Scholar]
- 3.Uphoff CC, Drexler HG. Detection of mycoplasma in leukemia-lymphoma cell lines using polymerase chain reaction. Leukemia. 2002;16:289–293. doi: 10.1038/sj.leu.2402365. [DOI] [PubMed] [Google Scholar]
- 4.Uphoff CC, Drexler HG. Comparative PCR analysis for detection of mycoplasma infections in continuous cell lines. In Vitro Cell. Dev. Biol. 2002;38:79–85. doi: 10.1290/1071-2690(2002)038<0079:CPAFDO>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 5.McGarrity GJ. Spread and control of mycoplasmal infection of cell cultures. In Vitro. 1976;12:643–648. doi: 10.1007/BF02797464. [DOI] [PubMed] [Google Scholar]
- 6.van Diggelen OP, Phillips DM, Shin SI. Endogenous HPRT activity in a cryptic strain of mycoplasma and its effect on cellular resistance to selective media in infected cell lines. Exp. Cell Res. 1977;106:191–203. doi: 10.1016/0014-4827(77)90256-7. [DOI] [PubMed] [Google Scholar]
- 7.van Diggelen OP, Shin SI, Phillips DM. Reduction in cellular tumorigenicity after mycoplasma infection and elimination of mycoplasma from infected cultures by passage in nude mice. Cancer Res. 1977;37:2680–2687. [PubMed] [Google Scholar]
- 8.Bolske G. Survey of Mycoplasma infections in cell cultures and a comparison of detection methods. Zentralbl. Bakteriol. Mikrobiol. Hyg. 1988;269:331–340. doi: 10.1016/s0176-6724(88)80176-7. [DOI] [PubMed] [Google Scholar]
- 9.Heberling RL, Barker ST, Kalter SS, Smith GC, Helmke RJ. Oncornavirus: isolation from a squirrel monkey (Saimiri sciureus) lung culture. Science. 1977;195:289–292. doi: 10.1126/science.63993. [DOI] [PubMed] [Google Scholar]
- 10.German Central Commission for Biological Safety (ZKBS). Position statement of the Central Biosafety Commission on the risk assessment of Squirrel Monkey Retrovirus according to § 5 Paragraph 1 of the Genetic Engineering Safety Regulations (GenTSV). . 2008 http://www.bvl.bund.de/cln_007/nn_496790/EN/06__Genetic__Engineering/ZKBS/01__Allg__Stellungnahmen/09__viruses/SMRV.html__nnn=true. [Google Scholar]
- 11.Sun R, Grogan E, Shedd D, Bykovsky AF, Kushnaryov VM, Grossberg SE, Miller G. Transmissible retrovirus in Epstein-Barr virus-producer B95-8 cells. Virology. 1995;209:374–383. doi: 10.1006/viro.1995.1269. [DOI] [PubMed] [Google Scholar]
- 12.Middleton PG, Miller S, Ross JA, Steel CM, Guy K. Insertion of SMRV-H viral DNA at the c-myc gene locus of a BL cell line and presence in established cell lines. Int. J. Cancer. 1992;52:451–454. doi: 10.1002/ijc.2910520320. [DOI] [PubMed] [Google Scholar]
- 13.Nelson-Rees WA, Flandermeyer RR. HeLa cultures defined. Science. 1976;191:96–98. doi: 10.1126/science.1246601. [DOI] [PubMed] [Google Scholar]
- 14.Chatterjee R. Cell biology. Cases of mistaken identity. Science. 2007;315:928–931. doi: 10.1126/science.315.5814.928. [DOI] [PubMed] [Google Scholar]
- 15.Masters JR, Thomson JA, Daly-Burns B, Reid YA, Dirks WG, Packer P, Toji LH, Ohno T, Tanabe H, Arlett CF, et al. Short tandem repeat profiling provides an international reference standard for human cell lines. Proc. Natl Acad. Sci. USA. 2001;98:8012–8017. doi: 10.1073/pnas.121616198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Helden PD, Wiid IJ, Albrecht CF, Theron E, Thornley AL, Hoal-van Helden EG. Cross-contamination of human esophageal squamous carcinoma cell lines detected by DNA fingerprint analysis. Cancer Res. 1988;48:5660–5662. [PubMed] [Google Scholar]
- 17.Drexler HG, Dirks WG, Matsuo Y, MacLeod RA. False leukemia-lymphoma cell lines: an update on over 500 cell lines. Leukemia. 2003;17:416–426. doi: 10.1038/sj.leu.2402799. [DOI] [PubMed] [Google Scholar]
- 18.ATCC. Cell line verification test recommendations. ATCC Technical Bulletin 8. 2007 [Google Scholar]
- 19.Thompson J, Higgins D, Gibson T. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4690. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalendar R, Lee D, Schulman AH. Genes, Genomes and Genomics. 2009. Review: FastPCR software for PCR primer and probe design and repeat search. [Google Scholar]
- 21.Schmitt M, Bravo IG, Snijders PJ, Gissmann L, Pawlita M, Waterboer T. Bead-based multiplex genotyping of human papillomaviruses. J. Clin. Microbiol. 2006;44:504–512. doi: 10.1128/JCM.44.2.504-512.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmitt M, Dondog B, Waterboer T, Pawlita M. Homogeneous amplification of genital human alpha papillomaviruses by PCR using novel broad-spectrum GP5+ and GP6+ primers. J. Clin. Microbiol. 2008;46:1050–1059. doi: 10.1128/JCM.02227-07. [DOI] [PMC free article] [PubMed] [Google Scholar]