Abstract
RNA secondary structure prediction using free energy minimization is one method to gain an approximation of structure. Constraints generated by enzymatic mapping or chemical modification can improve the accuracy of secondary structure prediction. We report a facile method that identifies single-stranded regions in RNA using short, randomized DNA oligonucleotides and RNase H cleavage. These regions are then used as constraints in secondary structure prediction. This method was used to improve the secondary structure prediction of Escherichia coli 5S rRNA. The lowest free energy structure without constraints has only 27% of the base pairs present in the phylogenetic structure. The addition of constraints from RNase H cleavage improves the prediction to 100% of base pairs. The same method was used to generate secondary structure constraints for yeast tRNAPhe, which is accurately predicted in the absence of constraints (95%). Although RNase H mapping does not improve secondary structure prediction, it does eliminate all other suboptimal structures predicted within 10% of the lowest free energy structure. The method is advantageous over other single-stranded nucleases since RNase H is functional in physiological conditions. Moreover, it can be used for any RNA to identify accessible binding sites for oligonucleotides or small molecules.
INTRODUCTION
RNA folds into well-defined tertiary structures that lead to diverse functions including regulation of gene expression (1–3), cellular localization (4), catalysis (5–10) and serving as a structural scaffold (11,12). In many cases, misfolding of an RNA leads to functional incompetence. For example, misfolding leads to catalytic inactivity in group I introns (13–17) and RNase P RNAs (18–20), and to the inability of rRNAs to form scaffolds for ribosomal assembly (21–24). Thus, determination of the secondary and tertiary structure of an RNA and how structure affects function is important not only in order to understand biological function but also to understand how to modulate it.
RNA tertiary structure is a composite of secondary structure elements such as paired regions and various types of single-stranded regions (internal loops, hairpin loops, bulges and multibranch loops). Approximately 46% of the bases in RNA are unpaired or non-canonically paired (25). Often these sites are of functional importance—forming tertiary contacts (26–28), forming scaffolds for binding of other biomolecules (29,30) and metabolites (2,3,31,32) and direct involvement in catalysis (8).
Despite the importance of RNA structure in cellular processes, there are relatively few NMR and X-ray crystal structures of RNA in the Protein Data Bank (PDB; http://www.pdb.org) (33) when compared to available protein structures. This is due to inherent difficulty in crystallizing RNA and the overlapping signals in NMR spectra of nucleic acids. Fortunately, RNA secondary structure can be predicted accurately from sequence by free energy minimization or phylogenetic comparison, and there are a variety of experimental methods that can be used to improve prediction. The program RNAstructure (34,35) uses free energy minimization to predict an ensemble of possible secondary structures for a given RNA sequence. On average, the lowest free energy structure outputted by the program contains 73% of the base pairs predicted correctly. In an ensemble of 1000 suboptimal structures within 10% free energy of the lowest free energy structure, one structure has at least 87% of the base pairs predicted correctly (35). Experimental constraints aid in selection of that structure. The secondary structure then constrains the possible tertiary structures the RNA can adopt. In fact, a recently reported program uses secondary structures as an essential starting point to predict tertiary structures (36).
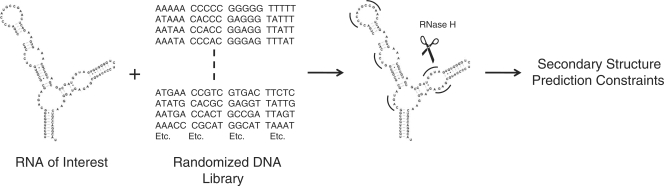
Insights into RNA secondary and tertiary structure can be gained by chemical modification and enzymatic mapping. In some cases, chemical modification and enzymatic mapping are conducted under conditions very different from those inside a cell. Such divergence could cause the RNA to fold into a non-native form, creating problems when trying to estimate in vivo structure. An alternative approach is to generate secondary structure constraints using a library of DNA oligonucleotides and RNase H, which cleaves DNA/RNA hybrids under a wide variety of conditions at pH 7 (Figure 1) (37). Previously, RNase H cleavage induced by binding of semi- (38,39) and fully randomized DNA libraries (40,41), semi-randomized DNA/2′–O–methyl RNA chimeras (42,43) and partially randomized tethered oligonucleotides (44) to RNAs have been used to design antisense oligonucleotides. In addition, designed oligonucleotides probes and RNase H cleavage have been used to identify accessible binding sites in whole cell extracts (45) and to study RNA folding pathways (20,46).
Figure 1.
Schematic of the general method used in this study. Randomized DNA oligonucleotides are incubated with an RNA of interest. Only DNAs complementary to single-stranded regions bind, inducing RNase H cleavage of the RNA strand. Nucleotides which are subject to RNase H cleavage are used as single-stranded constraints in RNA secondary structure prediction.
Herein, we describe a facile method to generate secondary structure constraints by identifying nucleotides that are subject to RNase H cleavage when bound to a member of fully randomized DNA libraries of different lengths (Figure 1). This method was applied to the 5S rRNA from Escherichia coli and yeast tRNAPhe. The secondary structure of the 5S rRNA is poorly predicted by free energy minimization (27% of base pairs) while the tRNA is well predicted (95% of base pairs). The single-stranded constraints generated by RNase H cleavage improved the prediction of the 5S rRNA (only the phylogenetic structure is predicted) while the accuracy of prediction for the tRNA is unchanged. The single-stranded constraints for the tRNA, however, eliminate all other suboptimal structures that ranged in accuracy from 29 to 76%. This method not only generates secondary structure restraints but also identifies accessible binding sites within an RNA that may be appropriate for design of antisense oligonucleotides or small molecules that modulate RNA function.
MATERIALS AND METHODS
General
All experiments were completed with diethyl pyrocarbonate (DEPC) -treated nanopure water. Randomized oligonucleotides were purchased from Integrated DNA Technologies. Yeast tRNAPhe was purchased from Sigma Aldrich.
RNA secondary structure prediction
All secondary structures (Figures 2 and 4) were predicted using the RNAstructure program (version 4.5 or 4.6) (34,35) and the following suboptimal structure parameters: max. % energy difference = 10, max. no. of structures = 20, window size = 3. These parameters were chosen such that no more than five structures were generated in order to simplify design of single sequences. It is assumed that a DNA oligonucleotide is unable to significantly invade fully paired RNA helices. Nucleotides where cleavage occurred were constrained as single-stranded in secondary structure predictions.
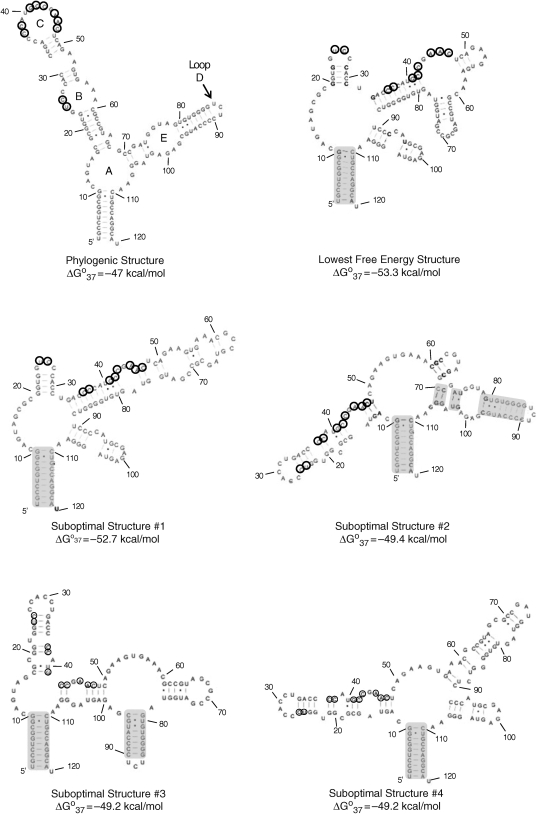
Figure 2.
Phylogenetic secondary structure, the predicted lowest free energy secondary structure, and the four suboptimal structures of E. coli 5S rRNA. Loops A–E are labeled in the phylogenetic structure. Base-paired regions that are predicted correctly in the suboptimal structures are shaded. Nucleotides cleaved by RNase H cleavage are circled. The lowest free energy structure only has 27% of the base pairs present in the phylogenetic structure.
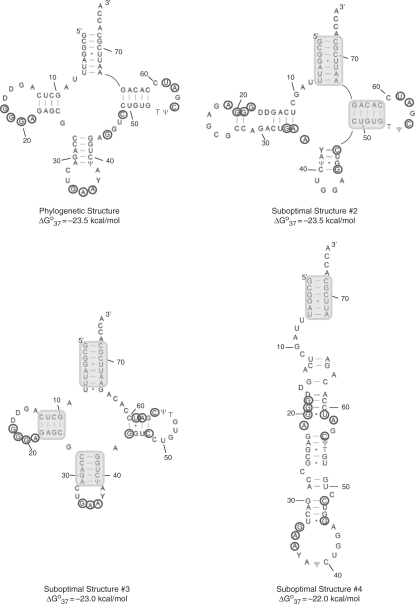
Figure 4.
Phylogenetic secondary structure and three predicted suboptimal structures of yeast tRNAPhe. The lowest free energy structure has 95% of the base pairs predicted correctly. Base paired regions that are predicted correctly in the suboptimal structures are shaded. Nucleotides cleaved by RNase H after 1 or 2 h incubation are circled. D denotes dihydrouracil while Y denotes wybutosine.
OligoWalk
The ΔG°binding values for oligonucleotides binding to the phylogenetic structures, lowest free energy structures and suboptimal structures (Tables 1 and 2) were predicted using the OligoWalk program (47,48), which is part of RNAstructure. The following parameters were used: Mode = Break Local Structure; Oligo Length = 5, 6, 7 or 8; Oligo Concentration = 5 µM (ΔG°binding values are independent of concentration); Oligomer Chemistry = DNA; and Target Structure Limits for Walk: Start = 1 and Stop = 120.
Table 1.
Oligonucleotide sequences and binding free energies to the phylogenetic structure, lowest free energy structure and two suboptimal structures
| Oligonucleotide sequence | Binding site | ΔG°binding to phylogenetic structure (kcal/mol) | ΔG°binding to lowest fee energy structure (kcal/mol) | ΔG°binding to suboptimal structure #1 (kcal/mol) | ΔG°binding to suboptimal structure #2 (kcal/mol) | ΔG°binding to suboptimal structure #3 (kcal/mol) | ΔG°binding to suboptimal structure #4 (kcal/mol) | RNase H cleavage observed using a single sequence? | RNase H cleavage observed in this region using randomized oligonucleotides? |
|---|---|---|---|---|---|---|---|---|---|
| 5-mers | |||||||||
| GTTTC | 56–60 | 4.1 | 3.2 | 1.7 | –0.4 | –1.9 | 3.9 | No | No |
| ATCGG | 70–74 | 1.0 | –0.5 | 3.2 | 1.1 | –0.5 | 3.4 | No | No |
| 6-mers | |||||||||
| GGGACC | 23–28 | –0.2 | –3.5 | –3.5 | 2.1 | 2.1 | 2.1 | No | Yes |
| ACTTCT | 50–54 | 5.5 | –2.3 | 3.1 | –3.5 | –3.5 | –3.5 | No | No |
| CTACCA | 74–79 | 2.1 | 10.0 | 2.5 | 2.1 | 9.7 | 0.2 | No | No |
| CTCTCG | 97–102 | 2.2 | –0.3 | –0.3 | 2.2 | 9.2 | –0.3 | No | No |
| 7-mers | |||||||||
| GCCAGGC | 2–8 | 15.6 | 15.6 | 15.6 | 15.6 | 15.6 | 15.6 | No | No |
| AGGTGGG | 26–32 | –0.3 | 1.7 | 1.7 | –4.0 | –2.3 | –4.0 | No | No |
| CGGCATG | 38–44 | –6.4 | 4.9 | 4.0 | 2.6 | 3.3 | 2.6 | Yes | Yes |
| AGTTCGG | 42–48 | –4.7 | 1.0 | 0.3 | –0.1 | 5.4 | 0.2 | Yes | Yes |
| 8-mers | |||||||||
| CATGGGGT | 34–41 | –7.4 | 14.5 | 14.5 | 10.0 | 4.8 | 10.0 | Yes | Yes |
‘Binding site’ indicates the nucleotides in the E. coli 5S rRNA to which the oligonucleotide probe is complementary. All ΔG°binding values were generated using the OligoWalk program in ‘break local’ structure mode. Observation of RNase H cleavage is indicated for each oligonucleotide.
Table 2.
Positions of RNase H cleavage of yeast tRNAPhe with randomized 5-mer oligonucleotides after a 1 or 2 h incubation and the corresponding ΔG°binding values for the phylogenetic and suboptimal structures
| Cleavage position | Phylogenetic structure ΔG°binding (kcal/mol) | Suboptimal structure #2 ΔG°binding (kcal/mol) | Suboptimal structure #3 ΔG°binding (kcal/mol) | Suboptimal structure #4 ΔG°binding (kcal/mol) |
|---|---|---|---|---|
| 18 | −4.1 | 2.9 | −4.1 | 1.7 |
| 19 | −5.2 | 0.4 | −5.2 | 0.6 |
| 20 | −1.1 | −2.0 | −1.1 | 6.9 |
| 21 | 2.0 | −0.4 | 2.0 | 6.3 |
| 34 | −1.9 | 10.1 | −1.3 | 3.7 |
| 35 | −2.2 | 8.6 | −2.2 | −0.4 |
| 36 | −1.9 | 9.2 | −1.9 | −1.9 |
| 45 | −0.1 | 1.7 | 5.8 | 2.6 |
| 48 | 2.1 | 8.8 | 3.8 | 9.6 |
| 56 | −1.2 | −1.2 | 4.4 | 6.4 |
| 58 | −1.9 | −1.9 | 3.1 | 7.4 |
| 59 | 0.5 | 0.5 | 2.7 | 5.7 |
The ΔG°binding values are for the oligonucleotides in which the nucleotide where cleavage occurs is the middle nucleotide of the resulting duplex.
Preparation of E. coli 5S rRNA
The E. coli 5S rRNA was prepared by overexpression from the pKK5-1 plasmid as previously reported (49), with the following modifications. The 5S rRNA was isolated by gel purification on a denaturing 8% polyacrylamide gel. The RNA was visualized by UV-shadowing, excised and extracted into 300 mM NaCl by tumbling overnight at 4°C. RNA was concentrated with 2-butanol and ethanol precipitated. Concentration was determined by measuring the absorbance at 260 nm and the corresponding extinction coefficient (1.19 × 106 M–1 cm–1). The extinction coefficient was determined by the HyTher program (50), which is based on nearest neighbor parameters (51).
5′-End labeling of RNAs
The 5S rRNA and tRNAPhe were 5′-end labeled with [γ-32P]ATP (PerkinElmer) and T4 polynucleotide kinase (New England BioLabs) as previously described (52). The RNA was purified and extracted as described above except the gel was exposed to a phosphor screen to visualize the RNA.
Design of DNA oligonucleotides for RNase H cleavage of E. coli 5S rRNA
Using the phylogenetic secondary structure of E. coli 5S rRNA and the secondary structures predicted by RNAstructure, DNA oligonucleotides were designed to distinguish between them. Negative controls were also designed that bind to a region that is double stranded in the phylogenetic structure and the predicted structures. These predictions were confirmed using OligoWalk.
RNase H cleavage experiments
The E. coli 5S rRNA was folded into Form A (53) in 50 µl of 1× assay buffer (150 mM NaCl, 4 mM MgCl2, 10 mM Tris–HCl, pH 7.4) by heating at 65°C for 5 min and slow cooling (∼0.5°C/min) to 37°C (53,54). For digestions using single sequences, DNA and DTT were added to final concentrations of 5 µM and 10 mM, respectively. For digestions with the randomized oligonucleotides, a final DNA concentration of 3.25 µM each possible 5-mer, 815 nM each possible 6-mer or 200 nM each possible 7-mer, and 10 mM DTT were used in a total volume of 150 µl. The RNA and DNA oligonucleotides were incubated for 15 min at 37°C. Then, 5 µl (5 units/µl) RNase H (New England BioLabs) were added, and the samples were incubated at 37°C for 10 h. The samples were ethanol precipitated and resuspended in 1× loading buffer (1 mM Tris, pH 7, 3.5 M urea and 1 mM EDTA). The products were separated on a denaturing 8% polyacrylamide gel. The gels were dried and exposed to a phosphor screen. Images were collected using a BioRad FX imaging system.
Yeast tRNAPhe was folded by heating at 95°C for 3 min in 1× assay buffer without MgCl2 and then placing the sample on ice for 10 min. Then, 10 mM MgCl2 and 200 nM final concentration of each possible 5-mer were added. The sample was allowed to equilibrate at 37°C for 15 min followed by addition of DTT to a final concentration of 10 mM and 25 units of RNase H. Time points were taken at 1, 2 and 4 h. The reaction was quenched by addition of 2.5 volumes of ethanol. Products were separated on a denaturing 10% polyacrylamide gel.
Hydrolysis and T1 ladders
The RNA was incubated in 5 µl of hydrolysis buffer (0.1 M NaHCO3, pH 10 and 1 mM EDTA) for 1 min at 95°C. To stop hydrolysis, 5 µl of 2× loading buffer were added and the samples were stored at –80°C until use. Guanosine residues were identified by T1 ribonuclease cleavage under denaturing conditions. The RNA was incubated in 10 µl of 1× T1 Buffer (25 mM sodium citrate, pH 5, 7 M urea and 1 mM EDTA) and 2.5 units/µl of T1 nuclease at 55°C for 10 min. The samples were stored at –80°C if they were not immediately subjected to gel electrophoresis.
RESULTS
The secondary structure of the E. coli 5S rRNA was predicted by free energy minimization using the program RNAstructure (34,35). The output of the program includes the lowest free energy structure and suboptimal structures that are within 10% free energy of the lowest free energy structure. Figure 2 shows the phylogenetic structure, and the lowest free energy structure and the four suboptimal structures that were predicted by the program. The five predicted structures were then compared to the phylogenetic (accepted) structure. Interestingly, the phylogenetic structure has the highest free energy value. The lowest free energy structure has only 27% of the base pairs predicted correctly while the suboptimal structures have between 27 and 59%. These percentages are quite low when considering on average the lowest free energy structure predicted by RNAstructure contains 73% of base pairs found in the corresponding phylogenetic structure (35).
On the basis of the differences in structure, 11 DNA oligonucleotide probes were designed to differentiate between the structures when subjected to an RNase H cleavage assay (Table 1 and Figure 2). RNase H cleaves the RNA phosphodieser backbone of DNA/RNA hybrids. The DNA oligonucleotide should only bind to regions in the RNA that are single stranded. We assumed that the short DNA probes would be unable to invade paired regions. (This assumption is correct based on experimental results.) The oligonucleotides are complementary to the following nucleotides within the E. coli 5S rRNA: 2–8 (designed to bind a paired region in all structures and serves as a negative control), 23–28 (designed to bind the phylogenetic structure, the lowest free energy structure, and suboptimal structure #1), 26–32 (designed to bind suboptimal structures #3–5), 34–41 (designed to bind the phylogenetic structure), 38–44 (designed to bind the phylogenetic structure), 42–48 (designed to bind the phylogenetic structure and the lowest free energy structure), 50–54 (designed to bind all structures), 56–60 (designed to bind the phylogenetic structure, the lowest free energy structure and suboptimal structures #2–4), 70–74 (designed to bind suboptimal structures #2 and 4), 74–79 (designed to bind the phylogenetic structure and suboptimal structures #2 and 3) and 97–102 (designed to bind all structures except suboptimal structure #4).
The DNA oligonucleotides were incubated individually with the E. coli 5S rRNA in order to determine which sites are accessible. The RNA was first folded into Form A as described previously (53) and then incubated with a DNA probe for 15 min at 37°C prior to addition of RNase H. Cleavage was only observed for three of the oligonucleotides—those that were complementary to nucleotides 34–41, 38–44 and 42–48 (Figure 3). Positions where cleavage occurred were entered as single-stranded constraints in secondary structure prediction by the RNAstructure program. This afforded only one structure, the phylogenetic structure; there were no other structures predicted within 10% free energy.
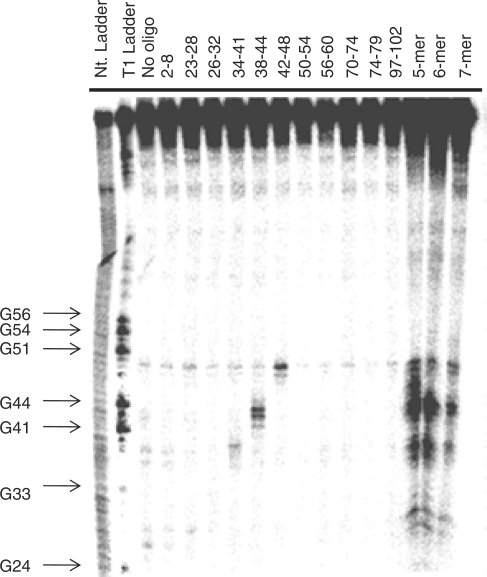
Figure 3.
Representative gel autoradiogram of RNase H cleavage experiments to identify single-stranded regions in E. coli 5S rRNA. The numbers above the lanes indicate to which nucleotides in the RNA the oligonucleotide probe is complementary.
The same RNase H cleavage assay was then used to determine if randomized DNA oligonucleotides can be used to generate secondary structure prediction constraints. The 5S rRNA was incubated with DNA libraries containing all possible 5-mers (1024 unique probes), 6-mers (4096 unique probes) or 7-mers (16 384 unique probes). A sufficiently high concentration of DNA library was used to ensure that all library members were present at a final concentration of ≥200 nM. All three lengths of randomized DNA probes identified four regions accessible to binding and subsequent RNase H cleavage (Figure 3). These regions correspond to nucleotides 26–27 (Loop B), 36–37 (Loop C), 41–44 (Loop C) and 46–48 (Loop C), and are similar to those identified using single DNA sequences. Each is consistent with the phylogenetic structure. As for the single sequences, the positions of RNase H cleavage were used as single-stranded constraints for secondary structure prediction, resulting in the phylogenetic structure.
Cleavage was not observed in Loops A, D or E. For Loop E, this is consistent with the absence of cleavage for the single sequences complementary to nucleotides 74–79 and 97–102. The lack of cleavage in Loop E is consistent with another study that used nucleases to determine the secondary structure of E. coli 5S rRNA (55). In that study, only one nucleotide in Loop E (U77) was subject to cleavage by Nuclease S1. In fact, another nucleotide in the loop (G100) was cleaved by the double-stranded Nuclease V1 (cobra venom nuclease). A crystal structure of Loop E has been solved and revealed that Loop E is a highly structured loop containing non-canonical pairs (56). Likewise, cleavage may not be observed in Loop A as nucleotides 11 and 12 are subjected to cleavage by Nuclease V1 although Nuclease S1 does cleave nucleotides 13–16 (55). Loop D is only a three nucleotide hairpin. The stem of this hairpin contains eight base pairs; the four pairs immediately adjacent to the hairpin are GC. Such a helix is likely difficult to invade with a DNA oligonucleotide.
The OligoWalk program (47,48) was then used to determine if RNase H cleavage correlates with accessibility of the corresponding site in the RNA as indicated by the ΔG°binding value. It was expected that the more negative the value of ΔG°binding, the more likely RNase H cleavage is. Large positive values for ΔG°binding should indicate a region where oligonucleotide binding would be unlikely. These results are summarized in Table 1. When individual sequences are used, cleavage is only observed if ΔG°binding was –4.7 kcal/mol or less, even if the region is predicted to be single stranded. The randomized oligonucleotides were able to induce cleavage when ΔG°binding was much higher, –0.2 kcal/mol (probe 23–28). It is interesting that the oligonucleotide that binds in the same region (26–32) has a similar ΔG°binding (−0.3 kcal/mol) but does not induce cleavage. However, the probe that binds nucleotides 23–28 forms one less base pair than the probe that binds nucleotides 26–32. It should be noted that 23–28 binds to four single-stranded nucleotides while 26–32 only binds two.
The secondary structure of yeast tRNAPhe (Figure 4) was also predicted using the RNAstructure program. In contrast to the E. coli 5S rRNA, tRNAPhe is well predicted with 95% of the base pairs in the phylogenetic structure present in the lowest free energy structure. The only difference between the phylogenetic structure and the lowest free energy structure is the formation of the U7–A66 base pair, which is not predicted by RNAstructure to form. Suboptimal structures #2, #3 and #4 contain 57, 76 and 29% of the base pairs in the phylogenetic structure, respectively (Figure 4). This RNA provides a test case to ensure that addition of constraints generated by RNase H cleavage does not negatively affect secondary structure prediction.
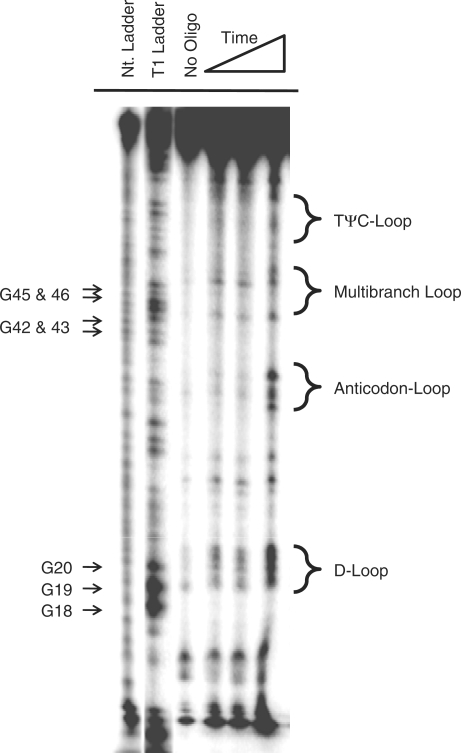
RNase H cleavage was observed for yeast tRNAPhe at the following nucleotides after a 1 or 2 h incubation when a DNA library of 5-mers was used (Figure 5): 18–21 (D-loop), 34–36 (anticodon-loop), 45 and 48 (multibranch loop) 56 and 58–59 (TΨC loop). These positions were then used as single-stranded constraints on secondary structure prediction. Interestingly, only one secondary structure was predicted—the lowest free energy structure, which has 95% of the base pairs predicted correctly. Although constraints do not improve prediction, importantly they do not decrease accuracy. It should be noted that cleavage at nucleotide C49 is also observed after a 4 h incubation period. This nucleotide forms a terminal GC pair (flanking the multibranch loop) and cleavage at this position was not expected. When included as a single-stranded restraint, only one structure is predicted. This structure is similar to the lowest free energy structure predicted without the use of experimental constraints except that the C49–G65 and the U51–A64 pairs are not predicted to form. Cleavage at this position could indicate that the structure of the terminal base pair is dynamic.
Figure 5.
Representative gel autoradiogram of RNase H cleavage experiments to identify single-stranded regions in yeast tRNAPhe with randomized 5-mer oligonucleotides.
As in the analysis of the position of RNase H cleavage for the E. coli 5S rRNA, the OligoWalk program was also used to estimate the ΔG°binding of oligonucleotides that induce cleavage in yeast tRNAPhe. As summarized in Table 2, these values are generally consistent with the phylogenetic structure. That is, if the position where cleavage is induced is assumed to be the middle nucleotide of the oligomer, most have negative ΔG° values. There are exceptions, however: the oligonucleotides where positions A21, C48 and U59 are the middle nucleotide. These oligonucleotides have ΔG°binding values of 2.0, 2.1 and 0.5 kcal/mol, respectively. These nucleotides are at or near the termini of helices. RNase H cleavage at the termini of helices has been observed previously in yeast tRNAasp (38). This suggests that loop dynamics, stacking of dangling ends (57) or coaxial stacking (58) could be important determinants of the sites accessible for binding DNA oligonucleotides (59).
DISCUSSION
The function of an RNA is intimately linked with its structure. Thus, having a reasonable estimate of RNA structure is important for understanding or predicting its function. Accurate structure prediction can be afforded by phylogenetic comparison if many related sequences are known. However, there are many cases in which a large data set is not available. In these cases, RNA secondary structure can be estimated via free energy minimization (35). The accuracy of secondary structure prediction can be improved if experimental constraints are used (35).
Many biochemical methods have been developed in order to gain insight into RNA secondary and tertiary structure including enzymatic mapping and chemical modification (60), equilibrium dialysis with short DNA oligonucleotides (61–63) and microarrays (54,64,65). One potential problem using enzymes is that some require ‘non-biological’ conditions that can affect RNA structure. For example, the optimal pH range for many nucleases specific for single-stranded regions is 4–5 or 9–10 (66). It is likely that a 2—3 unit pH difference (from pH 7) could cause the RNA to fold differently as the pKa's of some functional groups present in RNA bases can be in this range (67). Perturbations of RNA structures due to pH changes have been observed in the Hepatitis δ virus (68) and E. coli RNase P RNA (18). Many single-stranded nucleases, such as S1, Mung Bean and the single-stranded nucleases from Neurospora crassa and Schizopyllum commune, also require divalent transition metal ions such Zn2+ or Co2+ (66,69), which can also affect folding. In contrast, RNase H has optimal activity at pH 7.5–9.1, 100 mM monovalent metal ions, and 2–4 mM MgCl2, which are physiological (37).
Numerous methods have been developed to identify accessible binding sites using DNA oligonucleotides and RNase H cleavage (38–45). For example, a 2′-O-methyl RNA–DNA chimeric library of 11-mers and RNase H cleavage (43) have been used to identify accessible sites in the human multidrug resistance-1 mRNA (43) and the Hepatitis C Virus (HCV) RNA (40,70,71). Other studies have used accessible binding sites identified by RNase H cleavage to design effective antisense agents (42), tethered oligonucleotide probes to disrupt RNA–protein interactions (44) and ribozymes (41). The positions of cleavage were not used as constraints in RNA secondary structure prediction in any of these cases.
Equilibrium dialysis was used to study the binding of a library of 46 RNA trimers and 57 RNA tetramers to the E. coli 5S rRNA, all of which were fully complementary to the RNA sequence (61). The trimers and tetramers that bound the 5S rRNA and are consistent with the phylogenetic structure were complementary to nucleotides 11–14, 39–49, 68–70, 79–81 and 95–98. The probe complementary to nucleotides 68–70 is also complementary to nucleotides 17–19 and 61–63. Thus, although the sequence of accessible binding sites can be determined, the exact region in the structure to which a probe is binding cannot always be inferred due to the presence of multiple potential binding sites within the target RNA. The method reported herein can alleviate the problem of sequence degeneracy since the binding site is determined by RNase H cleavage.
A library of 7-mers has also been used to interrogate the structure of E. coli 5S rRNA via microarray (54). In these studies, every possible 2′-O-methyl 7-mer that could bind the target RNA (114 unique probes) was individually synthesized with a 5′-amino group and immobilized onto microarrays. The arrays were then hybridized with radioactively labeled RNA. In good agreement with the results present in this report, strong binding was observed to probes complementary to nucleotides: 24–30, 25–31, 26–32, 33–39 and 34–40. Medium binding was observed to probes complementary to nucleotides 27–33, 32–38, 35–41, 37–43, 38–44 and 41–47. It should also be noted that the sequence degeneracy problem present in the equilibrium dialysis studies also occurs with microarray methods. For example, the 7-mer probe that binds to nucleotides 35–41 can also bind to nucleotides 90–96 and the probe that binds to nucleotides 36–42 can also bind to nucleotides 91–97. Thus, our results are comparable to the microarray studies except that the binding sites for probes that have the potential to bind multiple regions within the RNA can be deconvoluted.
Binding of oligonucleotides to tRNAs has been studied by equilibrium dialysis (62,63), microarray (59,72) and RNase H cleavage (38). All revealed that there are four accessible sites in tRNAs—the acceptor arm, the anticodon-loop, the D-loop and variable-length region. Two different studies used microarrays to identifying accessible binding sites in yeast tRNAPhe (59,72). The binding patterns were similar despite the differences in the oligonucleotides used. The study reported by Mir and Southern used DNA oligonucleotides ranging in length from monomers to 12-mers (59), while the study reported by Jenek and Kierzek used isoenergetic LNA/2′-O-methyl 7-mers (72). Consistent with the data reported herein, binding was observed for oligonucleotides complementary to the D-loop, the variable loop/TΨC stem and the anticodon-loop. Binding was also observed to the double-stranded acceptor stem. This could be in part due to the concentration of salt in the buffers (1 M NaCl) or to the high local concentration of oligonucleotide immobilized on the array surface. Accessible binding sites in a related tRNA (yeast tRNAAsp) were also determined using RNase H cleavage and a semi-randomized DNA library. This library was generated by digesting the corresponding DNA template with DNase I. In good agreement with this report, strong cleavage sites were observed at nucleotides A21 (end of the D-loop), U35 (middle of the anticodon-loop), C56 (middle of the TΨC loop) and G73 (single-stranded region of acceptor stem).
Not all single-stranded regions in the E. coli 5S rRNA and yeast tRNAPhe were identified using this method. This is perhaps not unexpected based on previous reports in which the RNAs were probed for binding to libraries of oligonucleotides in solution (61) or displayed on a microarray surface (54,59,72). Important factors in oligonucleotide hybridization are the base composition (73), stacking at helix termini (dangling ends) (57) and coaxial stacking (58) among others. Coaxial stacking has been observed for both RNAs (74–77). Cleavage was also observed at longer incubation times (4 h) for one nucleotide that is not single stranded in the phylogenetic structure of yeast tRNAPhe, C49. Although this did not significantly affect secondary structure prediction accuracy, it is important to consider the relative amount of cleavage (weak or strong) and the incubation time required for RNase H cleavage to be observed.
Using the binding of randomized oligonucleotides and subsequent cleavage by RNase H to identify single-stranded regions is advantageous in many regards. For example, RNase H is active under conditions that are considered to mimic cellular pH and ionic strength. The randomized oligonucleotides can be used to map accessible sites in any RNA and does not require the individual syntheses of every possible DNA that might bind to the RNA of interest. The method is relatively fast and does not require specialized instrumentation such as an arrayer or microarray scanner. The instrumentation required for the RNase H method is common to most biochemistry laboratories.
Previous methods used to identify accessible sites in RNAs such as equilibrium dialysis (61–63) and microarrays (54,64) are able to determine which probes bind but they are not able to determine to which nucleotides. As mentioned previously, this can be problematic if a probe is complementary to more than one site as previously observed for the E. coli 5S rRNA using these methods (54,61). The RNase H method, however, also determines the binding site mitigating the inability to deconvolute sequence degeneracy.
Importantly, our method combines RNase H mapping with secondary structure prediction to improve prediction of both poorly predicted RNAs and to eliminate less accurate suboptimal structures from well predicted ones. As reported by other groups, the method can also be used to identify accessible sites for therapeutic intervention by oligonucleotides (antisense or RNA interference approaches) or small molecules. The use of a DNA library instead of designed sequences removes potential biases and prevents accessible sites not predicted by secondary structure prediction programs or programs that predict the stability of oligonucleotide binding from being overlooked. It is also likely that this approach can be applied to RNA folding studies. Previously, RNase H cleavage using single sequences has been used to study the folding of the Tetrahymena thermophila ribozyme (46) and the E. coli and Bacillus subtilis RNase P RNAs (20). By using randomized oligonucleotides in the form of DNA libraries, perhaps a clearer picture of RNA folding could be elucidated.
FUNDING
Grants from Canisius College and the Howard Hughes Medical Institute (institutional grant). Funding for open access charge: Canisius College.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Prof. Matthew Disney for helpful discussions and critical review of the manuscript.
REFERENCES
- 1.Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 2.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 3.Winkler WC, Cohen-Chalamish S, Breaker RR. An mRNA structure that controls gene expression by binding FMN. Proc. Natl Acad. Sci. USA. 2002;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keenan RJ, Freymann DM, Stroud RM, Walter P. The signal recognition particle. Annu. Rev. Biochem. 2001;70:755–775. doi: 10.1146/annurev.biochem.70.1.755. [DOI] [PubMed] [Google Scholar]
- 5.Zaug AJ, Cech TR. The intervening sequence RNA of Tetrahymena is an enzyme. Science. 1986;231:470–475. doi: 10.1126/science.3941911. [DOI] [PubMed] [Google Scholar]
- 6.Stark BC, Kole R, Bowman EJ, Altman S, Altman S. Ribonuclease P: an enzyme with an essential RNA component. Proc. Natl Acad. Sci. USA. 1978;75:3717–3721. doi: 10.1073/pnas.75.8.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.James BD, Olsen GJ, Liu JS, Pace NR. The secondary structure of ribonuclease P RNA, the catalytic element of a ribonucleoprotein enzyme. Cell. 1988;52:19–26. doi: 10.1016/0092-8674(88)90527-2. [DOI] [PubMed] [Google Scholar]
- 8.Qiao F, Cech TR. Triple-helix structure in telomerase RNA contributes to catalysis. Nat. Struct. Mol. Biol. 2008;15:634–640. doi: 10.1038/nsmb.1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeRose VJ. Two decades of RNA catalysis. Chem. Biol. 2002;9:961–969. doi: 10.1016/s1074-5521(02)00217-x. [DOI] [PubMed] [Google Scholar]
- 10.Fedor MJ, Williamson JR. The catalytic diversity of RNAs. Nat. Rev. Mol. Cell Biol. 2005;6:399–412. doi: 10.1038/nrm1647. [DOI] [PubMed] [Google Scholar]
- 11.Staley JP, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 12.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 13.Woodson SA, Cech TR. Alternative secondary structures in the 5′ exon affect both forward and reverse self-splicing of the Tetrahymena intervening sequence RNA. Biochemistry. 1991;30:2042–2050. doi: 10.1021/bi00222a006. [DOI] [PubMed] [Google Scholar]
- 14.Nikolcheva T, Woodson SA. Facilitation of group I splicing in vivo: misfolding of the Tetrahymena IVS and the role of ribosomal RNA exons. J. Mol. Biol. 1999;292:557–567. doi: 10.1006/jmbi.1999.3083. [DOI] [PubMed] [Google Scholar]
- 15.Pan J, Woodson SA. Folding intermediates of a self-splicing RNA: mispairing of the catalytic core. J. Mol. Biol. 1998;280:597–609. doi: 10.1006/jmbi.1998.1901. [DOI] [PubMed] [Google Scholar]
- 16.Emerick VL, Woodson SA. Self-splicing of the Tetrahymena pre-rRNA is decreased by misfolding during transcription. Biochemistry. 1993;32:14062–14067. doi: 10.1021/bi00213a040. [DOI] [PubMed] [Google Scholar]
- 17.Duncan CD, Weeks KM. SHAPE analysis of long-range interactions reveals extensive and thermodynamically preferred misfolding in a fragile group I intron RNA. Biochemistry. 2008;47:8504–8513. doi: 10.1021/bi800207b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Altman S, Guerrier-Takada C. M1 RNA, the RNA subunit of Escherichia coli ribonuclease P, can undergo a pH-sensitive conformational change. Biochemistry. 1986;25:1205–1208. doi: 10.1021/bi00354a002. [DOI] [PubMed] [Google Scholar]
- 19.Pan T, Sosnick TR. Intermediates and kinetic traps in the folding of a large ribozyme revealed by circular dichroism and UV absorbance spectroscopies and catalytic activity. Nat. Struct. Biol. 1997;4:931–938. doi: 10.1038/nsb1197-931. [DOI] [PubMed] [Google Scholar]
- 20.Zarrinkar PP, Wang J, Williamson JR. Slow folding kinetics of RNase P RNA. RNA. 1996;2:564–573. [PMC free article] [PubMed] [Google Scholar]
- 21.Hogan JJ, Gutell RR, Noller HF. Probing the conformation of 26S rRNA in yeast 60S ribosomal subunits with kethoxal. Biochemistry. 1984;23:3330–3335. doi: 10.1021/bi00309a033. [DOI] [PubMed] [Google Scholar]
- 22.Hogan JJ, Gutell RR, Noller HF. Probing the conformation of 18S rRNA in yeast 40S ribosomal subunits with kethoxal. Biochemistry. 1984;23:3322–3330. doi: 10.1021/bi00309a032. [DOI] [PubMed] [Google Scholar]
- 23.Hogan JJ, Noller HF. Altered topography of 16S RNA in the inactive form of Escherichia coli 30S ribosomal subunits. Biochemistry. 1978;17:587–593. doi: 10.1021/bi00597a005. [DOI] [PubMed] [Google Scholar]
- 24.Woese CR, Magrum LJ, Gupta R, Siegel RB, Stahl DA, Kop J, Crawford N, Brosius J, Gutell R, Hogan JJ, et al. Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res. 1980;8:2275–2293. doi: 10.1093/nar/8.10.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gutell RR, Weiser B, Woese CR, Noller HF. Comparative anatomy of 16-S-like ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- 26.Torres-Larios A, Swinger KK, Krasilnikov AS, Pan T, Mondragon A. Crystal structure of the RNA component of bacterial ribonuclease P. Nature. 2005;437:584–587. doi: 10.1038/nature04074. [DOI] [PubMed] [Google Scholar]
- 27.Kazantsev AV, Krivenko AA, Harrington DJ, Holbrook SR, Adams PD, Pace NR. Crystal structure of a bacterial ribonuclease P RNA. Proc. Natl Acad. Sci. USA. 2005;102:13392–13397. doi: 10.1073/pnas.0506662102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 29.Vila-Sanjurjo A, Ridgeway WK, Seymaner V, Zhang W, Santoso S, Yu K, Cate JH. X-ray crystal structures of the WT and a hyper-accurate ribosome from Escherichia coli. Proc. Natl Acad. Sci. USA. 2003;100:8682–8687. doi: 10.1073/pnas.1133380100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 31.Blount KF, Breaker RR. Riboswitches as antibacterial drug targets. Nat. Biotechnol. 2006;24:1558–1564. doi: 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- 32.Blount KF, Wang JX, Lim J, Sudarsan N, Breaker RR. Antibacterial lysine analogs that target lysine riboswitches. Nat. Chem. Biol. 2007;3:44–49. doi: 10.1038/nchembio842. [DOI] [PubMed] [Google Scholar]
- 33.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 35.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl Acad. Sci. USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Das R, Baker D. Automated de novo prediction of native-like RNA tertiary structures. Proc. Natl Acad. Sci. USA. 2007;104:14664–14669. doi: 10.1073/pnas.0703836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Berkower I, Leis J, Hurwitz J. Isolation and characterization of an endonuclease from Escherichia coli specific for ribonucleic acid in ribonucleic acid-deoxyribonucleic acid hybrid structures. J. Biol. Chem. 1973;248:5914–5921. [PubMed] [Google Scholar]
- 38.Matveeva O, Felden B, Audlin S, Gesteland RF, Atkins JF. A rapid in vitro method for obtaining RNA accessibility patterns for complementary DNA probes: correlation with an intracellular pattern and known RNA structures. Nucleic Acids Res. 1997;25:5010–5016. doi: 10.1093/nar/25.24.5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wrzesinski J, Legiewicz M, Ciesiolka J. Mapping of accessible sites for oligonucleotide hybridization on hepatitis delta virus ribozymes. Nucleic Acids Res. 2000;28:1785–1793. doi: 10.1093/nar/28.8.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lima WF, Brown-Driver V, Fox M, Hanecak R, Bruice TW. Combinatorial screening and rational optimization for hybridization to folded hepatitis C virus RNA of oligonucleotides with biological antisense activity. J. Biol. Chem. 1997;272:626–638. [PubMed] [Google Scholar]
- 41.Birikh KR, Berlin YA, Soreq H, Eckstein F. Probing accessible sites for ribozymes on human acetylcholinesterase RNA. RNA. 1997;3:429–437. [PMC free article] [PubMed] [Google Scholar]
- 42.Ho SP, Bao Y, Lesher T, Malhotra R, Ma LY, Fluharty SJ, Sakai RR. Mapping of RNA accessible sites for antisense experiments with oligonucleotide libraries. Nat. Biotechnol. 1998;16:59–63. doi: 10.1038/nbt0198-59. [DOI] [PubMed] [Google Scholar]
- 43.Ho SP, Britton DH, Stone BA, Behrens DL, Leffet LM, Hobbs FW, Miller JA, Trainor GL. Potent antisense oligonucleotides to the human multidrug resistance-1 mRNA are rationally selected by mapping RNA-accessible sites with oligonucleotide libraries. Nucleic Acids Res. 1996;24:1901–1907. doi: 10.1093/nar/24.10.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cload ST, Schepartz A. Selection of structure-specific inhibitors of the HIV Rev-Rev response element complex. J. Am. Chem. Soc. 1994;116:437–442. [Google Scholar]
- 45.Scherr M, Rossi JJ. Rapid determination and quantitation of the accessibility to native RNAs by antisense oligodeoxynucleotides in murine cell extracts. Nucleic Acids Res. 1998;26:5079–5085. doi: 10.1093/nar/26.22.5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zarrinkar PP, Williamson JR. Kinetic intermediates in RNA folding. Science. 1994;265:918–924. doi: 10.1126/science.8052848. [DOI] [PubMed] [Google Scholar]
- 47.Mathews DH, Burkard ME, Freier SM, Wyatt JR, Turner DH. Predicting oligonucleotide affinity to nucleic acid targets. RNA. 1999;5:1458–1469. doi: 10.1017/s1355838299991148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu ZJ, Mathews DH. OligoWalk: an online siRNA design tool utilizing hybridization thermodynamics. Nucleic Acids Res. 2008;36:W104–W108. doi: 10.1093/nar/gkn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moore PB, Abo S, Freeborn B, Gewirth DT, Leontis NB, Sun G. Preparation of 5S RNA-related materials for nuclear magnetic resonance and crystallography studies. Methods Enzymol. 1988;164:158–174. doi: 10.1016/s0076-6879(88)64041-9. [DOI] [PubMed] [Google Scholar]
- 50.Peyret N, Seneviratne PA, Allawi HT, SantaLucia J., Jr Nearest-neighbor thermodynamics and NMR of DNA sequences with internal A•A, C•C, G•G, and T•T mismatches. Biochemistry. 1999;38:3468–3477. doi: 10.1021/bi9825091. [DOI] [PubMed] [Google Scholar]
- 51.Puglisi JD, Tinoco I., Jr Absorbance melting curves of RNA. Methods Enzymol. 1989;180:304–325. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- 52.Disney MD, Testa SM, Turner DH. Targeting a Pneumocystis carinii group I intron with methylphosphonate oligonucleotides: backbone charge is not required for binding or reactivity. Biochemistry. 2000;39:6991–7000. doi: 10.1021/bi992937m. [DOI] [PubMed] [Google Scholar]
- 53.Ciesiollka J, Lorenz S, Erdmann VA. Different conformational forms of Escherichia coli and rat liver 5S rRNA revealed by Pb(II)-induced hydrolysis. Eur. J. Biochem. 1992;204:583–589. doi: 10.1111/j.1432-1033.1992.tb16671.x. [DOI] [PubMed] [Google Scholar]
- 54.Kierzek E, Kierzek R, Turner DH, Catrina IE. Facilitating RNA structure prediction with microarrays. Biochemistry. 2006;45:581–593. doi: 10.1021/bi051409+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Speek M, Lind A. Structural analyses of E. coli 5S RNA fragments, their associates and complexes with proteins L18 and L25. Nucleic Acids Res. 1982;10:947–965. doi: 10.1093/nar/10.3.947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lu M, Steitz TA. Structure of Escherichia coli ribosomal protein L25 complexed with a 5S rRNA fragment at 1.8-A resolution. Proc. Natl Acad. Sci. USA. 2000;97:2023–2028. doi: 10.1073/pnas.97.5.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Freier SM, Alkema D, Sinclair A, Neilson T, Turner DH. Contributions of dangling end stacking and terminal base-pair formation to the stabilities of XGGCCp, XCCGGp, XGGCCYp, and XCCGGYp helixes. Biochemistry. 1985;24:4533–4539. doi: 10.1021/bi00338a008. [DOI] [PubMed] [Google Scholar]
- 58.Walter AE, Turner DH, Kim J, Lyttle MH, Muller P, Mathews DH, Zuker M. Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc. Natl Acad. Sci. USA. 1994;91:9218–9222. doi: 10.1073/pnas.91.20.9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mir KU, Southern EM. Determining the influence of structure on hybridization using oligonucleotide arrays. Nat. Biotechnol. 1999;17:788–792. doi: 10.1038/11732. [DOI] [PubMed] [Google Scholar]
- 60.Ehresmann C, Baudin F, Mougel M, Romby P, Ebel JP, Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987;15:9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lewis JB, Doty P. Identification of the single-strand regions in Escherichia coli 5S RNA, native and A forms, by the binding of oligonucleotides. Biochemistry. 1977;16:5016–5025. doi: 10.1021/bi00642a012. [DOI] [PubMed] [Google Scholar]
- 62.Uhlenbeck OC. Complementary oligonucleotide binding to transfer RNA. J. Mol. Biol. 1972;65:25–41. doi: 10.1016/0022-2836(72)90489-5. [DOI] [PubMed] [Google Scholar]
- 63.Uhlenbeck OC, Baller J, Doty P. Complementary oligonucleotide binding to the anticodon loop of fMet-transfer RNA. Nature. 1970;225:508–510. doi: 10.1038/225508a0. [DOI] [PubMed] [Google Scholar]
- 64.Milner N, Mir KU, Southern EM. Selecting effective antisense reagents on combinatorial oligonucleotide arrays. Nat. Biotechnol. 1997;15:537–541. doi: 10.1038/nbt0697-537. [DOI] [PubMed] [Google Scholar]
- 65.Duan S, Mathews DH, Turner DH. Interpreting oligonucleotide microarray data to determine RNA secondary structure: application to the 3′ end of Bombyx mori R2 RNA. Biochemistry. 2006;45:9819–9832. doi: 10.1021/bi052618x. [DOI] [PubMed] [Google Scholar]
- 66.Desai NA, Shankar V. Single-strand-specific nucleases. FEMS Microbiol. Rev. 2003;26:457–491. doi: 10.1111/j.1574-6976.2003.tb00626.x. [DOI] [PubMed] [Google Scholar]
- 67.Moody EM, Brown TS, Bevilacqua PC. Simple method for determining nucleobase pK(a) values by indirect labeling and demonstration of a pK(a) of neutrality in dsDNA. J. Am. Chem. Soc. 2004;126:10200–10201. doi: 10.1021/ja047362h. [DOI] [PubMed] [Google Scholar]
- 68.Wadkins TS, Shih I, Perrotta AT, Been MD. A pH-sensitive RNA tertiary interaction affects self-cleavage activity of the HDV ribozymes in the absence of added divalent metal ion. J. Mol. Biol. 2001;305:1045–1055. doi: 10.1006/jmbi.2000.4368. [DOI] [PubMed] [Google Scholar]
- 69.Ando T. A nuclease specific for heat-denatured DNA in isolated from a product of Aspergillus oryzae. Biochim. Biophys. Acta. 1966;114:158–168. doi: 10.1016/0005-2787(66)90263-2. [DOI] [PubMed] [Google Scholar]
- 70.Smith RM, Wu GY. Secondary structure and hybridization accessibility of the hepatitis C virus negative strand RNA 5′-terminus. J. Viral. Hepat. 2004;11:115–123. doi: 10.1046/j.1365-2893.2003.00476.x. [DOI] [PubMed] [Google Scholar]
- 71.Smith RM, Walton CM, Wu CH, Wu GY. Secondary structure and hybridization accessibility of hepatitis C virus 3′-terminal sequences. J. Virol. 2002;76:9563–9574. doi: 10.1128/JVI.76.19.9563-9574.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jenek M, Kierzek E. Isoenergetic microarray mapping–the advantages of this method in studying the structure of Saccharomyces cerevisiae tRNAPhe. Nucleic Acids Symp. Ser. (Oxf.) 2008:219–220. doi: 10.1093/nass/nrn111. [DOI] [PubMed] [Google Scholar]
- 73.Ratmeyer L, Vinayak R, Zhong YY, Zon G, Wilson WD. Sequence specific thermodynamic and structural properties for DNA.RNA duplexes. Biochemistry. 1994;33:5298–5304. doi: 10.1021/bi00183a037. [DOI] [PubMed] [Google Scholar]
- 74.Robertus JD, Ladner JE, Finch JT, Rhodes D, Brown RS, Clark BF, Klug A. Structure of yeast phenylalanine tRNA at 3 Å resolution. Nature. 1974;250:546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- 75.Kim SH, Suddath FL, Quigley GJ, McPherson A, Sussman JL, Wang AH, Seeman NC, Rich A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974;185:435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- 76.Goringer HU, Wagner R. Does 5S RNA from Escherichia coli have a pseudoknotted structure? Nucleic Acids Res. 1986;14:7473–7485. doi: 10.1093/nar/14.18.7473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]