Abstract
H2A.Z is an evolutionarily conserved H2A variant that plays a key role in the regulation of chromatin transcription. To understand the molecular mechanism of H2A.Z exchange, we purified two distinct H2A.Z-interacting complexes termed the small and big complexes from a human cell line. The big complex contains most components of the SRCAP chromatin remodeling and TIP60 HAT complexes, whereas the small complex possesses only a subset of SRCAP and TIP60 subunits. Our exchange analysis revealed that both small and big complexes enhance the incorporation of H2A.Z-H2B dimer into the nucleosome. In addition, TIP60-mediated acetylation of nucleosomal H2A specifically facilitates the action of the small complex in the H2A.Z exchange reaction. Among factors present in the small complex, we determined that TIP48 and TIP49 play a major role in catalyzing H2A acetylation-induced H2A.Z exchange via their ATPase activities. Overall, our work uncovers the previously-unrecognized role of TIP48 and TIP49 in H2A.Z exchange and a novel epigenetic mechanism controlling this process.
INTRODUCTION
In eukaryotes, genomic DNA is packaged into a complex nucleoprotein structure called chromatin. The fundamental unit of chromatin is the nucleosome, which is composed of 147 bp of DNA wrapped around a histone octamer of two H2A-H2B heterodimers and a H3-H4 tetramer (1,2). Three major remodeling processes that regulate DNA accessibility in this repressive chromatin state are post-translational modifications of histones, ATP-dependent remodeling of chromatin, and incorporation of histone variants into chromatin (3–5). Recent studies demonstrated that histone variants play an important role in regulating gene expression and other DNA-templated cellular processes (6). H2A.Z is one of the evolutionarily conserved H2A variant and comprises about 5–10% of total cellular H2A (7). H2A.Z is expressed and integrated into chromatin independently of DNA replication and is essential for viability in many organisms, such as Tetrahymena, Drosophila, and mice (8–10).
The crystal structure of an H2A.Z-containing nucleosome indicates that H2A.Z confers a destabilization of interaction between the H2A.Z-H2B dimer and the H3–H4 tetramer (11). In agreement with these results, H2A.Z-containing nucleosomes exhibited more salt and heat-induced destabilization than canonical nucleosomes (12–14). However, more recent biophysical studies showed that integration of H2A.Z into nucleosomes has a positive effect on nucleosome stability (15,16). At the functional level, ChIP-chip analyses across the yeast genome indicated that H2A.Z is preferentially localized to specific sites within the promoter regions (12,17,18). This promoter localization of H2A.Z is remarkably specific, and appears to establish unique promoter architecture for transcription regulation (19–21). In this regard, a repressive role of H2A.Z in transcription is supported by its association with heterochromatin binding protein 1-α (HP1α) (22). However, the promoter-localized H2A.Z has also been implicated for gene activation by facilitating chromatin remodeling and recruitment of transcriptional machinery at gene promoter regions (23,24). A possible role of H2A.Z in transcription is further supported by the discovery that H2A.Z is dissociated from nucleosomes upon initiation of gene transcription in yeast and human cells (12,25).
Recent studies revealed that several chromatin remodeling activities have an ability to catalyze the replacement of H2A.Z for H2A within canonical nucleosomes. In yeast, the SWR1 complex containing swi2/snf2-related ATPase Swr1 has been shown to incorporate H2A.Z-H2B dimers into nucleosomes in an ATP-dependent manner (26–28). A subsequent study conducted in Drosophila showed that TIP60-mediated acetylation of nucleosomes plays a role in replacement of nucleosomal phospho-H2Av (the drosophila H2A.Z/H2A.X homolog) with unmodified H2Av (29). So far, two different activities were identified for H2A.Z exchange in human cells. SRCAP complex was purified and shown to include multiple subunits, one of which is Swr1-related chromatin remodeling protein SRCAP (30). In vitro exchange experiments indicated that the SRCAP complex is able to exchange H2A with H2A.Z in the reconstituted nucleosome (31). p400 is another human homologue of yeast Swr1 found in human TIP60 complex, and shown to have a catalytic activity for the exchange of H2A.Z into the promoter regions of p53 target genes (32). The fact that SRCAP and TIP60 complexes share several subunits (e.g. TIP48, TIP49, Actin, YL1) supports the participation of these components in regulating the H2A.Z exchange process.
To further understand the molecular mechanisms that regulate the incorporation of H2A.Z into the nucleosome, we purified and characterized two H2A.Z-associated complexes: the H2A.Z big complex containing most SRCAP and TIP60 subunits and the H2A.Z small complex containing only a subset of SRCAP and TIP60 subunits. In our exchange assays, we found that both small and big complexes can moderately promote the replacement of nucleosomal H2A with H2A.Z. Significantly, TIP60-mediated acetylation of nucleosomal H2A facilitates H2A.Z incorporation catalyzed by the small complex, but not by the big complex. More importantly, we show that TIP48 and TIP49 present in the small complex are sufficient to recapitulate H2A.Z exchange capabilities of the entire complex. These results provide new insight into the role of TIP48/TIP49 ATPase in the exchange of H2A.Z, which is facilitated by TIP60-mediated H2A acetylation.
MATERIALS AND METHODS
Purification and identification of H2A.Z complexes
The establishment of HeLa cell lines that stably express epitope-tagged H2A.Z (f/h:H2A.Z) and affinity purification of the H2A.Z complex were conducted by following essentially the same procedures as described in our recent study (33). The affinity-purified H2A.Z complex (0.3 ml) was further purified by 4.7 ml 15–40% glycerol gradient sedimentation as described (26). The purified H2A.Z complexes were identified by USC Mass Spectrometry and Proteomics Core Facility. Experimental details, including plasmid construction, cell line establishment and protein purification/identification, can be found in the Supplementary Data.
Preparation of recombinant histones and TIP48/TIP49
For the preparation of H2A.Z histone octamers and dimers, H2A.Z cDNA was PCR-amplified and inserted into the NdeI and BamHI sites of pET11a (for untagged H2A.Z), pET15b (for His-tagged H2A.Z/h:H2A.Z) or pET11d (for FLAG-tagged H2A.Z/f:H2A.Z). H2A.Z proteins were expressed in bacteria (Rosetta (DE3)pLysS, Novagen) and affinity-purified using Ni+-NTA (for h:H2A.Z) and M2 agarose (for f:H2A.Z) beads. For histone octamer preparation, untagged H2A.Z and canonical H2B, H3 and H4 were prepared as described (34,35), dissolved in 8 M guanidium solution by rotating at room temperature and combined for octameric reconstitutions by renaturation. For H2A.Z-H2B dimer preparation, His-tagged or FLAG-tagged H2A.Z was renatured together with H2B. The refolded histones were passed through Sephacryl S300 column (GE healthcare) to remove unreconstituted histones. H2A–H2B and H2A.X-H2B dimers were prepared essentially as described (34,36). To prepare recombinant TIP48 and TIP49, full-length cDNA sequences encoding human TIP48 and TIP49 were PCR amplified from 293T cell mRNA and subcloned into the NdeI and BamHI sites of pET11d together with a FLAG tag at their amino termini. FLAG-tagged TIP48 (f:TIP48) and TIP49 (f:TIP49) were expressed in bacteria and purified using M2 agarose beads. For reconstitution of TIP48/TIP49 complex, equal amounts of TIP48 and TIP49 were mixed at BC150 binding buffer and further purified by 15–40% glycerol gradient sedimentation.
Reconstitution of mononucleosomes
Mononucleosomes were reconstituted with 5′ biotinylated 207 bp DNA fragments containing 601 nucleosome positioning sequence (37,38) and either canonical or H2A.Z-containing histone octamers by salt dialysis method (39). The reconstituted nucleosomes were purified by sedimentation in a 5–30% (vol/vol) glycerol gradient to remove free DNA and core histones.
Histone acetyltransferase and ATPase assays
For histone acetyltransferase (HAT) assays, free histones (1 μg) or reconstituted mononucleosomes (0.5 μg) were incubated with the initial H2A.Z complex in the presence of 2.8 μM [3H] acetyl-CoA or 10 μM cold acetyl-CoA and analyzed by 15% SDS–PAGE and western blot analysis (34). All antibodies used for western blot analysis were from Millipore. ATPase assays were performed as described previously (40) by using the purified H2A.Z complexes (6 μl) or recombinant TIP48/TIP49 (150 ng) in the presence of 0.05 μCi of [γ-32P] ATP for 30 min at 30°C. Free phosphate and ATP were separated by 12% polyacrylamide gel (19:1) containing 7 M urea in a 1× TBE buffer.
Histone exchange assays
Exchange assays were performed as recently described (26,36) with minor modifications. Briefly, regular mononucleosomes (150 ng DNA equivalents) immobilized on streptavidin-conjugated Dynabeads M-280 (Dynal) were incubated with H2A.Z-S or H2A.Z-B complex, in exchange reaction buffer (25 mM HEPES, pH 7.6, 0.37 mM EDTA, 0.35 mM EGTA, 5 mM MgCl2, 1 mM DTT, 70 mM KCl, 10% glycerol, 0.02% NP-40 and 0.1 mg/ml BSA) for 60 min at 30°C in the absence or presence of 1 mM ATP. Note that both H2A.Z-S and H2A.Z-B complexes provide FLAG/HA-H2A.Z and H2B for exchange reactions. Beads were concentrated on a magnetic particle concentrator (Dynal) and washed three times with washing buffer (25 mM HEPES, pH 7.6, 0.1 mM EDTA, 0.5 mM EGTA, 5 mM MgCl2, 1 mM DTT, 70 mM KCl, 10% glycerol, 0.02% NP-40 and 0.1 mg/ml BSA). The supernatant was also collected from the initial exchange reaction for analysis of free unincorporated H2A.Z. Nucleosomal and free H2A.Z proteins collected from the exchange reaction were subjected to western blot analysis using anti-FLAG antibody (Sigma). Histone exchange assays using the acetylated nucleosomes were as described above, except that nucleosomes were pre-acetylated by the initial H2A.Z complex in HAT reaction buffer containing cold acetyl-CoA (10 μM, Sigma) for 1 h before their immobilization onto Dynabeads. Immobilized nucleosomes were washed extensively with 150 mM exchange buffer (25 mM HEPES, pH 7.6, 0.37 mM EDTA, 0.35 mM EGTA, 5 mM MgCl2, 1 mM DTT, 150 mM KCl, 10% glycerol, 0.02% NP-40 and 0.1 mg/ml BSA). For exchange reactions, immobilized nucleosomes was mixed with the TIP48/TIP49 complex (150 ng) in the exchange reaction buffer for 20 min at 30°C, and then incubated with h:H2A.Z-H2B, h:H2A.X-H2B or f:H2A-H2B dimers (400 ng each) for 60 min at 30°C. Nucleosomal proteins were analyzed by immunoblot using anti-His (Novagen) and anti-FLAG (Sigma) antibodies.
Chromatin isolation and mononucleosome immunoprecipitation
Chromatin was purified from 293T cells as previously described (41), but after expression of FLAG-tagged H2A.Z (f:H2A.Z). For preparation of H2A.Z mononucleosomes, nuclei of HeLa cells stably expressing f/h:H2A.Z were first digested with micrococcal nuclease (MNase) (0.6 U, Sigma), and preparation of mononucleosome fractions was confirmed by 2% agarose gel electrophoresis of nucleosomal DNA purified from the MNase digestion reaction. To isolate mononucleosomes containing f/h:H2A.Z, the entire mononucleosome fraction was further subjected to immunoprecipitation using M2-agarose beads. After overnight incubation, beads were washed five times with BC300, and bead-bound mononucleosomes were eluted from M2 agarose by using FLAG peptide (200 ng/ml). The eluted mononucleosomes were analyzed on 15% SDS-gel electrophoresis.
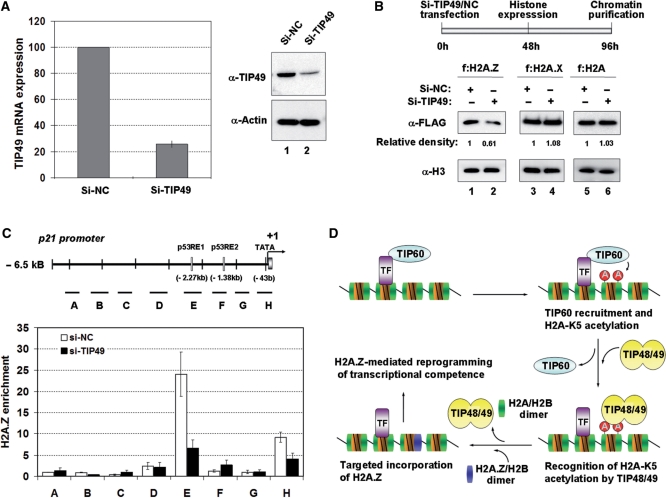
TIP49 depletion and RT–qPCR
For TIP49 depletion, 293T cells were transfected with 25 nM of the validated TIP49 siRNA (Ambion, siRNA ID #13702 or #13514) using siPORT NeoFX (Ambion) and incubated for 96 h. The expression level of TIP49 in siRNA-transfected cells was determined by western blot analysis using anti-TIP49 antibody (Santa Cruz). The real time quantitative PCR was performed using an iCycler iQ Real-Time Detection System with the iQ SYBR Green supermix (Bio-Rad). The primers used for qPCR are as listed in the Supplementary Data (Supplementary Table S1). The specificity of the amplification reactions were monitored by melting curve analysis and subsequently by agarose gel electrophoresis. The threshold cycle (Ct) value for each gene was normalized to the Ct value for β-actin. All samples were run in triplicate. For the analysis of histone incorporation on chromatin after TIP49 knockdown, the cells were initially transfected with TIP49 siRNA for 48 h and then retransfected with plasmids expressing f:H2A.Z, f:H2A.X, or f:H2A for another 48 h. Chromatin was isolated as described above, and incorporation of ectopic histones was analyzed by western blot analysis with anti-FLAG antibody.
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (42) with minor modifications. See the Supplementary Data for experimental details and primer sequences (Supplementary Table S2).
RESULTS
Free H2A.Z stably associates with components of SRCAP and TIP60 complexes
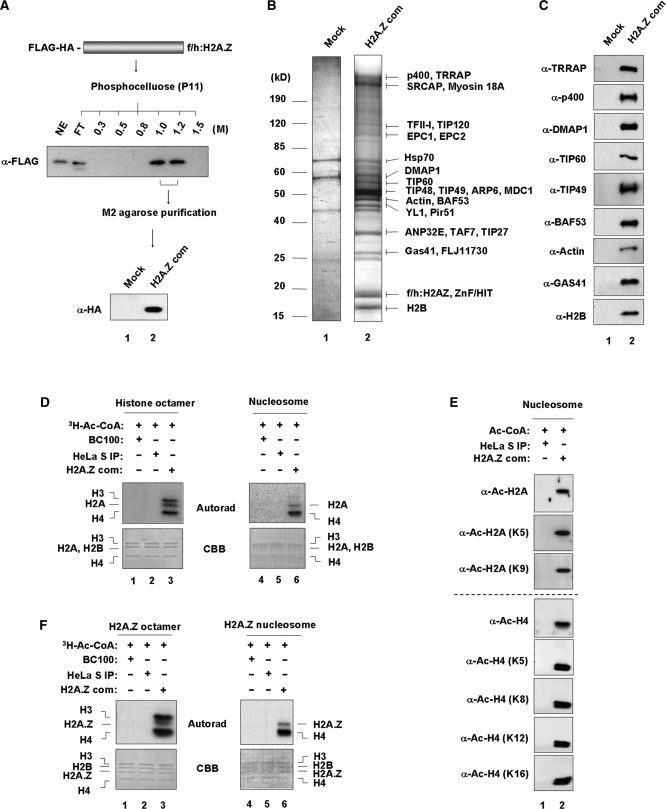
In an effort to enhance our understanding of H2A.Z exchange machinery, we have generated a cell line constitutively expressing the FLAG/HA-tagged version of H2A.Z (Figure 1A). Ectopic H2A.Z was found to be mainly localized in the nucleus as identified by immunofluorescence analysis with an anti-FLAG antibody (Supplementary Figure S1). Nuclear extracts were prepared from the cell line and initially fractionated on a phosphocellulose P11 column. The 1.0 M and 1.2 M KCl fractions containing ectopic H2A.Z were combined and further purified by M2-agarose affinity chromatography under stringent conditions (300 mM KCl, 0.1% Nonidet P-40) (Figure 1A). Coomassie blue stain analysis of elution fractions revealed the association of ectopic H2A.Z with 14 prominent bands that were not detectable with control HeLa nuclear extract (Figure 1B, lane 2). Mass spectrometric analysis of these 14 major bands identified a number of proteins, most of which are subunits of SRCAP chromatin remodeling and/or TIP60 HAT complexes as summarized in Figure 1B. We further confirmed the mass spectrometry results by western blot analysis using available antibodies (Figure 1C). Since the H2A.Z complex contains TIP60, which has been characterized as a HAT (43,44), we first asked whether the H2A.Z complex has HAT activity. To exclude the effect of any pre-existing modifications of native histones, we prepared recombinant histone octamers after expression of individual histones in bacteria for our HAT assays (Supplementary Figure S2, lanes 1–5). We found that the H2A.Z complex can acetylate H2A, H3 and H4 in H2A-containing histone octamers (Figure 1D, lane 3) and H2A.Z, H3 and H4 in H2A.Z-containing histone octamers (Figure 1F, lane 3). Since previous studies with TIP60 detected the same substrate specificity (43,44), these results strongly support that TIP60 in the complex is the responsible HAT for this modification.
Figure 1.
Purification of H2A.Z complex from human cells. (A) Schematic summary of purification of H2A.Z complex. Nuclear extracts from H2A.Z-expressing cells were first fractionated by P11 cation exchange column with BC buffer as indicated. The P11 1.0 and 1.2 M KCl fractions containing ectopic H2A.Z were combined and further purified with anti-FLAG affinity chromatography as described under ‘Materials and Methods’. The purified proteins were separated in 4–20% gradient SDS–PAGE and subjected to western blot analysis with anti-FLAG and -HA antibodies. Lane 1, mock purification with nuclear extracts prepared from regular HeLa cells; lane 2, FLAG/HA-H2A.Z immunoprecipitation with nuclear extracts prepared from H2A.Z expressing cells. (B) Mass spectrometric analysis of H2A.Z-associated polypeptides. After a large-scale isolation of H2A.Z complex, the purified polypeptides were resolved in 4–20% gradient SDS–PAGE. The protein bands, that were not present in the control lane, were excised, and identified by mass spectrometric analysis. Lane 1, the proteins purified from control HeLa cells; lane 2, the proteins purified from H2A.Z expressing cells. The positions of the molecular mass markers (in kDa) are indicated on the left. (C) Western blot analysis of the purified H2A.Z complex. H2A.Z complex was separated by 4–20% gradient SDS-PAGE, transferred to nitrocellulose, and probed by western analysis with antibodies as indicated on the left. Lane 1, mock-purified control; lane 2, H2A.Z complex. (D) Acetylation of H2A histone octamer and H2A nucleosome by H2A.Z complex. Recombinant histone octamer and nucleosome were subjected to HAT assay with [3H]-acetyl-CoA and the H2A.Z complex. HAT reactions were analyzed by 15% SDS–PAGE with subsequent fluorography. Lanes 1 and 4, buffer control; lanes 2 and 5, mock-purified control; lanes 3 and 6, H2A.Z complex. (E) H2A/H4-targeted acetylation of H2A nucleosome by H2A.Z complex. HAT assays were as in (D), but with nucleosomes and cold acetyl-CoA. HAT reactions were analyzed by western blot analysis with the indicated antibodies. Lane 1, mock-purified control; lane 2, H2A.Z complex. (F) Acetylation of H2A.Z histone octamer and H2A.Z nucleosome by H2A.Z complex. Assays were identical to (D), except that H2A.Z containing octamer and nucleosome were used as substrates. Lanes 1 and 4, buffer control; lanes 2 and 5, mock-purified control; lanes 3 and 6, H2A.Z complex.
To further analyze the ability of TIP60 in the H2A.Z complex to acetylate nucleosomes, we reconstituted nucleosomes with 207 bp DNA fragments containing 601 nucleosome positioning sequence and recombinant histone octamers by the method of salt gradient dialysis. The successful reconstitution of a nucleosome on the 601 DNA fragment was confirmed by appearance of a retarded band in nucleoprotein gels (Supplementary Figure S2, lanes 6 and 7). As shown in Figure 1D and F, TIP60 in the H2A.Z complex acetylates nucleosomal H2A, H2A.Z and H4 to a level comparable to that of free H2A, H2A.Z and H4 (lane 6 versus lane 3). However, in contrast to free H3 substrate, nucleosomal H3 was not acetylated by H2A.Z-associated TIP60 (lane 6 versus lane 3). Moreover, in western blotting to check acetylation of two lysine substrates (K5 and K9) in H2A tails and four lysine substrates (K5, K8, K12 and K16) in H4 tails, we detected acetylation of all these substrates in the nucleosome by H2A.Z-associated TIP60 (Figure 1E). Collectively, our results show that TIP60 present in the H2A.Z complex can acetylate the nucleosome with an intrinsic preference for H2A/H2A.Z and H4 tails.
Initial H2A.Z complex can be separated into two different subcomplexes
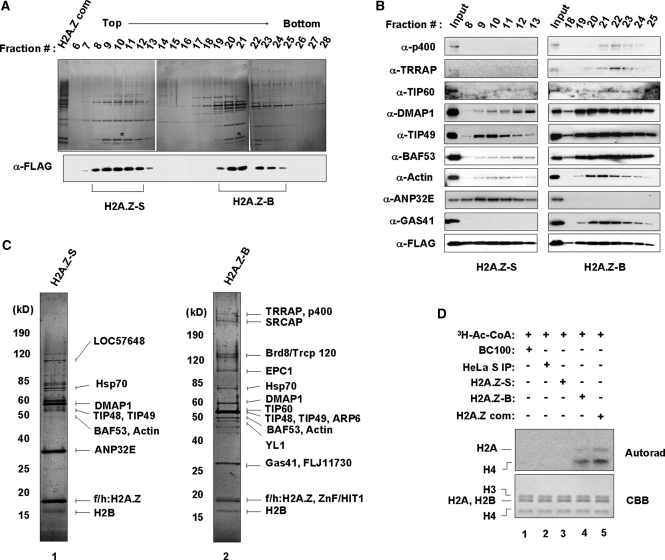
To determine whether the proteins co-purified with ectopic H2A.Z form a single distinct complex or multiple complexes, we further purified the initial H2A.Z complex by ultracentrifugation in a 15–40% glycerol gradient. Silver staining and western blot analysis of each gradient fraction revealed that the initial complex is primarily sedimented as two peaks in fractions 8–12 and 19–24 (Figures 2A and B), indicating the presence of two distinct subcomplexes in the initial complex. Mass spectrometry analysis of pooled 19–24 fractions revealed that most of SRCAP and TIP60 components are co-sedimented with H2A.Z in this fast-migrating peak (Figure 2C, lane 2). However, similar analysis of pooled 8–12 fractions detected only a subset of factors, including TIP48, TIP49, BAF53, DMAP1 and Actin, in this slow-sedimenting peak (Figure 2C, lane 1). We have termed two subcomplexes purified from fractions 8–12 and 19–24 as H2A.Z-S and H2A.Z-B, respectively, implying that they are small (S) and big (B) complexes. Consistent with results from western blot analysis of gradient fractions (Figure 2B), we detected HAT activity only in the big complex, which contains a high concentration of TIP60 (Figure 2D). To further confirm the presence of two distinct H2A.Z complexes in vivo, we fractionated the nuclear extracts prepared from the H2A.Z stable cell lines through a gel filtration column. Our western blot analysis of the fractions clearly showed that ectopic H2A.Z and its associated factors are eluted as two broad peaks corresponding to the small and big complexes (Supplementary Figure S3).
Figure 2.
Isolation of H2A.Z subcomplexes. (A) Glycerol gradient fractionation of H2A.Z subcomplexes. The H2A.Z complex was separated by glycerol gradient centrifugation as describe under ‘Materials and Methods’ section. Total 32 fractions were collected from the top to the bottom, and analyzed by 4–20% gradient SDS–PAGE and silver staining. Each fraction was also subjected to western blot analysis with anti-FLAG antibody. Asterisks indicate the ectopic H2A.Z. H2A.Z-S and H2A.Z-B indicate the H2A.Z-small (S) and H2A.Z-big (B) complexes, respectively. (B) Western blot analysis of the purified H2A.Z subcomplexes. Aliquots of fractions corresponding to H2A.Z-S and H2A.Z-B complexes were separated by 4–20% SDS–PAGE and analyzed by immunoblot with the indicated antibodies. The initial H2A.Z complex shown in the Figure 1B was used as input. (C) Mass spectrometric analysis of H2A.Z subcomplexes. H2A.Z-S and H2A.Z-B complexes were separated by 4–20% SDS–PAGE followed by silver staining and identification of the bands by tandem mass spectrometry. Protein size markers are indicated on the left. Lane 1, H2A.Z-S complex; lane 2, H2A.Z-B complex. (D) HAT assay of H2A.Z subcomplexes. Assays were identical to Figure 1D, except that H2A.Z-S (lane 3) and H2A.Z-B complexes (lane 4) described in (C) were used. Lane 1, buffer control; lane 3, mock-purified control; lane 5, initial H2A.Z complex.
Both small and big complexes mildly facilitate H2A.Z exchange
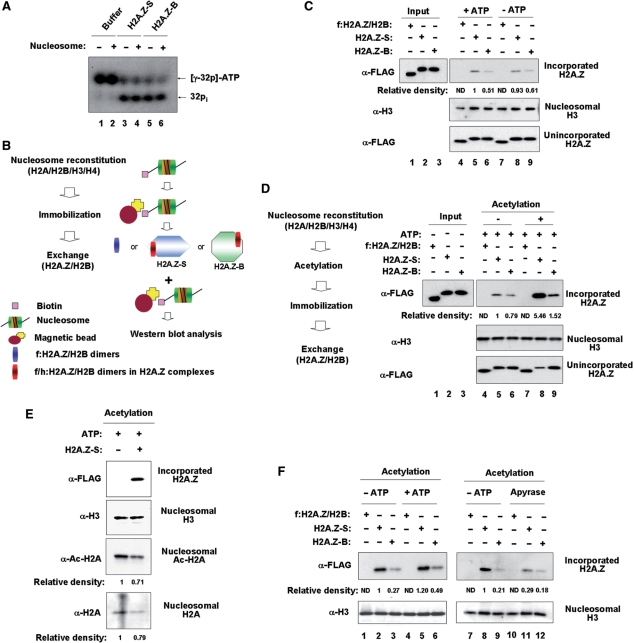
Considering that SRCAP complex and p400 have recently been shown to exchange H2A.Z/H2B dimers for nucleosomal H2A/H2B (31,32), we set out to determine whether the small and big complexes can catalyze the H2A.Z exchange through their ATP-dependent remodeling activities. The big complex contains four catalytic ATPase subunits, SRCAP, p400, TIP48 and TIP49, while the small complex contains only two ATPases, TIP48 and TIP49. Thus, we first checked the ability of the small and big complexes to hydrolyze ATP in the presence or absence of the 601 nucleosomes. This analysis revealed that both complexes have similar ATPase activities, and the ATPase activity of neither small nor big complex depends on the presence of nucleosomes (Figure 3A).
Figure 3.
Stimulatory effect of H2A acetylation on H2A.Z exchange. (A) ATP hydrolysis by H2A.Z-S and H2A.Z-B complexes. ATPase activities of the H2A.Z-S and H2A.Z-B complexes were checked with [γ-32P] ATP in the presence or absence of nucleosomes. ATPse assays were normalized to the level of FLAG/HA-H2A.Z in the complexes. Lanes 1 and 2, buffer control; lanes 3 and 4, H2A.Z-S complex; lanes 5 and 6, H2A.Z-B complex. (B) Schematic summary of H2A.Z exchange assay. Nucleosomes were reconstituted with 5′ biotinylated 207 bp 601 DNA fragments and immobilized on streptavidin-conjugated Dynabeads. Immobilized nucleosomes were incubated with either free FLAG-H2A.Z-H2B dimers or the H2A.Z subcomplexes containing FLAG/HA-H2A.Z-H2B dimers. Nucleosomal incorporation of H2A.Z was determined by western blot analysis with anti-FLAG antibody. (C) H2A.Z exchange by H2A.Z-S and H2A.Z-B complexes in vitro. Immobilized nucleosomes were incubated with either free FLAG-H2A.Z-H2B dimers (lanes 4 and 7) or the H2A.Z complexes containing FLAG/HA-H2A.Z-H2B dimers (lanes 5, 6, 8 and 9) in the presence or absence of ATP for 60 min. After extensive washing of the immobilized nucleosomes, H2A.Z incorporation was determined by western blot analysis with anti-FLAG antibody (first panel). Relative intensity of bands was quantified using Phosphorimager (Bio-Rad). For analysis of free unincorporated H2A.Z, supernatant was collected before the washing step and 25% of the supernatant was analyzed by western blot (third panel). Nucleosomal H3 was used as an internal loading control (second panel). The amount of H2A.Z-S and H2A.Z-B complexes was normalized based on the level of FLAG/HA-H2A.Z present in the complexes. Results shown are from a single experiment and are representative of three independent experiments. Lanes 1–3, input (50%) of free FLAG-H2A.Z-H2B dimer, FLAG/HA-H2A.Z-H2B dimer in H2A.Z-S complex and FLAG/HA-H2A.Z-H2B dimer in H2A.Z-B complex. (D) H2A.Z exchange into acetylated nucleosomes in vitro. Exchange assay was identical to (C), but nucleosomes were acetylated with the initial H2A.Z complex before immobilizing on magnetic beads and ATP was included in all reactions. (E) Preferential exchange of H2A.Z for acetylated H2A. After H2A.Z exchange reaction, equal amounts of nucleosomes were analyzed by western blot analyses with anti-FLAG (α-FLAG), anti-H3 (α-H3), anti-acetyl-H2A (α-Ac-H2A) and anti-H2A antibodies (α-H2A). Equal loading of nucleosomes was confirmed by western blotting for nucleosomal H3 (α-H3). (F) Requirement of ATP hydrolysis for H2A.Z exchange. In vitro exchange reactions were identical to lanes 7–9 in (D), but in the absence (lanes 1–3 and lanes 7–9) or presence (lanes 4–6) of ATP or pre-treatment of the H2A.Z complexes by 0.5 U apyrase (lane 10–12).
To analyze the ability of the small and big complexes to catalyze H2A.Z exchange, H2A-containing nucleosomes were reconstituted on 5′ biotinylated 207 bp DNA fragments containing 601 nucleosome positioning sequence with recombinant core histones (as in Supplementary Figure S2, see ‘Materials and Methods’ section for details). After immobilizing nucleosomes to paramagnetic beads, exchange assays were performed with small or big complex in the presence or absence of ATP as summarized in Figure 3B. Since both small and big complexes contain FLAG/HA-H2A.Z and H2B, we checked the incorporation of these dimers into immobilized nucleosomes without adding free H2A.Z-H2B dimers. As a gel loading control, we normalized for the levels of histone H3 on the immobilized nucleosomes (Figure 3C, Nucleosomal H3). Probing of the membranes with anti-FLAG antibody revealed that incubations of small and big complexes with H2A-containing nucleosomes induced transfer of about 10% of FLAG/HA-H2A.Z-H2B dimers into nucleosomes (Incorporated H2A.Z, lanes 5 and 6). Interestingly, similar experiments in the absence of ATP also showed nucleosomal incorporation of detectable amount of FLAG/HA-H2A.Z-H2B dimers from the complexes (Incorporated H2A.Z, lanes 8 and 9). Such H2A.Z transfer is most likely to be made by endogenous ATP co-purified with the complexes as revealed in recent studies (26,44). The effects of small and big complexes were specific, since the control reactions with free FLAG-H2A.Z-H2B dimers failed to show any H2A.Z incorporation (Incorporated H2A.Z, lanes 4 and 7). Anti-FLAG immunoblot of the supernatants collected from the exchange reactions showed no significant change in the level of unincorporated H2A.Z (Unincorporated H2A.Z), indicating moderate transfer of FLAG/HA-H2A.Z-H2B dimers into the nucleosome (note the faster mobility of FLAG-H2A.Z in the recombinant H2A.Z-H2B dimers compared to FLAG/HA-H2A.Z in the complexes).
Pre-acetylation of nucleosomal histones significantly facilitates the action of small complex
Because TIP60-mediated histone acetylation is known to facilitate ATP-dependent exchange of phosphorylated H2Av with unmodified H2Av in Drosophila (29), it is possible that TIP60 present in the purified H2A.Z complex cooperates with small and/or big complexes for H2A.Z exchange. Thus, having found that TIP60 in the initial H2A.Z complex can efficiently acetylate nucleosomal H2A/H4 (Figure 1D), we performed exchange assays after acetylation of the nucleosome by the initial H2A.Z complex. When nucleosomes were pre-acetylated by TIP60, big complex-induced incorporation of H2A.Z-H2B dimer was only slightly enhanced (Figure 3D, Incorporated H2A.Z, lane 9 versus lane 6). In striking contrast, identical assays using small complex showed about five-fold increase in H2A.Z exchange (Incorporated H2A.Z, lane 8 versus lane 5), indicating that TIP60-induced pre-acetylation is required for full action of small complex, but not of big complex. Since the pre-acetylated nucleosomes are free of the initial complex components (Supplementary Figure S4, lanes 1–4), the observed changes in H2A.Z incorporation is rather specific for the small complex and not due to the initial complex left from HAT reaction. To determine whether acetylated H2A proteins are preferentially replaced by H2A.Z, we also analyzed equivalent amounts of nucleosomes by western blot analysis after exchange reactions. As shown in Figure 3E, western blot intensities perceived for H2A and acetyl H2A are decreased to a comparable extent after exchange reactions, strongly supporting the selective dissociation of acetylated H2A upon H2A.Z incorporation.
We next tested whether ATP hydrolysis is required for acetylation-dependent incorporation of H2A.Z. As was the case for the acetylation-independent exchange reactions (Figure 3C), the small complex was capable of catalyzing acetylation-mediated H2A.Z exchange even in the absence of ATP, although a slight stimulation of H2A.Z exchange was observed by including free ATP (Figure 3F, Incorporated H2A.Z, lanes 2 and 5). Such H2A.Z exchange is most likely due to endogenous ATP copurified with the small complex, since the exchange reaction without exogenous ATP was significantly inhibited when apyrase was added in the beginning of the exchange assays (lane 8 versus lane 11). Big complex, having a much weaker activity in catalyzing acetylation-mediated H2A.Z exchange reaction, also showed a detectable inhibition on its activity upon apyrase treatment (lane 9 versus lane 12). When apyrase treatment was tried for acetylation-independent exchange reactions, we could also observe, albeit to a lesser extent, reduction of H2A.Z incorporation (Supplementary Figure S5).
Acetylation of H2A-K5 facilitates small complex-mediated H2A.Z exchange
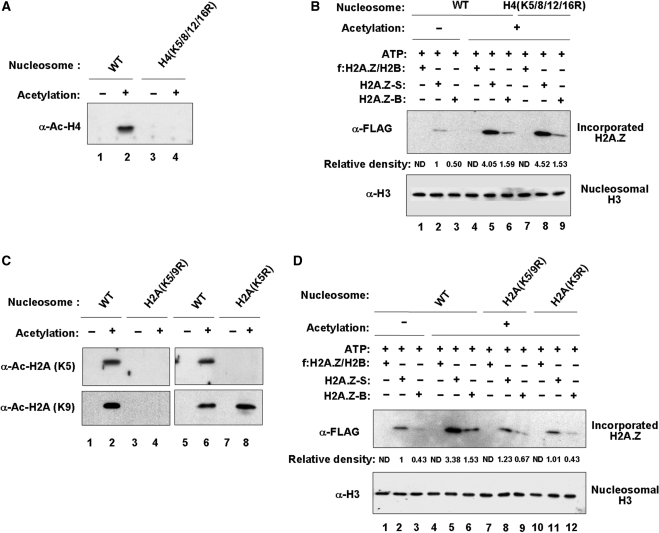
The aforementioned results showing the stimulatory effect of nucleosome acetylation on small complex-induced H2A.Z exchange suggest that acetylation of specific lysine substrates could be responsible for this stimulation. To address this issue, we repeated the exchange assays using nucleosomes reconstituted with mutant histones. Since TIP60 present in the initial H2A.Z complex has an intrinsic ability to acetylate nucleosomal H2A and H4 (Figure 1D and E), we mutagenized the major acetylation sites of H2A and H4 (K5 and K9 of H2A and K5, K8, K12 and K16 of H4). Western blot analyses confirmed that these mutations completely blocked acetylations of H2A-K5/K9 and H4-K5/K8/K12/K16 by TIP60 in the initial complex (Figure 4A and C). When the effect of H4 mutations on acetylation-mediated H2A.Z exchange was examined, the mutant H4 nucleosome showed H2A.Z incorporation comparable to that with the wild-type nucleosome (Figure 4B, lanes 5 and 6 versus lanes 8 and 9). In contrast, the exchange assay with the H2A-mutated nucleosome failed to show acetylation-facilitated incorporation of H2A.Z into the nucleosome (Figure 4D, lane 5 versus lane 8). More importantly, similar exchange reactions with the nucleosome containing K5 mutant form of H2A also showed significant repression in H2A.Z exchange (lane 5 versus lane 11), indicating that H2A-K5 acetylation is necessary for optimal incorporation of H2A.Z. The fact that the K5 mutation did not change the level of acetylation of K9 (Figure 4C, lane 8) strongly suggests that K9 acetylation alone is not sufficient for small complex-induced H2A.Z exchange. Overall, these results establish a primary role of acetylation of H2A-K5 in facilitating the action of small complex for H2A.Z exchange.
Figure 4.
Dominant role of H2A-K5 acetylation for H2A.Z exchange. (A) HAT assay with recombinant nucleosomes containing wild-type or mutant H4. HAT assay was identical to Figure 1E, but using nucleosomes reconstituted with intact (lanes 1 and 2) or lysine mutated (lanes 3 and 4) H4. Reaction products were subjected to western blot analysis with an antibody recognizing acetylated H4. (B) Minimal effect of H4 acetylation on H2A.Z exchange. In vitro exchange assay was identical to Figure 3D, but using nucleosomes reconstituted with intact (lanes 1–6) or lysine mutated (lanes 7–9) H4. Nucleosomes were unmodified (lanes 1–3) or acetylated (lanes 4–9) by TIP60 in the initial H2A.Z complex. Nucleosomal incorporation of H2A.Z was analyzed by western blot analysis with anti-FLAG antibody (α-FLAG). Equal loading of nucleosomes was confirmed by western blotting for nucleosomal H3 (α-H3). (C) HAT assay with recombinant nucleosomes containing wild-type or mutant H2A. HAT assay was identical to Figure 4A, but using nucleosomes reconstituted with intact (lanes 1, 2, 5 and 6), K5/K9 mutated (lanes 3 and 4) or K5 mutated (lanes 7 and 8) H2A. HAT reactions were subjected to immunoblot analysis with antibodies recognizing acetylated K5 or K9 of H2A. (D) Significant effect of H2A acetylation on H2A.Z exchange. In vitro exchange assay was identical to Figure 4B, but using nucleosomes containing intact (lanes 1–6), K5/K9 mutated (lanes 7–9) or K5 mutated (lanes 10–12) H2A. Again, histone H3 was used as loading control.
TIP48 and TIP49 are catalytic subunits of the small complex
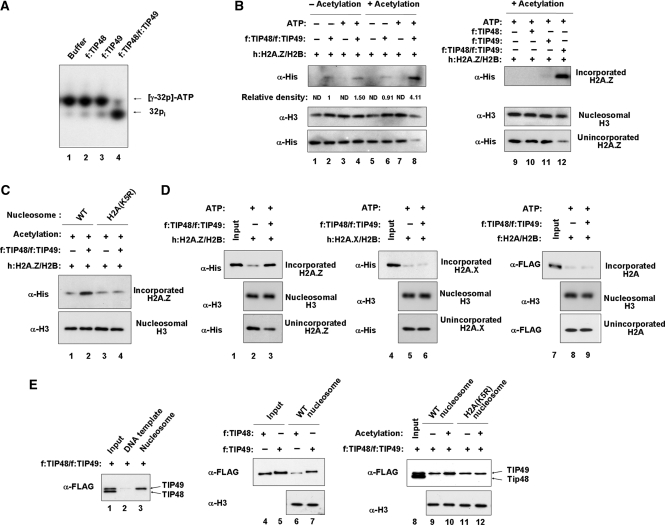
Although the exchange assays described above emphasize the specific role of the small complex in acetylation-facilitated H2A.Z exchange, it is unclear which factors within the complex are mainly responsible for the exchange reaction. Because ATP hydrolysis is necessary for the small complex-induced H2A.Z exchange (Figure 3F) and because TIP48 and TIP49 are the only ATPase activities of the small complex (Figure 2C), we speculated that TIP48 and TIP49 in the small complex are crucial for the acetylation-facilitated exchange reaction. We first conducted ATPase assays with recombinant TIP48 and TIP49 which were expressed in bacteria and purified using their FLAG epitope tags (Supplementary Figure S6A). In agreement with previous studies (45), when either TIP48 or TIP49 was incubated with ATP, they were unable to show catalytic activity for ATP hydrolysis (Figure 6A, lanes 2 and 3). However, when we used TIP48/TIP49 complex that was reconstituted and fractionated by glycerol gradient centrifugation (http://nar.oxfordjournals.org/cgi/content/full/gkp660/DC1B), the complex showed a strong ATPase activity (Figure 5A, lane 4), confirming that both proteins are required for ATP hydrolysis.
Figure 6.
Requirement of TIP48/TIP49 for H2A.Z deposition into chromatin in vivo. (A) Knockdown of TIP49 using TIP49 siRNA. 293T cells were transfected with TIP49-targeted siRNA (Si-Tip49) or negative control siRNA (Si-NC), and repression of TIP49 expression was confirmed by RT–qPCR and western blotting. The mRNA levels of TIP49 were normalized to the β-actin mRNA, which was unaffected by TIP49 siRNA transfection. Whole-cell lysates of the cells transfected with TIP49 or negative control siRNA were analyzed by immunoblot with anti-TIP49 and anti-actin antibodies. Error bars indicate the mean ± SD of results from three independent experiments. (B) Repressive effect of TIP49 knockdown on H2A.Z incorporation. 293T cells were initially depleted TIP49 for 48 h and transfected with FLAG-H2A.Z (f:H2A.Z, Lanes 1 and 2), FLAG-H2A.X (f:H2A.X, lanes 3 and 4) or FLAG-H2A (f:H2A, lanes 5 and 6) for 48 h. Chromatin was isolated from the transfected cells, and the amount of chromatin was normalized by western blot analysis of nucleosomal histone H3 (lower panel). Nucleosomal incorporations of f:H2A.Z, f:H2A.X and f:H2A were analyzed by western blot analysis with anti-FLAG antibody (α-FLAG). Results are representative of three independent experiments. (C) TIP48/TIP49-dependent incorporation of H2A.Z at the p21 promoter region. ChIP analysis of the p21 promoter was performed as described in ‘Materials and Methods’ section. Lines (A–H) under the diagram of p21 promoter region indicate segments used for qPCR. Error bars indicate the mean ± SD of results from three independent experiments. The white boxes and black box indicate two known p53 response elements (p53RE1 and p53RE2) and TATA box, respectively. (D) Summary of TIP48/TIP49-induced H2A.Z exchange. TIP60 is recruited to target gene promoters for localized acetylation of histone H2A at K5, and this acetylation mark acts as a signal for the recruitment of TIP48 and TIP49. TIP48 and TIP49 then facilitate promoter-targeted exchange of H2A.Z to establish platform that would favor the action of chromatin remodeling factors to regulate transcription.
Figure 5.
Exchange of H2A.Z-H2B dimer by TIP48 and TIP49. (A) ATPase activity of recombinant TIP48 and TIP49. ATPase assay was performed with recombinant TIP48/TIP49 and [γ-32P] ATP, and ATP hydrolysis was examined by 12% polyacrylamide gel (19:1) containing 7 M urea in a 1× TBE buffer. Lane 1, buffer control; lane2, recombinant TIP48; lane 3, recombinant TIP49; lane 4, recombinant TIP48/TIP49 complex. (B) H2A.Z exchange by TIP48, TIP49 or TIP48/TIP49 complexes in vitro. H2A.Z exchange activity of recombinant TIP48 and/or TIP49 was checked by using unmodified (lanes 1–4) or pre-acetylated (lanes 5–12) nucleosomes in the presence (lanes 3, 4, 7 and 8–12) or absence (lanes 1, 2, 5 and 6) of ATP as described in ‘Materials and Methods’ section. Nucleosomal incorporation of His-H2A.Z was analyzed by western blot analysis with anti-His antibody (α-His, upper panel). Free unincorporated H2A.Z was determined by western blot analysis of the supernatant (25%) with anti-His antibody (α-His, lower panel). Nucleosomal H3 was used as an internal loading control. Results are representative of three independent experiments. Lanes 1, 3, 5, 7 and 9–12 are reactions without TIP48 and/or TIP49. (C) Effect of H2A acetylation on H2A.Z exchange by TIP48/TIP49 complexes. In vitro exchange assay was identical to lanes 7–8 in Figure 6B, but using nucleosomes containing intact (lanes 1 and 2) or K5 mutated (lanes 3 and 4) H2A. Nucleosomal incorporation of His-H2A.Z was analyzed by western blot analysis with anti-His antibody (α-His). Nucleosomal H3 (α-H3) was used as an internal loading control. (D) Specific action of TIP48/TIP49 complex on H2A.Z exchange. In vitro exchange assays were performed with acetylated nucleosomes as in Figure 6B, using h:H2A.Z/H2B dimer (lanes 2 and 3), h:H2A.X/H2B dimer (lanes 5 and 6) and f:H2A/H2B dimer (lanes 8 and 9). Nucleosomal incorporation of dimers was analyzed by western blot analysis with the indicated antibodies (upper panel). Free unincorporated dimers were determined by western blot analysis of the supernatant (25%) with the indicated antibodies (lower panel). Nucleosomal H3 was used as an internal loading control (second panel). Results are representative of three independent experiments. Lanes 1, 4 and 7, input (25%) of free dimers. (E) Positive effect of acetylation of H2A-K5 on interaction of TIP49 with nucleosome. The unmodified (lanes 3, 6, 7, 9 and 11) or acetylated (lanes 10 and 12) nucleosomes containing wild type (lanes 6, 7, 9 and 10) or K5 mutated (lanes 11 and 12) H2A were immobilized on magnetic beads and incubated with FLAG-TIP48 (lane 6), FLAG-TIP49 (lane 7) or FLAG-TIP48/TIP49 complex (lanes 9–12). The interaction of TIP48/TIP49 with nucleosomes was analyzed by immunoblot with anti-FLAG antibody. The immobilized 5′ biotinylated 207 bp DNA fragments containing 601 nucleosome positioning sequence (lane 2) was also included to determine relative DNA binding affinity of TIP48/TIP49. Histone H3 was used as a loading control. Results are representative of three independent experiments. Lane 1, input of FLAG-TIP48; lane 2, input of FLAG-TIP49; lane 5, input of FLAG-TIP48/TIP49 complex.
We next prepared free H2A.Z-H2B dimers (Supplementary Figure S6C) and examined the H2A.Z exchange following their incubation with the immobilized nucleosomes in the presence of TIP48 and/or TIP49. Significantly, we found that TIP48 and TIP49 can closely recapitulate the action of the small complex in the H2A.Z exchange reaction by facilitating incorporation of H2A.Z-H2B dimers into acetylated nucleosomes (Figure 5B, lane 8). Addition of a nonhydrolyzable ATP analog ATP-γ-S reduced H2A.Z exchange activity of TIP48/TIP49 complex to the background level (Supplementary Figure S7, lane 4). Parallel exchange assays with TIP48 or TIP49 alone failed to show nucleosomal incorporation of H2A.Z-H2B dimers (Figure 5B, lanes 10 and 11), indicating that they play a combinatorial role in H2A.Z exchange. The finding that the stimulatory effect of pre-acetylation was lost upon mutation of H2A-K5 confirms a requirement of the modification, per se (Figure 5C, lane 4). To assess the specificity of action of TIP48 and TIP49, we also extended exchange assays to canonical H2A and another H2A variant H2A.X (Supplementary Figure S6C). In sharp contrast, TIP48 and TIP49 were unable to facilitate incorporations of H2A.X-H2B and H2A–H2B dimers into nucleosomes (Figure 5D), supporting highly selective action of TIP48 and TIP49 for H2A.Z exchange. Hence, we conclude that TIP48 and TIP49 in the small complex are sufficient to catalyze acetylation-facilitated incorporation of H2A.Z into the nucleosome.
Acetylation of H2A-K5 stabilizes the nucleosome-TIP49 interaction
While our results emphasize the critical role of H2A-K5 acetylation in H2A.Z-H2B dimer integration into nucleosomes, how this acetylation facilitates H2A.Z exchange reaction is unclear. One possibility is that K5 acetylation positively affects interaction of the nucleosome with TIP48/TIP49, which in turn allows a stable action of TIP48/TIP49 during H2A.Z exchange process. Thus, we analyzed the ability of TIP48 or TIP49 to interact with a fixed concentration of the H2A nucleosome immobilized to magnetic beads. As shown in Figure 5E, TIP48 and TIP49 both showed direct interactions with immobilized nucleosomes, although TIP49 appears to have much higher affinity to nucleosomes (lanes 6 and 7). To investigate cooperative binding of TIP48 and TIP49 to nucleosomes, we conducted identical binding assays with TIP48/TIP49 complex. As expected from individual binding assays, TIP49 was able to bind to nucleosomes, but somewhat surprisingly, TIP48 did not show any detectable binding to the nucleosome (lane 3). A possible explanation of these results is that TIP48 and TIP49 both bind to the same surface of the nucleosome. Thus, TIP48 interaction is less stable in the presence of its binding partner TIP49, which shows high affinity binding to nucleosomes. These results also hint at a transient action of TIP48 onto the TIP49-bound nucleosome in H2A.Z exchange reaction. Interestingly, the immobilized 601 DNA template also showed no detectable binding of TIP48 and TIP49 (lane 2), suggesting that TIP48 and TIP49 recognize unique platforms within the structural context of the nucleosome. Given the demonstrated role of pre-acetylation in H2A.Z exchange, we next assessed the interaction of TIP48/TIP49 following TIP60-mediated acetylation of nucleosomes. Significantly, there was an apparent increase in TIP49-nucleosome interaction after pre-acetylation of nucleosomes (lane 9 versus lane 10). Moreover, similar binding assays with nucleosomes bearing K5 mutant form of H2A failed to show stimulatory effects of TIP60 in TIP49-nucleosome interaction (lane 11 versus lane 12), indicating that H2A-K5 acetylation distinctly regulates TIP49 recruitment. Again, no stable interaction was observed for TIP48 in the reactions (lanes 9–12). These binding experiments were repeated three times and reproducibly showed at least 2-fold increase in TIP49-nucleosome interaction upon H2A-K5 acetylation. Taken together, these results suggest that acetylation of nucleosomal H2A at K5 is critical for a stable association of TIP49 during TIP48/TIP49-mediated H2A exchange process.
TIP48 and TIP49 are required for H2A.Z deposition into chromatin in vivo
To assess the in vivo relevance of our in vitro results, we asked whether H2A.Z exchange is altered when endogenous TIP48 and TIP49 are down-regulated using the siRNA approach. Since TIP48 or TIP49 alone has no ATPase activity and both proteins are required for H2A.Z exchange (Figure 5B), we knocked down only TIP49 to check the catalytic activity of TIP48/TIP49 complex for H2A.Z exchange. As confirmed by real-time PCR analysis, the transfection of cells with TIP49 siRNA suppressed TIP49 mRNA expression to levels equivalent to <30% of that observed in cells transfected with control siRNA (Figure 6A, left panel). Western blot analysis with anti-TIP49 antibody also verified a significant decrease in TIP49 expression, but no change in Actin expression, after TIP49 siRNA treatment (Figure 6A, right panel), confirming the specificity of siRNA-induced repression of TIP49. After 48 h depletion of TIP49, cells were transfected with FLAG-H2A.Z, -H2A.X or -H2A for another 48 h, and chromatin was isolated from cell nuclei. Our western blot analysis of chromatin prepared from cells transfected with the control siRNA confirmed the incorporation of ectopic H2A.Z into cellular nucleosomes (Figure 6B, lane 1). However, the nucleosomal incorporation of ectopic H2A.Z was significantly decreased in cells transfected with TIP49 siRNA (lane 2). Similar assays with ectopic H2A.X and H2A did not show any effect of TIP49 siRNA on their incorporation into cellular nucleosomes (lanes 3–6), underscoring the specific action of TIP48/TIP49 for H2A.Z exchange in living cells.
As H2A.Z is known to be localized at the p21 promoter (32), we next checked whether TIP48 and TIP49 are required for H2A.Z incorporation into this promoter region by ChIP analysis after siRNA-mediated depletion of cellular TIP49. Consistent with recent results (32), H2A.Z was enriched at the distal p53 response element as well as the TATA transcription initiator region, albeit a higher level of H2A.Z was detected in the distal response region (Figure 6C). However, in a parallel ChIP analysis after treatment of cells with TIP49 siRNA, a significant reduction of H2A.Z peaks at both p53 response element and transcription initiator regions was observed (Figure 6C). The entire experiment was repeated with a different siRNA and similar results were obtained (Supplementary Figure S8). Thus, although other factors may also contribute to cellular H2A.Z exchange, these results confirm that TIP49 is essential for cellular deposition of H2A.Z into distinct chromatin regions.
DISCUSSION
Recent studies in yeast revealed that H2A.Z can be incorporated into chromatin as H2A.Z-H2B dimers by the SWR1 complex, many subunits of which are homologous to the subunits of INO80 complex. The cellular requirement of SWR1 in H2A.Z exchange was further confirmed by gene expression studies that showed a considerable overlap of genes regulated by H2A.Z and SWR1 (26). In human cells, SRCAP and p400 ATP-dependent chromatin remodeling complexes were identified as a counterpart of yeast SWR1 and found to catalyze H2A.Z exchange reaction (31,32). However, these complexes seem mutually exclusive and exert their actions in distinct chromatin regions during specific cellular processes, as exemplified by p21 gene-specific action of p400 (32). Such a targeted action would predict that there might be additional activities and mechanisms to regulate the incorporation of H2A.Z into the nucleosome.
In this study, we have purified H2A.Z-containing complexes from a human cell line to dissect potential molecular mechanisms employed for the deposition of histone variant H2A.Z into the nucleosome. Taking advantage of our established purification method, we demonstrated that free H2A.Z can form two distinct complexes, which we named small and big complexes. The big complex consists of most components of SRCAP and TIP60 complexes, whereas the small complex contains only several components of SRCAP and TIP60 complexes. In comparison with a recent study indicating that free H2A.Z is associated primarily with SRCAP complex (30,31), it is somewhat surprising to find the TIP60 components in our purification. These differences may reflect the use of different cell types (HeLa versus 293/FRT cells) and the differences in nuclear extract preparation and purification procedures. That TIP60 components are bona fide subunits of the H2A.Z-containing big complex is further supported by (i) their copurification with H2A.Z via ion-exchange chromatography and immunoaffinity purification steps and (ii) their co-sedimentation with H2A.Z and SRCAP components of the purified big complex on glycerol gradients.
In accord with recent studies with SRCAP and p400, the big complex containing these activities facilitated incorporation of H2A.Z into the nucleosome. Since the small complex lacks SRCAP and p400, one would expect that the complex does not have function in H2A.Z incorporation. However, our exchange assays clearly indicate that the small complex can also exchange H2A.Z into the nucleosome by replacing canonical H2A. Importantly, a more prominent feature of the small complex is derived from our finding that TIP60-mediated acetylation of the nucleosomal H2A stimulates the action of the small complex, but not that of the big complex, in H2A.Z exchange reaction. This observation significantly enhances our understanding of the mechanism by which H2A.Z exchange process is regulated through epigenetic alterations in target nucleosomes. In fact, previous studies have also shown that the H2A.Z incorporation into specific genes or loci in yeast was facilitated by acetylation of H3 and H4 (12,46–48). Moreover, since other modifications, especially H3/H4 methylation, were also enriched in H2A.Z nucleosomes (49,50), it is not unlikely that these modifications represent another mechanism of regulation of H2A.Z exchange reaction. Bearing this possibility in mind, we may be able to take advantage of our in vitro assay system to identify other epigenetic marks and mechanisms of their action in the H2A.Z exchange pathway.
In search of which factors in the small complex play a major role in H2A acetylation-facilitated incorporation of H2A.Z, we focused on TIP48 and TIP49, since they are the only subunits with ATPase activity in the small complex. Consistent with published results (45), our ATPase assays with recombinant TIP48 and TIP49 clearly showed the requirement of both proteins for ATP hydrolysis. Further analysis of H2A.Z exchange revealed that TIP48 and TIP49 are the catalytic activity in the small complex responsible for H2A.Z exchange reaction. Although there have been reports linking TIP48/TIP49 to chromatin remodeling processes (51,52), these results provide the first direct connection between TIP48/TIP49 and histone variant exchange. There was no observable effect of TIP48/TIP49 on the incorporation of canonical histone H2A and histone variant H2A.X (Figure 5D), indicating H2A.Z-specific action of TIP48/TIP49. Moreover, the requirement of TIP48 and TIP49 in cellular H2A.Z exchange was supported by apparent decrease of the incorporation of ectopic H2A.Z into chromatin after RNAi-mediated depletion of TIP49 (Figure 6). Parallel ChIP experiments showed that TIP49 knockdown impaired H2A.Z deposition at the p21 promoter, specifically at the p53 response element and transcription initiator regions. Thus, although we cannot strictly rule out the possibility that SRCAP and p400 act in parallel on H2A.Z exchange pathways, such an impairment of H2A.Z exchange at a specific chromatin region upon TIP49 knockdown is indicative of the critical targeted action of TIP48 and TIP49 on H2A.Z exchange.
It should be noted that both small and big complexes possess TIP48 and TIP49, but acetylation of nucleosomal H2A promotes only small complex-induced incorporation of H2A.Z. Although the reason for these results is not clear at present, one possibility is that some proteins in big complex inhibit the activity of one or both of TIP48 and TIP49, causing H2A.Z exchange to be dependent solely on SRCAP and p400 ATPase activities. This negative regulator might have crucial functions for economizing the action of TIP48/TIP49 on the cellular H2A.Z exchange process. In agreement with this idea, the initial H2A.Z complex, albeit consisting of small and big complexes, induced only moderate transfer of H2A.Z-H2B dimers into pre-acetylated nucleosomes (data not shown). Further analysis, such as identification of these repressive factors and their functional interaction with TIP48/TIP49, would be helpful for understanding the mechanisms of regulation of H2A.Z exchange.
The most striking finding from our studies was the identification of H2A-K5 acetylation as a key regulator for the action of TIP48/TIP49 in the H2A.Z exchange process. Although there have been reports linking histone acetylation to H2A.Z exchange, our use of nucleosomes reconstituted with wild type and mutant recombinant histones allowed us to provide the first direct connection between H2A.Z exchange and site-specific H2A acetylation. An immediate question raised from these findings was how TIP60-mediated acetylation of nucleosomal H2A acts as a positive regulator for H2A.Z exchange. There are two potential explanations for the effect of H2A acetylation. One possibility implies that H2A acetylation destabilizes nucleosome structure by reducing the affinity of H2A-H2B dimer for the H3-H4 tetramer within the nucleosome and/or inducing a conformational change of the nucleosome. However, our structural analyses of unmodified and acetylated nuclesomes did not allow us to reveal any differences in their structural characteristics (data not shown). Another possibility is that H2A acetylation positively influences the association of TIP48/TIP49 with the N-terminal tail of histone H2A, thereby increasing the affinity of TIP48/TIP49 to the target nucleosome. An intriguing observation we made in this regard is that TIP49 preferentially interacts with nucleosome containing K5-acetylated H2A (Figure 5E). This result strongly suggests that H2A-K5 acetylation contributes to the action of TIP48/TIP49 during H2A.Z exchange process by stabilizing TIP49 interaction with nucleosomes. In addition, several recent studies have suggested that H2A.Z is specifically localized at gene promoter regions to exert its influence on transcription (53,54). A possible explanation for this promoter-targeted incorporation of H2A.Z is that recruitment of TIP60 (or other HATs) to gene promoters through its interaction with DNA binding factors results in increased level of H2A acetylation and H2A.Z exchange.
Thus, taking our in vitro and in vivo observations together with recent studies, we propose the following model for how TIP48/TIP49 ATPase activities and TIP60-mediated H2A acetylation promote H2A.Z exchange (Figure 6D). In the initial step, TIP60 is recruited to specific gene promoter regions likely through its interaction with sequence-specific DNA-binding factors (such as p53). Once recruited, TIP60 acetylates nucleosomal H2A localized in target promoters. This promoter-targeted H2A acetylation will in turn function as a marker to direct TIP48/TIP49-induced deposition of H2A.Z into the promoter regions. In this way, while TIP48/TIP49, SRCAP and p400 all have an intrinsic ability to exchange H2A.Z, TIP48/TIP49 can adopt a highly specific strategy for localizing H2A.Z at specific chromatin regions. It is also worth noting that TIP48 and TIP49 have been shown to interact with the RNA Polymerase II as well as transcription factors/coregulators (TBP, ATF2 and c-myc) (55–57). Thus, interaction of TIP48/TIP49 with transcription-related factors might also play a role in their initial localization to acetylated promoter regions. Further investigations of whether different genes employ different epigenetic signals and/or factors for H2A.Z deposition will be an exciting challenge for us in the future.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Margaret E. Early Medical Research Grant; the James H. Zumberge Research Grant; the National Institutes of Health (R01GM84209). Funding for open access charge: National Institutes of Health (R01GM84209).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank K. Luger (Colorado State University) for histone expression vectors, H. Willard (Duke University) for H2A.Z cDNA, D. Rhodes (MRC Laboratory of Molecular Biology) for DNA template containing 207 bp 601 nucleosome positioning sequence, and R. G. Roeder (The Rockefeller University) for anti-DMAP1, anti-BAF53 and anti-GAS41 antibodies. We also thank M. R. Stallcup for critical reading and helpful comments on the manuscript, and members of the An laboratory for instructive discussions and valuable suggestions.
REFERENCES
- 1.Harp JM, Hanson BL, Timm DE, Bunick GJ. Asymmetries in the nucleosome core particle at 2.5 A resolution. Acta Crystallogr. D. Biol. Crystallogr. 2000;56:1513–1534. doi: 10.1107/s0907444900011847. [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 3.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat. Rev. Genet. 2008;9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- 5.Wang GG, Allis CD, Chi P. Chromatin remodeling and cancer, Part II: ATP-dependent chromatin remodeling. Trends Mol. Med. 2007;13:373–380. doi: 10.1016/j.molmed.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamakaka RT, Biggins S. Histone variants: deviants? Genes Dev. 2005;19:295–310. doi: 10.1101/gad.1272805. [DOI] [PubMed] [Google Scholar]
- 7.Zlatanova J, Thakar A. H2A.Z: view from the top. Structure. 2008;16:166–179. doi: 10.1016/j.str.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 8.Faast R, Thonglairoam V, Schulz TC, Beall J, Wells JR, Taylor H, Matthaei K, Rathjen PD, Tremethick DJ, Lyons I. Histone variant H2A.Z is required for early mammalian development. Curr. Biol. 2001;11:1183–1187. doi: 10.1016/s0960-9822(01)00329-3. [DOI] [PubMed] [Google Scholar]
- 9.Clarkson MJ, Wells JR, Gibson F, Saint R, Tremethick DJ. Regions of variant histone His2AvD required for Drosophila development. Nature. 1999;399:694–697. doi: 10.1038/21436. [DOI] [PubMed] [Google Scholar]
- 10.Liu X, Li B, Gorovsky MA. Essential and nonessential histone H2A variants in Tetrahymena thermophila. Mol. Cell Biol. 1996;16:4305–4311. doi: 10.1128/mcb.16.8.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suto RK, Clarkson MJ, Tremethick DJ, Luger K. Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat. Struct. Biol. 2000;7:1121–1124. doi: 10.1038/81971. [DOI] [PubMed] [Google Scholar]
- 12.Zhang H, Roberts DN, Cairns BR. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell. 2005;123:219–231. doi: 10.1016/j.cell.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flaus A, Rencurel C, Ferreira H, Wiechens N, Owen-Hughes T. Sin mutations alter inherent nucleosome mobility. EMBO J. 2004;23:343–353. doi: 10.1038/sj.emboj.7600047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Abbott DW, Ivanova VS, Wang X, Bonner WM, Ausio J. Characterization of the stability and folding of H2A.Z chromatin particles: implications for transcriptional activation. J. Biol. Chem. 2001;276:41945–41949. doi: 10.1074/jbc.M108217200. [DOI] [PubMed] [Google Scholar]
- 15.Park YJ, Dyer PN, Tremethick DJ, Luger K. A new fluorescence resonance energy transfer approach demonstrates that the histone variant H2AZ stabilizes the histone octamer within the nucleosome. J. Biol. Chem. 2004;279:24274–24282. doi: 10.1074/jbc.M313152200. [DOI] [PubMed] [Google Scholar]
- 16.Thambirajah AA, Dryhurst D, Ishibashi T, Li A, Maffey AH, Ausio J. H2A.Z stabilizes chromatin in a way that is dependent on core histone acetylation. J. Biol. Chem. 2006;281:20036–20044. doi: 10.1074/jbc.M601975200. [DOI] [PubMed] [Google Scholar]
- 17.Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5' ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guillemette B, Bataille AR, Gevry N, Adam M, Blanchette M, Robert F, Gaudreau L. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keogh MC, Mennella TA, Sawa C, Berthelet S, Krogan NJ, Wolek A, Podolny V, Carpenter LR, Greenblatt JF, Baetz K, et al. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 2006;20:660–665. doi: 10.1101/gad.1388106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Millar CB, Xu F, Zhang K, Grunstein M. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 2006;20:711–722. doi: 10.1101/gad.1395506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Babiarz JE, Halley JE, Rine J. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev. 2006;20:700–710. doi: 10.1101/gad.1386306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fan JY, Rangasamy D, Luger K, Tremethick DJ. H2A.Z alters the nucleosome surface to promote HP1alpha-mediated chromatin fiber folding. Mol. Cell. 2004;16:655–661. doi: 10.1016/j.molcel.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 23.Adam M, Robert F, Larochelle M, Gaudreau L. H2A.Z is required for global chromatin integrity and for recruitment of RNA polymerase II under specific conditions. Mol. Cell Biol. 2001;21:6270–6279. doi: 10.1128/MCB.21.18.6270-6279.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santisteban MS, Kalashnikova T, Smith MM. Histone H2A.Z regulats transcription and is partially redundant with nucleosome remodeling complexes. Cell. 2000;103:411–422. doi: 10.1016/s0092-8674(00)00133-1. [DOI] [PubMed] [Google Scholar]
- 25.Farris SD, Rubio ED, Moon JJ, Gombert WM, Nelson BH, Krumm A. Transcription-induced chromatin remodeling at the c-myc gene involves the local exchange of histone H2A.Z. J. Biol. Chem. 2005;280:25298–25303. doi: 10.1074/jbc.M501784200. [DOI] [PubMed] [Google Scholar]
- 26.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 27.Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol. Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 28.Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J. A protein complex containing the conserved Swi2/Snf2-related ATPase Swr1p deposits histone variant H2A.Z into euchromatin. PLoS Biol. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kusch T, Florens L, Macdonald WH, Swanson SK, Glaser RL, Yates JR, 3rd, Abmayr SM, Washburn MP, Workman JL. Acetylation by Tip60 is required for selective histone variant exchange at DNA lesions. Science. 2004;306:2084–2087. doi: 10.1126/science.1103455. [DOI] [PubMed] [Google Scholar]
- 30.Cai Y, Jin J, Florens L, Swanson SK, Kusch T, Li B, Workman JL, Washburn MP, Conaway RC, Conaway JW. The mammalian YL1 protein is a shared subunit of the TRRAP/TIP60 histone acetyltransferase and SRCAP complexes. J. Biol. Chem. 2005;280:13665–13670. doi: 10.1074/jbc.M500001200. [DOI] [PubMed] [Google Scholar]
- 31.Ruhl DD, Jin J, Cai Y, Swanson S, Florens L, Washburn MP, Conaway RC, Conaway JW, Chrivia JC. Purification of a human SRCAP complex that remodels chromatin by incorporating the histone variant H2A.Z into nucleosomes. Biochemistry. 2006;45:5671–5677. doi: 10.1021/bi060043d. [DOI] [PubMed] [Google Scholar]
- 32.Gevry N, Chan HM, Laflamme L, Livingston DM, Gaudreau L. p21 transcription is regulated by differential localization of histone H2A.Z. Genes Dev. 2007;21:1869–1881. doi: 10.1101/gad.1545707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choi J, Kim B, Heo K, Kim K, Kim H, Zhan Y, Ranish JA, An W. Purification and characterization of cellular proteins associated with histone H4 tails. J. Biol. Chem. 2007;282:21024–21031. doi: 10.1074/jbc.M703883200. [DOI] [PubMed] [Google Scholar]
- 34.An W, Roeder RG. Reconstitution and transcriptional analysis of chromatin in vitro. Methods Enzymol. 2004;377:460–474. doi: 10.1016/S0076-6879(03)77030-X. [DOI] [PubMed] [Google Scholar]
- 35.Dyer PN, Edayathumangalam RS, White CL, Bao Y, Chakravarthy S, Muthurajan UM, Luger K. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 36.Heo K, Kim H, Choi SH, Choi J, Kim K, Gu J, Lieber MR, Yang AS, An W. FACT-mediated exchange of histone variant H2AX regulated by phosphorylation of H2AX and ADP-ribosylation of Spt16. Mol. Cell. 2008;30:86–97. doi: 10.1016/j.molcel.2008.02.029. [DOI] [PubMed] [Google Scholar]
- 37.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 38.Robinson PJ, Fairall L, Huynh VA, Rhodes D. EM measurements define the dimensions of the “30-nm” chromatin fiber: evidence for a compact, interdigitated structure. Proc. Natl Acad. Sci. USA. 2006;103:6506–6511. doi: 10.1073/pnas.0601212103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hamiche A, Sandaltzopoulos R, Gdula DA, Wu C. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell. 1999;97:833–842. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- 40.Dignam JD. Preparation of extracts from higher eukaryotes. Methods Enzymol. 1990;182:194–203. doi: 10.1016/0076-6879(90)82017-v. [DOI] [PubMed] [Google Scholar]
- 41.Wysocka J, Reilly PT, Herr W. Loss of HCF-1-chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol. Cell Biol. 2001;21:3820–3829. doi: 10.1128/MCB.21.11.3820-3829.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saramaki A, Banwell CM, Campbell MJ, Carlberg C. Regulation of the human p21(waf1/cip1) gene promoter via multiple binding sites for p53 and the vitamin D3 receptor. Nucleic Acids Res. 2006;34:543–554. doi: 10.1093/nar/gkj460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamamoto T, Horikoshi M. Novel substrate specificity of the histone acetyltransferase activity of HIV-1-Tat interactive protein Tip60. J. Biol. Chem. 1997;272:30595–30598. doi: 10.1074/jbc.272.49.30595. [DOI] [PubMed] [Google Scholar]
- 44.Ikura T, Ogryzko VV, Grigoriev M, Groisman R, Wang J, Horikoshi M, Scully R, Qin J, Nakatani Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell. 2000;102:463–473. doi: 10.1016/s0092-8674(00)00051-9. [DOI] [PubMed] [Google Scholar]
- 45.Puri T, Wendler P, Sigala B, Saibil H, Tsaneva IR. Dodecameric structure and ATPase activity of the human TIP48/TIP49 complex. J. Mol. Biol. 2007;366:179–192. doi: 10.1016/j.jmb.2006.11.030. [DOI] [PubMed] [Google Scholar]
- 46.Durant M, Pugh BF. NuA4-directed chromatin transactions throughout the Saccharomyces cerevisiae genome. Mol. Cell Biol. 2007;27:5327–5335. doi: 10.1128/MCB.00468-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shia WJ, Li B, Workman JL. SAS-mediated acetylation of histone H4 Lys 16 is required for H2A.Z incorporation at subtelomeric regions in Saccharomyces cerevisiae. Genes Dev. 2006;20:2507–2512. doi: 10.1101/gad.1439206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Auger A, Galarneau L, Altaf M, Nourani A, Doyon Y, Utley RT, Cronier D, Allard S, Cote J. Eaf1 is the platform for NuA4 molecular assembly that evolutionarily links chromatin acetylation to ATP-dependent exchange of histone H2A variants. Mol. Cell Biol. 2008;28:2257–2270. doi: 10.1128/MCB.01755-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Viens A, Mechold U, Brouillard F, Gilbert C, Leclerc P, Ogryzko V. Analysis of human histone H2AZ deposition in vivo argues against its direct role in epigenetic templating mechanisms. Mol. Cell Biol. 2006;26:5325–5335. doi: 10.1128/MCB.00584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarcinella E, Zuzarte PC, Lau PN, Draker R, Cheung P. Monoubiquitylation of H2A.Z distinguishes its association with euchromatin or facultative heterochromatin. Mol. Cell Biol. 2007;27:6457–6468. doi: 10.1128/MCB.00241-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Diop SB, Bertaux K, Vasanthi D, Sarkeshik A, Goirand B, Aragnol D, Tolwinski NS, Cole MD, Pradel J, Yates J.R., 3rd, et al. Reptin and Pontin function antagonistically with PcG and TrxG complexes to mediate Hox gene control. EMBO Rep. 2008;9:260–266. doi: 10.1038/embor.2008.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gallant P. Control of transcription by Pontin and Reptin. Trends Cell Biol. 2007;17:187–192. doi: 10.1016/j.tcb.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 53.Brickner DG, Cajigas I, Fondufe-Mittendorf Y, Ahmed S, Lee PC, Widom J, Brickner JH. H2A.Z-mediated localization of genes at the nuclear periphery confers epigenetic memory of previous transcriptional state. PLoS Biol. 2007;5:e81. doi: 10.1371/journal.pbio.0050081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zanton SJ, Pugh BF. Full and partial genome-wide assembly and disassembly of the yeast transcription machinery in response to heat shock. Genes Dev. 2006;20:2250–2265. doi: 10.1101/gad.1437506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cho SG, Bhoumik A, Broday L, Ivanov V, Rosenstein B, Ronai Z. TIP49b, a regulator of activating transcription factor 2 response to stress and DNA damage. Mol. Cell Biol. 2001;21:8398–8413. doi: 10.1128/MCB.21.24.8398-8413.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wood MA, McMahon SB, Cole MD. An ATPase/helicase complex is an essential cofactor for oncogenic transformation by c-Myc. Mol. Cell. 2000;5:321–330. doi: 10.1016/s1097-2765(00)80427-x. [DOI] [PubMed] [Google Scholar]
- 57.Kanemaki M, Makino Y, Yoshida T, Kishimoto T, Koga A, Yamamoto K, Yamamoto M, Moncollin V, Egly JM, Muramatsu M, et al. Molecular cloning of a rat 49-kDa TBP-interacting protein (TIP49) that is highly homologous to the bacterial RuvB. Biochem. Biophys. Res. Commun. 1997;235:64–68. doi: 10.1006/bbrc.1997.6729. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.