Abstract
Potent antiviral RNAi can be induced by intracellular expression of short hairpin RNAs (shRNAs) and artificial microRNAs (miRNAs). Expression of shRNA and miRNA results in target mRNA degradation (perfect base pairing) or translational repression (partial base pairing). Although efficient inhibition can be obtained, error-prone viruses such as human immunodeficiency virus type 1 (HIV-1) can escape from RNAi-mediated inhibition by mutating the target sequence. Recently, artificial miRNAs have been shown to be potent RNAi inducers due to their efficient processing by the RNAi machinery. Furthermore, miRNAs may be more proficient in suppressing imperfect targets than shRNAs. In this study, we tested the knockdown efficiency of miRNAs and shRNAs against wild-type and RNAi-escape HIV-1 variants with one or two mutations in the target sequence. ShRNAs and miRNAs can significantly inhibit the production of HIV-1 variants with mutated target sequences in the open reading frame. More pronounced mutation-tolerance was measured for targets in the 3′ untranslated region (3′ UTR). Partially complementary sequences within the 3′ UTR of the HIV-1 RNA genome efficiently act as target sites for miRNAs and shRNAs. These data suggest that targeting imperfect target sites by antiviral miRNAs or shRNAs provides an alternative RNAi approach for inhibition of pathogenic viruses.
INTRODUCTION
RNAi against chronic virus infections requires a stable gene therapy to durably protect cells against virus replication (1–3). Such a gene therapy involves vector-mediated RNAi that can be induced by intracellular expression of antiviral microRNA (miRNA) mimics or short hairpin RNAs (shRNAs). miRNA mimics are expressed as primary miRNA transcripts (pri-miRNAs) in the nucleus that are cleaved by the RNAse III-like endonuclease Drosha and its cofactor DGCR8 into precursor miRNAs (pre-miRNAs), which are hairpin RNAs of ∼70 nucleotides (nts) (4). The pre-miRNA is transported to the cytoplasm by Exportin-5 and further processed by the RNAse III-like enzyme Dicer into an imperfect ∼22 nt miRNA duplex (5,6). The single stranded mature miRNA is incorporated into the RNA-induced silencing complex (RISC) and directs the complex to complementary mRNA sequences (7,8) to cause mRNA cleavage or translational repression depending on the complementarity between the miRNA and the mRNA target (7,8). ShRNAs are perfect hairpins of 19–29 bp with a small apical loop and a 3′ UU overhang. Such shRNA constructs are usually expressed in the nucleus from a polymerase III promoter (9,10). The shRNA is translocated to the cytoplasm by Exportin-5 and processed by Dicer into functional 21-nt small interfering RNA duplexes (siRNAs) with 2-nt 3′ overhangs. The siRNA duplex is loaded into RISC. The passenger strand of the siRNA is cleaved, released and degraded (11–13), whereas the guide strand directs RISC to cleave the perfectly complementary mRNA (14).
It has been previously shown by us and others also that potent and specific inhibition of the human immunodeficiency virus type 1 (HIV-1) can be obtained using artificial antiviral miRNAs or shRNAs (15–18). However, prolonged culturing of HIV-1 infected human T cells that express an antiviral shRNA resulted in the emergence of RNAi-escape viruses (19–21). These escape viruses have acquired deletions or point mutations in the target sequence that prevent RNAi-mediated inhibition. Even when highly conserved HIV-1 sequences are targeted in essential open reading frames, the virus can escape by selecting silent codon changes, thus leaving the encoded protein unaltered (22,23). To avoid viral escape, one can use combinatorial RNAi that involves targeting of multiple HIV-1 sequences simultaneously (15,16,24,25). An alternative would be to test whether an escape-proof RNAi-mediated inhibition strategy can be designed, which should remain active on targets with one or two mutations. While shRNAs are usually designed to cleave a perfect complementary mRNA target, miRNAs can translationally repress a target mRNA by pairing with their ‘seed region’ (nucleotide positions 2–8) to multiple imperfect targets in the 3′ UTR (26–30). Recent studies confirmed that cellular miRNAs can inhibit viral gene expression by directing RISC to partially complementary viral sequences (31–33). For example, a set of cellular miRNAs are involved in the establishment of HIV-1 latency by targeting partially complementary sequences in the 3′ UTR of the viral RNA genome (31,34). These miRNAs also appear to determine the non-susceptibility of resting peripheral blood monocytes for a productive HIV-1 infection by establishing a latent provirus (35).
In this study, we tested the inhibitory effect of miRNAs and shRNAs on imperfect HIV-1 target sequences in the open reading frames and 3′ UTR of the mRNAs. We showed that shRNAs and miRNAs can significantly inhibit the production of HIV-1 variants with mutated target sequences in the open reading frame. Furthermore, shRNAs and miRNAs can inhibit multiple imperfect targets in the 3′ UTR of the HIV-1 RNA genome. These data suggest that targeting of partially complementary sequences can provide an alternative RNAi approach for durable inhibition of error-prone viruses.
MATERIALS AND METHODS
DNA constructs
Hairpin RNA constructs used in this study were described previously (15,24). The pLAI plasmid encoding the HIV-1 isolate LAI (36) was used to produce virus upon transfection of the human embryonic kidney (HEK) 293T cells. For the A mutants, a single (8A or 15A) or double (8A and 15A) mutation was introduced in the target sequence of pLAI. For the D mutants, single mutations were introduced at position 6 or 11 and double mutations were introduced at position 8/11 and 13/15 in the target sequence. These mutations have been previously observed in a large-scale HIV-1 escape study (22). Mutations were introduced by fusion PCR using an oligonucleotide containing the mutations.
The previously described firefly luciferase expression vector pGL3-Nef (21) was used to construct the luciferase reporter constructs Luc-I, Luc-II and Luc-III. An HIV-1 fragment of approximately 600, 1000 and 1600 bp was PCR amplified using the full-length molecular clone pLAI as template with primers Luc-I f: GGAATTCATTATCGTTTCAGACCCACCTC and Luc-I r: AACTGCAGGCTGCCTTGTAAGTCATTGGTC; Luc-II f: GGAATTCCACACCTCAGGTACCTTTAAGAC and Luc-II r: AACTGCAGTCACCAGCGTTTCTGGGTGAGC; Luc-I f and Luc-III r. The PCR fragments were purified from agarose gel, digested with EcoRI and PstI and inserted into the corresponding sites of the pGL3-Nef. The luciferase reporter constructs encoding the ACDE wild-type targets ACDEwt and the mutated targets ACDEm1 and ACDEm2 were constructed by annealing oligonucleotides and inserting into the EcoRI and PstI sites of the pGL3 vector. The luciferase reporter ACDEm1 with the ACDE target was altered by the introduction of observed viral escape mutations (22): G8A in target A and D, A6G in target C, and a G6A mutation in target E. Luciferase reporter ACDEm2 encodes the ACDE target with a point mutation at position 15 in each target.
Cell culture and transfection
The HEK 293T cell line was cultured in Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA, USA) containing 10% fetal calf serum (FCS), 100 U/ml penicillin, 100 U/ml streptomycin and minimal essential medium non-essential amino acids (DMEM/10% FCS) at 37°C and 5% CO2. For luciferase and HIV-1 inhibition assays, HEK 293T cells were seeded in 24-well plates at a density of 1.3 × 105 cells per well in 0.5 ml DMEM/10% FCS without antibiotics. The next day, transfections were performed using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions.
Luciferase assays
HEK 293T cells were co-transfected with 100 ng of firefly luciferase reporter plasmids (pGL3; Promega, Madison, WI, USA), 1 ng of renilla luciferase expression plasmid (pRL-CMV) and different amounts of hairpin RNA expression constructs. The renilla plasmid served as an internal control for cell viability and transfection efficiency. To obtain equal DNA concentrations, pBluescript SK− (pBS) (Promega) was added to the transfection mixtures. Two days post-transfection, luciferase and renilla expression were measured with the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's protocol. The relative luciferase activity was calculated as the ratio between the firefly and renilla luciferase expression and corrected for between-session variation (37).
HIV-1 inhibition assays
Inhibition of virus production was determined by co-transfection of 250 ng HIV pLAI, 1 ng pRL-CMV and different amounts of hairpin RNA constructs into HEK 293T cells. We added pBS to obtain equal DNA concentrations in all transfections. Two days post-transfection, the CA-p24 levels in the culture supernatant was determined by ELISA as described previously (38). The transfected cells were lysed and renilla luciferase expression was determined using 10 μl of the lysates with the Renilla Luciferase Assay System (Promega) according to the manufacturer's instructions. Relative virus production was calculated as the ratio between the CA-p24 level and the renilla luciferase activity, and corrected for between-session variation (37).
In silico analysis of miRNA targets in the HIV-1 RNA genome
To screen for putative target sites for miRNAs in the 3′ UTR of the HIV-1 RNA genome, we used the miRanda algorithm. The identification was performed for the sense and antisense sequences of the miRNA constructs A, C, D and E versus the HIV-1 molecular clone LAI. The miRanda scoring matrix used for this analysis allows G–U ‘wobble’ bp, which are important for the accurate detection of RNA : RNA duplexes. Complementarity-scoring for individual positions was: +5 for G–C, +5 for A–U, +2 for G–U and −3 for all other nt pairs. Affine penalties were used for gap-opening (−8) and gap-extension (−2). The threshold was set at a cut-off value of 90. The algorithm applied the following empirical rules (with position 1 defined as the 5′ end of the miRNA): (i) no mismatches at positions 2 to 5; (ii) fewer than five mismatches between positions 3–12; (iii) at least one mismatch between positions 9 and L-5 (where L is total alignment length); and (iv) fewer than two mismatches in the last five positions of the alignment. The analysis of possible miRNA target sites in the resulting candidate sequences were tested with the new rna22 algorithm for miRNA heteroduplex prediction (39).
RESULTS
Inhibition of HIV-1 by miRNA and shRNA
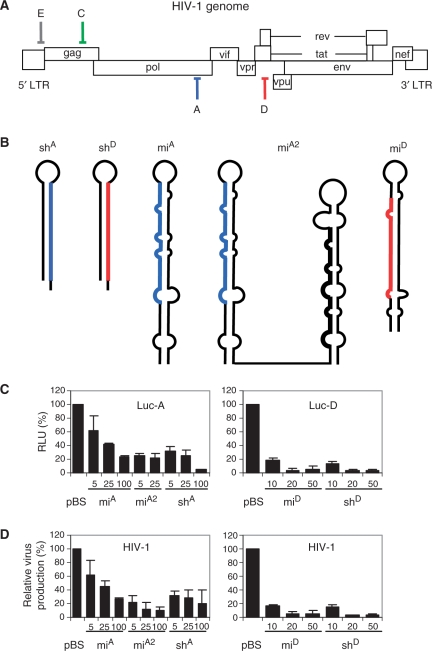
We induced antiviral RNAi with vectors expressing an shRNA or miRNA that target the pol or tat/rev genes of HIV-1 (target A or D in Figure 1A). Both types of constructs were designed to encode the same guide strand RNA. The ∼50 nt shRNAs are characterized by a perfect hairpin stem of 19 or 21 bp and a small loop, whereas the ∼70 nt miRNAs contain multiple mismatches in the hairpin stem (Figure 1B). We previously analysed the shRNA-derived siRNAs and the processed mature miRNAs by Northern blotting and showed that guide RNAs of similar sizes were expressed (15). For the miRNA against the A target, we tested two variations; as single hairpin miA or double hairpin transcript miA2. The miA2 construct encodes miA and an irrelevant miRNA in a single transcript. We first determined the knockdown efficiency of these inhibitors on wild-type, fully complementary mRNA targets in a luciferase reporter (Figure 1C). Luciferase expression in the presence of the pBS control was set at 100%. Profound inhibition was observed for miA and shA on Luc-A. The miA2 construct was previously shown to be more effective than the single miA construct (15), a result that was confirmed in this study (Figure 1C). Inhibition observed in the Luc-D system was even more potent, both for miD and shD. We also tested the ability of the antiviral constructs to inhibit HIV-1 production by co-transfection with the molecular clone pLAI (Figure 1D). Virus production in the presence of pBS was set at 100%. Dose-dependent inhibition of HIV-1 production was observed, and the constructs against target D were again more potent. In general, inhibition by the antiviral shRNAs and miRNAs was equally effective, both with luciferase reporters and in the HIV-1 production assay.
Figure 1.
Inhibition of HIV-1 using antiviral miRNA and shRNA constructs. (A) The HIV-1 genome showing the positions of the target sites for the inhibitors used in this study: A in pol (blue), C in gag (green), D in r/t (red) and E in the untranslated leader (ldr) sequence (grey). (B) Structure of the shRNA and miRNA transcripts. The guide strand siRNA or mature miRNA against HIV-1 is marked in blue (A) and red (D). (C) Inhibition of luciferase expression. HEK 293T cells were co-transfected with 100 ng of the luciferase reporter construct, 1 ng of pRL and the indicated amount of hairpin RNA constructs. Two days post-transfection cells were lysed and the lysates were used to measure firefly and renilla luciferase expression. Normalized luciferase expression in the absence of inhibitor was set at 100%. (D) Inhibition of HIV-1 production. HEK 293T cells were co-transfected with 250 ng HIV-1 molecular clone pLAI, 1 ng pRL and the indicated amount of hairpin RNA constructs. CA-p24 levels in the culture supernatant and renilla luciferase expression in cells were measured at 2 days post-transfection. Normalized CA-p24 expression in the absence of inhibitor was set at 100%. The mean values and SDs were calculated based on seven independent transfections.
Inhibition of RNAi-escape mutants by shRNA and miRNA
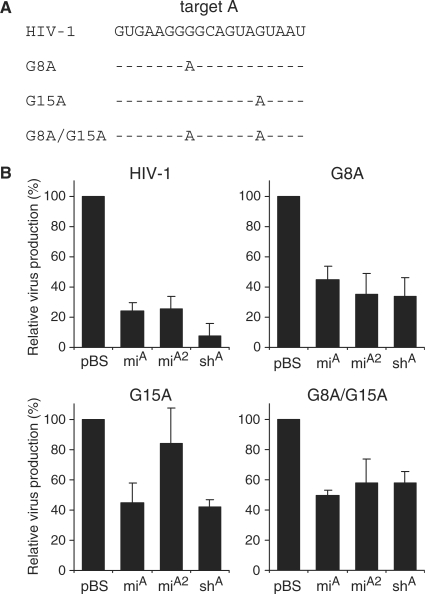
To test the ability of antiviral miRNAs and shRNAs to inhibit targets that are not perfectly complementary, we analyzed the inhibitory effect of miA and shA on the production of HIV-1 RNAi-escape mutants. We introduced point mutations in the HIV-1 genome that were selected in previous virus evolution experiments with the shA inhibitor: G8A, G15A and the double mutant G8A/G15A (22) (Figure 2A). We tested the ability of miA, miA2 and shA to inhibit the mutant viruses, and the wild-type HIV-1 construct was included for comparison (Figure 2B). As expected, we measured potent knockdown of wild-type HIV-1 with miA, miA2 and shA. A significant reduction of 20–50% in knockdown efficiency was observed for all three inhibitors on the two HIV-1 genomes with a point mutation in the target sequence. The knockdown efficiency of miA and shA was still ∼60% for the escape viruses with a single mutation (G8A and G15A) and approximately 40% for the escape virus with two point mutations (G8A/G15A). To test whether the reduction in viral production was due to degradation of HIV-1 mRNAs, we performed Northern blots analyses. We observed a significant reduction in the amount of the full-length 9-kb HIV-1 genomic RNA in the presence of the antiviral miRNA or shRNA (results not shown).
Figure 2.
Inhibition of HIV-1 escape variants by artificial miRNAs. (A) Sequence of the HIV-1 pol target A and RNAi-escape viruses with the mutations G8A, G15A or G8A/G15A. (B) Inhibition of HIV-1 variants by miA, miA2 and shA. HEK 293T cells were co-transfected with 250 ng HIV-1 molecular clone pLAI, 1 ng pRL and 10 ng of the hairpin RNA constructs. CA-p24 levels in the culture supernatant and renilla luciferase expression in cells were measured at 2 days post-transfection. Normalized CA-p24 expression in the presence of pBS was set at 100%. The mean values and SDs were derived from seven independent transfections.
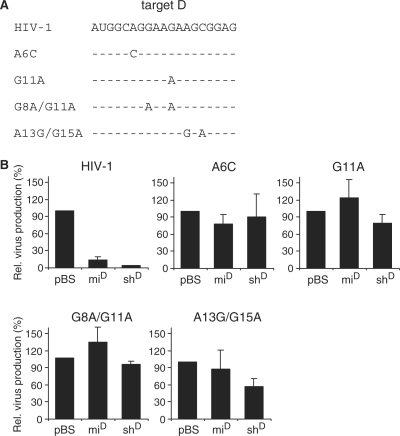
A similar strategy was used for target D, in which we introduced single point mutations (A6C or G11A) or double mutations (G8A/G11A and A13G/G15A) that were detected in previous virus evolution experiments (22) (Figure 3A). Efficient HIV-1 inhibition was scored for miD and shD (Figure 3B). Single and double point mutations in the target sequence resulted in a loss of inhibition. Only shD retained a 40% inhibition efficiency on the A13G/G15A escape virus. Taken together, these results demonstrate that introduction of one or two point mutations in a target can lead to a loss in knockdown efficiency by both the miRNA and the shRNA inhibitors. However, depending on the actual target sequence, still up to 60% inhibition of virus production can be obtained even if the target contains two point mutations. Thus, both shRNAs and artificial miRNA mimics show a variable degree of mutation tolerance when targeting HIV-1 escape viruses.
Figure 3.
Inhibition of HIV-1 escape variants by artificial miRNAs targeting the r/t region. (A) Sequence of the HIV-1 r/t target D and RNAi-escape variants with the mutations A6C, G11A, G8A/G11A or A13G/G15A. (B). Inhibition of mutant HIV-1 variants by miD and shD. HEK 293T cells were co-transfected with 250 ng HIV-1 molecular clone pLAI, 1 ng pRL and 10 ng of the hairpin RNA constructs. CA-p24 levels in the culture supernatant and renilla luciferase expression in cells were measured at 2 days post-transfection. Normalized CA-p24 expression in the presence of pBS was set at 100%. The mean values and SDs are based on four independent experiments.
Knockdown of reporter constructs with multiple imperfect targets in the 3′ UTR
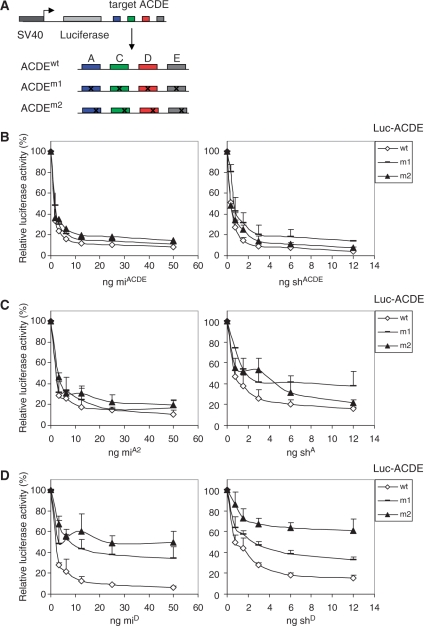
Cellular miRNAs generally regulate gene expression by targeting multiple sites within the 3′ UTR of gene transcripts. Similarly, mutation-tolerant RNAi against HIV-1 may require multiple targets in the 3′ UTR of the transcript. To test this scenario, we constructed luciferase reporters with four 3′ UTR targets: ACDE, each with and without point mutations (Figure 4A). The new targets C and E correspond to HIV-1 gag and untranslated leader (ldr) sequences (Figure 1A). The introduced point mutations mimic the sequence variation observed in RNAi-escape virus variants (22). Each target in ACDEm1 has a single point mutation either at position 6 or 8, which does not affect base pairing with the miRNA ‘seed region’. Each target in ACDEm2 has a point mutation at position 15, which is in the centre of the ‘seed region’. This set of reporters was subsequently tested with the antiviral miRNA polycistron miACDE that encodes four active miRNAs against wild-type HIV-1 (15). For comparison, we used the construct shACDE that encodes four shRNAs against the same targets (24). We titrated the amount of miRNA and shRNA constructs in co-transfection with the ACDEwt reporter in HEK 293T cells to obtain similar inhibitory activities (Figure 4B). The normalized firefly luciferase expression in the absence of inhibitor was set at 100%. We observed potent inhibition of ACDEwt by miACDE and only a slightly reduced inhibition of the mutant reporters ACDEm1 and ACDEm2 (Figure 4B, left panel). Similarly, we observed potent inhibition of the ACDEwt and mutant ACDEm1 and ACDEm2 reporters by shACDE (Figure 4B, right panel). These findings show that artificial antiviral miRNAs and shRNAs can inhibit the expression of luciferase reporters that contain multiple partially complementary sequences within the 3′ UTR.
Figure 4.
Inhibition of reporter constructs with multiple imperfect targets in the 3′ UTR. (A) The luciferase reporter constructs encoding four different target sequences, with or without point mutations, are depicted in different colors. ACDEwt is the luciferase reporter with four perfectly complementary targets. The targets in ACDEm1 have a point mutation either at position 6 or 8, the ACDEm2 reporter targets have a point mutation at position 15. (B) Inhibition of ACDEwt, ACDEm1 and ACDEm2 by miACDE and shACDE. Normalized luciferase expression in the presence of the mock plasmid pBS was set at 100%. (C) Inhibition of ACDEwt, ACDEm1 and ACDEm2 by a single miRNA or shRNA against target A. Inhibition of the three luciferase constructs by miA2 and shA was determined by co-transfection. Luciferase expression in the presence of pBS was set at 100%. (D) Inhibition of ACDEwt, ACDEm1 and ACDEm2 by a single miRNA or shRNA against target D. Inhibition of luciferase constructs by miD and shD was determined by co-transfection as described above. The averages and SDs of four independent transfections are shown.
We then wondered whether miRNA-induced silencing of a single target is differently affected by a point mutation when it is located in the 3′ UTR. To test this, we co-transfected the same set of wild-type and mutant luciferase reporters (ACDEwt, mutants ACDEm1 and ACDEm2) with miA2 or shA into HEK 293T cells. We again titrated the amount of miRNA and shRNA construct to obtain equal inhibitory activities on the wild-type reporter. Normalized firefly luciferase expression in the absence of inhibitor was set at 100%. Potent inhibition of the wild-type luciferase reporter was observed for the miA2 construct. Luciferase silencing was only marginally affected by mutation of target A at position 8 or 15 in mutant reporter ACDEm1 or ACDEm2 (Figure 4C, left panel). Inhibition by shA seemed more sensitive to the introduced target mutations, especially for mutant m1 (Figure 4C, right panel). In fact, this trend was also witnessed in the shACDE context (Figure 4B, right panel). We next examined whether the same trend could be observed using an miRNA and shRNA against target D. Luciferase silencing was significantly affected by mutation of target D at position 8 in the mutant reporter ACDEm1 and in particular by the mutation at position 15 in mutant reporter ACDEm2 (Figure 4D). The knockdown efficiencies were similarly affected by the miD and shD inhibitors. Consistent with the target A results, partial inhibition was retained upon mutation of the target sequence suggesting that shRNAs and artificial miRNAs can exhibit mutation-tolerance when targeting multiple as well as single sites within the 3′ UTR of mRNAs.
Targets in the 3′ UTR of the HIV-1 RNA genome for miACDE and shACDE
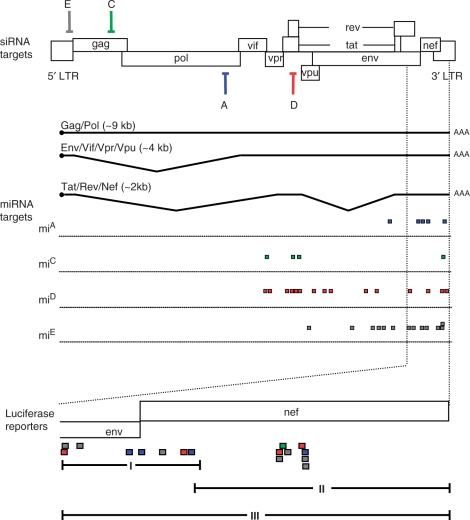
Our findings suggest that designed miRNAs can function like cellular miRNAs by partial base pairing with multiple targets in the 3′ UTR. We therefore wondered whether the antiviral mi/siRNAs derived from the miACDE and shACDE constructs could repress HIV-1 gene expression by targeting putative miRNA target sites within the HIV-1 3′ UTR. To study this, we used the miRanda algorithm to predict potential target sites in the 3′ UTRs of all spliced HIV-1 transcripts for each individual siRNA expressed from these constructs (39). The perfect complementary siRNA targets for miACDE and shACDE are depicted in the HIV-1 genome (Figure 5, upper part). Interestingly, the miRanda algorithm found multiple potential target sites in the 3′ UTR for each individual siRNA, depicted as colored squares in Figure 5. We focused on the 3′ UTR domain that is shared by all HIV-1 transcripts (40), marked in Figure 5, and blown up in the lower panel.
Figure 5.
Putative miRNA target sites in the HIV-1 3′ UTR for miACDE and shACDE. The HIV-1 genome is shown with the perfect target sites for miACDE and shACDE depicted in different colors. Putative miRNA target sites (color-coded) were predicted by computational analysis. Three luciferase reporter constructs were generated with part of all putative 3′ UTR targets.
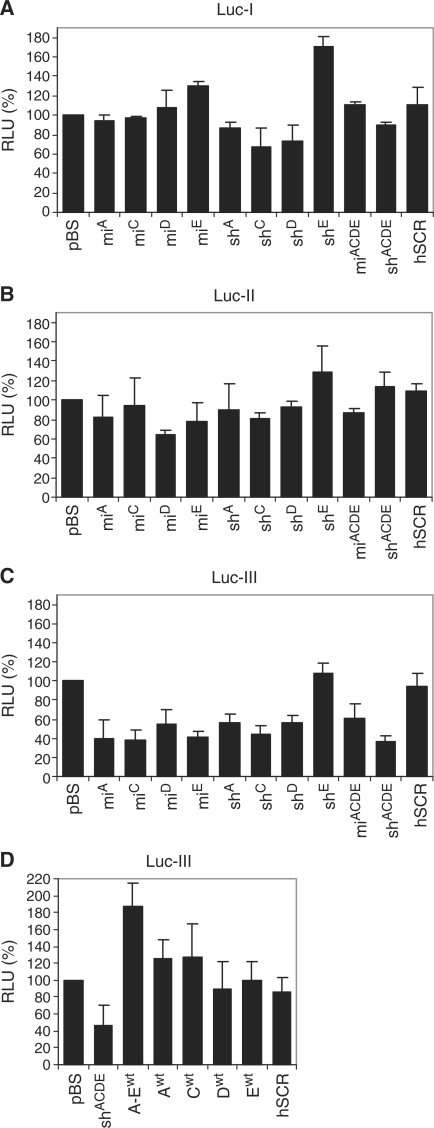
To determine the functionality of the putative miRNA targets, we cloned these sequences as 3′ UTR in luciferase reporter constructs. To study the impact of the multitude of target sites on the knockdown efficiency of the antiviral miRNA polycistron, we constructed three luciferase reporter constructs with different 3′ UTR domains: Luc-I, Luc-II and the combined segment Luc-III (Figure 5, lower panel). Luc-I and Luc-II contain approximately the same number of target sites, but the targets in Luc-I are dispersed, whereas the target sites in Luc-II are clustered. Luc-III combines the putative target sites encoded by Luc-I and Luc-II. We co-transfected the three luciferase reporter constructs with the antiviral miACDE and shACDE constructs into HEK 293T cells. As controls, we included the individual miRNA or shRNA constructs. Normalized firefly luciferase expression in the absence of inhibitor (pBS control) was set at 100%. As a negative control, we used a construct expressing a scrambled hairpin (hSCR). We did not detect knockdown of the Luc-I reporter construct by the individual miRNA or shRNA constructs (Figure 6A). Targeting of multiple targets by miACDE or shACDE also did not result in knockdown of luciferase expression. Similarly, we did not detect significant inhibition of the Luc-II reporter by miACDE or shACDE (Figure 6B).
Figure 6.
Validation of putative 3′ UTR targets for miACDE and shACDE. (A) HEK 293T cells were co-transfected with 100 ng Luc-I reporter, 1 ng pRL and 100 ng of the hairpin RNA constructs. Luciferase expression in the presence of pBS was set at 100%. The scambled hairpin hSCR was included as a negative control. (B, C). As described for (A), but instead Luc-II or Luc-III was used. (D) As described for (C), but now wild-type miRNA controls were used (Awt, Bwt, Cwt, Dwt and Ewt and Aw–Ewt) to test the sequence-specificity. As a positive control, the shACDE was used. Averages and SDs represent four independent transfections.
Finally, we assessed whether the antivirals were able to inhibit Luc-III that encodes all putative 3′ UTR targets of HIV-1 RNA. We observed up to 60% inhibition of reporter gene expression by all single miRNA constructs and all shRNA constructs except shE (Figure 6C). The combined expression of multiple antiviral miRNAs by miACDE did not further enhance the inhibition of Luc-III compared to the single miRNA constructs. In contrast, shACDE resulted in enhanced knockdown of Luc-III compared to the individual shRNA constructs. To test whether inhibition of Luc-III was sequence-specific, we performed the same experiment with single wild-type miRNA controls (Awt,Bwt,Cwt,Dwt,Ewt and Awt-Ewt (Figure 6D). The scrambled hairpin hSCR was used as negative control and shACDE as positive control that yielded ∼50% knockdown. We did not observe any significant knockdown of the Luc-III reporter with the individual wild-type miRNAs and the wild-type Awt-Ewt cluster.
DISCUSSION
A major challenge to obtain durable HIV-1 inhibition by means of induced RNAi is the prevention of viral escape via the selection of RNAi-resistant virus variants. Both shRNAs and artificial miRNAs have been used to induce antiviral RNAi. These antiviral molecules are usually designed to cleave fully complementary viral target mRNAs. In this study, we set out to test whether artificial miRNAs and shRNAs can repress HIV-1 gene expression by targeting partially complementary viral sequences. Cellular miRNAs allow mismatches with their 3′ UTR targets for translational repression. Therefore, translational inhibition of viral gene expression by artificial miRNAs may be sustained on viral escape variants that have acquired point mutations within the target sequence. To test if a durable therapy can be designed in this manner, we tested inhibition of wild-type and RNAi-escape HIV-1 variants by miRNA and shRNA constructs (15,16).
Depending on the target sequence, we observed a relatively strong (up to 60%) RNAi-mediated inhibition of HIV-1 escape viruses by both the miRNAs and shRNAs that target well-conserved sequences in viral open reading frames, despite the use of low amounts of inhibitory constructs. Although RNAi-escape viruses were previously shown to replicate in the presence of the specific RNAi inducer, their replication is in some cases delayed (21). Moreover, escape viruses that have acquired a single point mutation can also acquire additional mutations over time (21). These results suggest that partially complementary sequences are still targeted and that the virus needs to acquire additional mutations to optimize its resistance phenotype. Consistent with these results, other studies have reported that mismatched siRNAs or miRNAs resulted in a significant degree of translational repression when mRNAs contain target sequences in the coding region of non-viral genes (41–43). One study reported that reduced protein expression was still observed when up to four clustered mismatches were present in the centre of the duplex between the siRNA and the target (41). Another possibility is that HIV-1 can tolerate RNAi pressure, despite the fact that partially complementary target sequences are significantly targeted. This may be due to the expression of the viral RNAi suppressor protein Tat (44–46). However, it is currently unknown to what extent RNAi suppressors interfere with RNAi therapeutics (1).
Efficient miRNA-mediated inhibition requires targeting of multiple partially complementary sites within the 3′ UTR of mRNAs. Therefore, we also tested the miRNA- and shRNA-mediated inhibition on luciferase reporters with multiple target sites in the 3′ UTR. We found that shRNA- and miRNA-induced gene repression was only slightly reduced when a point mutation was introduced in the target sites. We observed similar effects when the luciferase reporters were attacked by a single inhibitor. These combined results indicate that designed miRNAs and shRNAs can function like cellular miRNAs by acting on imperfect targets in the 3′ UTR. Although miRNAs and shRNAs have been shown to be functionally interchangeable for inducing translational repression or mRNA cleavage (47,48), miRNAs also have greater inhibitory efficiency compared to conventional shRNAs (15,17,49,50). This is mainly due to the fact that miRNAs are better substrates for the RNAi machinery than shRNAs (50). Another possibility is that miRNA mimics are more efficient in activating translational repression because they are processed via the natural miRNA pathway, whereas shRNA processing skips the Drosha cleavage step. Furthermore, there may be functional differences between a RISC complex loaded with a miRNA or siRNA. Cellular miRNAs and endogenous siRNAs can interact with one or more of the four Argonaute proteins (Ago1–4) (51,52), and it currently remains unclear whether these inhibitors show distinct binding preferences (53).
In general, we observed more potent inhibition when the target was located in the 3′ UTR of a reporter transcript. However, it is difficult to directly compare the levels of inhibition when targeting an open reading frame versus the same target sequence within the 3′ UTR. Consistent with our results, a recent report showed that the knockdown efficiency is significantly reduced when a miRNA partially base pairs with a target in the open reading frame compared to the regular 3′ UTR position (54). Interestingly, the same study demonstrated that the knockdown efficiency of a perfectly base paired miRNA is not affected by the target position. These results indicate that the effectivity of miRNA-mediated gene repression is dependent on the location of the target.
Via computational analysis, we identified multiple putative miRNA targets within the HIV-1 3′ UTR for our combination inhibitory molecules, the four shRNA construct shACDE (24) and the miACDE polycistron that encode four antiviral siRNAs or miRNAs (15). Using luciferase reporters with these 3′ UTR targets, we found that the reporter with the maximum number of targets was inhibited up to 60% by both antiviral miRNAs and shRNAs. In contrast, reporters bearing less target sites could not be inhibited by the miRNAs or shRNAs. This finding suggests that the presence of multiple target sites in the complete 3′ UTR of HIV-1 may allow additional inhibition by the antiviral miRNA molecules. It is important to note that there are other critical factors that influence the inhibition by miRNAs and siRNAs, including RISC accessibility of the targets and the distance between the target sites (55–59). Thus, the presence of multiple partially complementary target sites in the 3′ UTR of mRNAs does not necessarily result in knockdown of these mRNAs. It is therefore needed to experimentally validate the targets. Nonetheless, these results suggest that potential off-target effects on sites of bystander mRNAs is a genuine concern for the development of any RNAi-based gene therapy. This off-targeting risk should be critically assessed in relevant in vivo models prior to an eventual clinical application (1).
It has recently been shown that cellular miRNAs can target and inhibit the expression of viral transcripts. Although these miRNAs are only partially complementary to the viral mRNAs, they have a profound inhibitory effect on viral gene expression and consequently viral replication (31–33). In case of HIV-1, multiple cellular miRNAs were shown to contribute to viral latency by targeting multiple sites in the 3′ UTR (31). Furthermore, HIV-1 is able to change (i.e. reduce) the miRNA expression profile of infected cells (60). In this study, we show that antiviral miRNAs and shRNAs can effectively target putative miRNA targets in the HIV-1 3′ UTR, resulting in reduced gene expression. Naturally occurring virus-encoded miRNA target sites are retained within the viral genome despite the fact that they contribute to reduced viral replication. Similarly, therapeutic miRNAs may provide a modest inhibition of viral gene expression that does not easily trigger the emergence of escape variants, allowing a sustained attenuation of virus replication over an extended period. For the design of such an miRNA therapy, one should preferentially target multiple sequences within the 3′ UTR.
In conclusion, we present data that both antiviral miRNAs and shRNAs can repress gene expression via partially complementary target sequences in the 3′ UTR and to a lesser extent in the open reading frame. Potent knockdown was observed for miRNAs and shRNAs that bind to a single target or multiple targets with one or two mismatches in the 3′ UTR of a reporter transcript. Multiple imperfect target sites in the 3′ UTR are required to obtain a significant knockdown efficiency of miRNAs and shRNAs. These findings indicate that designed miRNAs and shRNAs can function like cellular miRNAs on imperfect targets in the 3′ UTR. Targeting multiple viral sequences in the 3′ UTR by antiviral miRNAs or shRNAs may provide a successful alternative RNAi strategy for durable suppression of escape-prone viruses.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Netherlands Organisation for Health Research and Development (ZonMw; VICI grant); The Netherlands Organisation for Scientific Research (NWO-CW; TOP grant). Funding for open access charge: Deutscher Akademischer Austausch Dienst (DAAD) fellowship.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Stephan Heynen for performing CA-p24 ELISA.
REFERENCES
- 1.Haasnoot J, Westerhout EM, Berkhout B. RNA interference against viruses: strike and counterstrike. Nat. Biotechnol. 2007;25:1435–1443. doi: 10.1038/nbt1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haasnoot PCJ, Berkhout B. Handbook of Experimental Pharmacology. Vol. 173. Berlin, Heidelberg: Springer; 2006. Its use as antiviral therapy; pp. 117–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim DH, Rossi JJ. Strategies for silencing human disease using RNA interference. Nat. Rev. Genet. 2007;8:173–184. doi: 10.1038/nrg2006. [DOI] [PubMed] [Google Scholar]
- 4.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.Filipowicz W. RNAi: the nuts and bolts of the RISC machine. Cell. 2005;122:17–20. doi: 10.1016/j.cell.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 9.Paddison PJ, Caudy AA, Bernstein E, Hannon GJ, Conklin DS. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 11.Leuschner PJ, Ameres SL, Kueng S, Martinez J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- 13.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–629. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 14.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 15.Liu YP, Haasnoot J, Ter Brake O, Berkhout B, Konstantinova P. Inhibition of HIV-1 by multiple siRNAs expressed from a single microRNA polycistron. Nucleic Acids Res. 2008;36:2811–2824. doi: 10.1093/nar/gkn109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ter Brake O, Konstantinova P, Ceylan M, Berkhout B. Silencing of HIV-1 with RNA interference: a multiple shRNA approach. Mol. Ther. 2006;14:883–892. doi: 10.1016/j.ymthe.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 17.Boden D, Pusch O, Silbermann R, Lee F, Tucker L, Ramratnam B. Enhanced gene silencing of HIV-1 specific siRNA using microRNA designed hairpins. Nucleic Acids Res. 2004;32:1154–1158. doi: 10.1093/nar/gkh278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lo HL, Chang T, Yam P, Marcovecchio PM, Li S, Zaia JA, Yee JK. Inhibition of HIV-1 replication with designed miRNAs expressed from RNA polymerase II promoters. Gene Ther. 2007;14:1503–1512. doi: 10.1038/sj.gt.3303011. [DOI] [PubMed] [Google Scholar]
- 19.Das AT, Brummelkamp TR, Westerhout EM, Vink M, Madiredjo M, Bernards R, Berkhout B. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J. Virol. 2004;78:2601–2605. doi: 10.1128/JVI.78.5.2601-2605.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boden D, Pusch O, Lee F, Tucker L, Ramratnam B. Human immunodeficiency virus type 1 escape from RNA interference. J. Virol. 2003;77:11531–11535. doi: 10.1128/JVI.77.21.11531-11535.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westerhout EM, Ooms M, Vink M, Das AT, Berkhout B. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 2005;33:796–804. doi: 10.1093/nar/gki220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.von Eije KJ, Ter Brake O, Berkhout B. Human immunodeficiency virus type 1 escape is restricted when conserved genome sequences are targeted by RNA interference. J. Virol. 2008;82:2895–2903. doi: 10.1128/JVI.02035-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.ter Brake O, von Eije KJ, Berkhout B. Probing the sequence space available for HIV-1 evolution. AIDS. 2008;22:1875–1877. doi: 10.1097/QAD.0b013e328309efe3. [DOI] [PubMed] [Google Scholar]
- 24.ter Brake O, ‘t Hooft K, Liu YP, Centlivre M, von Eije KJ, Berkhout B. Lentiviral vector design for multiple shRNA expression and durable HIV-1 Inhibition. Mol. Ther. 2008;16:557–564. doi: 10.1038/sj.mt.6300382. [DOI] [PubMed] [Google Scholar]
- 25.Liu YP, Haasnoot J, Berkhout B. Design of extended short hairpin RNAs for HIV-1 inhibition. Nucleic Acids Res. 2007;35:5683–5693. doi: 10.1093/nar/gkm596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 28.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 29.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 30.Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L, Linsley PS. Widespread siRNA “off-target” transcript silencing mediated by seed region sequence complementarity. RNA. 2006;12:1179–1187. doi: 10.1261/rna.25706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang J, Wang F, Argyris E, Chen K, Liang Z, Tian H, Huang W, Squires K, Verlinghieri G, Zhang H. Cellular microRNAs contribute to HIV-1 latency in resting primary CD4(+) T lymphocytes. Nat. Med. 2007;13:1241–1247. doi: 10.1038/nm1639. [DOI] [PubMed] [Google Scholar]
- 32.Lecellier CH, Dunoyer P, Arar K, Lehmann-Che J, Eyquem S, Himber C, Saib A, Voinnet O. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 33.Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ahluwalia JK, Khan SZ, Soni K, Rawat P, Gupta A, Hariharan M, Scaria V, Lalwani M, Pillai B, Mitra D, et al. Human cellular microRNA hsa-miR-29a interferes with viral nef protein expression and HIV-1 replication. Retrovirology. 2008;5:117. doi: 10.1186/1742-4690-5-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Ye L, Hou W, Zhou Y, Wang YJ, Metzger DS, Ho WZ. Cellular microRNA expression correlates with susceptibility of monocytes/macrophages to HIV-1 infection. Blood. 2009;113:671–674. doi: 10.1182/blood-2008-09-175000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peden K, Emerman M, Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology. 1991;185:661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- 37.Ruijter JM, Thygesen HH, Schoneveld OJ, Das AT, Berkhout B, Lamers WH. Factor correction as a tool to eliminate between-session variation in replicate experiments: application to molecular biology and retrovirology. Retrovirology. 2006;3:1–8. doi: 10.1186/1742-4690-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeeninga RE, Hoogenkamp M, Armand-Ugon M, de Baar M, Verhoef K, Berkhout B. Functional differences between the long terminal repeat transcriptional promoters of HIV-1 subtypes A through G. J. Virol. 2000;74:3740–3751. doi: 10.1128/jvi.74.8.3740-3751.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 40.Purcell DFJ, Martin MA. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 1993;67:6365–6378. doi: 10.1128/jvi.67.11.6365-6378.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saxena S, Jonsson ZO, Dutta A. Small RNAs with imperfect match to endogenous mRNA repress translation. Implications for off-target activity of small inhibitory RNA in mammalian cells. J. Biol. Chem. 2003;278:44312–44319. doi: 10.1074/jbc.M307089200. [DOI] [PubMed] [Google Scholar]
- 42.Kloosterman WP, Wienholds E, Ketting RF, Plasterk RH. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res. 2004;32:6284–6291. doi: 10.1093/nar/gkh968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holen T, Amarzguioui M, Wiiger MT, Babaie E, Prydz H. Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res. 2002;30:1757–1766. doi: 10.1093/nar/30.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bennasser Y, Le SY, Benkirane M, Jeang KT. Evidence that HIV-1 encodes an siRNA and a suppressor of RNA silencing. Immunity. 2005;22:607–619. doi: 10.1016/j.immuni.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 45.Haasnoot J, de Vries W, Geutjes EJ, Prins M, de Haan P, Berkhout B. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog. 2007;3:e86. doi: 10.1371/journal.ppat.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qian S, Zhong X, Yu L, Ding B, de Haan P, Boris-Lawrie K. HIV-1 Tat RNA silencing suppressor activity is conserved across kingdoms and counteracts translational repression of HIV-1. Proc. Natl Acad. Sci. USA. 2009;106:605–610. doi: 10.1073/pnas.0806822106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc. Natl Acad. Sci. USA. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qu J, Ye J, Fang R. Artificial microRNA-mediated virus resistance in plants. J. Virol. 2007;81:6690–6699. doi: 10.1128/JVI.02457-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, et al. Second-generation shRNA libraries covering the mouse and human genomes. Nat. Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 51.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 52.Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Aronautes in microRNA silencing. Genes Dev. 2009;23:304–317. doi: 10.1101/gad.1749809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 54.Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nat. Struct. Mol. Biol. 2009;16:144–150. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saetrom P, Heale BS, Snove O, Jr., Aagaard L, Alluin J, Rossi JJ. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 2007;35:2333–2342. doi: 10.1093/nar/gkm133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown KM, Chu CY, Rana TM. Target accessibility dictates the potency of human RISC. Nat. Struct. Mol. Biol. 2005;12:469–470. doi: 10.1038/nsmb931. [DOI] [PubMed] [Google Scholar]
- 57.Shao Y, Chan CY, Maliyekkel A, Lawrence CE, Roninson IB, Ding Y. Effect of target secondary structure on RNAi efficiency. RNA. 2007;13:1631–1640. doi: 10.1261/rna.546207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rudnick SI, Swaminathan J, Sumaroka M, Liebhaber S, Gewirtz AM. Effects of local mRNA structure on posttranscriptional gene silencing. Proc. Natl Acad. Sci. USA. 2008;105:13787–13792. doi: 10.1073/pnas.0805781105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Obernosterer G, Tafer H, Martinez J. Target site effects in the RNA interference and microRNA pathways. Biochem. Soc. Trans. 2008;36:1216–1219. doi: 10.1042/BST0361216. [DOI] [PubMed] [Google Scholar]
- 60.Houzet L, Yeung ML, de Lame V, Desai D, Smith SM, Jeang KT. MicroRNA profile changes in human immunodeficiency virus type 1 (HIV-1) seropositive individuals. Retrovirology. 2008;5:118. doi: 10.1186/1742-4690-5-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.