Abstract
Gene transfer to the corneal endothelium has potential in preventing corneal transplant rejection. In this study, we transfected mouse corneal endothelial cells (MCEC) with a class of novel arginine-rich oligopeptides. The peptides featured a tri-block design and mediated reporter gene expression in MCEC more efficiently than the commercial polyethylenimine standard. The functionality of each block was demonstrated to critically influence the performance of the peptide. Results from confocal imaging and flow cytometry then showed that energy-dependent endocytosis was the dominant form of uptake and multiple pathways were involved. Additionally, uptake was strongly dependent on interactions with cell-surface heparan sulphate. Fluorescence resonance energy transfer studies revealed that the peptide/DNA entered cells as an associated complex and some will have dissociated by 8.5 h. Large-scale accumulation of uncondensed DNA within the nucleus can also be observed by 26 h. Finally, as a proof of biological relevance, we transfected MCEC with plasmids encoding for the functional indoleamine 2,3-dioxygenase (IDO) enzyme. We then demonstrated that the expressed IDO could catalyse the degradation of l-tryptophan, which in turn suppressed the growth of CD4+ T-cells in a proliferation assay.
INTRODUCTION
The cornea is an immune privileged tissue, in large part due to its avascular nature and the scarcity of dendritic cells in the corneal graft, although other factors also contribute to its status (1,2). This privilege, while not absolute, explains the high success rate that first-time corneal grafts routinely enjoy even in the absence of systemic immunosuppression (2). Nonetheless, allograft failure still occurs and around a quarter of grafts will fail within 5 years (3,4). Immune rejection is the biggest cause for this graft loss and is especially prevalent among recipients with previous episodes of graft rejection, inflammation or neovascularization (5). There is therefore an urgent need for novel forms of therapy and the ex vivo delivery of immunomodulatory genes into the cornea prior to transplantation represents one promising strategy to allow long term survival of the graft (6,7).
The cornea, specifically, its posterior endothelial monolayer which fulfils the critical function of maintaining corneal transparency (8), is particularly amenable to such a genetic approach because of its accessibility, simple anatomy and its ability to be stored and manipulated ex vivo for up to a month before surgery (8). The ex vivo immunomodulation of corneal transplantation is thus a realistic goal as it avoids many of the challenges associated with gene delivery at the systemic level. To achieve this objective, viral vectors, for example, adenovirus (9,10), lentivirus (11), equine infection anaemia virus (12), recombinant adeno-associated virus and herpes simplex virus (13), among others, have been explored actively. The biggest advantage of viral mediated gene delivery is the natural efficiency of viruses at infecting and transferring genetic cargoes into cells. However, most viral systems are limited by their innate immunogenicity, proinflammatory nature, cytotoxicity, limited cargo loading capacity and the complexity of industrial scale production (5). As a result, non-viral vectors may be a safer alternative for clinical applications (14), although their efficiency is lower and the gene expression they mediate is more transient than some viral vectors.
Oligopeptides as a class of non-viral vector has recently gained prominence. Advantages of such a system include their biodegradability, biocompatibility and ease of production and compositional control. Current oligopeptide designs are based mainly on cationic arginine (15) or lysine (16) to achieve DNA binding effects. To further improve transfection efficiency, either hydrophobic moieties (17–19) or endosomolytic agents (20–23) are frequently incorporated.
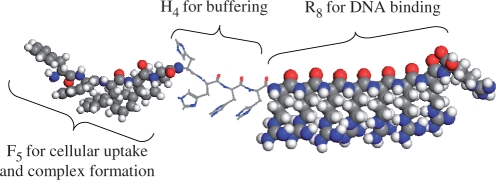
Recently, we explored a novel class of non-viral vector made from natural amino acids which combined all the above features (24). The peptides had a tri-block design comprising of arginine for DNA binding and membrane penetration, histidine for buffering effects and a segment of hydrophobic residues for enhancing cellular uptake and DNA binding ability (Figure 1). We demonstrated that they could bind and protect DNA from enzymatic degradation. Importantly, the peptides mediated efficient reporter gene expression in a variety of cell lines and, more significantly, following injection into mice bearing solid tumours. While the systemic application of these peptides is limited due to the large size (up to 50 µm) of the peptide/DNA complexes (24), they can still have a role in the ex vivo manipulation of corneas and other tissues or cells.
Figure 1.
Design features of the oligopeptide vectors with (1) octaarginine for DNA binding and membrane penetration, (2) histidine residues for buffering capacity and (3) a hydrophobic segment (shown as F5) for enhancing cellular uptake and DNA complex formation. Colour scheme: red, O; white, H; grey, C; blue, N.
In this study, we explored the possibility of using these peptides to transfect mouse corneal endothelial cells (MCEC). We showed that the peptides were able to transfect MCEC more efficiently than polyethylenimine (PEI), a commercially available transfection reagent (25). We systematically demonstrated that the functionality of each block in the peptide design was essential to achieve high transfection. We then investigated the pathways of endocytosis and intracellular trafficking followed by these vectors. Fluorescence resonance energy transfer (FRET) was also used to investigate the events occurring between early internalization and eventual nuclear localization. Finally, to demonstrate the biological relevance of this system, we transfected MCEC with plasmids encoding for the functional indoleamine 2,3-dioxygenase (IDO) enzyme. We showed in a functional assay that the expressed IDO could catalyse the breakdown of tryptophan, an amino acid T-cells critically depend on for growth. Consequently, their growth was retarded in a proliferation assay involving CD4+ T-cells and IDO-transfected MCEC. This study therefore provides a useful platform from which we can develop the ex vivo manipulation of whole corneas.
MATERIALS AND METHODS
Materials
Nocodazole, cytochalasin D, dynasore, sodium azide, 2-deoxyglucose, methyl-β-cyclodextrin, filipin III, 5-(N-ethyl-N-isopropyl)amiloride (EIPA), heparin, chloroquine, branched polyethylenimine (Mw ∼25 000 g/mol) and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazoliumbromide (MTT) were all purchased from Sigma (Singapore) and were of the highest purity available. Transferrin-AlexaFluor488, cholera toxin-AlexaFluor488, dextran-Texas Red (10 000 g/mol, neutral overall charge) and Hoechst 34580 were from Invitrogen (Singapore). Label IT® Cy™5, Cy™3 and TM-rhodamine labelling kits were from Mirus (Madison, WI, USA). Reporter genes used were either the 6.4 kb firefly luciferase (pCMV-luciferase VR1255C) driven by the cytomegalovirus promoter (Carl Wheeler, Vical, San Diego, CA, USA) or the 4.7 kb GFPmut1 variant (pEGFP-C1) driven by the SV40 early promoter (Clontech, Palo Alto, CA, USA). For a therapeutic gene, the 8.5 kb pSMART-2G plasmid was used encoding IDO under control of the cytomegalovirus promoter. The cell line used was the SV40-immortalized BALB/c strain MCEC (12). All peptides were synthesized as reported previously with a purity >96% according to high performance liquid chromatography (HPLC) (24).
In vitro luciferase and GFP expression
MCEC was maintained in Dulbecco's modified Eagle's medium (DMEM) completed with 10% (v/v) fetal bovine serum, 1% (v/v) non-essential amino acids, 100 µg/ml of streptomycin and 100 U/ml of penicillin at 37°C with 5% CO2. Cells were seeded 1 day before experiments. For the luciferase transfection assay, 0.5 ml of medium containing 6.0 × 104 cells were seeded into each well of a 24-well plate, while for the GFP transfection assay, 1.0 ml of medium containing 1.8 × 105 cells were pipetted into each well of a 12-well plate.
Peptide complexes were formed at various N/P ratios (defined here as the molar ratio of arginine to the DNA phosphorus content) by the drop-wise addition of equivolume solutions of peptide into DNA. Ten millimolar phosphate buffer (pH 7.0) was used to form the particles for all experiments unless otherwise stated. The desired N/P ratios were achieved by fixing the amount of DNA used and varying the amount of peptide. The mixture was then vortexed for ∼10 s and left to stand for ∼30 min before use. PEI/DNA complexes were similarly formed either in water or 10 mM phosphate buffers. However, only data for PEI/complexes formed in water were presented as this formulation gave higher luciferase expression results. All media was replaced before the solution containing the complexes was added drop-wise into each well. Full-serum conditions were always maintained throughout transfection unless otherwise stated. The media was again replaced after 4 h of incubation.
Analysis for luciferase expression was carried out after 3 days. Cells were washed with 0.5 ml of PBS before the addition of 0.2 ml of ‘Reporter Lysis Buffer’ (Promega, Madison, WI, USA). Membrane lysis was completed with two freeze–thaw cycles and manual cell scratching between the cycles. The cell suspension was centrifuged at 20 800 g for 10 min. The supernatant (20 μl) was removed and mixed with 100 µl of a luciferase assay reagent (Promega). The relative light unit (RLU) was measured with a luminometer (Lumat LB 9507, Mandel Scientific, Ontario, Canada) and protein concentration with a bicinchoninic acid protein assay kit (Pierce, Rockfort, IL, USA). Luciferase activity was expressed as RLU per milligram protein ± SD of at least four samples.
For the GFP transfection assay, cells were harvested after 2 days, washed three times with 1.0 ml of PBS and detached with 0.3 ml of trypsin (0.5 g/l in PBS). The cell suspension was then centrifuged at 20 800g for 10 min, followed by resuspension in a dilute trypsin solution (0.2 g/l) to further digest and remove any surface-bound peptide complexes. The cell pellet was again collected and finally resuspended in PBS for analysis using a flow cytometer (FACSCalibur, BD Bioscience, San Jose, CA, USA). A total of 10 000 events were recorded and percentage of MCEC expressing GFP was reported as a mean ± SD of at least four samples. Cells were defined to be GFP positive by setting the gating region to include 1% of cells mock transfected with plasmids encoding for luciferase.
Uptake studies
Uptake of complexes into MCEC was either measured using flow cytometry or imaged by confocal microscopy. In both cases, plasmids encoding for luciferase were labelled with Cy5 according to the manufacturer's recommendation, followed by ethanol precipitation. The labelling density was determined to be about 1 Cy5 dye per 135 base pairs by absorbance at 649 and 260 nm.
One day before the experiment, cells were either seeded into 12-well plates at 2.2 × 105 cells per well or confocal chambers (Nunc, Rochester, NY, USA) at 1.2 × 105 cells per chamber. Peptide complexes were then formed with the Cy5-labelled DNA and added to the cells. After 3 h of incubation at 37°C, cells were washed thoroughly and either harvested for flow cytometry analysis (3-laser LSR II, BD Bioscience) as described above, or stained with the Hoechst nuclear dye for confocal imaging (LSM 5 DUO, Carl Zeiss, Jena, Germany). The parameter used to quantify uptake in flow cytometry was the mean fluorescence intensity from 10 000 gated cells. Cells transfected with unlabelled DNA were used as negative controls.
Inhibitor studies
The effects of various inhibitors were assessed at both the uptake and luciferase expression level. Depending on the experiment, cells were pre-incubated with one of the following inhibitors for 1 h before transfection: nocodazole (4.5 µg/ml) (26), cytochalasin D (5.0 µg/ml) (27), dynasore (6.8 µg/ml) (28), sodium azide (0.5 mg/ml) with 2-deoxyglucose (4.1 mg/ml) (26), methyl-β-cyclodextrin (5.2 mg/ml) (29), filipin III (1.0 µg/ml) (26), EIPA (25 µg/ml) (29). Inhibitors that had limited water solubility were first dissolved in DMSO to make a stock solution. The cells were then replaced with fresh growth media containing the respective inhibitors before addition of peptide complexes (N/P 20) containing either fluorescently labelled or unlabelled DNA encoding for luciferase. No pre-incubation was carried out for experiments involving heparin (50 µg/ml) (30) or chloroquine (100 µM) (31), which was present in the media only during transfection. All inhibitor solutions were 0.22 µm filtered before use. Analyses by flow cytometry and confocal microscopy were performed 3 h later, while luciferase expression was quantified 72 h later, as described above. All data were presented relative to control cells transfected in the presence of vehicle control (DMSO or water) alone (100%). Toxicity induced by the various inhibitors was accounted for in preliminary experiments where a suitable working concentration was established for which cell death was not evident.
Co-localization with endocytosis markers
Cells were concurrently incubated with peptide complexes (N/P 20) containing DNA labelled with TM-rhodamine and one of the following markers for specific endocytosis pathways: transferrin-AlexaFluor488 (25 µg/ml), cholera toxin-AlexaFluor488 (5 µg/ml) and dextran-Texas Red (10 µM). After 30–40 min, the cells were extensively rinsed with PBS, stained with Hoechst and immediately imaged using confocal microscopy. Z-stacking was done to confirm that signals originate from within the cells in all confocal experiments in this study.
FRET
Plasmids encoding for luciferase were double-labelled with Cy3 and Cy5 (approximately 1.5 Cy3 dye: 1 Cy5 dye: 100 base pairs of the plasmid). On the day of experiment, peptide complexes (N/P 20) containing the double-labelled DNA were incubated with the cells for 2 h at 37°C, followed by extensive washing with PBS. The cells were then returned to the incubator followed by live cell analysis at selected time points. During imaging, Cy3 was excited with the 543 nm laser. Cy5 would only be excited if there was FRET. Both the emissions of Cy3 (560-615 nm band-pass) and Cy5 (650 nm long-pass) were collected in the experiments. FRET images were then quantified with Matlab R2006 ver7.2.0.232 by binary-scoring each pixel as ± for fluorescence. The ratio of Cy5-positive to Cy3-positive pixels was used to indicate the extent of FRET for that image.
Heparin displacement assays
Peptide complexes were first formed at N/P ratio 20 and then incubated with heparin (0.1 to 20.0 mg/ml) for 1 h at room temperature (22°C). Agarose gel electrophoresis (80 V, 50 min) with ethidium bromide staining was used to detect DNA released by heparin treatment. DNA still bound to peptide was either seen within the loading well or was not visible due to the inability of ethidium bromide to intercalate condensed DNA.
Titration experiments
To compare the relative buffering capacity of the peptides, titration was carried out with an automatic potentiometric titrator (AT-610, Kyoto Electronics Manufacturing, Tokyo, Japan) against 0.01 M NaOH. The temperature was maintained at 25°C by a circulating water bath and the electrodes were calibrated with standards before use. A 16 ml of peptide solution (0.5 mg/ml) was used for all titration experiments.
IDO mRNA expression
The expression of IDO was first quantified at the mRNA level using RT–PCR. MCEC cultured in 12-well plates were exposed to either peptide F or PEI complexed to plasmids encoding for IDO for 4 h, as per earlier experiments. Lipofectamine™2000 (Invitrogen) was also used at a ratio of 1 : 2.5 (DNA : lipofectamine™2000) according to the manufacturer's protocol. Cells were harvested 48 h post-transfection and their RNA isolated for reverse-transcription into cDNA. Samples were then analysed using the Sybr-Green PCR Mastermix (Applied Biosystem, Warrington, UK) and a 7900 HT Fast Real Time PCR system (Applied Biosystems). The thermal profile used was: stage 1, initial denaturation, 95°C for 10 min; stage 2, amplification, 95°C for 15 s, 60°C for 1 min, 72°C for 30 s for 40 cycles and stage 3, dissociation, 95°C for 15 s, 60°C for 15 s, 95°C for 15 s. Specificity of the amplification was confirmed by the observation of a single dissociation peak from the samples. IDO mRNA expression was normalized to hypoxanthine phosphoribosyl transferase (HPRT) mRNA. Primers used for IDO and HPRT have been described elsewhere (12).
Western blot
Cells were transfected in 24-well plates and then lysed 48 h later by resuspension in Laemmli reducing sample buffer (Sigma, UK). Protein lysates were resolved on a 12% SDS–polyacrylamide gel and then blotted onto a nitrocellulose membrane. Anti-mouse IDO mAb (MAB5412, Chemicon, CA, USA) or anti-β-actin mAb (Sigma) was used as a primary probe, followed by HRP-conjugated rabbit anti-mouse antibody (P0161, DakoCytomation, UK). Blots were developed using the Lumigen PS-3 (Lumigen, MI, USA) system.
l-kynurenine functional assay
The biological activity of the expressed IDO was determined by colorimetrically measuring the levels of l-kynurenine, the metabolic product of tryptophan. Cells were cultured in 24-well plates and the media was supplemented with 200 µM of tryptophan. Cells were either transfected with peptide F (IDO), mock-transfected (GFP) or untransfected for 4 h. Where appropriate, 1-methyl-dl-tryptophan (1-MT) (Sigma) was added at a final concentration of 0.75 mM to inhibit the activity of IDO. The level of l-kynurenine in the culture supernatant was then quantified 48-h post-transfection. Briefly, 100 µl of culture supernatant was incubated with 100 µl of 30% trichloroacetic acid (Sigma) at 50°C for 30 min in a 96-well plate. The precipitated protein was then spun down and 100 µl of the clear supernatant was transferred to a new 96-well plate for incubation with 100 µl of freshly prepared Ehrlich's reagent at 65°C for 15 min. Absorbance was then measured at 492 nm with a spectrophotometer (Anthos Labtec HT 2, Salzburg, Austria). A calibration curve (R2 > 0.99) was generated with samples of known l-kynurenine concentration.
T-cell proliferation assay
CD4+ responder T-cells were freshly isolated from the spleens of C3H mice using procedures described elsewhere (12). 105 T-cells (stimulated with anti-CD3 and CD28 beads) were then added to each well of a 96-well plate already containing 5 × 104 MCEC (treated with 40 µg/ml of mitomycin C for 30 min) which were either untransfected or transfected with peptide F complexed with plasmids encoding for IDO or GFP. Where appropriate, 1-MT was added at a final concentration of 0.75 mM. After 3 days of co-incubation, 30 µl of [3H]thymidine solution (1 µCi/well, Amersham Pharmacia Biotech, UK) was added. The amount of incorporated radioactivity was measured 16 h later with a 1450 MicroBeta TriLux (PerkinElmer, Shelton, CT, USA).
RESULTS AND DISCUSSION
The peptides have a tri-block design (Figure 1) in which the arginine sequence was used for DNA binding, the histidine block for buffering capacity and the hydrophobic segment for enhancing uptake and DNA-binding ability. We carried out initial experiments to determine if each of these blocks was necessary.
The octaarginine block serves to bind DNA and therefore is a necessary component of the design. Furthermore, the length of eight repeat units had previously been optimized in terms of its ability to deliver the reporter gene encoding for luciferase into COS-7 cells (17).
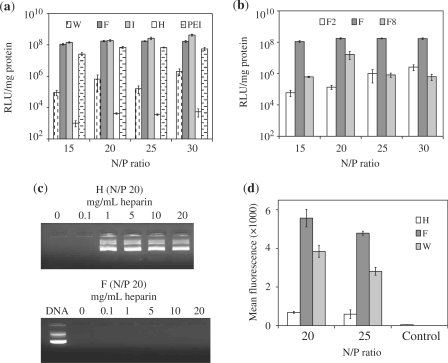
To determine if the hydrophobic block was important, we synthesized a series of peptides (Table 1), each containing a hydrophobic block made up of amino acids with varying degree of hydrophobicity: NH2-I5H4R8-CONH2 (I), NH2-F5H4R8-CONH2 (F), NH2-W5H4R8-CONH2 (W), arranged in order of decreasing hydrophobicity (32,33). As a control, we also removed the hydrophobic segment in one of the sequences, i.e. NH2-H4R8-CONH2 (H), and then evaluated all peptides in terms of their ability to mediate luciferase expression in MCEC. As shown in Figure 2a, there was a dependence on N/P ratio for expression efficiency. Removing the hydrophobic block resulted in a drastic reduction (more than four orders) in the amount of luciferase expressed by the cells across the entire range of N/P ratios tested. This clearly demonstrates the importance of the hydrophobic block in achieving high transfection levels. Additionally, significant differences in expression level were also observed by varying the degree of hydrophobicity of the peptides. W, the least hydrophobic, resulted in the least expression, while F and I performed comparably within the range of N/P ratios tested. Encouragingly, both F and I were superior to PEI—widely regarded as one of the most efficient non-viral vectors available (25)—in terms of luciferase expression. The length of the hydrophobic block was also optimized as peptide F mediated significantly more luciferase expression then complexes formed with NH2-F2H4R8-CONH2 (F2) or NH2-F8H4R8-CONH2 (F8) (Figure 2 b). Lower N/P ratios were excluded from the diagrams because of poor efficiency. For instance, the expression levels achieved for F at N/P 5 and 10 were 1% and 5% of that achieved at N/P 15, respectively.
Table 1.
A summary of the nature of hydrophobic segment and length of hydrophobic histidine, glycine or arginine blocks used for all peptides in this study
| Peptide | Hydrophobic block | Hydrophobic (length) | Histidine (length) | Arginine (length) |
|---|---|---|---|---|
| I | Isoleucine | 5 | 4 | 8 |
| F | Phenylalanine | 5 | 4 | 8 |
| W | Tryptophan | 5 | 4 | 8 |
| H | – | 0 | 4 | 8 |
| F2 | Phenylalanine | 2 | 4 | 8 |
| F8 | Phenylalanine | 8 | 4 | 8 |
| H8 | Isoleucine | 5 | 8 | 8 |
| G4 | Isoleucine | 5 | 4 (Glycine) | 8 |
Figure 2.
Effects of the hydrophobic block on the properties of the oligopeptide vectors. (a) Luciferase expression levels in MCEC transfected with W5H4R8 (W), F5H4R8 (F), I5H4R8 (I), H4R8 (H) or PEI complexes and (b) F2H4R8 (F2) F5H4R8 (F) or F8H4R8 (F8) complexes. (c) Heparin displacement assay with H/DNA and F/DNA complexes incubated with increasing concentrations of heparin for 1 h at room temperature. Naked DNA served as the positive control. (d) Relative uptake quantified with flow cytometry after MCEC have been transfected with either peptide H, F or W complexed to Cy5-labelled DNA. The negative control refers to MCEC transfected with F/unlabelled DNA at N/P 20. All error bars represent SD.
The presence of hydrophobic groups within a vector has been reported to increase transfection (25,34), and one reason proposed may be due to the stabilization of the complex (35,36). To test for this, we conducted a heparin displacement assay in which H and F complexes were first formed at N/P 20 and then incubated with a range of heparin concentrations for 1 h. Displacement of DNA by the anionic heparin was detected by gel electrophoresis (37,38). As shown in Figure 2c, a heparin concentration of 1 mg/ml was sufficient to completely displace the DNA condensed by H. F complexes, on the other hand, remained stable up till a concentration of 20 mg/ml. Similarly, complexes of I and W were also stable up to 20 mg/ml of heparin (data not shown), showing that the presence of a hydrophobic block results in complexes that are more resistant to heparin displacement. It should be noted that while a more stable complex would be better at protecting the DNA from degradation, it may also prevent intracellular release of the DNA thus limiting its bioavailability. However, the latter possibility does not seem to be of a major concern as F complexes still mediated significantly higher expression levels than H.
The presence of hydrophobic moieties has also been reported to improve cellular uptake (39,40). To investigate this, H and F peptides were used to deliver Cy5-labelled DNA encoding for luciferase into MCEC. For this experiment, cells were exposed to the respective complexes for 3 h at 37°C before washing and analysis. Using the mean fluorescence intensity as a parameter, results from flow cytometry showed that F complexes were internalized about eight times more efficiently than H complexes (Figure 2d). This trend was also consistent with our previous results which indicated that F complexes were internalized about 13 times more efficiently than H complexes by HEK293 cells (24). Additionally, F complexes were also more efficiently internalized than W complexes and this may be one possible factor for the higher luciferase expression achieved with the former. As a negative control, cells were also transfected with F complexes (N/P 20) containing unlabelled plasmids to show that the difference in mean fluorescence detected was due to DNA uptake and not novel protein expression or other nonspecific effects of transfection. The adequate removal of surface-adsorbed particles was proved by incubating the cells with sodium azide/2-deoxyglucose (NaN3/DOG), which inhibited energy-dependent endocytosis but not surface binding (41). This reduced the mean fluorescence near to background (see later results).
The observation that F complexes were more readily internalized than H complexes was further corroborated with confocal observations where brightly fluorescent cells were detected upon exposure to F complexes containing Cy5-labelled DNA (Supplementary Figure S1). In sharp contrast, little fluorescence was detected in cells exposed to H complexes. As above, the addition of NaN3/DOG blocked the internalization of F complexes (see later results). Z-stacking also confirmed the absence of signals throughout the entire thickness of the cell layer, demonstrating the efficient removal of surface-bound particles.
While it is clear that the hydrophobic group provided for greater complex stability, cellular penetration and transfection, the actual mechanism responsible for these effects remains to be explained. We note that the size of F complexes upon DNA binding was 15–50 µm [peptides with other hydrophobic groups, e.g. I, W and G showed similar size distribution up to N/P 30 (data not shown)], while H/DNA complexes were 0.8–2.0 µm (24). It is possible that the hydrophobic moieties may have caused the aggregation and precipitation of complexes onto the cell layer for a more direct contact. In turn, this may boost the internalization and transfection achieved with F complexes compared to H complexes. Although the contribution of precipitation cannot presently be ascertained, it is still noteworthy that the luciferase expression level mediated by F complexes was on average more than two orders higher than that achieved using calcium phosphate (Supplementary Figure S2), a system known to efficiently transfect cells by precipitation (42). This justifies the further development of these peptides. Moreover, the transfection mediated by F complexes was also more than two orders higher than that achieved with W complexes (Figure 2a), despite both having comparable size. This argues that precipitation alone is not sufficient to ensure high transfection and that other aspects of the molecular design are important. Current efforts in our lab are on the elucidation of the effects of the hydrophobic block.
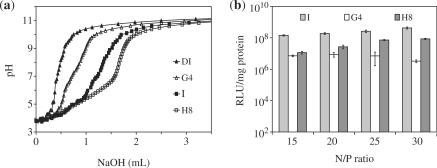
We next investigated whether the buffering histidine block is a useful component of the design. The imidazole sidegroup of histidine has a pKa ∼6.0 and as such, is suited to disrupt endosomal vesicles by proton scavenging effects under the proton sponge hypothesis (31,43,44). To directly test for the influence of the histidine block on the buffering capacity, we synthesized additional peptide sequences in which the length of the histidine block was either increased to 8, i.e. NH2-I5H8R8-CONH2 (H8), or totally replaced with non-functional glycine spacers, i.e. NH2-I5G4R8-CONH2 (G4). We then compared the pH profile of peptides I, H8 and G4 upon titration against a standard solution of dilute sodium hydroxide (0.01 M). Figure 3a indicates that a larger amount of base was needed to cause a unit increase in pH of H8, followed by I and then by G4. This confirms that a larger buffering capacity can be achieved with a longer histidine block and most probably explains why I transfected significantly better than G4 (Figure 3b). The presence of a buffering histidine block is thus an important design consideration.
Figure 3.
Effects of the buffering histidine block on the properties of the oligopeptide vectors. (a) Titration experiments with 16 ml of de-ionized water (DI) containing 0.5 mg/ml peptide (G4, I or H8) or nothing (DI) against 0.01 M NaOH. (b) Luciferase expression levels in MCEC transfected with peptide/DNA complexes with varying length and nature of the buffering block. Error bars represent SD.
However, as also observed by others (45), a larger buffering capacity does not necessarily result in greater transfection ability. As shown in Figure 3b, I still mediated higher luciferase expression than H8, despite the latter having a greater buffering capacity. This may be because the process of endosomal escape, albeit important, is just one of the several possible limiting steps encountered by the DNA complex en route to the nucleus. DNA release, for instance, may be another potential limitation. To test this, we conducted FRET experiments comparing peptides I and H8. For this study, plasmids encoding for luciferase were double-labelled with the FRET pair Cy3 (donor) and Cy5 (acceptor). During live-cell imaging, only Cy3 was excited but emissions from both Cy3 and Cy5 were collected. When the DNA is condensed, Cy3 and Cy5 are close enough for FRET to occur (46). Upon complex dissociation and DNA uncondensation, FRET is reduced and only Cy3 is detected. Each image was binary-scored according to the number of pixels positive for Cy 3 (green) or Cy5 (false-coloured red). The ratio of red to green pixels (R/G) was then used to indicate the extent of FRET.
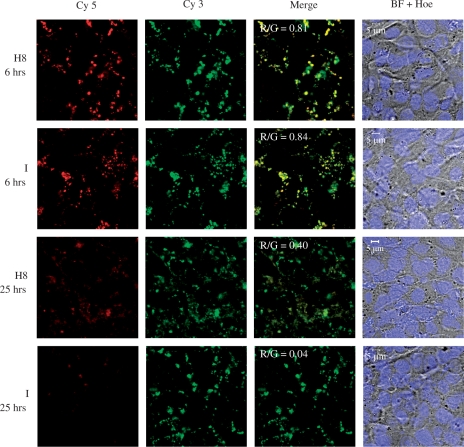
Figure 4 shows that at 6 h post-exposure, strong co-localization of red and green signals were observed in cells transfected with either H8 (R/G = 0.81) or I (R/G = 0.84). This suggests that the DNA remained condensed, presumably by the peptides up to this point. After 25 h, however, FRET was largely unobservable in cells transfected with I (R/G = 0.04), whereas significant FRET can still be seen in cells transfected with H8 (R/G = 0.40). This suggests that DNA release from H8 complexes is indeed more difficult compared to I complexes and may contribute to the lower luciferase expression achieved with H8.
Figure 4.
MCEC were exposed to peptide I or H8/DNA complexes (N/P 20) formed at pH 7.0. FRET was used to show if the DNA was still condensed at selected time points. The DNA had been double-labelled with both Cy5 and Cy3. During imaging, only Cy3 was excited. If there was FRET, Cy5 (false-coloured red) can also be detected, which will then co-localize with Cy3 (green) to give yellow signals in the merged image. All cells were stained for their nuclei with Hoechst (Hoe) and presented with the bright field (BF) images.
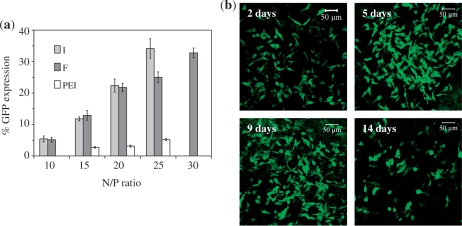
Based on luciferase expression results, two of the best peptides (F and I) were selected to deliver plasmids encoding for GFP into MCEC. Flow cytometry was then used to quantify the percentage of cells that expressed GFP. As shown in Figure 5a, F and I were generally comparable and more efficient (>5×) than PEI in inducing GFP expression in MCEC. However, although peptide I seemed to perform better than F at N/P 25, the toxicity of I complexes was higher, as shown in Supplementary Figure S3. At N/P 20 and 25, for instance, the viability of cells transfected with I complexes was ∼70% and ∼65%, respectively, compared to viabilities of ∼90% and ∼80% with F complexes. Hence, peptide F at N/P 20 (peptide concentration in medium ∼32 µM) was selected for all further work.
Figure 5.
(a) Percentage of MCEC expressing GFP 2 days post-transfection with peptides I, F or PEI. (b) Confocal images showing MCEC expressing GFP up to 14 days post-transfection with F/DNA complexes (N/P 20). Each image is a merger of several z-stack images acquired as the cell layers became overly confluent with the long incubation period. Cells also started to detach from the culture chambers after prolonged incubation, explaining the apparent reduction in cells expressing GFP on day 14.
GFP expression can be observed in MCEC up to a period of 14 days post-transfection with peptide F at N/P 20 (Figure 5b). The number of GFP positive cells (but not necessarily the proportion) appeared to increase and then decrease, consistent with cell proliferation and death. Due to technical limitations, however, it was not possible to quantify the percentage of cell population expressing GFP upon prolonged incubation.
The presence of serum during transfection significantly augmented both luciferase (Supplementary Figure S4a) and GFP (Supplementary Figure S4b) expression mediated by peptide F, despite reports to the contrary with other arginine-rich vectors (47). This may be due to the enhanced viability of cells in the presence of serum (Supplementary Figure S4a). This finding is particularly useful for our future application as it is preferable to culture whole corneas in the presence of serum throughout the ex vivo manipulation. Additionally, we observed that the expression level can be further increased with prolonged exposure of MCEC to the complexes. For instance, increasing the incubation period from 4 to 6 h produced a 2.5 times increment in the amount of luciferase expressed (Supplementary Figure S5).
The pH used for complex formation also influenced the properties of the vector. It was observed that DNA binding became increasingly efficient as pH was reduced from 7.0 to 5.0, as shown by an ethidium bromide exclusion assay (Supplementary Figure S6a). This was consistent with the more extensive protonation of histidine at a lower pH. However, the level of luciferase expression mediated by complexes formed at pH 7.0 was significantly higher than those formed at pH 6.3 or 5.0 (Supplementary Figure S6b). FRET experiments then indicated that decondensation of DNA within the cells was slower with complexes formed at pH 5.0 (R/G = 0.70 at 25 h post-transfection) compared to those formed at pH 7.0 (R/G = 0.28 at 25 h post-transfection), possibly contributing to their lower transfection ability (Supplementary Figure S7).
Having justified the tri-block design and demonstrated the ability of the peptides to deliver genetic cargoes efficiently into MCEC, we next investigated the mechanism(s) by which the peptide complexes were internalized by MCEC. The pathways of cellular uptake can be broadly divided into those that are energy dependent or independent. Endocytosis, which is energy-dependent, is by far the best understood and can be further subcategorized into clathrin-mediated endocytosis, caveolae/lipid raft-mediated endocytosis (simply referred to as caveolae-mediated endocytosis hereon), macropinocytosis and clathrin- and caveolae-independent endocytosis (48).
The exact internalization mechanism of arginine-rich peptides (including HIV-Tat derived peptides) is controversial. Energy dependent (29,30,49–52), energy independent (53,54) and a combination of energy-dependent and -independent pathways (depending on the concentration of peptide present) (55), have all been implicated as possible internalization pathways for arginine-rich peptides. In fact, just within the realm of endocytosis, the clathrin-mediated pathway (52), caveolae-mediated pathway (30) and macropinocytosis (29) had each been individually identified as the main route of entry, while others claim that a combination of several pathways was more likely (50). Reasons for the apparent discrepancy include factors such as differing experimental conditions, sequence of peptide used and lineage of cells studied. Care should also be taken when drawing conclusions based on inhibitors that may affect more than one pathway concurrently. We thus investigated the possible route(s) by which MCEC internalize the peptide/DNA complexes.
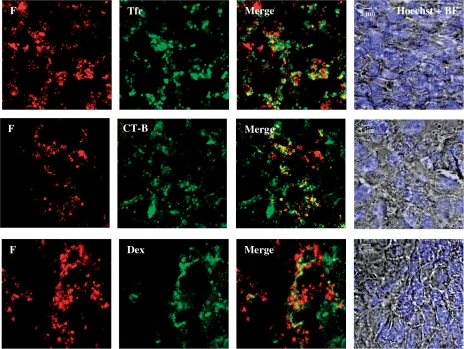
We co-cultured the cells with peptide F complexes (N/P 20) containing rhodamine-labelled DNA and either one of the fluorescently labelled makers for specific pathways of endocytosis: namely, transferrin, cholera toxin or dextran, which are known to be internalized exclusively via the clathrin-mediated pathway (49,56,57), caveolae-mediated pathway (27) and macropinocytosis (55,58), respectively. After a brief incubation, cells were washed extensively with PBS and observed for any co-localization of the complexes and endocytosis markers. Cells were observed live to avoid artefacts due to fixation (49). Interestingly, we observed that the complexes exhibited partial co-localization with all three endocytosis markers (Figure 6). This suggests that all three mechanisms could contribute to the uptake of peptide complexes by MCEC, which seems reasonable in light of the heterogeneity of the complex particles.
Figure 6.
Co-localization (yellow) of F peptide complexed with DNA (red) at N/P 20 with classical markers for specific endocytosis pathways like transferrin (Tfr, green) for clathrin-mediated endocytosis, cholera toxin B (CT-B, green) for caveolae/lipid raft-mediated endocytosis and dextran (Dex, false-coloured green) for macropinocytosis. Cells were all stained with the Hoechst nuclear dye (blue) and presented with the bright field (BF) images. Imaging was done 40–90 min post-exposure to peptide/DNA complexes.
Involvement of the clathrin-mediated pathway—whose vesicles undergo acidification (48,58,59), unlike caveosomes (30,40,58,60) and macropinosomes (29,58,59) which remain neutral—was also supported by the observation that chloroquine increased luciferase expression by 3.5- to 7.6-fold (Supplementary Figure S8). This is consistent with the ability of chloroquine to promote endosomal rupture and cytosolic escape by the prevention of acidification (31,44).
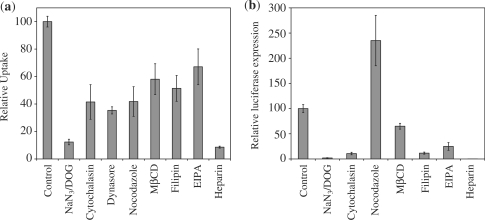
To provide further evidence for the involvement of different endocytotic pathways, we quantified the effects of various inhibitors on MCEC at both the uptake (Figure 7a) and gene expression (Figure 7b) level. This then allows discrimination of which pathway is most biologically relevant, as efficient internalization may not necessarily lead to high expression. All data shown were normalized to cells transfected in the presence of vehicle control (DMSO or water) alone.
Figure 7.
Effects of various inhibitors on the (a) uptake and (b) gene expression of MCEC transfected with F complexes (N/P 20). Cells were pre-incubated with inhibitors for 1 h prior to exposure to F complexes containing Cy5-labelled DNA (for uptake quantification using flow cytometry) or unlabelled DNA (for gene expression). Values were all referenced to control cells normally transfected with F complexes without any inhibitors (assigned as 100%). In cases whereby the inhibitors had to be dissolved in DMSO, control cells were also transfected in the presence of the same volume of pure DMSO. Error bars represent SD.
The combination of NaN3/DOG blocks intracellular ATP synthesis and inhibits all forms of energy-dependent translocation (41). Since incubating the cells with NaN3/DOG drastically reduced their ability to endocytose (∼87.7% inhibition) and express the gene (∼98.2% inhibition), this confirms that energy-dependent endocytosis is the main form of uptake relevant to this system. Live-cell confocal observations further demonstrated the impaired uptake upon NaN3/DOG treatment (Supplementary Figure S9). Results from both flow cytometry and confocal microscopy also show that surface-adsorbed particles have been adequately removed in all experiments.
The actin cytoskeleton (required during the early budding process of vesicles near the cell surface), GTPase dynamin (known to mediate the budding of pits) and microtubules (required for the subsequent trafficking of budded vesicles) are all reported to be important for the trafficking of endosomes, caveosomes and macropinosomes (27,48,61,62). Furthermore, both the clathrin- and caveolae-mediated pathways have been shown to be sensitive to cholesterol depletion (52,62,63). Treatment of the cells with cytochalasin [which disrupts the actin cytoskeleton (56)], dynasore [which inhibits the GTPase activity of dynamin (28)], nocodazole [which disrupts microtubule depolymerization (26)] and methyl-β-cyclodextrin [MβCD, which extracts cholesterol from cell membrane (63)] significantly reduced uptake by 58.5%, 64.7%, 58.2% and 41.9%, respectively. Gene expression also dropped by 89.3% and 35.0% after cytochalasin and MβCD treatment, respectively. The effects of dynasore on gene expression cannot be examined due to its pronounced toxicity during the longer incubation period used for gene expression assays. Nocodazole, on the other hand, increased gene expression 2.35 times. A possible reason may be due to the disruption of the microtubule network and therefore, late-stage trafficking. For instance, it was reported that in nocodazole-treated cells, internalized molecules remained sequestered longer within early endosomes (64). The delay in maturation into digestive lysosomes may mean that the complexes have more opportunity to escape into the cytosol, in turn increasing the chance of their genetic cargoes being expressed.
The involvement of both the caveolae-mediated pathway and macropinocytosis is given further credit by using inhibitors that shuts down the respective pathways. Filipin is an inhibitor of the caveolae-mediated pathway (26,27,54,57,63) and treatment with this drug reduced uptake and gene expression by 48.6% and 88.4%, respectively. Similarly, 5-(N-ethyl-N-isopropyl)amiloride (EIPA), an inhibitor of macropinocytosis (29,50,55), reduced uptake and expression by 32.9% and 75.1%, respectively, though it is recognized that the action of these inhibitors may not be totally specific.
Cell-surface proteoglycans, in particular, heparan sulphate (HS) have been reported to be key mediators in the initial binding events leading to internalization (58,65,66). Our results suggest that interaction with HS indeed seems pivotal for internalization as the presence of heparin, which competes with HS for binding with the complexes, dramatically reduced uptake (of which it may the binding process that is blocked, and not internalization) and luciferase expression by 91.4% and 99.9%, respectively. The concentration of heparin used for this experiment (50 µg/ml) is not expected to affect the stability of the complexes (Figure 2c).
Recently, it was demonstrated that particles as large as 3 µm can still be internalized by non-phagocytic cells using endocytotic pathways (41), suggesting that the actual size cut-off limit for internalization may be larger than previously thought. Although the peptide F complexes are larger than 3 µm, it is possible that their internalization involves fragmentation into smaller ‘bite-size’ pieces. This fragmentation process has been observed during macropinocytosis and the pinching-off of membrane vesicles (67).
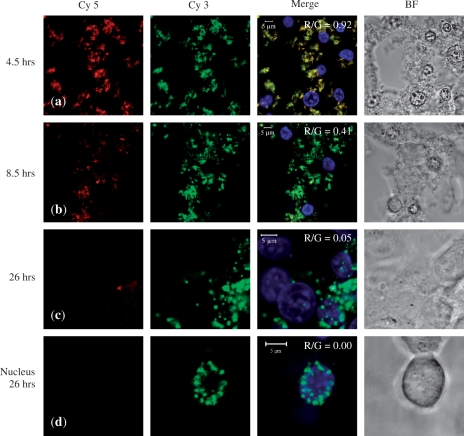
While the previous confocal studies determined the locality of the DNA, they gave no information as to whether the DNA remained condensed. To address this, we conducted FRET experiments, this time involving peptide F and MCEC using earlier time points and analysing more closely cellular localization. Figure 8a shows that strong co-localization of Cy3 and Cy5 (R/G = 0.92) can be observed up to 4.5 h after transfection, indicating that the DNA was still condensed, presumably by the peptide carrier. There was also evidence of early stage nuclear entry of condensed DNA, as indicated by the co-localization of yellow and blue Hoechst signals. This suggests that the peptide/DNA entered the nucleus as an associated complex. Similar nuclear entry of PEI/DNA complexes has been previously reported, albeit at different time-points post-transfection (68).
Figure 8.
FRET study. Confocal images taken of live MCEC at row (a) 4.5 h, row (b) 8.5 h and row (c) 26 h after initial exposure to peptide F/DNA complexes at N/P 20. The DNA had been double-labelled with both Cy5 and Cy3. During imaging, only Cy3 was excited. If there was FRET, Cy5 (false-coloured red) can also be detected, which will then co-localize with Cy3 (green) to give yellow signals in the merged image. Row (d) A nucleus at 26 h showing large scale accumulation of uncondensed DNA. All cells were stained for their nuclei with Hoechst (blue) and presented with the bright field (BF) images.
By 8.5 h, however, there was evidence of DNA uncondensing, as shown by a decrease in the red Cy 5 signal (Figure 8b). The merged image therefore appears more green than yellow (R/G = 0.41). By 26 h, large amount of DNA had accumulated within the nuclei which showed little FRET signal (R/G = 0.05) and so was presumably not condensed (Figure 8c). Figure 8d focuses on a nucleus at 26 h exhibiting large-scale accumulation of uncondensed DNA (R/G = 0.00). Finally, to ensure that the disappearance of Cy5 signals was due to a lost of FRET and not simply an artefact of photobleaching or degradation, we excited with the Cy5 wavelength alone, which resulted in the complete disappearance of Cy3 signals and appearance of Cy5 fluorescence (data not shown).
To demonstrate biological relevance, we next used the peptides to deliver plasmids encoding for IDO into MCEC. IDO is a cytosolic haem-containing enzyme which catalyses the biodegradation of tryptophan down the kynurenine pathway (69). Since the growth of T lymphocytes are critically dependent on tryptophan—without which they get arrested in their G1 phase (70)—it has been shown that IDO overexpression can suppress T-cell responses (71,72). In the context of corneal transplantation, the induction of IDO expression in the corneal endothelium prolongs allograft survival by locally suppressing T-cell activity (12).
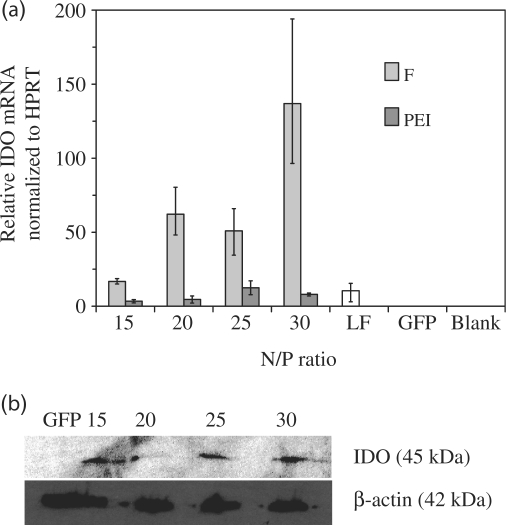
We first transfected MCEC with peptide F complexed with plasmids encoding for IDO. IDO mRNA expression normalized to the housekeeping HPRT gene was then quantified using RT–PCR (Figure 9a). As can be seen, there was a strong increase (>105 fold) in the level of IDO mRNA following peptide-mediated transfection, compared to untransfected and mock-transfected (GFP) MCEC. More significantly, the levels of IDO mRNA expression induced by peptide F were also higher (by up to 17-fold) than those induced by PEI or lipofectamine™2000. IDO protein was also detected by western blotting after cells have been transfected with peptide F/IDO complexes (Figure 9b).
Figure 9.
(a) IDO mRNA expression in MCEC transfected with peptide F, PEI or lipofectamine™2000 (LF). Negative controls include untransfected MCEC (Blank) or mock transfection with F/GFP complexes (GFP) at N/P 30. Error bars represent standard deviation. (b) IDO protein expression detected by western blot following transfection with peptide F at N/P ratios 15–30. Mock transfection with F/GFP complexes (GFP) at N/P 30 served as the negative control.
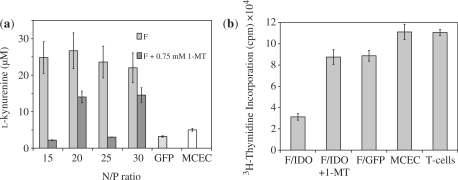
To show that the IDO expressed by the cells retained functional activity, the amount of l-kynurenine, the main breakdown product of tryptophan (73), in the culture supernatant was measured (Figure 10a). As can be seen, the levels of l-kynurenine detected in the supernatant of MCEC transfected with peptide F were elevated compared to untransfected and mock-transfected controls. The IDO produced by the cells was therefore biologically functional. Further, the levels of l-kynurenine detected were reduced significantly, though to a variable extent, by the addition of 1-MT, an inhibitor of IDO (70–72). This showed that the effect of l-kynurenine production was due to IDO activity.
Figure 10.
IDO functional assays. (a) Levels of l-kynurenine detected in the supernatants of MCEC transfected with F/IDO complexes, F/GFP complexes (GFP) at N/P 30 or untransfected (MCEC). The addition of the specific IDO antagonist, 1-MT, reduced the catalytic activity of IDO and the amount of l-kynurenine detected by varying degrees. (b) CD4+ T-cells were either incubated alone (T-cells) or with MCEC in a 3-day proliferation assay. The proliferation rate of CD4+ T-cells was then assessed by [3H]thymidine incorporation. MCEC used were either untransfected (MCEC), or transfected with IDO (F/IDO) or GFP (F/GFP) using peptide F at N/P 30. The addition of 0.75 mM of 1-MT (F/IDO+1-MT) reversed the inhibiting effects of IDO activity. All error bars represent SD.
CD4+ T-cells are the chief mediator of ocular inflammation (2). It was therefore important to evaluate the effect of IDO production on the proliferation of CD4+ T-cells. To achieve this, peptide-transfected MCEC was incubated with CD4+ responder T cells freshly isolated from the spleens of C3H mice and stimulated with anti-CD3 and CD28 beads. The proliferation of the T cells was then measured by [3H]thymidine incorporation after a 3-day period. It was observed that the proliferative ability of T-cells incubated with IDO-transfected MCEC, but not GFP-transfected MCEC, was reduced by up to 70% compared to the untransfected MECE control (Figure 10b). Further, the inhibitory effect was reversed by the addition of 1-MT. These data showed that the suppressed growth of T cells was specifically due to the effects of IDO activity, offering us a promising strategy to apply in the ex vivo manipulation of whole corneas for our future work.
CONCLUSION
In summary, we have demonstrated that the tri-block arginine-rich oligopeptides were more effective than PEI in mediating the expression of reporter (luciferase and GFP) and therapeutic (IDO) genes in MCEC. The presence of a hydrophobic block and a buffering component within the tri-block design were systematically shown to be important in influencing the transfection performance of the peptide. We elucidated the pathways of internalization and showed that the MCEC mainly utilized energy-dependent endocytosis where internalization was heavily dependent on interactions with cell-surface HS. Additionally, distinct pathways such as the clathrin-mediated endocytosis, caveolae-mediated endocytosis and macropinocytosis all contributed to the eventual uptake, although none of the pathways seemed to dominate. FRET studies demonstrated that the DNA entered cells as a condensed complex. Events after uptake, for instance, DNA uncondensation (by 8.5 h) and the accumulation of free DNA within nuclei (by 26 h) were demonstrated. We have also shown that we can use the peptides to deliver plasmids encoding for a biologically relevant protein, IDO, that has previously been shown to prolong corneal allograft survival (12). IDO expression was then detected at the mRNA and protein level and shown to be functionally active and capable of inhibiting T-cell proliferation. Although transfection efficiency may depend on the size and nature of plasmids used, we have shown that the peptides were able to deliver plasmids ranging in size from 4.7 to 8.5 kb. This class of non-viral vector therefore promises to be a versatile vehicle for the immunomodulation of corneal transplantations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Institute of Bioengineering and Nanotechnology, Agency for Science; Technology and Research, Singapore and the Wellcome Trust; NIHR Biomedical Research Centre 5 funding scheme (to A.J.T.G.). Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Xin Dong Guo (School of Chemistry and Chemical Engineering, South China University of Technology) for simulating the molecular structure of the peptide and Bai Xue Zheng (National University of Singapore) for assistance in the quantification of FRET images.
REFERENCES
- 1.Niederkorn JY. The immune privilege of corneal allografts. Transplantation. 1999;67:1503–1508. doi: 10.1097/00007890-199906270-00001. [DOI] [PubMed] [Google Scholar]
- 2.Niederkorn JY. The immune privilege of corneal grafts. J. Leukoc. Biol. 2003;74:167–171. doi: 10.1189/jlb.1102543. [DOI] [PubMed] [Google Scholar]
- 3.Gong N, Pleyer U, Volk H-D, Ritter T. Effects of local and systemic viral interleukin-10 gene transfer on corneal allograft survival. Gene Ther. 2007;14:484–490. doi: 10.1038/sj.gt.3302884. [DOI] [PubMed] [Google Scholar]
- 4.Jun AS, Larkin DFP. Prospects for gene therapy in corneal disease. Eye. 2003;17:906–911. doi: 10.1038/sj.eye.6700565. [DOI] [PubMed] [Google Scholar]
- 5.George AJT, Arancibia-Cárcamo CV, Awad HM, Comer RM, Fehevari Z, King WJ, Kadifachi M, Hudde T, Kerouedan-Lebossé C, Mirza F, et al. Gene delivery to the corneal endothelium. Am. J. Respir. Crit. Care Med. 2000;162:S194–S200. doi: 10.1164/ajrccm.162.supplement_3.15tac11. [DOI] [PubMed] [Google Scholar]
- 6.Larkin DFP, Oral HB, Ring CJA, Lemoine NR, George AJT. Adenovirus-mediated gene delivery to the corneal endothelium. Transplantation. 1996;61:363–370. doi: 10.1097/00007890-199602150-00005. [DOI] [PubMed] [Google Scholar]
- 7.Klausner EA, Peer D, Chapman RL, Multack RF, Andurkar SV. Corneal gene therapy. J. Control Rel. 2007;124:107–133. doi: 10.1016/j.jconrel.2007.05.041. [DOI] [PubMed] [Google Scholar]
- 8.George AJT, Larkin DFP. Corneal transplantation: the forgotten graft. Am. J. Transplant. 2004;4:678–685. doi: 10.1111/j.1600-6143.2004.00417.x. [DOI] [PubMed] [Google Scholar]
- 9.Borras T, Tamm ER, Zigler JS., Jr. Ocular adenovirus gene transfer varies in efficiency and inflammatory response. Invest. Ophthalmol. Vis. Sci. 1996;37:1282–1293. [PubMed] [Google Scholar]
- 10.Klebe S, Sykes PJ, Coster DJ, Krishnan R, Williams KA. Prolongation of sheep corneal allograft survival by ex vivo transfer of the gene encoding interleukin-10. Transplantation. 2001;71:1214–1220. doi: 10.1097/00007890-200105150-00006. [DOI] [PubMed] [Google Scholar]
- 11.Wang X, Appukuttan B, Ott S, Patel R, Irvine J, Song J, Park J-HC, Smith R, Stout JT. Efficient and sustained transgene expression in human corneal cells mediated by a lentiviral vector. Gene Ther. 2000;7:196–200. doi: 10.1038/sj.gt.3301075. [DOI] [PubMed] [Google Scholar]
- 12.Beutelspacher SC, Pillai R, Watson MP, Tan PH, Tsang J, McClure MO, George AJT, Larkin DFP. Function of indoleamine 2,3-dioxygenase in corneal allograft rejection and prolongation of allograft survival by over-expression. Eur. J. Immunol. 2006;36:690–700. doi: 10.1002/eji.200535238. [DOI] [PubMed] [Google Scholar]
- 13.Hudde T, Rayner SA, Alwis MD, Thrasher AJ, Smith J, Coffin RS, George AJT, Larkin DFP. Adeno-associated and herpes simplex viruses as vectors for gene transfer to the corneal endothelium. Cornea. 2000;19:369–373. doi: 10.1097/00003226-200005000-00022. [DOI] [PubMed] [Google Scholar]
- 14.Luo D, Saltzman WM. Synthetic DNA delivery systems. Nat. Biotechnol. 2000;18:33–37. doi: 10.1038/71889. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell DJ, Kim DT, Steinman L, Fathman CG, Rothbard JB. Polyarginine enters cells more efficiently than other polycationic homopolymers. J. Peptide Res. 2000;56:318–325. doi: 10.1034/j.1399-3011.2000.00723.x. [DOI] [PubMed] [Google Scholar]
- 16.Zauner W, Ogris M, Wagner E. Polylysine-based transfection systems utilizing receptor-mediated delivery. Adv. Drug Deliv. Rev. 1998;30:97–113. doi: 10.1016/s0169-409x(97)00110-5. [DOI] [PubMed] [Google Scholar]
- 17.Futaki S, Ohashi W, Suzuki T, Niwa M, Tanaka S, Ueda K, Harashima H, Sugiura Y. Stearylated arginine-rich peptides: a new class of transfection systems. Bioconjugate Chem. 2001;12:1005–1011. doi: 10.1021/bc015508l. [DOI] [PubMed] [Google Scholar]
- 18.Wadhwa MS, Collard WT, Adami RC, McKenzie DL, Rice KG. Peptide-mediated gene delivery: influence of peptide structure on gene expression. Bioconjugate Chem. 1997;8:81–88. doi: 10.1021/bc960079q. [DOI] [PubMed] [Google Scholar]
- 19.McKenzie DL, Smiley E, Kwok KY, Rice KG. Low molecular weight disulfide cross-linking peptides as nonviral gene delivery carriers. Bioconjugate Chem. 2000;11:901–909. doi: 10.1021/bc000056i. [DOI] [PubMed] [Google Scholar]
- 20.Yu W, Pirollo KF, Yu B, Rait A, Xiang L, Huang W, Zhou Q, Ertem G, Chang EH. Enhanced transfection efficiency of a systemically delivered tumor-targeting immunolipoplex by inclusion of a pH-sensitive histidylated oligolysine peptide. Nucleic Acids Res. 2004;32:e48. doi: 10.1093/nar/gnh049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyer M, Philipp A, Oskuee R, Schmidt C, Wagner E. Breathing life into polycations: functionalization with pH-responsive endosomolytic peptides and polyethylene glycol enables siRNA delivery. J. Am. Chem. Soc. 2008;130:3272–3273. doi: 10.1021/ja710344v. [DOI] [PubMed] [Google Scholar]
- 22.Curiel DT, Agarwal S, Wagner E, Cotten M. Adenovirus enhancement of transferrin-polylysine-mediated gene delivery. Proc. Natl Acad. Sci. 1991;88:8850–8854. doi: 10.1073/pnas.88.19.8850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wagner E, Plank C, Zatloukal K, Cotten M, Birnstiel ML. Influenza virus hemagglutinin HA-2 N-terminal fusogenic peptides augment gene transfer by transferrin-polylysine-DNA complexes: toward a synthetic virus-like gene-transfer vehicle. Proc. Natl Acad. Sci. 1992;89:7934–7938. doi: 10.1073/pnas.89.17.7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seow WY, Yang YY. A class of cationic triblock amphiphilic oligopeptides as efficient gene-delivery vectors. Adv. Mater. 2009;21:86–90. [Google Scholar]
- 25.Putnam D. Polymers for gene delivery across length scales. Nat. Mater. 2006;5:439–451. doi: 10.1038/nmat1645. [DOI] [PubMed] [Google Scholar]
- 26.Gratton SEA, Napier ME, Ropp PA, Tian S, DeSimone JM. Microfabricated particles for engineered drug therapies: elucidation into the mechanisms of cellular internalization of PRINT particles. Pharm. Res. 2008;25:2845–2852. doi: 10.1007/s11095-008-9654-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torgersen ML, Skretting G, Deurs BV, Sandvig K. Internalization of cholera toxin by different endocytic mechanisms. J. Cell Sci. 2001;114:3737–3747. doi: 10.1242/jcs.114.20.3737. [DOI] [PubMed] [Google Scholar]
- 28.Macia E, Ehrlich M, Massol R, Boucrot E, Brunner C, Kirchhausen T. Dynasore, a cell-permeable inhibitor of dynamin. Dev. Cell. 2006;10:839–850. doi: 10.1016/j.devcel.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 29.Wadia JS, Stan RV, Dowdy SF. Transducible TAT-HA fusogenic peptide enhances escape of TAT-fusion proteins after lipid raft macropinocytosis. Nat. Med. 2004;10:310–315. doi: 10.1038/nm996. [DOI] [PubMed] [Google Scholar]
- 30.Ferrari A, Pellegrini V, Arcangeli C, Fittipaldi A, Giacca M, Beltram F. Caveolae-mediated internalization of extracellular HIV-1tat fusion proteins visualized in real time. Mol. Ther. 2003;8:284–294. doi: 10.1016/s1525-0016(03)00122-9. [DOI] [PubMed] [Google Scholar]
- 31.Akinc A, Thomas M, Klibanov AM, Langer R. Exploring polyethylenimine-mediated DNA transfection and the proton sponge hypothesis. J. Gene Med. 2005;7:657–663. doi: 10.1002/jgm.696. [DOI] [PubMed] [Google Scholar]
- 32.Janin J. Surface and inside volumes in globular proteins. Nature. 1979;277:491–492. doi: 10.1038/277491a0. [DOI] [PubMed] [Google Scholar]
- 33.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 34.Lin C, Zhong Z, Lok MC, Jiang X, Hennink WE, Feijen J, Engbersen JFJ. Novel bioreducible poly(amido amine)s for highly efficient gene delivery. Bioconjugate Chem. 2007;18:138–145. doi: 10.1021/bc060200l. [DOI] [PubMed] [Google Scholar]
- 35.Breitenkamp RB, Emrick T. Pentalysine-grafted romp polymers for DNA complexation and delivery. Biomacromolecules. 2008;9:2495–2500. doi: 10.1021/bm800511p. [DOI] [PubMed] [Google Scholar]
- 36.Jadhav V, Maiti S, Dasgupta A, Das PK, Dias RS, Miguel MG, Lindman B. Effect of the head-group geometry of amino acid-based cationic surfactants on interaction with plasmid DNA. Biomacromolecules. 2008;9:1852–1859. doi: 10.1021/bm8000765. [DOI] [PubMed] [Google Scholar]
- 37.Rimann M, Lühmann T, Textor M, Guerino B, Ogier J, Hall H. Characterization of PLL-g-PEG-DNA nanoparticles for the delivery of therapeutic DNA. Bioconjugate Chem. 2008;19:548–557. doi: 10.1021/bc7003439. [DOI] [PubMed] [Google Scholar]
- 38.Koping-Hoggard M, Varum KM, Issa M, Danielsen S, Christensen BE, Stokke BT, Artursson P. Improved chitosan-mediated gene delivery based on easily dissociated chitosan polyplexes of highly defined chitosan oligomers. Gene Ther. 2004;11:1441–1452. doi: 10.1038/sj.gt.3302312. [DOI] [PubMed] [Google Scholar]
- 39.Duncan R. Polymer conjugates as anticancer nanomedicine. Nat. Rev. Cancer. 2006;6:688–701. doi: 10.1038/nrc1958. [DOI] [PubMed] [Google Scholar]
- 40.Brooks H, Lebleu B, Vive. E. Tat peptide-mediated cellular delivery: back to basics. Adv. Drug Deliv. Rev. 2005;57:559–577. doi: 10.1016/j.addr.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 41.Gratton SEA, Ropp PA, Pohlhaus PD, Luft JC, Madden VJ, Napier ME, DeSimone JM. The effect of particle design on cellular internalization pathways. Proc. Natl Acad. Sci. 2008;105:11613–11618. doi: 10.1073/pnas.0801763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pouton CW, Seymour LW. Key issues in non-viral gene delivery. Adv. Drug Deliv. Rev. 2001;46:187–203. doi: 10.1016/s0169-409x(00)00133-2. [DOI] [PubMed] [Google Scholar]
- 44.Gabrielson NP, Pack DW. Acetylation of polyethylenimine enhances gene delivery via weakened polymer/DNA interactions. Biomacromolecules. 2006;7:2427–2435. doi: 10.1021/bm060300u. [DOI] [PubMed] [Google Scholar]
- 45.Liu Y, Reineke TM. Poly(glycoamidoamine)s for gene delivery. Structural effects on cellular internalization, buffering capacity, and gene expression. Bioconjugate Chem. 2007;18:19–30. doi: 10.1021/bc060029d. [DOI] [PubMed] [Google Scholar]
- 46.Liu X, Yang JW, Lynn DM. Addition of “charge-shifting” side chains to linear poly(ethyleneimine) enhances cell transfection efficiency. Biomacromolecules. 2008;9:2063–2071. doi: 10.1021/bm800291v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moulton HM, Nelson MH, Hatlevig SA, Reddy MT, Iversen PL. Cellular uptake of antisense morpholino oligomers conjugated to arginine-rich peptides. Bioconjugate Chem. 2004;15:290–299. doi: 10.1021/bc034221g. [DOI] [PubMed] [Google Scholar]
- 48.Conner SD, Schmid SL. Regulated portals of entry into the cell. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 49.Richard JP, Melikov K, Vives E, Ramos C, Verbeure B, Gait MJ, Chernomordik LV, Lebleu B. Cell-penetrating peptides: a reevaluation of the mechanism of cellular uptake. J. Biol. Chem. 2003;278:585–590. doi: 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
- 50.Nakase I, Niwa M, Takeuchi T, Sonomura K, Kawabata N, Koike Y, Takehashi M, Tanaka S, Ueda K, Simpson JC, et al. Cellular uptake of arginine-rich peptides: roles for macropinocytosis and actin rearrangement. Mol. Ther. 2004;10:1011–1022. doi: 10.1016/j.ymthe.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 51.Fawell S, Seery J, Daikh Y, Moore C, Chen LL, Pepinsky B, Barsoum J. Tat-mediated delivery of heterologous proteins into cells. Proc. Natl Acad. Sci. 1994;91:664–668. doi: 10.1073/pnas.91.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Richard JP, Melikov K, Brooks H, Prevot P, Lebleu B, Chernomordik LV. Cellular uptake of unconjugated tat peptide involves clathrin dependent endocytosis and heparan sulfate receptors. J. Biol. Chem. 2005;280:15300–15306. doi: 10.1074/jbc.M401604200. [DOI] [PubMed] [Google Scholar]
- 53.Rothbard JB, Jessop TC, Lewis RS, Murray BA, Wender PA. Role of membrane potential and hydrogen bonding in the mechanism of translocation of guanidinium-rich peptides into cells. J. Am. Chem. Soc. 2004;126:9506–9507. doi: 10.1021/ja0482536. [DOI] [PubMed] [Google Scholar]
- 54.Eguchi A, Akuta T, Okuyama H, Senda T, Yokoi H, Inokuchi H, Fujita S, Hayakawa T, Takeda K, Hasegawa M, et al. Protein transduction domain of HIV-1 tat protein promotes efficient delivery of DNA into mammalian cells. J. Biol. Chem. 2001;276:26204–26210. doi: 10.1074/jbc.M010625200. [DOI] [PubMed] [Google Scholar]
- 55.Duchardt F, Fotin-Mleczek M, Schwarz H, Fischer R, Brock R. A comprehensive model for the cellular uptake of cationic cell-penetrating peptides. Traffic. 2007;8:848–866. doi: 10.1111/j.1600-0854.2007.00572.x. [DOI] [PubMed] [Google Scholar]
- 56.Sahay G, Batrakova EV, Kabanov AV. Different internalization pathways of polymeric micelles and unimers and their effects on vesicular transport. Bioconjugate Chem. 2008;19:2023–2029. doi: 10.1021/bc8002315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rejman J, Oberle V, Zuhorn IS, Hoekstra D. Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis. Biochem. J. 2004;377:159–169. doi: 10.1042/BJ20031253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lundin P, Johansson H, Guterstam P, Holm T, Hansen M, Langel Ü, Andaloussi SEL. Distinct uptake routes of cell-penetrating peptide conjugates. Bioconjugate Chem. 2008;19:2535–2542. doi: 10.1021/bc800212j. [DOI] [PubMed] [Google Scholar]
- 59.Zauner W, Kichler A, Schmidt W, Mechtler K, Wagner E. Glycerol and polylysine synergize in their ability to rupture vesicular membranes: a mechanism for increased transferrin–polylysine-mediated gene transfer. Exp. Cell Res. 1997;232:137–145. doi: 10.1006/excr.1997.3486. [DOI] [PubMed] [Google Scholar]
- 60.Pelkmans L, Helenius A. Endocytosis via caveolae. Traffic. 2002;3:311–320. doi: 10.1034/j.1600-0854.2002.30501.x. [DOI] [PubMed] [Google Scholar]
- 61.Pelkmans L, Puntener D, Helenius A. Local actin polymerization and dynamin recruitment in SV40-induced internalization of caveolae. Science. 2002;296:535–539. doi: 10.1126/science.1069784. [DOI] [PubMed] [Google Scholar]
- 62.Mayor S, Pagano RE. Pathways of clathrin-independent endocytosis. Nature Rev. Mol. Cell Biol. 2007;8:603–612. doi: 10.1038/nrm2216. [DOI] [PubMed] [Google Scholar]
- 63.Rodal SK, Skretting G, Garred Ø, Vilhardt F, Deurs BV, Sandvig K. Extraction of cholesterol with methyl-β-cyclodextrin perturbs formation of clathrin-coated endocytic vesicles. Mol. Biol. Cell. 1999;10:961–974. doi: 10.1091/mbc.10.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gruenberg J, Griffiths G, Howell KE. Characterization of the early endosome and putative endocytic carrier vesicles in vivo and with an assay of vesicle fusion in vitro. J. Cell Biol. 1989;108:1301–1316. doi: 10.1083/jcb.108.4.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wadia JS, Dowdy SF. Transmembrane delivery of protein and peptide drugs by TAT-mediated transduction in the treatment of cancer. Adv. Drug Deliv. Rev. 2005;57:579–596. doi: 10.1016/j.addr.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 66.Ziegler A, Nervi P, Durrenberger M, Seelig J. The cationic cell-penetrating peptide cpptat derived from the HIV-1 protein tat is rapidly transported into living fibroblasts: optical, biophysical, and metabolic evidence. Biochemistry. 2005;44:138–148. doi: 10.1021/bi0491604. [DOI] [PubMed] [Google Scholar]
- 67.Geng Y, Dalhaimer P, Cai S, Tsai R, Tewari M, Minko T, Discher DE. Shape effects of filaments versus spherical particles in flow and drug delivery. Nat. Nanotechnol. 2007;2:249–255. doi: 10.1038/nnano.2007.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Godbey WT, Wu KK, Mikos AG. Tracking the intracellular path of poly(ethylenimine)/DNA complexes for gene delivery. Proc. Natl Acad. Sci. 1999;96:5177–5181. doi: 10.1073/pnas.96.9.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Batabyal D, Yeh SR. Human tryptophan dioxygenase: a comparison to indoleamine 2,3-dioxygenase. J. Am. Chem. Soc. 2007;129:15690–15701. doi: 10.1021/ja076186k. [DOI] [PubMed] [Google Scholar]
- 70.Uyttenhove C, Pilotte L, Theate I, Stroobant V, Colau D, Parmentier N, Boon T, Eynde BJVD. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2,3-dioxygenase. Nat. Med. 2003;9:1269–1274. doi: 10.1038/nm934. [DOI] [PubMed] [Google Scholar]
- 71.Mellor AL, Baban B, Chandler P, Marshall B, Jhaver K, Hansen A, Koni PA, Iwashima M, Munn DH. Induced indoleamine 2,3 dioxygenase expression in dendritic cell subsets suppresses T cell clonal expansion. J. Immunol. 2003;171:1652–1655. doi: 10.4049/jimmunol.171.4.1652. [DOI] [PubMed] [Google Scholar]
- 72.Munn DH, Zhou M, Attwood JT, Bondarev I, Conway SJ, Marshall B, Brown C, Mellor AL. Prevention of allogeneic fetal rejection by tryptophan catabolism. Science. 1998;281:1191–1193. doi: 10.1126/science.281.5380.1191. [DOI] [PubMed] [Google Scholar]
- 73.Muller AJ, Prendergast GC. Indoleamine 2,3-dioxygenase in immune suppression and cancer. Curr. Cancer Drug Tar. 2007;7:31–40. doi: 10.2174/156800907780006896. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.