Abstract
RNA aptamers offer a potential therapeutic approach to the competitive inhibition of DNA-binding transcription factors. In previous reports we described in vitro selection and characterization of anti-NF-κB p50 and p65 RNA aptamers. We now describe the further characterization of these aptamers in vitro and in vivo. We show that sub-saturating concentrations of certain anti-p50 RNA aptamers promote complex formation with NF-κB p50 tetramers, whereas anti-p65 R1 RNA aptamers bind NF-κB dimers under all conditions tested. Yeast three-hybrid RNA aptamer specificity studies corroborate previous in vitro results, verifying that anti-p50 and anti-p65 R1 RNA aptamers are highly specific for NF-κB p502 and p652, respectively. These studies introduce a novel T-cassette RNA transcript that improves RNA display from a four-way RNA junction. Mutagenesis of the anti-p65 R1 aptamer reveals tolerated substitutions, suggesting a complex tertiary structure. We describe in vivo selections from a yeast three-hybrid RNA library containing sequences present early in the R1 SELEX process to identify novel anti-p65 RNA aptamers, termed Y1 and Y3. These aptamers appear to be compact bulged hairpins, reminiscent of anti-p50. Y1 competitively inhibits the DNA-binding domain of NF-κB p652 in vitro.
INTRODUCTION
The mammalian NF-κB–signaling cascade is activated in response to inputs from many receptors (e.g. TNF, Toll, cytokine, integrin and EGF), canonical and non-canonical downstream–signaling molecules (e.g. IKKα/β/γ, NIK), and crosstalk from other pathways [e.g. PI3K, AKT, CK2, etc. (1–3)]. These inputs convey information sensing infection, adhesion and DNA damage. Depending on cell type and both extra- and intracellular conditions, such signals act in concert to activate NF-κB responses. NF-κB transcription factors are key regulators of hundreds of genes whose products induce specific immune responses, proliferation, or promote cell survival by evasion of apoptosis (4–6).
When proper NF-κB signaling is disrupted, NF-κB-induced gene products (e.g. TNFα, cyclinD, IL-6, IL-8, c-IAPs, Bcl family members, c-FLIP) contribute to disease pathogenesis (7,8). There is strong experimental support for the hypothesis that constitutive NF-κB activation in cancer directly contributes to drug resistance and a malignant phenotype (9–17). Mechanisms of NF-κB constitutive activation vary. Activation may result from upstream receptor overexpression and/or aberrant kinase signaling (18–23). Mutation of genes encoding NF-κB transcription factors is unusual, making NF-κB transcription factors attractive targets for therapeutic inhibition.
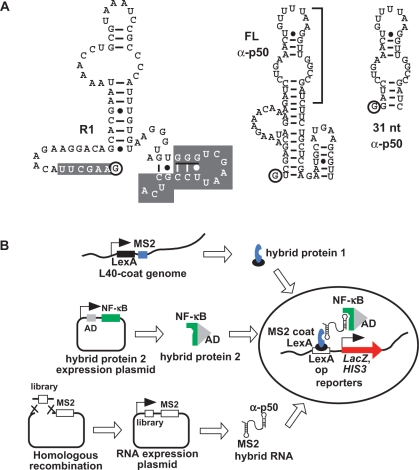
Previously, we used a SELEX approach to identify anti-NF-κB p50 and p65 aptamers (Figure 1A) that bind to the DNA-binding domain of NF-κB transcription factors (24,25). These RNA aptamers were shown to act as decoys that competitively inhibit DNA binding in vitro. We exploited the yeast three-hybrid system (Y3H; Figure 1B) to characterize and optimize anti-p50 RNA aptamers (26,27). Others have also reported the utility of the Y3H in mapping aptamer/protein-binding sites (28,29).
Figure 1.
(A) Predicted anti-NF-κB RNA aptamer secondary structures. Full-length R1, full-length anti-p50, and 31-nt version of anti-p50 (bracket) are depicted with 5′ terminal nt circled and nt of the non-randomized regions of R1 shaded. The trinucleotide G67–69 feature of R1 is underlined. (B) Screening RNA libraries using the Y3H. Transcription of lacZ and HIS3 reporter genes (filled arrow at right) depends on a tri-molecular interaction between integrated hybrid protein 1 (LexA/MS2 coat protein fusion, top), a hybrid protein 2 (GAL4AD/NF-κB family member, middle), and a hybrid RNA composed of the MS2 recognition sequence and an early round or degenerate R1 library sequence cloned into the MS2 plasmid by homologous recombination (bottom).
Our long-term interests include understanding the basis for, and specificity of, RNAs that bind to DNA-binding proteins. Here we further characterize an anti-NF-κB p65 RNA aptamer termed R1 (25) and compare its specificity to the anti-NF-κB p50 RNA aptamer that has previously been studied in detail (24,26,27,30). Figure 1A depicts a predicted secondary structure of R1 (25). In previous biochemical experiments (25), truncation analysis suggested that 3′-terminal sequences of R1 are important for high-affinity binding to homodimeric murine p65 (mp652). When 25 nt at the 3′ terminus of R1 were replaced by A25, affinity for mp652 was reduced 100-fold. An important feature within this 3′ sequence is the G67–69 trinucleotide (Figure 1A, underlined). Mutation of G67–69 to A67–69 resulted in a 20-fold reduction in affinity for mp652. These and other results of preliminary mutational analysis suggested that, unlike the much more compact 31-nt anti-p50 RNA aptamer (24), both 5′ and 3′ R1 terminal sequences are necessary for high affinity binding to mp652 (25). One goal of the present work was to identify anti-p65 RNA aptamers with simpler structures that might facilitate in vivo applications.
We report experiments in vitro and in the Y3H to characterize the interaction between anti-NF-κB RNA aptamers and NF-κB proteins including (i) NF-κB multimer preference (ii) NF-κB dimer specificity, (iii) mutagenesis data and (iv) selection of new anti-p65 RNA aptamers.
MATERIALS AND METHODS
NF-κB protein expression and purification
The Rel homology region of human p652 was cloned into a pET-15b derivative encoding a Tev protease cleavage site and protein expressed in Escherichia coli strain BL21(DE3). Cells were resuspended in 50 mM sodium phosphate buffer (pH 7.5) containing 300 mM NaCl (binding buffer), lysed with a high-pressure microfluidizer Emulsiflex C-5 (Avestin), subjected to centrifugation at 20 000× g for 30 min, and the supernatant loaded onto a nickel-NTA column (Qiagen), which was washed and then eluted with aliquots of binding buffer containing 20 mM and 500 mM imidazole, respectively. The hexahistidine tag was then cleaved with TEV protease overnight at room temperature. The protein was purified by Superdex 75 chromatography (Amersham Biosciences). Untagged recombinant murine p652 and human p502 proteins were expressed and purified as described (25).
Native electrophoretic RNA titration analysis of aptamer/NF-κB complexes
Binding preferences for R1 RNA/p652 and full-length or 31-nt anti-p50 RNA/p502 complexes were investigated by incubating a constant concentration of p652 or p502 protein (100 nM or 250 nM) in 20 µl-binding reactions (20 mM Tris–HCl, pH 8.0, 50 mM NaCl, 1 mM MgCl2) with increasing concentrations of [32P]-pCp-labeled RNA. Reactions contained 0.25-, 0.5-, 1.0-, 1.5-, 2.0-, 3.0-, 4.0- or 5.0-fold molar excess RNA relative to 250 nM protein, or 0.025-, 0.05-, 0.1-, 0.25-, 0.5-, 1.0 or 2.0-fold molar excess RNA relative to 100 nM protein. Components were incubated for 20 min and electrophoresed through native polyacrylamide gels in 0.25× TBE buffer. Complexes were detected and analyzed by storage phosphor imaging.
Glutaraldehyde crosslinking analysis of aptamer/NF-κB complexes
Binding reactions were performed as described for native RNA titration analysis except in binding buffer lacking BSA and salmon sperm DNA. Reactions were then incubated in the presence of 0.02% glutaraldehyde for 30 min at room temperature. Cross-linking reactions were quenched with 0.25 M glycine for 10 min, heat denatured in 2% SDS, and analyzed by electrophoresis through 10% Bis–Tris NuPage polyacrylamide gels (Invitrogen). Proteins were stained with Sypro Ruby (Invitrogen) and imaged by UV transillumination.
Electrophoretic mobility shift assays and binding affinity estimates
RNA molecules were 3′ radiolabeled by ligation of [32P]-pCp using T4 RNA ligase. Affinities for p652 protein were determined by incubating 1 nM radiolabeled RNA with increasing concentrations of p652 in 20 μl-binding reactions containing 20 mM Tris–HCl, pH 8.0, 50 mM NaCl, 1 mM MgCl2, 1 μg poly(dI·dC), 0.5 μg tRNA, 0.25 mg/ml BSA, 5% (v/v) glycerol and 1 mM DTT. Binding reactions were incubated for 20 min at room temperature and electrophoresed through 29 : 1 acrylamide:bisacrylamide native polyacrylamide gels in 0.25× TBE buffer. Complexes were detected and analyzed by storage phosphor imaging. Affinity estimates were obtained by fitting the fractional saturation (θ) of the radiolabeled nucleic acid target to the total protein concentration (L):
| 1 |
where Kd is the equilibrium dissociation constant.
Hybrid protein constructs
Yeast GAL4 transcriptional activation domain fusions with NF-κB proteins were generated by appending 40-bp homology extensions to PCR products encoding the appropriate NF-κB Rel homology regions and cloning into expression vector pGAD424 (Clontech) via yeast homologous recombination at the EcoRI site.
Yeast reporter gene assays
For quantitative lacZ reporter assays, yeast strains were grown to mid-log phase, lysed by three liquid N2 freeze-thaw cycles, and exposed to buffered CPRG or ONPG substrate as described (27).
Cloning of anti-p65 RNAs present early in the R1 SELEX process
RNA sequences present at round 6 of the prior 12-round in vitro selection that yielded the R1 aptamer (25) were reverse-transcribed, PCR-amplified, and cloned into pJ713, a version of pIIIA/MS2–2 (31), via yeast homologous recombination.
Synthesis of degenerate R1 RNA library
A degenerate R1 DNA oligonucleotide library was synthesized by phosphoramidite methodology in the Mayo Molecular Biology Core Facility. Phosphoramidite mixtures were adjusted to yield ∼3% random mutagenesis at each position.
T-cassette construction
A four-way RNA junction was designed to isolate the secondary and tertiary structures of modular RNA domains required for the Y3H. A pair of oligonucleotides was extended into a duplex by overlap extension PCR. This cassette was cloned into the pIIIA/MS2-2 RNA expression plasmid (pJ207) using yeast homologous recombination (32) to produce plasmid pJ1582. EcoRI sites correspond to cloning sites in pIIIA/MS2-2, SphI, SmaI and XhoI sites were introduced. In the present experiments, MS-2 hairpin sequences where cloned at the SmaI site and RNA aptamers at the SphI site. In some cases random sequences were cloned at the XhoI site. All cloning was performed by yeast homologous recombination. The DNA sequence corresponding to the T-cassette transcript (restriction sites in bold, aptamer insertion sites in italics) is:
GA2CGA3CTCTG3AGCTGCGAT2G2CAGA2T2CCGT2AGCA2G2C2GCAG2ACT2GCATGCT2ATC2TGCG2CGCG3CGCGT3 C3G3T2ACGCGC3GC2T2A2GTGT3CTCGAGT2G2CACT2A2GCT2GCTA2CGGA2T2CC4ATATC2A2CT2C2A2T3A2TCT3CT6A2T4CACT2AT3GCG
Y3H screening
The Y3H host strain L40–coat (31) was simultaneously transformed with the GAL4AD/zebrafish p652 (zp652) expression plasmid (pJ1596), a linearized MS2 RNA expression plasmid (pJ713), and the PCR-amplified candidate aptamer library flanked with 40 bp of homology to the MS2 RNA expression plasmid. Alternatively, the transformation contained a GAL4AD/murine p652 expression plasmid (pJ1448), the linearized T-cassette RNA expression plasmid (pJ1582), and PCR-amplified candidate aptamer library flanked with 40 bp of homology to the T-cassette RNA expression plasmid. Transformation mixtures were plated on media selective for the protein and RNA expression plasmids as well as a selective level of 3-AT, and the plates were incubated at 30°C for 5–8 days. Large colonies were replica-plated onto selective media and checked for lacZ reporter gene activity by filter assay. RNA expression plasmids were then isolated from lacZ positive transformants and sequenced.
Northern blot analysis of RNA accumulation in yeast
Total RNA was extracted from Y3H strains as described (33). Northern blot analysis was conducted as described (26), simultaneously probing with radiolabeled oligonucleotides complementary to the RNase P RPR1 leader (5′-AGCAC2ACAGCGTAC2ATGT) contained in both the MS2 cassette and T-cassette, and to the U6 small nuclear RNA (snRNA) (5′-TC2T2ATGCAG4A2CTGC). Signals were detected and analyzed by storage phosphor technology.
RESULTS AND DISCUSSION
R1 and anti-p50 RNA aptamer binding to dimer and tetramer forms of NF-κB
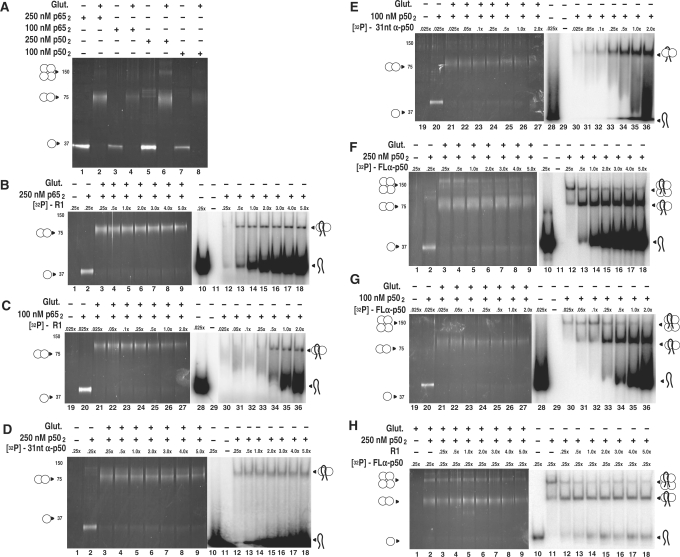
The availability of anti-NF-κB p50 and p65 RNA aptamers allows for exploration of aptamer specificity in vitro and in the cell nucleus. Because NF-κB proteins participate in various homo-multimerization equilibria (34) we began by studying the in vitro specificity of anti-NF-κB RNA aptamers for NF-κB protein multimers. Previously (25) we reported the titration of R1 RNA over a range of concentrations relative to a constant concentration of p652. Analysis of the resulting complexes by native gel electrophoresis showed that the anti-NF-κB p65 R1 RNA binds to p652 with a stoichiometry of one RNA aptamer to one p65 dimer (p652), even at high RNA excess. Likewise, a single anti-p50 RNA binds to p502 in solution (30), although the crystal complex studied by X-ray diffraction shows two anti-p50 RNAs bound to the p502 dimer (35). We analyzed R1 binding to p652 and anti-p50 binding to p502 over a range of protein and RNA concentrations where different multimeric protein species are possible. To clarify the NF-κB multimer state (e.g. monomer, dimer, tetramer) in RNA complexes, we compared electrophoretic gel mobility shift assays with denaturing SDS polyacrylamide gel electrophoresis after glutaraldehyde cross-linking.
Glutaraldehyde cross-linking was first performed in the absence of aptamers to determine the distribution of NF-κB p50 and p65 monomers, homodimers and homotetramers at nominal dimer concentrations of 100 nM or 250 nM (Figure 2A). The results show that both recombinant p50 and p65 NF-κB proteins are readily cross-linked as homodimers with no free monomer detected. Some p50 homotetramer is evident under these conditions (Figure 2A, lanes 6 and 8). We then titrated radiolabeled R1 over a wide concentration range relative to p652 (Figure 2B and C). Consistent with previous results (25), only a single RNA:protein complex was observed regardless of the p652 concentration or the relative concentration of R1 (Figure 2B, lanes 12–18; Figure 2C, lanes 30–36). The shifted species involves only p652 as shown by glutaraldehyde cross-linking (Figure 2B, lanes 3–9; Figure 2C, lanes 21–27). The presence of a single complex independent of relative concentrations of RNA and protein monomer confirms a 1 : 2 RNA:protein stoichiometry for R1:p65.
Figure 2.
RNA aptamer specificities for different NF-κB multimers. (A) Glutaraldehyde cross-linking of p50 and p65 proteins at 100 nM and 250 nM nominal dimer concentrations. (B) and (C): R1 RNA titration of p652/R1 RNA complexes. Increasing concentrations of labeled R1 RNA (0.25-, 0.5-, 1-, 2-, 3-, 4-, 5-fold relative to 250 nM p652) were incubated with p652 and electrophoresed on native gels (lanes 1–7) or cross-linked with 0.03% glutaraldehyde (Glut.) and electrophoresed on denaturing SDS gels (lanes 8–14). The analysis was repeated for 100 nM p652. (D) and (E): 31-nt anti-p50 RNA titration of p502/31-nt anti-p50 RNA complexes, performed as in (A). (F) and (G): Full-length anti-p50 RNA titration of p502/full-length anti-p50 RNA complexes, performed as in (A). (H) Competition assay of full-length anti-p50/p504 complex. Labeled full-length anti-p50 RNA was incubated at a concentration of 0.25-fold relative to 250 nM p652, with or without 0.03% glutaraldehyde (lanes 1 and 9, respectively) or in the presence of unlabeled R1 RNA competitor (non-specific for p50; 0.25-, 0.5-, 1-, 2-, 3-, 4-, 5-fold) relative to 250 nM p652, and electrophoresed on a native gel (lanes 2–8), or incubated with 0.03% glutaraldehyde and electrophoresed on a denaturing SDS gel. (lanes 10–16).
We repeated these experiments for complexes of anti-p50 and NF-κB p502 protein, using either a 31-nt truncated version of anti-p50 (Figure 2D and E) similar to the RNA that had been studied by X-ray crystallography (35), or the full-length (79 nt) anti-p50 molecule identified from SELEX (Figure 2F and G) (24). The 31-nt anti-p50 aptamer complex with p50 appears as a single species for all RNA:p50 ratios with p502 at 250 nM or 100 nM (Figure 2D, lanes 12–18; Figure 2E, lanes 30–36). Glutaraldehyde cross-linking shows the p502 protein form to predominate (Figure 2D, lanes 3–9; Figure 2E, lanes 21–27). These results are consistent with our previous studies of the binding of the 31-nt anti-p50 RNA aptamer to p502 in solution (30).
Testing of the full-length anti-p50 RNA aptamer with NF-κB p50 protein unexpectedly revealed two RNA:protein complexes (Figure 2F and G). A slow-migrating p50:anti-p50 complex was prevalent in the presence of sub-stoichiometric RNA concentrations relative to p502 (Figure 2F, lane 12, Figure 2G, lane 30). Increasing RNA excess shifted the distribution to favor a complex of higher mobility (Figure 2F, lanes 13–18, Figure 2G, lanes 31–36). Glutaraldehyde cross-linking studies revealed the basis for these two complexes (Figure 2F, lanes 3–9, Figure 2G, lanes 21–27). As previously shown in Figure 2A, 100 nM or 250 nM dimer concentrations of recombinant p50 (but not p65) protein include a significant fraction of tetramer. The full-length (79 nt) anti-p50 RNA aptamer (but not the 31-nt truncated version) shows a strong binding preference for p504, favoring this species even though p502 is the major protein form present (Figure 2F, compare lanes 3 and 12; Figure 2G, compare lanes 21 and 30). We hypothesize that the 79-nt anti-p50 RNA aptamer is sufficiently long that the RNA makes specific contacts with one p50 subunit and simultaneous non-specific contacts with an unoccupied monomer within the same tetramer. This cooperative binding enhances the stability of the complex. As the concentration of full-length anti-p50 RNA is increased, p504 becomes saturated and excess RNA aptamers bind to the p502 form of the protein (Figure 2G, lanes 33–36).
To test that it is non-specific interactions that enhance binding of full-length anti-p50 RNA aptamers to p504, we created binding reactions with a sub-saturating concentration of anti-p50, resulting in a predominant anti-p50:p504 complex (Figure 2H, lanes 2 and 11). We then added increasing amounts of an unlabeled non-specific RNA (in this case R1, which has low p50 affinity) to determine if the non-specific single-stranded RNA could compete the labeled RNA aptamer into p502 complexes. The results show that the non-specific RNA partially competes with the specific full-length anti-p50/p504 complex (Figure 2H, compare lanes 11 and 12). These results demonstrate the existence of full-length anti-p50/p50 complexes with stoichiometries of both 1 : 2 and 1 : 4. The latter are stabilized by both specific and non-specific aptamer/protein interactions.
RNA aptamer specificity for NF-κB proteins
Results of previous in vitro studies suggest that anti-p50 and anti-p65 R1 aptamers are highly specific for NF-κB p502 and p652, respectively (25,35). To test anti-p65 R1 binding to p652 in vivo in the context of the yeast nucleus we employed the Y3H [Figure 1B; (26,27,31,32)]. NF-κB proteins were cloned and expressed as Gal4 activation domain fusions in Y3H strains also expressing different tethered RNA aptamers in the context of the original MS2 RNA expression plasmid. LacZ reporter gene activation was monitored by β-galactosidase assay, and scores were normalized to those from the strain expressing MS2-R1 and Gal4AD-murine p652 (Table 1 ). Anti-p50 recognized p502 but not p652 or Drosophila NF-κB homologs tested in the Y3H. Similarly, low lacZ reporter gene expression was detected for R1 with all NF-κB family members except murine p652 (Table 1). These results corroborate in vitro analyses, confirming that the anti-p50 and R1 aptamers are highly selective for their original SELEX NF-κB targets, over all other NF-κB family members tested in the yeast assay.
Table 1.
Binding activities of RNA aptamers in yeast and in vitro
| Aptamera | Proteinb | β-Galactosidasec (MS2 cassette) | β-Galactosidasec (T-cassette) | Kd, nMd |
|---|---|---|---|---|
| – | AD | 0.3 ± 0.3 | 1.0 ± 0.6 | – |
| R1 | AD | 2.2 ± 0.3 | 3.3 ± 2.3 | – |
| R1 | AD-mp502 | 1.7 ± 0.1 | 4.8 ± 3.3 | – |
| R1 | AD-mp652 | 100 | 476 ± 56 | 38 ± 20 |
| R1 | AD-hp652 | 4.4 ± 0.4 | – | – |
| R1 | AD-zp652 | 4.5 ± 0.3 | – | – |
| R1 | AD-dorsal2 | 1.7 ± 0.1 | 4.8 ± 3.3 | – |
| R1 | AD-dif2 | 2.1 ± 0.1 | 5.2 ± 3.3 | – |
| anti-p50 | AD | 1.9 ± 0.1 | 8.5 ± 0.5 | – |
| anti-p50 | AD-mp502 | 254 ± 14 | 576 ± 30 | – |
| anti-p50 | AD-mp652 | 2.5 ± 0.6 | 7.1 ± 1.0 | – |
| anti-p50 | AD-dorsal2 | 1.1 ± 0.1 | 5.2 ± 0.5 | – |
| anti-p50 | AD-dif2 | 0.9 ± 0.1 | 2.4 ± 0.5 | – |
| Y1 | AD | 1.5 ± 0.8 | – | – |
| Y1 | AD-mp652 | 375 ± 8 | 556 ± 90 | 74 ± 2 |
| Y1 | AD-hp652 | 38 ± 9 | – | – |
| Y1 | AD-zp652 | 175 ± 8 | – | – |
| Y1-1 | AD-mp65 | 113 ± 16 | 552 ± 90 | 86 ± 10 |
| Y1-1T | AD-mp652 | 161 ± 26 | – | 101 ± 50 |
| Y3 | AD | 1.1 ± 0.7 | – | – |
| Y3 | AD-mp652 | 73 ± 10 | 456 ± 142 | 94 ± 31 |
| Y3 | AD-hp652 | 8.3 ± 2.6 | – | – |
| Y3 | AD-zp652 | 80 ± 30 | – | – |
| Y3-1 | AD-mp652 | 86 ± 33 | 618 ± 123 | >2500 |
| Y3-1T | AD-mp652 | 90 ± 25 | – | >2500 |
Dash indicates not tested.
aRNA aptamers cloned in MS2 cassette, T-cassette, or tested in isolation in vitro. R1: 83 nt; anti-p50: 31 nt; Y1: 86 nt; Y1-1: 40 nt; Y1-1T: 39 nt; Y3: 83 nt; Y3-1: 32 nt; Y3-1T: 33 nt.
bAD: GAL4 activation domain.
cβ-galactosidase activities normalized to positive control combination MS2-R1-mp652 set at 100. Averages represent at least three independent trials for each transformant.
dEquilibrium dissociation constant from electrophoretic gel mobility shift data fit to Equation (1) for target protein without activation domain. Mean and standard deviation are shown based on at least two repeats.
An unexpected difference in Y3H signal between murine and human p65 (Supplementary Table S1) prompted further consideration. Miyakawa et al. (36) recently reported that an anti-human IgG RNA aptamer was selective for IgG from human versus other vertebrates. Thus, RNA aptamers have the potential for species selectivity. Previous filter binding experiments with the anti-p50 RNA aptamer suggested no obvious preference for murine p502 versus human p502 (24). The murine p652 ortholog was used in the original SELEX experiment that identified R1 (25). The DNA recognition residues of p65 Loop L1 are highly conserved among p65 orthologs, suggesting that R1 should bind multiple forms of p652. We performed electrophoretic gel mobility shift experiments comparing purified human and murine p652 orthologs to estimate equilibrium dissociation constants. Indeed, R1 displayed similar affinities for both murine p652 and human p652 in these experiments (Supplementary Figure S1A). Y3H experiments suggested R1 discrimination among murine, human, and zebrafish p652 orthologs, although all produced lacZ reporter gene activities above background (Supplementary Table S1). Western blotting showed that similar levels of murine and human target proteins accumulate in yeast (Supplementary Figure S1B). Hybrid p652 proteins used to determine specificity in the Y3H were also tested in the yeast one-hybrid system (Y1H) with three copies of the NF-κB DNA consensus sequence ([5′-G4ACT3C2]) integrated upstream of lacZ and HIS3 reporter genes. The results (Supplementary Table S2) confirm that cloned Gal4AD-p652 fusion proteins are functional in yeast. The lack of correlation between in vitro and in vivo R1 interaction with human p652 is likely related to reduced human p65 protein function detected in the Y1H. The ∼2-fold lower activity of human p652 relative to murine p652 observed in the Y1H is apparently amplified in the Y3H assay. We conclude that anti-NF-κB RNA aptamers demonstrate specificity for their targets in vivo.
Novel T-cassette improves Y3H RNA display and facilitates R1 mutation screening
Protein:aptamer interactions in the Y3H system depend on at least three factors: (i) intrinsic affinity of the isolated RNA insert for the target Gal4AD fusion protein, (ii) retention of required RNA insert tertiary structure when expressed within a larger transcript and (iii) accessibility of the RNA insert to the target protein. The combination of these three factors influences aptamer/target protein interaction and reporter gene readout. RNA ‘display’ describes the tertiary structure and accessibility of an RNA aptamer in the Y3H.
Unlike the compact anti-p50 RNA hairpin, R1 contains 5′ and 3′ terminal sequences required for p652 recognition. This may complicate preservation of R1 tertiary structure in the context of Y3H RNA display transcripts. To optimize R1 display in the conventional Y3H MS2 cassette we initially undertook systematic folding predictions to identify flanking sequences that maximize RNA aptamer autonomy from the surrounding transcript context. Computational approaches were used to predict the folding autonomy of defined or random RNA inserts within the conventional Y3H display transcript for all possible 6-bp (4096) or 10-bp (1048576) complementary sequences that could form base-paired stems flanking the R1 aptamer. Optimal R1 stem sequences were then tested in the Y3H in the presence of murine p652. In the case of the optimized 6-bp stem, Y3H readout for the R1 aptamer was marginally improved relative to a non-optimal stem, but aptamer function remained sensitive to details of the stem junction (data not shown). Several optimized 10-bp stems were then included flanking a 60-nt random sequence library adjacent to a functional RNA aptamer. When expressed with the target protein, aptamers with random inserts flanked by sequences for optimized stems were twice as likely to produce detectable Y3H readouts. However, even in this case Y3H readouts in the presence of an adjacent 60-nt random sequence averaged only 3% of readouts without the random sequence (data not shown). Thus, optimally designed 6- or 10-bp GC-rich stems did not assure preservation of RNA aptamer tertiary structure.
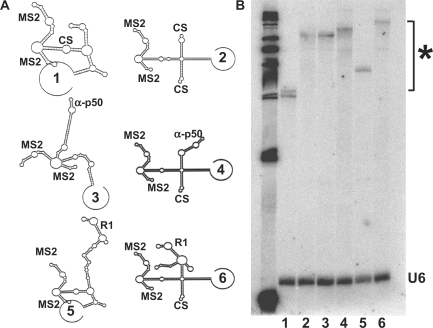
We therefore created a ‘T-cassette’ RNA transcript to stabilize modular RNA aptamer folding in the Y3H system. The structure of the expressed RNA is designed to isolate the aptamer domain from other RNA domains emanating from a four-way helical junction. Figure 3A compares the predicted secondary structures of the original yeast RNAs (odd numbers) with the corresponding T-cassette forms (even numbers). One arm of the T-cassette transcript is occupied by a pair of MS2 RNA hairpins, two arms are available for displaying experimental RNAs, and the fourth arm carries stabilizing yeast RNAse P sequences derived from the original hybrid RNA expression cassette (Figure 3A, Construct 2). To test the T-cassette context for display of aptamers R1 and anti-p50, we measured β-galactosidase activities for Y3H strains expressing either T-cassette RNAs or MS2-RNAs in the presence of the appropriate NF-κB family member (Table 1). Strains with T-cassette RNAs showed consistently higher reporter gene activation. A 5-fold increase was observed for T-R1 versus MS2-R1, and a ∼2-fold increase for T-anti-p50 versus MS2-anti-p50 strain (Table 1). To confirm equal accumulation of T-cassette and MS2-RNAs in the Y3H strain, we harvested total RNA and performed northern blot analysis (Figure 3B). MS2-RNAs (lanes 1, 3 and 5) are expressed at levels comparable to T-cassette RNAs (lanes 2, 4 and 6). Thus, the higher reporter gene activity induced by T-cassette RNAs may result from improved aptamer folding and accessibility. Importantly, the 5-fold increase in β-galactosidase readout for the T-R1 strain versus the MS2-R1 strain significantly improves the dynamic range available for mutation analysis.
Figure 3.
Optimizing RNA display in Y3H. (A) Predicted secondary structure of original MS2 and T-cassette transcripts alone (RNAs 1 and 2), with 31-nt anti-p50 (RNAs 3 and 4), and with R1 (RNAs 5 and 6) for RNA display in Y3H. (B) Northern blot comparison of T-cassette and MS2 RNA transcript accumulation in test strains discussed in Table 1. Note that the highly-structured T-cassette RNAs resist complete denaturation, causing retarded mobility (*).
R1 mutation analysis and secondary structure prediction
To improve our understanding of the R1 aptamer, tolerated mutations were mapped in its primary sequence. A random library of R1 mutants was created in the context of the T-cassette and subjected to selection for aptamer function in the Y3H with murine p652 as bait. R1 variants were scored for in vivo function in the Y3H by β-galactosidase assay. Twenty-four unique functional variants were included in the analysis (Table 2). These sequences carried an average of three mutations per R1 molecule. Measured β-galactosidase activities displayed an 11-fold range relative to the original R1 aptamer. Interestingly, none of the 24 functional R1 mutants contained mutations near the 3′ terminus of the RNA. These results suggest that 3′ sequences are important for R1 binding to p652 (25).
Table 2.
Functional mutants of RNA aptamer R1 defined in yeast
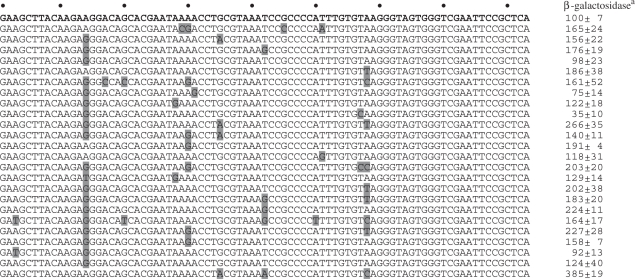 |
aβ-galactosidase units normalized to positive control combinations set at 100.
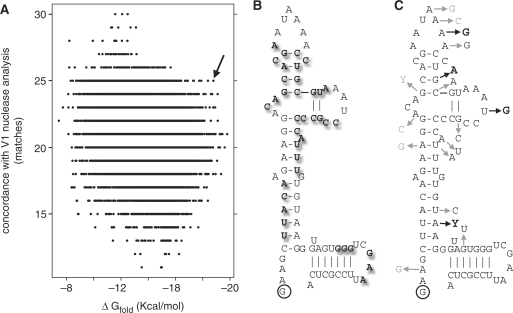
V1 nuclease susceptibility results for R1 (25) were used to generate a revised R1 secondary structure prediction for mapping tolerated mutations. The result is shown in Figure 4. Panel A plots predicted R1 secondary structures in terms of free energy change associated with folding and concordance with V1 nuclease analysis (25). The arrow indicates a favorable example. Panel B shows this fold overlaid with V1 nuclease susceptibility. Panel C indicates tolerated nucleotide substitutions, with changes that strongly improve yeast three-hybrid function shown in bold. Analysis of these mutagenesis data by three available consensus structure prediction software programs was not conclusive (Supplementary Figure S2).
Figure 4.
Anti-p65 R1 RNA aptamer improved secondary structure model. (A) Plot of optimal and sub-optimal predicted R1 secondary structures and concordance with prior V1 nuclease susceptibility data (25). Arrow indicates candidate fold. (B) Depiction of prior V1 nuclease susceptibility data (bold) in this context. (C) Tolerated R1 mutations (Table 1) are indicated by arrows. Mutations that enhanced R1 function in the Y3H are indicated in bold.
Yeast selection of anti-p65 aptamers from early in the SELEX process
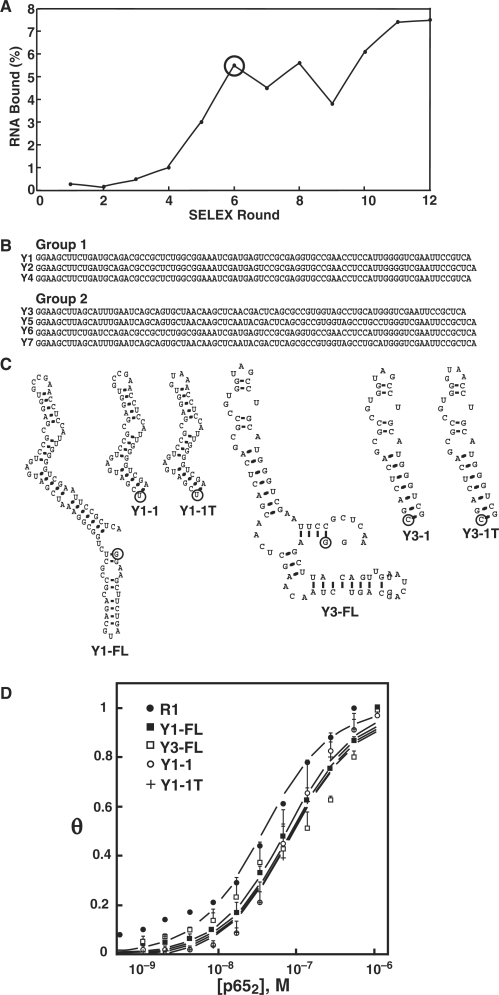
While the R1 aptamer may be valuable in many settings, we sought smaller anti-p65 RNA aptamers with compact structures to facilitate RNA display in in vivo experimental settings. We used the Y3H to screen a library of RNA sequences present at round 6 of the original murine p652 SELEX experiment that eventually gave rise to the R1 aptamer at round 12 (25) (Figure 5A). This approach takes advantage of the RNA diversity present before convergence of the SELEX process (27). The Round 6 RNA library was cloned and expressed in the context of the original MS2 RNA cassette. The Y3H strain expressed GAL4AD-zebrafish p652 (zp652), a related vertebrate NF-κB target to select anti-p65 RNA aptamers with broad specificity. Approximately 50 000 RNA sequences were screened for strong HIS3 activation (40 mM 3-AT as selective agent). Seven RNAs induced strong expression of both HIS3 and lacZ (Figure 5B). Representative full length (FL) aptamers Y1 and Y3 and derivatives (Figure 5C) were inserted into MS2 RNA and/or T-cassette contexts and tested in the Y3H. Table 1 summarizes aptamer interactions in yeast. Strikingly, the Y1/mp652, Y1/hp652 and Y1/zp652 strains displayed 4-, 8- and 39-fold higher lacZ reporter gene activation relative to the corresponding yeast strains expressing R1. Our previous studies demonstrated that R1 could not be readily truncated (25). In contrast, Y1-1 and Y3-1 truncations proved to be functional in the Y3H, activating lacZ by 33 and 120% of full-length Y1 and Y3 RNAs, respectively (Table 1). Truncated variants of Y1 and Y3 containing tetraloop sequences were at least as effective (Table 1). The same constructs were also evaluated in the improved T-cassette context (Table 1). The results again indicate superior aptamer presentation in the T-cassette with reporter gene activation often 5- to 7-fold higher than for aptamers presented from the conventional MS2 cassette. Interestingly, Y1 and Y3 aptamers and derivatives were all comparable to R1 in binding mp652 in the T-cassette context (Table 1).
Figure 5.
Y3H selection of anti-p65 RNA aptamers from early SELEX rounds. (A) Percent RNA recovered during original SELEX that yielded the anti-p65 RNA aptamer R1 (25). (B) Candidate anti-p65 RNA aptamers from SELEX round 6. (C) Predicted secondary structures of Y1, Y3, and truncation variants. The 5′ terminal nt of each RNA is circled. (D) Electrophoretic gel mobility shift binding isotherms for murine p652 in the presence of R1 (closed circles), Y1 (closed squares), Y3 (open squares), truncation Y1-1 (open circles), or truncation Y1-1T with substituted tetraloop (plus).
We tested the anti-NF-κB p65 Y1, Y3 aptamers and derivatives for binding to murine p652 in vitro. Full-length Y1 and Y3 bound murine p652 in vitro with affinities 2- to 3-fold lower than for the R1 aptamer (Figure 5D). Y1-1 and Y1-1T aptamers displayed in vitro affinities similar to full-length Y1. In contrast to derivatives of Y-1, the Y3-1 and Y3-1T truncations that functioned well in the Y3H did not bind mp652 in vitro (Table 1). These results emphasize that aptamer function in the context of a larger stabilizing RNA scaffold (Y3H) may not reliably predict behavior of an isolated aptamer in vitro. Because 2- to 3-fold lower affinities were measured for Y series aptamers versus R1 in vitro (Table 1), this suggests that aptamer folding is particularly stabilized in the T-cassette. We conclude that the Y1 anti-NF-κB p65 RNA aptamer shows particular promise because of its compact folded structure and relaxed species specificity relative to R1.
Y1 and Y1-1 aptamers as decoys for NF-κB
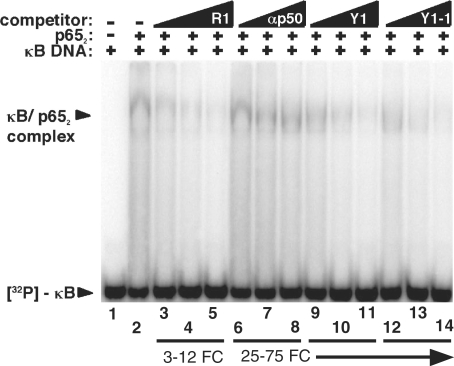
We have previously shown that anti-p65 R1 competes with κB DNA for p652 protein binding in vitro (25). We used competition gel mobility shift assays to test the ability of the newly identified anti-p65 full length and truncated Y1 RNA aptamers to compete with κB DNA for binding to p652 (Figure 6). Radiolabeled κB DNA was incubated in the absence of p652 (lane 1), in the presence of p652 alone (lane 2) or together with various unlabeled competitors: R1 (lanes 3–5), anti-p50 (lanes 6–8), full-length Y1 (lanes 9–11), and truncated Y1 (lanes 12–14). Increasing unlabeled competitor nucleic acid concentrations are indicated: 3-, 6-, or 12-fold molar excess (lanes 3–5) or 25-, 50- or 75-fold molar excess (lanes 6–14) relative to p652. Full length and truncated Y1 RNAs compete with κB DNA for binding to the DNA-binding domain of murine p652, suggesting that, like R1, the binding site of these RNA aptamers overlaps with the electrostatically favorable DNA-binding domain of p65.
Figure 6.
Y1 and Y1-1 RNA aptamers as decoys for NF-κB. Labeled κB DNA (1 nM) was incubated alone (lanes 1 and 15) or in the presence of p652 (20 nM) alone (lane 2) or together with various unlabeled RNA competitors (lanes 3–14): R1 RNA (lanes 3–5), anti-p50 RNA (lanes 6–8), Y1 RNA (lanes 9–11), and truncated Y1-1 (lanes 12–14), Triangles indicate increasing unlabeled molar excess competitor concentration [3-, 6- or 12-fold competitor (FC; lanes 3–5)] and [25-, 50- or 75-fold competitor (lanes 6–14)] relative to p652 concentration.
Summary and future prospects
There have been interesting recent efforts to develop transcription factor decoys based on DNA analogs [e.g. NF-κB; (37)] or RNA molecules [e.g. heat shock factor; (38) and β-catenin; (39)]. The work described here constitutes additional progress toward the goal of developing a ‘toolbox’ of anti-NF-κB RNA aptamers for competitive inhibition of p50 and p65 homo- and heterodimers. Native gel electrophoresis and glutaraldehyde crosslinking studies revealed a 1 : 2 stochiometry for both short anti-p50 and anti-p65 aptamers interacting with NF-κB subunits. This result confirms the dimeric status of the transcription factors under our chosen in vitro conditions, and confirms that a single aptamer binds each dimer under both limiting and excess RNA concentrations. This result confirms and extends the previous stoichiometry study of the anti-p50 aptamer (30), and indicates that the 2:2 stoichiometry observed in the X-ray crystal structure is a special feature of that complex (35,40). Work reported here also confirms that the Y3H system provides powerful verification that the anti-NF-κB RNA aptamers we have identified are specific for their intended targets in an in vivo setting.
Future efforts will be directed to achieve stable transgenic expression of RNA aptamers in model organisms and cultured cells. The goal is to characterize the ability of anti-NF-κB aptamers to artificially regulate gene transcription by acting as molecular decoys for NF-κB transcription factors.
The therapeutic potential of NF-κB RNA aptamers will depend on development of delivery technologies allowing RNA aptamers to reach high intra-nuclear concentrations within specific organs and/or tissue types. Anti-NF-κB RNA aptamer delivery via a viral vector strategy is an option (41).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Mayo Foundation; National Institutes of Health (grant GM68128 to L.J.M.); post-baccalaureate fellowship through National Institutes of Health (grant GM7514 to Y.F.H.). Funding for open access charge: Mayo Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Gouri Ghosh for assistance with protein purification, and Matilda Sprung, Temilola Abdul, Kasandra Riley, Whyte Owen, Cris Charlesworth and past and present members of the Maher lab for technical assistance.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint first authors.
REFERENCES
- 1.Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 2.Luo JL, Kamata H, Karin M. IKK/NF-kappaB signaling: balancing life and death – a new approach to cancer therapy. J. Clin. Invest. 2005;115:2625–2632. doi: 10.1172/JCI26322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Waes C. Nuclear factor-kappaB in development, prevention, and therapy of cancer. Clin. Cancer Res. 2007;13:1076–1082. doi: 10.1158/1078-0432.CCR-06-2221. [DOI] [PubMed] [Google Scholar]
- 4.Chen FE, Ghosh G. Regulation of DNA binding by Rel/NF-kappaB transcription factors: structural views. Oncogene. 1999;18:6845–6852. doi: 10.1038/sj.onc.1203224. [DOI] [PubMed] [Google Scholar]
- 5.Fujita T, Nolan GP, Ghosh S, Baltimore D. Independent modes of transcriptional activation by the p50 and p65 subunits of NF-kappa B. Genes Dev. 1992;6:775–787. doi: 10.1101/gad.6.5.775. [DOI] [PubMed] [Google Scholar]
- 6.Verma IM, Stevenson JK, Schwarz EM, Van Antwerp D, Miyamoto S. Rel/NF-kappaB/I kappaB family: intimate tales of association and dissociation. Genes Dev. 1995;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 7.Dutta J, Fan Y, Gupta N, Fan G, Gelinas C. Current insights into the regulation of programmed cell death by NF-kappaB. Oncogene. 2006;25:6800–6816. doi: 10.1038/sj.onc.1209938. [DOI] [PubMed] [Google Scholar]
- 8.Karin M, Lin A. NF-kappaB at the crossroads of life and death. Nat. Immunol. 2002;3:221–227. doi: 10.1038/ni0302-221. [DOI] [PubMed] [Google Scholar]
- 9.Abdel-Latif MM, O'Riordan J, Windle HJ, Carton E, Ravi N, Kelleher D, Reynolds JV. NF-kappaB activation in esophageal adenocarcinoma: relationship to Barrett's metaplasia, survival, and response to neoadjuvant chemoradiotherapy. Ann. Surg. 2004;239:491–500. doi: 10.1097/01.sla.0000118751.95179.c6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bargou RC, Emmerich F, Krappmann D, Bommert K, Mapara MY, Arnold W, Royer HD, Grinstein E, Greiner A, Scheidereit C, et al. Constitutive nuclear factor-kappaB-RelA activation is required for proliferation and survival of Hodgkin's disease tumor cells. J. Clin. Invest. 1997;100:2961–2969. doi: 10.1172/JCI119849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kojima M, Morisaki T, Sasaki N, Nakano K, Mibu R, Tanaka M, Katano M. Increased nuclear factor-kB activation in human colorectal carcinoma and its correlation with tumor progression. Anticancer Res. 2004;24:675–681. [PubMed] [Google Scholar]
- 12.Mukhopadhyay T, Roth JA, Maxwell SA. Altered expression of the p50 subunit of the NF-kappaB transcription factor complex in non-small cell lung carcinoma. Oncogene. 1995;11:999–1003. [PubMed] [Google Scholar]
- 13.Nair A, Venkatraman M, Maliekal TT, Nair B, Karunagaran D. NF-kappaB is constitutively activated in high-grade squamous intraepithelial lesions and squamous cell carcinomas of the human uterine cervix. Oncogene. 2003;22:50–58. doi: 10.1038/sj.onc.1206043. [DOI] [PubMed] [Google Scholar]
- 14.Ondrey FG, Dong G, Sunwoo J, Chen Z, Wolf JS, Crowl-Bancroft CV, Mukaida N, Van Waes C. Constitutive activation of transcription factors NF-(kappa)B, AP-1, and NF-IL6 in human head and neck squamous cell carcinoma cell lines that express pro-inflammatory and pro-angiogenic cytokines. Mol. Carcin. 1999;26:119–129. doi: 10.1002/(sici)1098-2744(199910)26:2<119::aid-mc6>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 15.Sovak MA, Bellas RE, Kim DW, Zanieski GJ, Rogers AE, Traish AM, Sonenshein GE. Aberrant nuclear factor-kappaB/Rel expression and the pathogenesis of breast cancer. J. Clin. Invest. 1997;100:2952–2960. doi: 10.1172/JCI119848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suh J, Payvandi F, Edelstein LC, Amenta PS, Zong WX, Gelinas C, Rabson AB. Mechanisms of constitutive NF-kappaB activation in human prostate cancer cells. Prostate. 2002;52:183–200. doi: 10.1002/pros.10082. [DOI] [PubMed] [Google Scholar]
- 17.Wang W, Abbruzzese JL, Evans DB, Larry L, Cleary KR, Chiao PJ. The nuclear factor-kappaB RelA transcription factor is constitutively activated in human pancreatic adenocarcinoma cells. Clin. Cancer Res. 1999;5:119–127. [PubMed] [Google Scholar]
- 18.Gasparian AV, Yao YJ, Kowalczyk D, Lyakh LA, Karseladze A, Slaga TJ, Budunova IV. The role of IKK in constitutive activation of NF-kappaB transcription factor in prostate carcinoma cells. J. Cell Sci. 2002;115:141–151. doi: 10.1242/jcs.115.1.141. [DOI] [PubMed] [Google Scholar]
- 19.Madrid LV, Mayo MW, Reuther JY, Baldwin AS., Jr Akt stimulates the transactivation potential of the RelA/p65 Subunit of NF-kappaB through utilization of the IkappaB kinase and activation of the mitogen-activated protein kinase p38. J. Biol. Chem. 2001;276:18934–18940. doi: 10.1074/jbc.M101103200. [DOI] [PubMed] [Google Scholar]
- 20.Madrid LV, Wang CY, Guttridge DC, Schottelius AJ, Baldwin A.S., Jr., Mayo MW. Akt suppresses apoptosis by stimulating the transactivation potential of the RelA/p65 subunit of NF-kappaB. Mol. Cell Biol. 2000;20:1626–1638. doi: 10.1128/mcb.20.5.1626-1638.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nikolopoulos SN, Blaikie P, Yoshioka T, Guo W, Giancotti FG. Integrin beta4 signaling promotes tumor angiogenesis. Cancer Cell. 2004;6:471–483. doi: 10.1016/j.ccr.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 22.Romieu-Mourez R, Landesman-Bollag E, Seldin DC, Traish AM, Mercurio F, Sonenshein GE. Roles of IKK kinases and protein kinase CK2 in activation of nuclear factor-kappaB in breast cancer. Cancer Res. 2001;61:3810–3818. [PubMed] [Google Scholar]
- 23.Tamatani T, Azuma M, Aota K, Yamashita T, Bando T, Sato M. Enhanced IkappaB kinase activity is responsible for the augmented activity of NF-kappaB in human head and neck carcinoma cells. Cancer Lett. 2001;171:165–172. doi: 10.1016/s0304-3835(01)00611-5. [DOI] [PubMed] [Google Scholar]
- 24.Lebruska LL, Maher LJ., 3rd. Selection and characterization of an RNA decoy for transcription factor NF-kappaB. Biochemistry. 1999;38:3168–3174. doi: 10.1021/bi982515x. [DOI] [PubMed] [Google Scholar]
- 25.Wurster SE, Maher LJ., 3rd. Selection and characterization of anti-NF-kappaB p65 RNA aptamers. RNA. 2008;14:1037–1047. doi: 10.1261/rna.878908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cassiday LA, Maher LJ., 3rd. In vivo recognition of an RNA aptamer by its transcription factor target. Biochemistry. 2001;40:2433–2438. doi: 10.1021/bi002376v. [DOI] [PubMed] [Google Scholar]
- 27.Cassiday LA, Maher LJ., 3rd. Yeast genetic selections to optimize RNA decoys for transcription factor NF-kappaB. Proc. Natl Acad. Sci. USA. 2003;100:3930–3935. doi: 10.1073/pnas.0736013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konig J, Julius C, Baumann S, Homann M, Goringer HU, Feldbrugge M. Combining SELEX and the yeast three-hybrid system for in vivo selection and classification of RNA aptamers. RNA. 2007;13:614–622. doi: 10.1261/rna.334307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang B, Kraemer B, SenGupta D, Fields S, Wickens M. Yeast three-hybrid system to detect and analyze interactions between RNA and protein. Methods Enzymol. 1999;306:93–113. doi: 10.1016/s0076-6879(99)06007-3. [DOI] [PubMed] [Google Scholar]
- 30.Cassiday LA, Lebruska LL, Benson LM, Naylor S, Owen WG, Maher LJ., 3rd. Binding stoichiometry of an RNA aptamer and its transcription factor target. Anal. Biochem. 2002;306:290–297. doi: 10.1006/abio.2002.5710. [DOI] [PubMed] [Google Scholar]
- 31.SenGupta DJ, Zhang B, Kraemer B, Pochart P, Fields S, Wickens M. A three-hybrid system to detect RNA-protein interactions in vivo. Proc. Natl Acad. Sci. USA. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.SenGupta DJ, Wickens M, Fields S. Identification of RNAs that bind to a specific protein using the yeast three-hybrid system. RNA. 1999;5:596–601. doi: 10.1017/s1355838299002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caponigro G, Muhlrad D, Parker R. A small segment of the MAT alpha 1 transcript promotes mRNA decay in Saccharomyces cerevisiae: a stimulatory role for rare codons. Mol. Cell Biol. 1993;13:5141–5148. doi: 10.1128/mcb.13.9.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sengchanthalangsy LL, Datta S, Huang DB, Anderson E, Braswell EH, Ghosh G. Characterization of the dimer interface of transcription factor NFkappaB p50 homodimer. J. Mol. Biol. 1999;289:1029–1040. doi: 10.1006/jmbi.1999.2823. [DOI] [PubMed] [Google Scholar]
- 35.Huang DB, Vu D, Cassiday LA, Zimmerman JM, Maher LJ, 3rd., Ghosh G. Crystal structure of NF-kappaB (p50)2 complexed to a high-affinity RNA aptamer. Proc. Natl Acad. Sci. USA. 2003;100:9268–9273. doi: 10.1073/pnas.1632011100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miyakawa S, Nomura Y, Sakamoto T, Yamaguchi Y, Kato K, Yamazaki S, Nakamura Y. Structural and molecular basis for hyperspecificity of RNA aptamer to human immunoglobulin G. RNA. 2008;14:1154–1163. doi: 10.1261/rna.1005808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.King DJ, Bassett SE, Li X, Fennewald SA, Herzog NK, Luxon BA, Shope R, Gorenstein DG. Combinatorial selection and binding of phosphorothioate aptamers targeting human NF-kappa B RelA(p65) and p50. Biochemistry. 2002;41:9696–9706. doi: 10.1021/bi020220k. [DOI] [PubMed] [Google Scholar]
- 38.Zhao X, Shi H, Sevilimedu A, Liachko N, Nelson HC, Lis JT. An RNA aptamer that interferes with the DNA binding of the HSF transcription activator. Nucleic Acids Res. 2006;34:3755–3761. doi: 10.1093/nar/gkl470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choi YS, Hur J, Lee HK, Jeong S. The RNA aptamer disrupts protein-protein interaction between beta-catenin and nuclear factor-kappaB p50 and regulates the expression of C-reactive protein. FEBS Lett. 2009;583:1415–1421. doi: 10.1016/j.febslet.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 40.Ghosh G, Huang DB, Huxford T. Molecular mimicry of the NF-kappaB DNA target site by a selected RNA aptamer. Curr. Opin. Struct. Biol. 2004;14:21–27. doi: 10.1016/j.sbi.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 41.Mi J, Zhang X, Liu Y, Reddy SK, Rabbani ZN, Sullenger BA, Clary BM. NF-kappaB inhibition by an adenovirus expressed aptamer sensitizes TNFalpha-induced apoptosis. Biochem. Biophys. Res. Commun. 2007;359:475–480. doi: 10.1016/j.bbrc.2007.05.125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.