Abstract
The Sm-like protein Hfq promotes the association of small antisense RNAs (sRNAs) with their mRNA targets, but the mechanism of Hfq's RNA chaperone activity is unknown. To investigate RNA annealing and strand displacement by Hfq, we used oligonucleotides that mimic functional sequences within DsrA sRNA and the complementary rpoS mRNA. Hfq accelerated at least 100-fold the annealing of a fluorescently labeled molecular beacon to a 16-nt RNA. The rate of strand exchange between the oligonucleotides increased 80-fold. Therefore, Hfq is very active in both helix formation and exchange. However, high concentrations of Hfq destabilize the duplex by preferentially binding the single-stranded RNA. RNA binding and annealing were completely inhibited by 0.5 M salt. The target site in DsrA sRNA was 1000-fold less accessible to the molecular beacon than an unstructured oligonucleotide, and Hfq accelerated annealing with DsrA only 2-fold. These and other results are consistent with recycling of Hfq during the annealing reaction, and suggest that the net reaction depends on the relative interaction of Hfq with the products and substrates.
INTRODUCTION
In bacteria, small antisense RNAs (sRNAs) participate in regulatory networks that control the response to environmental stress such as low temperature (1), low iron (2) or oxidative stress (3). Many sRNAs activate or repress the translation of their target messenger RNAs (mRNAs) by base pairing with complementary sequences in the mRNA (4,5). sRNAs can also target the mRNA for degradation, as in the case of the RyhB sRNA and sodB mRNA (2,6). Despite the importance of these regulatory interactions, however, the dynamics of base pairing between sRNAs and their mRNA targets remain poorly understood.
The RNA-binding protein Hfq binds many sRNAs in vivo and is necessary for their function (5,7,8). Hfq is a 11-kDa homolog of the eukaryotic Sm proteins (9–11) that forms ring-shaped homohexamers. Hfq preferentially binds single-stranded A/U-rich sequences adjacent to stem-loops (9,12–14), and facilitates the association of sRNAs with their mRNA targets. Because Hfq is not required to stabilize the sRNA–mRNA complex once it is formed, Hfq is considered a chaperone (9). Hfq binding also protects sRNAs and some mRNAs from degradation (15,16).
Several mechanisms for Hfq ‘chaperoning’ of base pairs between sRNAs and mRNAs base pairing have been proposed (4). Hfq may unfold or weaken RNA secondary structures, allowing sRNAs to access their targets (17,18). In addition, Hfq forms stable ternary complexes with sRNAs and mRNAs, that may facilitate annealing of complementary strands by increasing the local concentration of each strand (9,10). The Escherichia coli Hfq hexamer is proposed to simultaneously bind U-rich and A-rich strands, bringing them together (19). There is also evidence that Hfq hexamers can form higher-order complexes (20), although the functional importance of such complexes remains unclear.
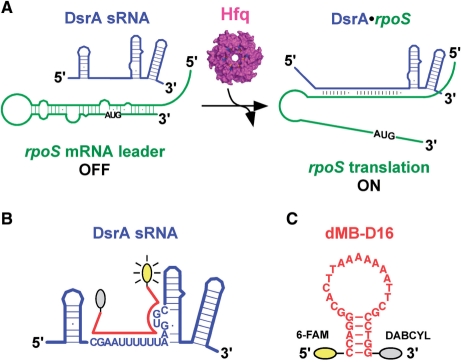
We have used the well-characterized interactions between DsrA sRNA and rpoS mRNA to study the function of Hfq in riboregulation and how it facilitates base pairing between complementary RNAs. In E. coli, the 87-nt sRNA DsrA upregulates the translation of rpoS (1,21,22), which encodes the stationary phase transcription factor σS (23). The ribosome-binding site of the rpoS mRNA is normally blocked by a stem–loop within the 5′ leader, inhibiting translation initiation (24). DsrA sRNA activates rpoS translation by base pairing to the 5′ leader and opening up its inhibitory stem–loop (Figure 1A) (21), in a manner that depends on Hfq protein (15,25).
Figure 1.
DsrA regulation of rpoS. (A) DsrA sRNA (blue) binds to the leader region of rpoS mRNA (green), exposing the Shine–Dalgarno sequence and initiation codon (AUG) (see text for details). Hfq (pink) binds DsrA and rpoS mRNA, and is required for DsrA regulation of rpoS in vivo. (B) Molecular beacon (MB; red) targeted against the U-rich Hfq binding site in DsrA sRNA. Letters indicate the sequence of the D16 oligomer. (C) Sequence of dMB-D16, conjugated with fluorescein 6-FAM (5′) and DABCYL (3′). rMB-D16 has the same sequence except U's replace T's. dMB-R16 is the complement of dMB-D16.
Hfq binds both DsrA and rpoS mRNA in vitro, and the formation of stable ternary complexes between Hfq, DsrA and rpoS mRNA correlates with faster association of the two RNAs (14,26,27). Arluison et al. (28) used FRET to suggest that an Hfq hexamer bound to both DsrA and rpoS slowly melts the rpoS secondary structure and promotes DsrA and rpoS binding by increasing the local concentration of the two RNAs. They proposed that Hfq uses strand-exchange to open the rpoS leader and promote DsrA annealing. This is consistent with biochemical data showing that Hfq partially destabilizes the secondary structure of the full-length rpoS leader (27). In contrast, Rajkowitsch and Schroeder (29) suggested Hfq functions mainly through RNA annealing rather than through strand exchange. The difference between the results of these two studies may be a result of the different RNA substrates used by each group. Arluison et al. (28) used RNAs that contained some secondary structure, whereas Rajkowitsch and Schroeder (29) used short, unstructured oligonucleotides.
To examine the ability of Hfq to accelerate annealing and strand exchange of structured and unstructured RNAs, we designed fluorescently labeled ‘molecular beacons’ to base pair with a 16-nt sequence that encompasses the Hfq-binding site within DsrA (Figure 1B). The use of molecular beacons in place of rpoS mRNA allowed us to detect the intrinsic annealing and exchange activities of Hfq, while avoiding the complication of multiple Hfq binding sites within the natural rpoS leader. The beacons also sidestepped the requirement to unfold the secondary structure of the rpoS mRNA. The thermodynamics and kinetics of reactions between the beacon and a 16-mer target RNA were compared with similar reactions containing full-length DsrA sRNA. We found that Hfq increased the rates of annealing and strand exchange as much as 100-fold. However, this activity depends strongly on the ionic strength of the buffer and the secondary structure of the substrates.
MATERIALS AND METHODS
Molecular beacons and oligonucleotides
Synthetic oligodeoxynucleotides (Invitrogen) were used without further purification: MB-D16: 5′ dCCAGGGCACTTAAAAAATTCGCCTGG; MB-D16s: 5′ dCCCGGGCACTTAAAAAATTCGCCGGG; nf-MB-R16: 5′ dCCCCTCGAATTTTTTAAGTGCAGGGG. Oligoribonucleotides (Invitrogen) were purified by 8% PAGE, eluted and resuspended in water. D16: 5′ CGAAUUUUUUAAGUGC and R16: 5′ GCACUUAAAAAAUUCG. Concentrations were determined by absorption at 260 nm: D16 ϵ260: 172.3 OD/μmol; R16 ϵ260: 187.7 OD/μmol; dMB-D16: ϵ260: 291.5 OD/μmol; dMB-D16s: ϵ260: 286.1 OD/μmol; nf-MB-R16: ϵ260: 277.9 OD/μmol.
Molecular beacons were designed with the help of MFOLD (30). Dye-conjugated oligomers were synthesized and purified by reverse phase HPLC (Trilink Biotechnologies): MB-D16: 5′ (6-FAM) CCAGGGCACTTAAAAAATTCGCCTGG (C6-NH-DABCYL) 3′; MB-R16: 5′ (6-FAM) CCCCTCGAATTTTTTAAGTGCAGGGG (C6-NH- DABCYL) 3′; MB-D16 (RNA): 5′ (6-FAM) CCAGGGCAC UUAAAAAAUUCGCCUGG (C6-NH- DABCYL) 3′. Complementary residues that form the hairpin of the beacon are underlined. The concentrations were determined by absorption at 260 nm: dMB-D16: ϵ260: 289.6 OD/μmol; dMB-R16: ϵ260: 283.4 OD/μmol; rMB-D16: ϵ260: 295.6 OD/μmol.
Preparation of DsrA RNA
DsrA (87 nt) was transcribed with T7 RNA polymerase from pUCT7DsrA and purified as previously described (26).
Purification of Hfq
His-tagged Hfq was expressed from a plasmid kindly provided by A. Feig (Wayne State University), and purified as previously described (19). Hfq stocks were stored at –80°C. Some experiments in TNKM, TN50 and TN500 buffers used natural Hfq, which was purified as previously described (9). Both preparations had similar activities on oligonucleotide substrates.
Native PAGE
Oligomers were 5′ labeled with 32P according to standard protocols and passed through a size-exclusion column (BD Sciences). Samples (10 µl) containing beacon, D16 RNA, and Hfq as stated in the text were incubated 30 min at 30°C prior to loading on 8% polyacrylamide gels containing 1× TBE. Gels were maintained at 4–10°C during electrophoresis. Experiments were carried out in TNK buffer (10 mM Tris–HCl pH 7.5, 50 mM NaCl, 50 mM KCl) or Hfq storage buffer (HB; 10 mM Tris–HCl pH 7.5, 50 mM NH4Cl, 0.2 mM EDTA pH 8.5, 2% glycerol) as stated in the text.
Fluorescence experiments
Equilibrium fluorescence intensity measurements (515-nm emission) were made using a Perkin-Elmer LS-50B Luminescence Spectrometer, with excitation at 496 nm. To measure the equilibrium binding constant (Kd), the fluorescence intensity of 2 nM dMB-D16 was recorded in TNK or HB buffer at 30°C, with 0–1.5 μM oligonucleotide target. The beacon and target were equilibrated 5 min after each addition. The relative change in fluorescence intensity ΔF was assumed to be proportional to the fraction of bound beacon and was fit to a single-site binding isotherm. Where applicable, Hfq was added to the beacon before the addition of target. Hfq did not change the baseline fluorescence of DNA beacons, but the baseline fluorescence of rMB-D16 increased slightly. Experiments with DsrA RNA (300 nM) were done as above, except DsrA was incubated briefly at 65°C before use.
Fluorescence anisotropy measurements were made using a Fluorolog-3 spectrofluorometer (Horiba) in L format with single excitation and emission monochromators at 495 nm and 515 nm, respectively. D16 RNA (5 nM) conjugated with fluorescein (6-FAM) in TNK or HB buffer was titrated with Hfq. The sample was incubated 5 min after each addition before the anisotropy was measured. For titrations, the fractional binding was obtained from the change in anisotropy, 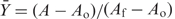 , in which A is the anisotropy at each Hfq concentration, Ao in the anistropy of D16-FAM without Hfq and Af is the anisotropy at saturation. The fractional binding was fit to a two-site isotherm,
, in which A is the anisotropy at each Hfq concentration, Ao in the anistropy of D16-FAM without Hfq and Af is the anisotropy at saturation. The fractional binding was fit to a two-site isotherm, 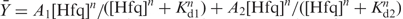 , in which A1 and A2 are the anisotropies of complexes 1 and 2, Kd1 and Kd2 are the binding constants and n = 2.3 is the cooperativity of forming each complex (26). The salt dependence of binding was measured by incubating 5 nM D16-FAM with 150 nM Hfq6 in HB, and then adding NaCl to the complex. Samples were incubated 5 min after each addition before the anistropy was measured.
, in which A1 and A2 are the anisotropies of complexes 1 and 2, Kd1 and Kd2 are the binding constants and n = 2.3 is the cooperativity of forming each complex (26). The salt dependence of binding was measured by incubating 5 nM D16-FAM with 150 nM Hfq6 in HB, and then adding NaCl to the complex. Samples were incubated 5 min after each addition before the anistropy was measured.
Stopped-flow spectroscopy
The hybridization kinetics was measured in HB or TNK buffer at 30°C using an Applied Photophysics SX 18MV stopped-flow spectrometer. Reactions contained 50 nM molecular beacon (final), 100 nM target RNA and 50 nM Hfq6 unless stated otherwise in the text. The beacon and RNA oligomers were mixed with a dead time of 1.8 ms. Hfq was typically added to the syringe containing the beacon, although the order of mixing did not affect the results. The sample was excited at 496 nm and the emission intensity was measured using a 515-nm cutoff filter. At least three separate experiments were performed at each oligonucleotide concentration. The data were fit to a double exponential rate equation and the observed rate constants for three or more trials were averaged.
RESULTS
Beacon design
To study the RNA annealing activity of Hfq, we synthesized minimal RNA and DNA substrates that mimic functional sequences in DsrA sRNA. A molecular beacon (MB-D16) was designed to complement a 16-nt sequence in DsrA sRNA containing the U-rich Hfq-binding site in DsrA (Figure 1B and C). Experiments were carried out with DsrA or with a 16-nt RNA containing just the target sequence (D16). The length of the target sequence and its position relative to the Hfq-binding site were chosen to maximize the stability of the antisense complex. The stability of the hairpin stem was also varied to optimize binding while maintaining a low baseline of fluorescence (31).
In our initial experiments, we used a DNA molecular beacon (dMB-D16). Since Hfq does not bind DNA strongly (32), we anticipated that Hfq would not unfold the hairpin of the molecular beacon in the absence of complementary target. We also expected ternary complexes in which the two substrates are bridged by Hfq to be unstable. Thus, any increase in the rate of hybridization should be due to enhanced base pairing between dMB-D16 and D16 RNA. To test the importance of interactions between Hfq and the beacon, we also obtained an RNA version of the beacon, rMB-D16, as well as a beacon with the complementary loop sequence (dMB-R16).
Beacon–target binding by native PAGE
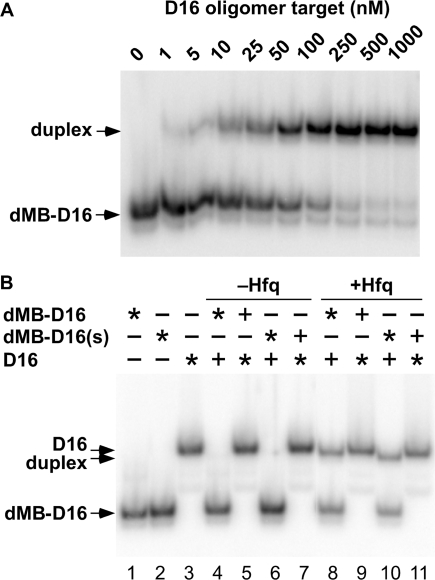
Molecular beacons were initially tested for the ability to bind D16 RNA by native PAGE. Radiolabeled dMB-D16 was incubated in TNK plus 1 mM MgCl2 (TNKM) at 30°C with increasing concentrations of D16 target (Figure 2A). As expected, the two complementary oligomers formed a stable hybrid duplex that migrated differently than the individual oligomers (Kd = 33 nM). Similar results were obtained in gel mobility shift experiments with dMB-R16, which base pairs with R16, the A-rich complement of D16 (data not shown).
Figure 2.
Hybridization of molecular beacons by native PAGE. (A) Molecular beacon dMB-D16 (without fluorophores) was 32P-labeled, and incubated in TNKM buffer for 15 min at 30°C with D16 RNA before native 8% PAGE; Kd = 33 nM. (B) Hfq stimulates base pairing in low salt buffer (HB), as indicated by the appearance of duplex (lanes 8 and 10). Trace radiolabeled (asterisk) and 50 nM unlabeled (+) dMB-D16 and D16 were co-incubated 15 min, with or without 83 nM Hfq6. dMB-D16(s) is the same as dMB-D16 except a C-G pair replaces the A-T pair in the hairpin stem. The similar mobilities of the duplex and D16 made it difficult to resolve the duplex when D16 is labeled (lanes 9 and 11).
The effect of Hfq on the association of these oligomers was also tested by native PAGE (Figure 2B). In TNKM buffer, Hfq had only a small effect on oligonucleotide binding (data not shown). When similar reactions were carried out in 50 mM NH4Cl (HB), however, 83 nM Hfq6 caused much more of the labeled oligonucleotide to shift into the duplex band, even when the stem of the beacon was strengthened by switching the AT pair to CG (dMB-D16s; Figure 2B). In HB buffer, almost no duplex was formed in the absence of Hfq.
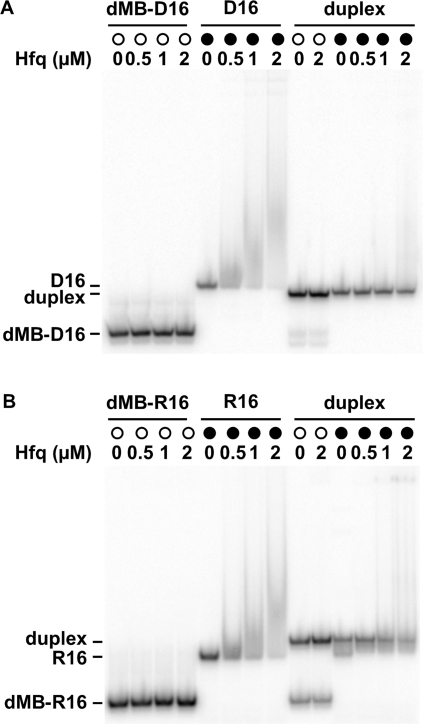
We next investigated whether Hfq formed stable complexes with the oligonucleotides. When the concentration of Hfq was increased from 0 to 330 nM (Hfq6), the gel mobility of dMB-D16 or dMB-R16 did not change (Figure 3, left lanes), confirming that Hfq does not interact strongly with DNA molecular beacons. Hfq was able to bind D16 or R16 RNA, although the complexes were too labile to produce a strong gel mobility shift (Figure 3, middle lanes). Hfq bound the hybrid DNA–RNA duplex much less strongly than D16 or R16 RNA (Figure 3, right lanes). In fact, the small amount of shifted material in these lanes is likely due to interactions between Hfq and single strands. Thus, Hfq was able to promote the association of the oligonucleotides but did not form a stable ternary complex with the base paired product.
Figure 3.
Hfq binding by native PAGE. Hfq binds the oligonucleotides weakly, as indicated by the absence of strong gel mobility shifts. 32P-labeled MB DNA (open circle), target RNA (closed circle), or their double-stranded complex (open or closed circles, respectively) were titrated with 0–2 µM Hfq monomer at 30°C in TNKM. (A) dMB-D16 and D16; (B) dMB-R16 and R16.
Binding of MB-D16 to its target RNA by fluorescence
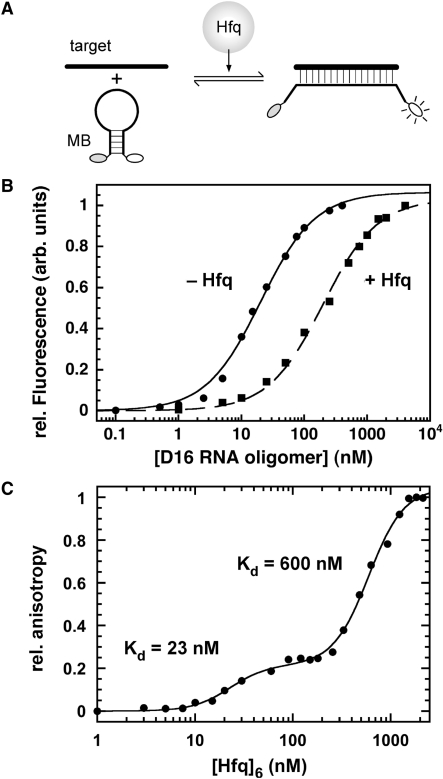
To quantify the effect of Hfq on the stability of the beacon–RNA complex, base pairing of dMB-D16 was examined using fluorescence spectroscopy. The MB-D16 beacon was conjugated with fluorescein on its 5′ end and DABCYL at the 3′ end (Figure 1C). In the absence of target, the hairpin stem of the beacon brings the fluorophore and quencher together, so that the baseline fluorescence is low (33). When base paired with a target RNA, separation of the fluorophore and quencher greatly increases the fluorescence intensity (Figure 4A).
Figure 4.
Hfq destabilizes the hybrid duplex. (A) Target RNA increases the fluorescence of molecular beacons. (B) Titration of dMB-D16 (5 nM) with D16 RNA in TNK buffer at 30°C. The extent of binding was measured from the relative fluorescence intensity at 515 nm. Circles, no Hfq, Kd = 19.6 ± 0.7 nM; squares, 330 nM Hfq6, Kd = 260 ± 63 nM. See Table 1 for additional data. (C) Hfq binding to D16-FAM measured by fluorescence anisotropy. Fractional binding in TNK was fit to a two-binding site isotherm with Kd1 = 23 ± 4 nM (Hfq6), Kd2 = 600 ± 25 nM.
When titrated with D16 RNA, the fluorescence of dMB-D16 beacon increased as expected (Figure 4B). In TNK buffer, the dissociation constant for D16 RNA and MB-D16 was 19.6 ± 0.7 nM at 30°C (Table 1), close to the estimate from native PAGE. In HB buffer, the Kd for the antisense complex was 79 ± 7 nM (Table 1). When the binding equilibrium between the beacon and RNA target was measured in the presence of 330 nM Hfq6, the Kd for hybridization of the beacon and the target increased 10-fold, to 260 ± 60 nM in TNK (Figure 4B; Table 1). This concentration of Hfq is above the Kd for single-stranded RNA (see below) and equivalent to the highest concentration of D16 RNA in our titration. The simplest explanation for these results is that Hfq destabilizes the beacon–target complex by binding single-stranded D16 RNA more strongly than the hybrid duplex. Hfq did not increase the fluorescence intensity of dMB-D16 in the absence of target, suggesting that it does not destabilize the DNA hairpin, consistent with the lack of a gel mobility shift in Figure 3.
Table 1.
Base pairing of molecular beacons with D16 RNA at 30°C
| Beacon | TNKa |
HBb |
||
|---|---|---|---|---|
| Kd (nM) | kobs (s–1)c | Kd (nM) | kobs (s–1)d | |
| dMB-D16 | ||||
| –Hfq | 19.6 ± 0.7 | 0.0052 ± 0.0005 | 79 ± 7 | 0.0080 ± 0.002 |
| +Hfq | 260 ± 63 | 0.58 ± 0.24 | – | 2.11 ± 0.96 |
| rMB-D16 | ||||
| –Hfq | 13.7 ± 1.0 | 0.005 ± 0.001 | 74 ± 4 | 0.005 ± 0.001 |
| +Hfq | 174 ± 29 | 0.12 ± 0.04 | – | 0.059 ± 0.0002 |
Error is the SD from at least three trials.
aTNK: 10 mM Tris–HCl pH 7.5; 50 mM NaCl; 50 mM KCl.
bHB: 10 mM Tris–HCl pH 7.5; 50 mM NH4Cl; 0.2 mM EDTA pH 8.5; 2% glycerol.
cObserved rate constant for binding of 50 nM beacon, 100 nM D16 oligoribonucleotide, in the presence or absence of 50 nM Hfq in the buffer specified at 30°C.
d300 nM beacon.
To determine the affinity of Hfq for single-stranded D16 RNA alone, we measured the change in fluorescence anisotropy of D16 conjugated to fluorescein (D16-FAM) (34). When D16-FAM was titrated with Hfq in TNK buffer at 30°C, we observed an increase in anisotropy indicative of two Hfq complexes with binding constants of 23 ± 4 nM and 600 ± 25 nM (Hfq6), respectively (Figure 4C). These are similar to previously reported dissociation constants for His6–Hfq binding to DsrA (14) and slightly lower than that of the wild type protein at 37°C (26). These results also showed that Hfq associates with D16 tightly enough to compete with base pairing between D16 and dMB-D16.
Hfq greatly increases the RNA annealing rate
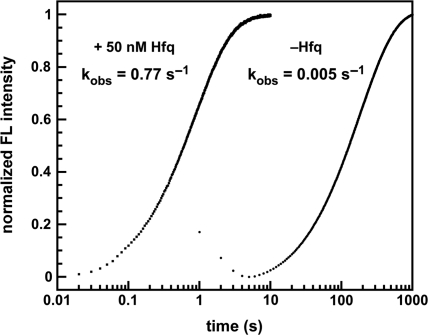
The association kinetics of the beacon and target were examined using stopped-flow spectroscopy (Figure 5). In the absence of Hfq, the observed hybridization rate of 50 nM MB-D16 to 100 nM D16 RNA was 0.0052 ± 0.0005 s–1 at 30°C. In the presence of 50 nM Hfq, the hybridization rate increased 100-fold, to an average value of 0.58 ± 0.24 s–1 (Table 1). In HB buffer, the effect of Hfq was even stronger, increasing the rate constant 260-fold to 2.1 s–1 (Table 1). Thus, Hfq strongly accelerated the base pairing kinetics of these oligonucleotides, demonstrating that it has an intrinsic strand-annealing activity. This stimulation depended on the RNA-binding activity of Hfq, because the amino acid substitution K56A in the U-rich RNA binding site of Hfq (19) resulted in ∼10-fold lower annealing rate under the same conditions (35). In addition, oligonucleotides completely lacking U- or A-rich sequences were poor substrates for Hfq (data not shown).
Figure 5.
Hfq increases the rate of beacon-target annealing. The hybridization rate of MB-D16 (50 nM) with 100 nM D16 RNA was measured by stopped-flow fluorescence in TNK buffer at 30°C. Hfq6 (50 nM) increases the binding rate 150-fold to kobs = 0.77 s–1 (64%). The initial burst phase yielded kobs = 2.1 s–1 (36%). In the absence of Hfq, kobs = 0.005 s–1. See Table 1 for further data.
Hfq increases the rate of strand exchange
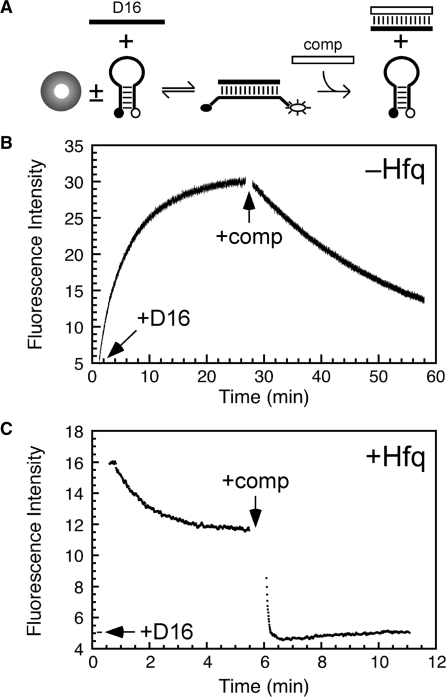
We next tested whether Hfq also accelerates strand exchange under these conditions (Figure 6A). The D16 RNA oligonucleotide was mixed with dMB-D16 for 20 min, until the fluorescence intensity reached a plateau, signaling that the beacon was base paired with RNA (Figure 6B). Next, an excess of complementary R16 RNA was added to the cuvette, trapping D16 as it dissociated from the dMB-D16 beacon. As the beacon was competed off the RNA, the fluorescence decreased with an observed rate constant of 0.18 min–1.
Figure 6.
Hfq accelerates strand exchange. (A) dMB-D16 was equilibrated in TNK buffer at 30°C before addition of 50 nM D16 target RNA. After the fluorescence stabilized, 50 nM R16 competitor was added to trap free D16. (B) Change in fluorescence as in (A), without Hfq. Strand association, kobs = 0.18 min–1; strand exchange kobs = 0.038 min–1. Arrows indicate the additions of target and competitor. (C) Change in fluorescence as in (A), with 200 nM Hfq6. The beacon was equilibrated with Hfq before the addition of D16; the baseline fluorescence before the addition of D16 is indicated by the arrow. Strand exchange, kobs = 14 min–1; slow decrease in dMB-D16·D16 fluorescence, kobs = 0.8 min–1. The rate of strand association was too fast to measure using a conventional spectrometer.
When the experiment was repeated in the presence of 200 nM Hfq6, the initial beacon–RNA complex formed very quickly, as expected (Figure 6C). This was followed by a slow decrease in fluorescence whose origin we do not yet understand. More importantly, the strand-exchange reaction after the addition of competitor occurred within 5–10 s (∼14 min–1). This corresponded to a 80-fold increase in the rate of strand displacement, comparable to the influence of Hfq on the rate of strand association. Thus, Hfq accelerates helix formation and helix dissociation (strand exchange) under the same experimental conditions.
Salt inhibits the annealing activity of Hfq
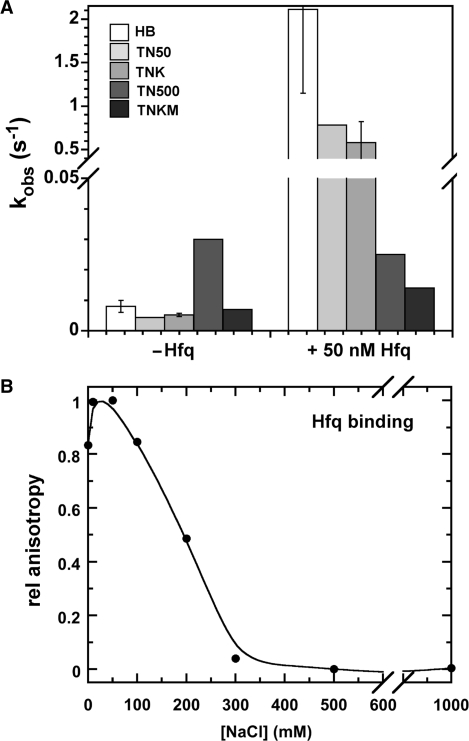
Since Hfq was more active in HB buffer than TNK buffer, next we investigated the effect of salt on Hfq activity. Salt increases the stability of nucleic acid duplexes, but often lowers the affinity of proteins for DNA or RNA (36). When the salt concentration was raised from 50 to 500 mM, the rate of protein-independent annealing increased from 0.0044 to 0.03 s–1 (Figure 7A), as expected from early work (37). In contrast, salt reduced the rate of RNA association in the presence of Hfq from 2.1 ± 1.0 s–1 in HB to 0.025 s–1 in TN500 (Figure 7A). Thus, Hfq has no net effect on the RNA annealing rate in 500 mM salt. This loss of activity corresponded to a steep decline in Hfq binding to D16 RNA over this salt concentration range (Figure 7B). Hfq's annealing activity was inhibited by 1 mM MgCl2 more than predicted by the increase in bulk ionic strength (compare TNK and TNKM; Figure 7A). Thus, the ability of Hfq to facilitate the formation and exchange of base pairs is extremely sensitive to the electrostatic environment around the RNA.
Figure 7.
Salt inhibits annealing by Hfq. (A) Observed rate constants for annealing with or without 50 nM Hfq (100 nM D16 and 50 nM MB-D16, 30°C). Salt lowered the Hfq-dependent annealing rate. Reactions in 10 mM Tris–HCl pH 7.5 plus: HB, 50 mM NH4Cl, 2% glycerol; TN50, 50 mM NaCl; TN500, 500 mM NaCl; TNK, 50 mM NaCl, 50 mM KCl; TNKM, 50 mM NaCl, 50 mM KCl, 1 mM MgCl2. Error bars represent the SD of three independent trials. (B) Effect of salt on Hfq binding to D16 RNA. Salt was added to complexes formed in HB (150 nM Hfq6 and 5 nM D16-FAM) and the extent of Hfq binding measured by fluorescence anisotropy. Error bars are about the size of the data points.
U-rich and A-rich RNAs are similar
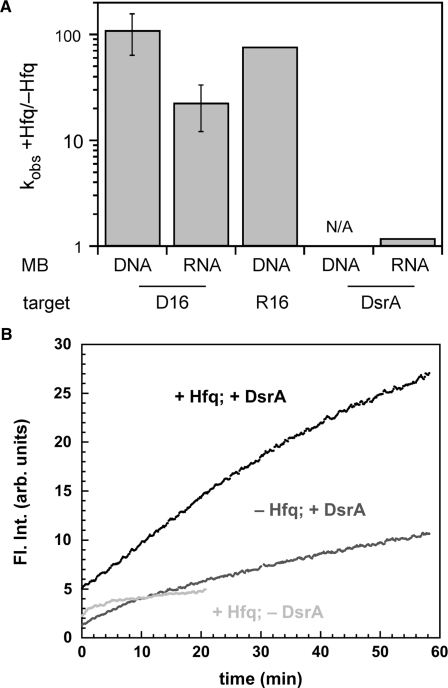
The inner surface of the Hfq hexamer binds both A- and U-rich RNAs, while the distal face is proposed to contain an A-specific binding site (19). To test the ability of Hfq to bind different RNA targets, annealing reactions were carried out with an A-rich RNA (R16) that is the complement of D16. The R16 RNA (100 nM) and dMB-R16 base paired with each other at a rate of 0.01 s–1 in the absence of protein. In the presence of 50 nM Hfq6, the association rate increased to 0.74 s–1 at 30°C in HB buffer. This 74-fold increase in the hybridization rate is less than the 260-fold effect of Hfq on the U-rich D16 RNA and dMB-D16 in HB buffer, but still substantial (Figure 8A). Thus, the activity of Hfq on these substrates depends little on whether the RNA strand is U- or A-rich.
Figure 8.
Substrate-dependence of Hfq annealing activity. (A) Ratio of annealing rate constants with and without Hfq (50 nM molecular beacon, 100 nM 16-mer or 300-nM DsrA and ±50 nM Hfq6). Rates for R16 are in HB buffer; all others are in TNK. Error bars represent the SD of three independent experiments. R16 is the average of multiple shots within the same experiment. (B) DsrA sRNA (300 nM) binding to rMB-D16 RNA (50 nM) at 30°C in TNK. Fluorescence increased more rapidly with 50 nM Hfq6 (black; kobs ≈ 0.014 min–1) than without Hfq (medium gray; kobs ≈ 0.004 min–1). A small rise in fluorescence was observed with Hfq and no target (light gray).
An RNA beacon results in slower hybridization rates
To determine whether Hfq accelerates the annealing of two RNAs similarly to an RNA and DNA strand, the same experiments were carried out with an RNA version of MB-D16. The equilibrium dissociation constants for binding of rMB-D16 and D16 RNA in TNK were 13.7 ± 1.0 nM (no Hfq) and 174 ± 29 nM (with Hfq) (Table 1). These binding constants are similar to those for the DNA beacon and D16. Thus, Hfq destabilizes RNA and hybrid duplexes to a comparable degree.
In contrast, Hfq accelerated the annealing of two RNA oligomers less than annealing of an RNA and DNA oligomer. In the absence of Hfq, the RNA and DNA beacons base paired with D16 RNA at the same rate (0.005 ± 0.001 s–1; Table 1). In the presence of 50 nM Hfq6, however, the annealing rate of rMB-D16 was 0.12 ± 0.04 s–1, about 5-fold lower than that of dMB-D16 (Table 1; Figure 8A). Since the annealing kinetics of the RNA and DNA beacons are identical in the absence of Hfq, it is unlikely that this difference comes from the stability of the stem or loop of the RNA beacon. As discussed below, it is more likely that the observed kinetics depend on the interactions of Hfq with the substrates and product.
Structure of DsrA inhibits hybridization
Previous studies showed that Hfq binding depends on RNA structure as well as sequence (9,14). To compare Hfq activity on oligonucleotides with a natural sRNA substrate, we carried out similar experiments with 300 nM DsrA RNA at 30°C. Under these conditions, the DNA beacon dMB-D16 was virtually unable to bind DsrA sRNA. This was ameliorated only slightly by the addition of 50 nM Hfq6 (data not shown). The RNA beacon rMB-D16 was able to slowly bind DsrA in the absence of Hfq (0.004 min–1) (Figure 8B). In the presence of 50 nM Hfq6, its fluorescence increased about three times faster (0.014 min–1). A slight increase in the baseline fluorescence when Hfq was added to rMB-D16 alone suggested that Hfq also unfolds the free RNA beacon to some extent (Figure 8B). These results not only show that DsrA is less available to pair with a complementary RNA strand, but that the sequence or secondary structure of DsrA attenuates the intrinsic capacity of Hfq to stimulate RNA helix formation.
DISCUSSION
The Sm-like protein Hfq is essential for the function of many sRNAs, and can help sRNAs base pair with their target mRNAs (7). Because Hfq is not required to stabilize the sRNA–mRNA complex once it is formed, Hfq acts as a chaperone for RNA interactions (9). Here, we show that Hfq has a strong intrinsic strand-annealing activity on substrates with minimal secondary structure. It accelerates the hybridization of two complementary oligonucleotides ≥100-fold, which is even larger than the 30–50-fold increase in the rate of DsrA binding to long rpoS leaders by Hfq (27,38). However, the magnitude of the annealing enhancement depends on the substrates used. The 100-fold effect of Hfq on oligonucleotides is much larger than the 2-fold rate increase for DsrA binding to a minimal rpoS leader (26) and the 4-fold effect previously reported for association of 37-nt RNA substrates (28). Moreover, we find that Hfq speeds up strand exchange about 80-fold, in qualitative agreement with previous reports (28,29).
Although the mechanism by which Hfq facilitates helix formation and strand exchange is not understood, one plausible scenario is that Hfq favors nucleation of the intermolecular helix, which is often the rate-determining step for the association of antisense RNAs (39). Helix nucleation requires that Hfq interacts at least transiently with one or both strands of the helix. Although Hfq has a relaxed binding specificity, oligomers without a U- or A-rich sequence were poor substrates in our annealing reactions, indicating that some sequence recognition is needed.
Unsurprisingly for a basic protein, the ability of Hfq to accelerate the association of two oligonucleotides depends strongly on salt concentration. In the absence of protein, salt increased the base-pairing rate, consistent with the expected reduction in the free energy barrier for helix nucleation (40). In contrast, 0.3 M salt strongly inhibited binding of Hfq to the D16 RNA and consequently eliminated the ability of Hfq to promote helix formation. Our results show that the concentrations and types of ions must be accounted for when comparing Hfq activity in different assays.
Since Hfq promotes both helix formation and exchange under similar conditions, the net flux of the annealing reaction will depend on the relative stabilities of the products and reactants. The fact that Hfq shifts the equilibrium away from the duplex is consistent with its stronger affinity for single-stranded RNA than double-stranded RNA apparent from the gel-mobility shift experiments. Thus, a small amount of Hfq might favor the antisense complex by increasing the association rate of the two RNAs, while too much Hfq will drive the reaction backwards by stabilizing the unpaired RNA. In preliminary experiments (data not shown), 50 nM Hfq6 destabilized the antisense complex only slightly, while 300 nM Hfq6 had a 10-fold effect (Figure 3A). The action of HIV nucleocapsid protein, another RNA chaperone, has been shown to be sensitive to the stabilities of the initial and final RNA structures and the affinity of nucleocapsid for each (41,42).
We find that Hfq is 10–100 times more active on a 16-nt RNA than on DsrA sRNA, despite the fact that it binds the two RNAs with similar affinity. Based on the annealing rates in the absence of Hfq, we estimate that the target sequence within DsrA is ∼100 times less accessible to an antisense probe than an unstructured oligonucleotide. Alternative secondary structures have been proposed for DsrA (43,44), and formation of such structures could mask our target sequence. We also observed, however, that Hfq is five times more active on a DNA molecular beacon than an RNA beacon of the same sequence. This is not due to differences in the stability of the RNA and DNA beacons, as the strand association rates were the same without Hfq.
The need to recycle Hfq during the annealing reaction could explain why it appears more active on weakly bound oligonucleotide substrates such as the DNA beacon. Previously, we observed that high occupancy by Hfq inhibits base pairing between DsrA and the rpoS leader, and proposed that Hfq must dissociate from its binding site on DsrA during the reaction cycle (26). Poor interactions with one or both strands would favor release of Hfq during the reaction cycle. By contrast, stable interactions with substrates could inhibit the release of Hfq and ‘zippering’ of the antisense helix. The natural rpoS leader may overcome this problem by offering a strong binding site for Hfq upstream of DsrA that transfers the protein away from the region of the intermolecular duplex (27).
An alternative explanation for our results is that Hfq interacts with single-stranded oligomers in a different mode than natural sRNAs, altering its activity. The U-rich RNA oligomer is expected to occupy the binding pockets which encircle the inner pore of Hfq, as shown in the crystal structure of the Staphylococcus aureus Hfq–RNA complex (45). However, recognition of A-rich RNAs has been linked to residues on the rim and distal face of Hfq, opposite the U-rich binding site (19,34). The outer surface of the Hfq hexamer has also been proposed to interact with double-stranded RNA (14). Additional interactions with A-rich sequences or double helices, which could be supplied by natural substrates but not a short oligonucleotide, could conceivably perturb the binding of single-stranded residues to the inner surface of Hfq. Further work is needed to test these possibilities.
In conclusion, the results presented here demonstrate that Hfq has potent annealing and strand-displacement activities. However, these intrinsic activities are modulated by its interactions with the substrates and the electrostatic environment, which in turn determine the net reaction flux. Further experiments to investigate the annealing mechanism are in progress.
FUNDING
The National Institutes of Health (R01 GM46686).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank T. Soper, D. Toptygin and L. Brand for their assistance and A. Feig for the gift of the His6-Hfq expression plasmid.
Footnotes
Present address: Julia F. Hopkins, Department of Biochemistry and Molecular Biology, Dalhousie University, 5850 College St, Halifax, NS B3H 1X5, Canada.
REFERENCES
- 1.Sledjeski DD, Gupta A, Gottesman S. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 1996;15:3993–4000. [PMC free article] [PubMed] [Google Scholar]
- 2.Masse E, Gottesman S. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl Acad. Sci. USA. 2002;99:4620–4625. doi: 10.1073/pnas.032066599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell. 1997;90:43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- 4.Storz G, Opdyke JA, Zhang A. Controlling mRNA stability and translation with small, noncoding RNAs. Curr. Opin. Microbiol. 2004;7:140–144. doi: 10.1016/j.mib.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 5.Gottesman S, McCullen CA, Guillier M, Vanderpool CK, Majdalani N, Benhammou J, Thompson KM, FitzGerald PC, Sowa NA, FitzGerald DJ. Small RNA regulators and the bacterial response to stress. Cold Spring Harbor Symp. Quant. Biol. 2006;71:1–11. doi: 10.1101/sqb.2006.71.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masse E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 8.Aiba H. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 2007;10:134–139. doi: 10.1016/j.mib.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 9.Zhang A, Wassarman KM, Ortega J, Steven AC, Storz G. The Sm-like Hfq protein increases OxyS RNA interaction with target mRNAs. Mol. Cell. 2002;9:11–22. doi: 10.1016/s1097-2765(01)00437-3. [DOI] [PubMed] [Google Scholar]
- 10.Moller T, Franch T, Hojrup P, Keene DR, Bachinger HP, Brennan RG, Valentin-Hansen P. Hfq: a bacterial Sm-like protein that mediates RNA–RNA interaction. Mol. Cell. 2002;9:23–30. doi: 10.1016/s1097-2765(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 11.Sun X, Zhulin I, Wartell RM. Predicted structure and phyletic distribution of the RNA-binding protein Hfq. Nucleic Acids Res. 2002;30:3662–3671. doi: 10.1093/nar/gkf508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Senear AW, Steitz JA. Site-specific interaction of Qbeta host factor and ribosomal protein S1 with Qbeta and R17 bacteriophage RNAs. J. Biol. Chem. 1976;251:1902–1912. [PubMed] [Google Scholar]
- 13.Vytvytska O, Moll I, Kaberdin VR, von Gabain A, Blasi U. Hfq (HF1) stimulates ompA mRNA decay by interfering with ribosome binding. Genes Dev. 2000;14:1109–1118. [PMC free article] [PubMed] [Google Scholar]
- 14.Brescia CC, Mikulecky PJ, Feig AL, Sledjeski DD. Identification of the Hfq-binding site on DsrA RNA: Hfq binds without altering DsrA secondary structure. RNA. 2003;9:33–43. doi: 10.1261/rna.2570803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sledjeski DD, Whitman C, Zhang A. Hfq is necessary for regulation by the untranslated RNA DsrA. J. Bacteriol. 2001;183:1997–2005. doi: 10.1128/JB.183.6.1997-2005.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Folichon M, Arluison V, Pellegrini O, Huntzinger E, Regnier P, Hajnsdorf E. The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res. 2003;31:7302–7310. doi: 10.1093/nar/gkg915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geissmann TA, Touati D. Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. EMBO J. 2004;23:396–405. doi: 10.1038/sj.emboj.7600058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moll I, Leitsch D, Steinhauser T, Blasi U. RNA chaperone activity of the Sm-like Hfq protein. EMBO Rep. 2003;4:284–289. doi: 10.1038/sj.embor.embor772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat. Struct. Mol. Biol. 2004;11:1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arluison V, Mura C, Guzman MR, Liquier J, Pellegrini O, Gingery M, Regnier P, Marco S. Three-dimensional structures of fibrillar Sm proteins: Hfq and other Sm-like proteins. J. Mol. Biol. 2006;356:86–96. doi: 10.1016/j.jmb.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 21.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl Acad. Sci. USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lease RA, Cusick ME, Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl Acad. Sci. USA. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lange R, Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol. Microbiol. 1991;5:49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- 24.Brown L, Elliott T. Mutations that increase expression of the rpoS gene and decrease its dependence on hfq function in Salmonella typhimurium. J. Bacteriol. 1997;179:656–662. doi: 10.1128/jb.179.3.656-662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muffler A, Traulsen DD, Fischer D, Lange R, Hengge-Aronis R. The RNA-binding protein HF-I plays a global regulatory role which is largely, but not exclusively, due to its role in expression of the sigmaS subunit of RNA polymerase in Escherichia coli. J. Bacteriol. 1997;179:297–300. doi: 10.1128/jb.179.1.297-300.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lease RA, Woodson SA. Cycling of the Sm-like protein Hfq on the DsrA small regulatory RNA. J. Mol. Biol. 2004;344:1211–1223. doi: 10.1016/j.jmb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 27.Soper TJ, Woodson SA. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA. 2008;14:1907–1917. doi: 10.1261/rna.1110608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arluison V, Hohng S, Roy R, Pellegrini O, Regnier P, Ha T. Spectroscopic observation of RNA chaperone activities of Hfq in post-transcriptional regulation by a small non-coding RNA. Nucleic Acids Res. 2007;35:999–1006. doi: 10.1093/nar/gkl1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajkowitsch L, Schroeder R. Dissecting RNA chaperone activity. RNA. 2007;13:2053–2060. doi: 10.1261/rna.671807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mathews DH, Sabina J, Zuker M, Turner DH. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 31.Hopkins JF, Woodson SA. Molecular beacons as probes of RNA unfolding under native conditions. Nucleic Acids Res. 2005;33:5763–5770. doi: 10.1093/nar/gki877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franze de Fernandez MT, Hayward WS, August JT. Bacterial proteins required for replication of phage Q ribonucleic acid. Purification and properties of host factor I, a ribonucleic acid-binding protein. J. Biol. Chem. 1972;247:824–831. [PubMed] [Google Scholar]
- 33.Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 34.Sun X, Wartell RM. Escherichia coli Hfq binds A18 and DsrA domain II with similar 2:1 Hfq6/RNA stoichiometry using different surface sites. Biochemistry. 2006;45:4875–4887. doi: 10.1021/bi0523613. [DOI] [PubMed] [Google Scholar]
- 35.Hopkins JF. Ph.D. Thesis. Baltimore: Johns Hopkins University; 2007. Secondary structure rearrangements monitored by fluorescent probes in RNA folding and RNA-protein interactions. [Google Scholar]
- 36.Record MT, Zhang WT, Anderson CF. Analysis of effects of salts and uncharged solutes on protein and nucleic acid equilibria and processes: a practical guide to recognizing and interpreting polyelectrolyte effects, Hofmeister effects, and osmotic effects of salts. Adv. Protein Chem. 1998;51:281–353. doi: 10.1016/s0065-3233(08)60655-5. [DOI] [PubMed] [Google Scholar]
- 37.Porschke D, Uhlenbec OC, Martin FH. Thermodynamics and kinetics of helix-coil transition of oligomers containing Gc base pairs. Biopolymers. 1973;12:1313–1335. [Google Scholar]
- 38.Updegrove T, Wilf N, Sun X, Wartell RM. Effect of Hfq on RprA-rpoS mRNA pairing: Hfq-RNA binding and the influence of the 5′ rpoS mRNA leader region. Biochemistry. 2008;47:11184–11195. doi: 10.1021/bi800479p. [DOI] [PubMed] [Google Scholar]
- 39.Wagner EG, Altuvia S, Romby P. Antisense RNAs in bacteria and their genetic elements. Adv. Genet. 2002;46:361–398. doi: 10.1016/s0065-2660(02)46013-0. [DOI] [PubMed] [Google Scholar]
- 40.Porschke D, Eigen M. Co-operative non-enzymic base recognition. 3. Kinetics of the helix-coil transition of the oligoribouridylic–oligoriboadenylic acid system and of oligoriboadenylic acid alone at acidic pH. J. Mol. Biol. 1971;62:361–381. doi: 10.1016/0022-2836(71)90433-5. [DOI] [PubMed] [Google Scholar]
- 41.Heilman-Miller SL, Wu T, Levin JG. Alteration of nucleic acid structure and stability modulates the efficiency of minus-strand transfer mediated by the HIV-1 nucleocapsid protein. J. Biol. Chem. 2004;279:44154–44165. doi: 10.1074/jbc.M401646200. [DOI] [PubMed] [Google Scholar]
- 42.Vo MN, Barany G, Rouzina I, Musier-Forsyth K. HIV-1 nucleocapsid protein switches the pathway of transactivation response element RNA/DNA annealing from loop–loop “kissing” to “zipper”. J. Mol. Biol. 2009;386:789–801. doi: 10.1016/j.jmb.2008.12.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lease RA, Belfort M. A trans-acting RNA as a control switch in Escherichia coli: DsrA modulates function by forming alternative structures. Proc. Natl Acad. Sci. USA. 2000;97:9919–9924. doi: 10.1073/pnas.170281497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones AM, Goodwill A, Elliott T. Limited role for the DsrA and RprA regulatory RNAs in rpoS regulation in Salmonella enterica. J. Bacteriol. 2006;188:5077–5088. doi: 10.1128/JB.00206-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schumacher MA, Pearson RF, Moller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]