Abstract
Numerous cellular factors belonging to the DNA repair machineries, including RAD18, RAD52, XPB and XPD, have been described to counteract human immunodeficiency virus type 1 (HIV-1) replication. Recently, Uracil DNA glycosylase 2 (UNG2), a major determinant of the uracil base excision repair pathway, was shown to undergo rapid proteasome-dependent degradation following HIV-1 infection. However, the specific role of intracellular UNG2 depletion during the course of HIV-1 infection is not clearly understood. Our study shows for the first time that overexpression of UNG2 inhibits HIV-1 replication. We demonstrate that this viral inhibition is correlated with a marked decrease in transcription efficiency as shown by monitoring HIV-1 LTR promoter activity and quantification of HIV-1 RNA levels. Interestingly, UNG2 inhibits LTR activity when stimulated by Tat transactivator or TNFα, while barely affected using Phorbol ester activation. Mutational analysis of UNG2 indicates that antiviral activity may require the integrity of the UNG2 catalytic domain. Altogether, our data indicate that UNG2 is likely to represent a new host defense factor specifically counteracted by HIV-1 Vpr. The molecular mechanisms involved in the UNG2 antiviral activity still remain elusive but may rely on the sequestration of specific cellular factor(s) critical for viral transcription.
INTRODUCTION
Multiple cellular DNA repair enzymes have been described as potential cellular cofactors required for human immunodeficiency virus type 1 (HIV-1) integration. These cofactors include components of the base excision repair (BER), the homologous recombination (HR) and the non-homologous end joining DNA repair pathways (1). In contrast, multiple DNA repair components have also been shown to counteract HIV-1 replication. For instance, RAD18, a cellular protein implicated in post-replication DNA repair, decreases the susceptibility of target cells to MLV and HIV-1 infection, probably by targeting the incoming viral DNA (2). The HR molecule RAD52 has also been shown to reduce retroviral infection by competing with active integration complexes (3). Finally, the human TFIIH complex proteins XPB and XPD, two DNA helicases with opposite polarity, play a critical role in the degradation of the retroviral DNA (4). To establish a productive infection, HIV-1 must be able to overcome these cellular DNA damage response machineries.
In this report, we investigated the role of the human Uracil DNA glycosylase 2 (UNG2) in the HIV-1 life cycle. Nuclear UNG2 and mitochondrial UNG1 isoforms are DNA repair enzymes that act in removing uracil bases from the sugar backbone of genomic and mitochondrial DNA respectively, leaving abasic sites and initiating the uracil BER pathway (5). Particularly, UNG2 activity is crucial for rapid removal of dUMP residues incorporated during genomic DNA replication (6). During HIV-1 infection, UNG2 was initially reported to be specifically packaged into virions via direct interaction with the viral integrase (IN) (7,8) or the Vpr regulatory protein (9). When packaged into HIV-1 particles, UNG2 was described to be essential for efficient viral replication by preventing dUMP misincorporation into the nascent viral DNA during the reverse transcription step (10,11). This role was proposed to be specific for HIV-1 since neither the related HIV-2 nor SIV retroviruses were found able to incorporate UNG2 into cell free particles (12). However, the contribution of UNG2 in the HIV-1 life cycle is highly debated. A recent report suggested that virion-associated UNG2 is dispensable for an efficient HIV-1 replication (13). Moreover, in the context of HIV-1 infected cells, UNG2 complexes with HIV-1 Vpr (14). This UNG2-Vpr interaction was recently shown to trigger the degradation of UNG2 in a proteasome-dependent manner through the specific recruitment of the damage-specific DNA-binding protein 1 (DDB1) by HIV-1 Vpr (15,16).
In this context, the aim of our study was to decipher the complex relationship that exists between UNG2 and HIV-1. First, we show that UNG2 overexpression inhibits HIV-1 RNA synthesis and viral particles production. Furthermore, we determine that depletion of endogenous UNG2, following RNA interference, promotes Tat-mediated activation of HIV-1 LTR promoter. UNG2 overexpression also inhibits TNFα-induced HIV-1 transcription but barely affects PMA-induced-LTR activation. Mutation of residues Q153D154 in UNG2 catalytic domain altered UNG2 anti-transcriptional activity. Testing UNG2 effects on a vast variety of promoters from cellular or viral origin put in evidence that UNG2 harbors a wide anti-transcriptional effect, suggesting that this activity may rely on the inhibition of cellular factor(s) critical for transcriptional regulation of multiple cellular and viral genes. Altogether, these data show for the first time that UNG2 harbors an antiviral activity. In addition, we do confirm that endogenous UNG2 is degraded in the presence of HIV-1 Vpr but is barely affected in cells infected with the related HIV-2 retrovirus. Therefore, these results support the hypothesis that the Vpr-mediated degradation of UNG2 may specifically protect HIV-1 from a negative regulatory effect of UNG2 on viral transcription.
MATERIALS AND METHODS
Reagents and antibodies
The following antibodies were used: rabbit polyclonal anti-UNG2 clone PU059 (17) (from Geir Slupphaug, Trondheim, Norway); this antibody cross reacts with human UNG1 (31 kDa), UNG2 (37 kDa) and phosphorylated isoforms of UNG2 (38–40 kDa); mouse monoclonal (mAb) anti-actin (C4, MP Biomedicals, France); mouse mAb anti-p24 ab9069 (Abcam, France); goat polyclonal anti-p24 (AbD serotec, France); Alexa fluor-594 F(ab’)2 fragment of goat anti-rabbit IgG and Alexa fluor-488 F(ab’)2 fragment of goat anti-mouse IgG (Invitrogen, France). PMA and TNF-α were obtained from Sigma-Aldrich (France).
Plasmids
The following plasmids encoding for viral molecular clones were used: pNLAD8, pNL4.3, pNL4.3-VprW54G, pNL4.3-INLK172/173AA (from J. Sire, Marseille, France), pNL4.3ΔVpr (from W.C. Greene, San Francisco, USA) and pROD10 (from K. Peden, Bethesda, USA). pUNG2-EGFP-N1 (here termed pUNG2-GFP) has been obtained from Geir Slupphaug. pcDNA3.1-UNG2 (here termed pUNG2) and the UNG2 catalytic mutant QD153-154LE were from J. Sire. pAS1B-Vpr was from Serge Bénichou (Paris, France). The pC53SN3, pG13PY-Luc, pcDNA-c-Jun and the pCollagenase-Luc plasmids have been kindly provided by Jean-Michel Mesnard (Montpellier, France).
Cell culture
Immortalized CEM T cells were maintained in RPMI 1640 medium. HEK 293T, MAGIC-5B (from M. Tatsumi, Tokyo, Japan) and Hela-LTRHIV-1-Luc cells (from S. Emiliani, Paris, France) were maintained in DMEM medium. Complete media were supplemented with penicillin-streptomycin and 10% heat-inactivated fetal bovine serum (FBS) (Cambrex, France).
Viral stocks production
Viral stocks were produced by calcium phosphate transfection of HEK 293T cells. Briefly, cells were transfected with the proviral DNA constructs for 8 h, washed with pre-warmed DMEM-10% SVF medium. For VSV-G pseudotyped HIV-1 production, the proviral DNA was cotransfected with pHEF-VSV-G as previously described (18). Viral supernatants were collected 48 h post-transfection, filtered and frozen in aliquots at –80°C. Viral stocks were titered using either an HIV-1 p24 Enzyme-linked immunosorbent assay (ELISA) kit (Beckman Coulter, France) or a reverse transcriptase (RT) assay as previously described (19).
Enzymatic assays
Plasmid transfections into Hela-LTRHIV-1-Luc and MAGIC-5B cells were carried out using cationic polymer (JetPei, Polyplus, France). Two days post-transfection or infection, cell extracts of Hela-LTRHIV-1-Luc were analyzed for luciferase (Luc) activity according to the manufacturer’s protocol (Yelen Corp., France). Cell extracts from MAGIC-5B cells were analyzed for β-galactosidase (β-Gal) activity, used as an index of HIV-1 LTR activity. Reporter activities were normalized to total protein contents.
Fluorescence microscopy
Infected MAGIC-5B cells were fixed in 3.7% paraformaldehyde for 10min, washed, permeabilized with 0.1% triton X-100 for 20min. Then, cells were incubated with mouse anti-p24 (1/300) and rabbit anti-UNG1/2 primary antibodies (1/300), washed, and incubated with Alexa-Fluor conjugated secondary antibodies (1/1000). Cells were mounted onto glass slides covered with anti-fade medium (Hardset Vectashield, Clinisciences, France). Two-color images were obtained with a light microscope, Leica DC250 (Leica, France) with a Plan Apo 63 × /1.32-0.6 oil-immersion objective lens. Digital images were processed with Adobe Photoshop.
Reverse transcription-quantitative PCR experiments
Quantification of viral genomic RNA was assessed by amplifying a 235-bp fragment from the HIV-1 gag gene (20) and quantification of LTR-luciferase mRNA was assessed by amplifying a 145-bp fragment from the promoter-proximal region (21). Briefly, total cellular RNA was extracted with Tri-Reagent (Sigma, France). Oligo(dT) was used as RT-primer and subsequent qPCR was conducted with a SYBR Green Kit (Roche, France) on the RotorGene system (Labgene, France). A standard curve was generated from 50 to 500 000 copies of pNL4.3 plasmid. Each assay was accompanied by controls without reverse transcriptase (DNA contamination levels < 1% of HIV-1 RNA). The copy numbers of HIV-1 genomic RNA or LTR-luciferase mRNA were normalized to that of the GAPDH mRNA quantified in parallel as endogenous control.
Western blot experiments and analysis
Proteins were separated by SDS–PAGE with 12% gels, transferred to a nitrocellulose membrane (Millipore, france) and immunoblotted with the appropriate primary and horseradish peroxidase–conjugated secondary antibodies (Immunotech, France). Autoradiographic films were scanned and quantified using Image J (NIH) software.
Stealth RNAi transfection
The Stealth RNAi sequence duplexes directed against UNG2 are as follows: sense, 5′-CGCGUUCGCUGCCUCCUCAGCUCCA-3′ and antisense, 5′-UGGAGCUGAGGAGGCAGCGAACGCG-3′. This target sequence corresponds to the 5′ untranslated region of UNG2 mRNA (positions 44–68 according to NM_080911 entry). As a negative control, we used the Stealth RNAi negative control duplexes with high GC content (from Invitrogen, France). Stealth RNAi duplexes (20 µM) were transfected using Lipofectamine 2000 following manufacturer’s instructions (Invitrogen). To obtain optimal conditions for endogenous UNG2 expression, cells were maintained at low density during the entire experiment. Forty-eight hours after transfection, cells were harvested and cell lysates were analyzed for their content in β-Gal activity used as an index of HIV-1 LTR activity.
RESULTS
Different capacity to inhibit intracellular UNG2 expression between HIV-1 and HIV-2
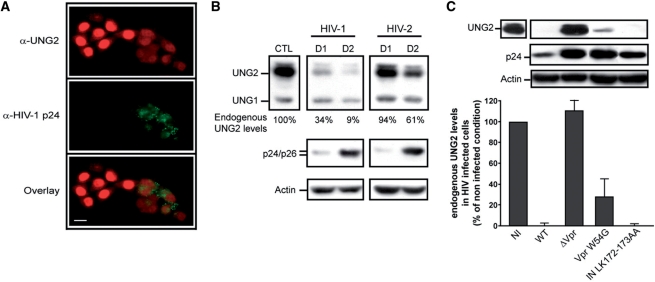
Conflicting results have been reported in the literature, on the fate of UNG2 following HIV-1 expression, raising either its possible incorporation into viral particles (8), or its targeting to the proteasome in cells expressing the Vpr accessory HIV-1 protein (15,16). Hence, we reexamined the expression of endogenous UNG2 in cells infected with HIV-1. Immunofluorescence experiments were performed with HIV-1-infected MAGIC-5B cells dually stained with antibodies raised against UNG1/2 and HIV-1 p24 antigen. As shown in Figure 1A, both mitochondrial UNG1 and nuclear UNG2 isoforms are recognized by anti-UNG antibody. Interestingly, endogenous UNG2 is highly expressed in the nucleus of p24-negative cells (left of the picture). In contrast, UNG2 is barely detected in HIV-1 positive cells (right of the picture), while UNG1 remains detectable into the cytoplasm of these cells.
Figure 1.
Endogenous UNG2 expression profile in HIV-infected cells. (A) Immunofluorescence of 5 × 104 MAGIC-5B cells infected with HIV-1NLAD8 (100 ng/ml of Gag p24) for 48 h. After fixation and permeabilization, cells are labeled for HIV-1 p24 antigen (Green) and mitochondrial UNG1 and nuclear UNG2 (Red). Scale bar, 20 µm (B) Cell lysates of immortalized CEM T cells (2 × 106) infected with HIV-1NL4.3 or HIV-2ROD (RT of viral input 4 × 105 cpm) were analyzed by immunoblot using anti-UNG1/2 or anti-p24 or anti-actin mAbs, either 1 day (D1) or 2 days (D2) after infection. These data are representative of two experiments (C) Infection in triplicate of CEM T cells with wild type NL4.3 (WT), Vpr-deleted NL4.3 (ΔVpr), Vpr mutant NL4.3-VprW54G and Integrase mutant NL4.3-INLK172-173AA as in B. Forty-eight hours after infection, cells were analyzed for endogenous UNG2 expression by immunoblot (upper panel). Expression of p24 and actin are shown. In lower panel, densitometry scanning of immunoblots obtained with anti-UNG2 serum are expressed as the percentage of UNG2 expression observed in non-infected (NI) cells. Values are the mean of three separate experiments with standard deviations.
We confirm these data using immunoblotting experiments of HIV-1-infected CEM T cells (Figure 1B). Increasing expression of HIV-1 p24 antigen was observed from total cell lysates at Days 1 and 2 post-infection. Concomitantly, expression of UNG2, detected as a 37 kDa, protein, was found decreased by 66 and 91% at Days 1 and 2 post-infection, respectively, as compared with basal expression level determined from uninfected cells. The UNG1 isoform was simultaneously detected from the same immunoblotting experiment as a 31 kDa protein. No significant variation of UNG1 levels was observed when uninfected and HIV-1-infected samples were compared (Figure 1B).
The present analysis was further extended to the study of HIV-2ROD, a close relative to HIV-1 that was previously reported unable to package UNG2, suggesting major differences between these related lentiviruses (12). Hence, we investigated whether endogenous UNG2 expression is affected in HIV-2-infected CEM cells. As shown in Figure 1B, UNG2 expression was found slightly decreased at Day 1 post-infection and was maintained around 60% at Day 2 post-infection. No significant variation in expression levels of UNG1 was observed in HIV-2-infected cells. Altogether these data indicate that the pool of cellular UNG2, but not UNG1, is strongly depleted following HIV-1 infection. In contrast, HIV-2 strain barely affects expression of endogenous UNG2, indicating that UNG2 depletion in infected cells is variable depending on the HIV strain.
Depletion of endogenous UNG2 is proteasome dependent and requires a direct interaction with Vpr but not with Integrase
UNG2 has been found to specifically interact with the Vpr accessory protein (14) and the IN domain of HIV-1 Gag-Pol precursor (8). Therefore, we investigated the contribution of both Vpr and IN proteins in UNG2 depletion observed in infected cells. When CEM cells are infected with HIV-1 deleted for the Vpr gene (ΔVpr) and processed for immunoblotting experiments, UNG2 is detected at levels comparable to those detected in uninfected cells (Figure 1C). In similar conditions, HIV-1 expressing VprW54G, a Vpr mutant deficient for UNG2 binding (22), induces an intermediate phenotype corresponding to the partial depletion of endogenous UNG2. In these cells, UNG2 expression levels persist on average at 27% compared with uninfected control cells. In addition, we found that endogenous UNG2 is fully depleted in cells infected with HIV-1 coding for a LK172-173AA Integrase mutant, that was reported to abolish incorporation of UNG2 into HIV-1 particles (23) (Figure 1C). Altogether, our data indicate that in HIV-1 infected T cells, interaction of Vpr with UNG2 is required to induce the depletion of the intracellular pool of UNG2. In contrast, the direct recruitment of UNG2 by IN is dispensable for UNG2 depletion.
We further extended these observations by analyzing the role of a proteasome-dependent process in UNG2 depletion (Supplementary Figure 1). We show that Vpr expression strongly decreases the intracellular expression level of UNG2 fused to the GFP protein (UNG2-GFP) (24) in a dose-dependent manner. When cells are maintained in the presence of MG132, a specific proteasome inhibitor, UNG2-GFP levels were strongly restored in cells co-transfected with a Vpr expression vector. These data confirm that Vpr-directed depletion of cellular UNG2 is mainly mediated by the proteasome as initially reported by Schröfelbauer et al. (15,16).
UNG2 overexpression inhibits HIV-1 particles production
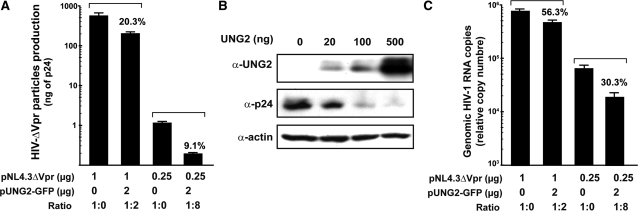
The cellular APOBEC3G cytidine deaminase was previously identified as an innate immune factor against retroviral infection (25). Concomitantly, the HIV-1 Vif protein was reported to counteract this antiviral activity by targeting APOBEC3G to the proteasome (26). In light of this recent description, we speculate that the depletion of intracellular UNG2 observed in presence of Vpr could represent a mechanism of protection against an antiviral activity harbored by UNG2. To study this hypothesis, we evaluated the production level of Vpr deficient HIV-1 particles in presence of UNG2. 293T cells were transfected with an HIV-1 proviral DNA lacking the Vpr coding sequence (pNL4.3ΔVpr) and either the pGFP or the pUNG2-GFP vectors that direct expression of GFP or UNG2-GFP, respectively. As shown in Figure 2A, we observed a strong decrease in the amount of viral particles released in the culture supernatants of UNG2-GFP positive cells, as measured by HIV-1 p24 ELISA. We have also determined, using immunoblotting experiments, that intracellular p24 expression levels are dramatically reduced in cells cotransfected with pNL4.3ΔVpr and increasing amounts of pUNG2 expression vector (Figure 2B). DNA repair mechanisms and cell cycle control are intimately linked (27). Therefore, we excluded any effect of UNG2 transfection on the cell cycle progress that could explain the antiviral activity using propidium iodide labeling experiments (Supplementary Figure 2).
Figure 2.
UNG2 overexpression impairs HIV-1 particles production and full length HIV-1 RNA transcription. (A) 293T cells were cotransfected with 2 µg of pUNG2-GFP and either 1 µg of pNL4.3ΔVpr (pNL4.3ΔVpr:pUNG2-GFP ratio of 1 : 2) or 0.25 µg of pNL4.3ΔVpr (pNL4.3ΔVpr:pUNG2-GFP ratio of 1 : 8). In absence of pUNG2-GFP (1 : 0), the transfection mixture was complemented with pGFP empty vector. Forty-eight hours post-transfection, viral production was monitored using an HIV-1 p24 ELISA kit. Experiment was carried out in triplicate and results are represented as the percentage of the pGFP control condition with standard deviations. Transfection efficiency has been controlled by flow cytometry analysis (86 ± 1% and 85 ± 2% of GFP and UNG2-GFP positive cells respectively). (B) 293T cells were cotransfected with 1 µg of pNL4.3ΔVpr and increasing amounts of pUNG2 as indicated. Forty-eight hours later, equal amounts of cell lysates were immunobloted for total UNG2, p24 and actin protein content. This result is representative of two different experiments. (C) Aliquots of 293T cells used in panel A were analyzed by RT-QPCR for their intracellular amount of genomic HIV-1 RNA as described in material and methods. Copy numbers of HIV-1 genomic RNA from three different experiments are represented as percentage values of pGFP control conditions with standard deviations.
UNG2 exerts its antiviral activity at the transcriptional level
Next, we hypothesized that inhibition of HIV-1 expression observed in presence of UNG2 could result from a decrease in viral gene transcription. Thus, we designed a Reverse transcription-quantitative PCR (RT-QPCR) assay to monitor the level of intracellular full length HIV-1 transcripts produced in infected cells overexpressing or not UNG2 (20). As shown in Figure 2C, when values are normalized to GAPDH mRNA levels, UNG2 overexpression reduces HIV-1 RNA levels in a dose dependent manner (for copy number data, see Supplementary Figure 3A). A 3-fold decrease in HIV-1 genomic RNA levels was observed when a 1 : 8 pNL4.3ΔVpr:UNG2 ratio was used. Interestingly, such modulation at the RNA level may account for the 10-fold inhibition of viral particles production observed in Figure 2A. Indeed, this observation is reminiscent of previously published data indicating that a siRNA directed against HIV-1 gag can lead simultaneously to a 10-fold reduction in full length viral transcripts and to a 25-fold decrease in viral particles production (28).
Overexpressed UNG2 negatively regulates HIV-1 LTR transcription
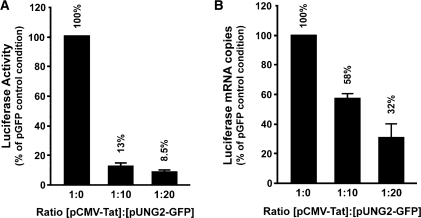
An efficient transcription of the HIV-1 genome requires the interaction of the viral transactivating protein Tat with the viral LTR promoter. Thus, we analyzed whether UNG2 interferes with viral transcription at the level of Tat-mediated LTR promoter activation. We used the reporter cell line Hela LTRHIV-1-Luc stably transfected with a single copy of the luciferase gene under the control of the entire HIV-1BRU LTR promoter (29). UNG2 overexpression in these cells strongly inhibited Tat-mediated luciferase expression, as shown by luciferase reporter assay (Figure 3A). These results were confirmed using Hela MAGIC-5B cells, which contain a stably integrated lacZ gene under the control of the HIV-1 LTR promoter (data not shown). To determine whether this inhibition of luciferase expression is the consequence of a transcriptional repression, luciferase mRNA transcripts have been quantified in Tat-expressing HeLa LTRHIV-1-Luc cells using a specific RT-QPCR assay. As shown in Figure 3B, luciferase mRNA levels, normalized to endogenous GAPDH mRNA levels, are diminished by 68% in cells expressing a 1 : 20 Tat to UNG2 ratio (for copy number data, see Supplementary Figure 3B). We do not observe a linear correlation between the repression level of the luciferase mRNA transcription and translation. Such discrepancy may result from a nonlinear process, typical in biological systems (30). However, we do not exclude the existence of an additional post-transcriptional antiviral effect of UNG2 exerted on HIV-1.
Figure 3.
UNG2 overexpression inhibits Tat-mediated stimulation of HIV-1 LTR transcription. (A) Hela LTRHIV-1-Luc cells were transfected with pCMV-Tat (0.25 µg) and either 2.5 or 5 µg of pUNG2-GFP (ratio 1 : 10 and 1 : 20, respectively). In absence of pUNG2-GFP, the transfection mixture was complemented with pGFP empty vector. Two days post-transfection, cell extracts were analyzed for content in luciferase activity. Experiment was carried out in duplicate and results are represented as the percentage of the pGFP control condition (1 : 0) with standard deviations. Transfection efficiency has been controlled by flow cytometry analysis (30 ± 1% of GFP (1 : 0), 37 ± 1% of UNG2-GFP (1 : 10) and 38 ± 3% of UNG2-GFP (1 : 20) positive cells). (B) Aliquots of Hela LTRHIV-1-Luc cells used in the panel A were analyzed by RT-QPCR for their intracellular amount of luciferase mRNA. Copy numbers of luciferase mRNA from two different experiments are represented as percentage values of GFP control condition (1 : 0) with standard deviations.
The antiviral activity of UNG2 requires the integrity of its catalytic domain
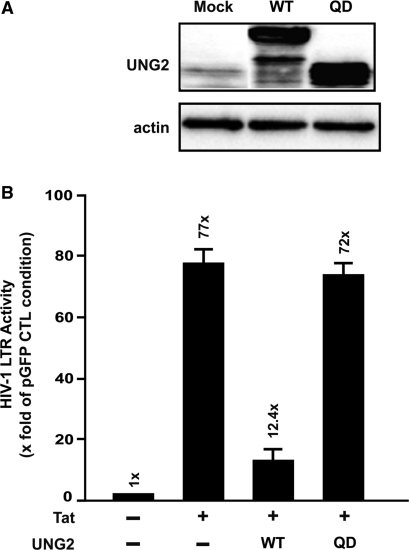
To define more precisely whether the catalytic domain of UNG2 is involved in the inhibition of HIV transcription, we analyzed the activity of the LTR promoter in presence of a UNG2 catalytic mutant (UNG2QD153-154LE) (31). First, expression of UNG2 constructs was controlled. 293T cells were transfected with vectors encoding either the GFP-fused UNG2 protein or the well characterized UNG2QD153-154LE mutant previously described by Priet et al. (11). Forty-eight hours post-transfection, similar expression levels were observed for both UNG2 constructs using immunoblotting experiments (Figure 4A). Subsequently, LTR-driven transcription was investigated by cotransfection of 293T cells with UNG2 constructs, a LTR-luciferase construct and a Tat-expressing vector. Luciferase activity was then measured 48 h post-transfection and used as an index of LTR activity (Figure 4B). The strong inhibition of Tat-induced LTR transcription observed in presence of wild-type UNG2 (12.4-fold compared with 77-fold LTR activity in absence of UNG2) was abolished in cells expressing the UNG2QD153-154LE catalytic mutant (72-fold LTR activity). This result suggests that the antiviral activity of UNG2 may require its glycosylase activity.
Figure 4.
Consequences of UNG2 mutations on inhibition of LTR-driven transcription. (A) 293T cells were transfected with equivalent amounts of pcDNA-UNG2-GFP or pcDNA-UNG2QD153-154LE (QD) or left untransfected (Mock). Expression of UNG2 constructs in the corresponding cell extracts was controlled using anti-UNG2 immunoblots (upper panel). Actin immunoblots were used as protein loading controls (lower panel). (B) 293T cells (8 × 104) were cotransfected with 0.2 µg of HIVLTR-Luc and 0.1 µg of pCMV-Tat plasmids or with 0.2 µg pcDNA-UNG2-GFP or 0.2 µg pcDNA-UNG2QD153-154LE (QD). Two days after transfection, cells were analyzed in triplicate for luciferase expression used as an index of HIV-1 LTR activity. Data are represented as the fold of LTR promoter activity observed in absence of Tat expression.
Effect of UNG2 overexpression on PMA and TNF-induced HIV-1 LTR transcription
In order to assess whether UNG2 is acting specifically on Tat-induced LTR transcription, LTR activity has been stimulated 24 h into HeLa LTRHIV-1-Luc cells with extracellular effectors and LTR activity has been monitored in absence or presence of UNG2 as described above. As shown in Figure 5, UNG2 harbors a modest inhibitory effect on the basal transcriptional activity of HIV-1 LTR promoter. Furthermore, we observed a 5.6-fold increase of LTR activity in cells stimulated with tumor necrosis factor alpha (TNF-α) compared with basal transcriptional activity of HIV-1 LTR. This activity was strongly inhibited in presence of UNG2-GFP, close to the background (1.4-fold). This result is similar to the one observed for Tat-induced LTR transcription in presence of UNG2 (decreased LTR activity from a 104- to 14-fold activity). Interestingly, LTR activity induced by an extracellular mitogen like the phorbol myristate acetate (PMA), a strong LTR activator (15.6-fold) is barely affected by UNG2 (11.9-fold). This result highlights the specificity of UNG2 acting on HIV-1 LTR activity through the inhibition of specific signaling pathways that are still to be identified.
Figure 5.
Consequences of UNG2 overexpression on PMA- and TNF-α-induced HIV-1 LTR transactivation. Hela LTRHIV-1-Luc cells were transfected with 0.8 µg of pGFP or pUNG2-GFP and cultured for 24 h in medium alone (CTL), or supplemented with PMA (20 ng/ml) or TNF-α (50 ng/ml). Subsequently, cell extracts were analyzed for content in luciferase activity. Experiment was carried out in triplicate and results are represented as the fold of activation observed in the pGFP control condition normalized to one. As a control, some Hela LTRHIV-1-Luc cells were co-transfected with 0.1 µg pCMV-Tat and 8 µg of pGFP.
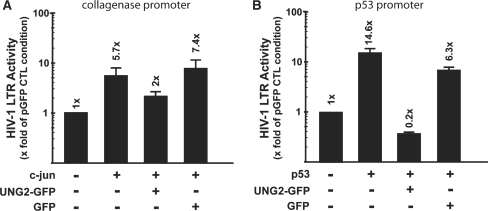
Effects of UNG2 overexpression on cellular promoters transactivation
Analysis of the UNG2 inhibitory effect on gene transcription was further investigated using different cellular promoters. 293T cells were cotransfected with DNA encoding the transcription factors c-jun or p53 and their specific DNA target, consisting of the luciferase gene under the control of either a collagenase promoter or repeated p53 binding sites, respectively (Figure 6A and B). Data from these experiments revealed that UNG2 overexpression repressed transactivation of these promoters to different extent. While a significant repression of the p53 promoter was observed in cells expressing UNG2, the collagenase promoter containing AP1 recognition sites transactivated by c-Jun was only mildly affected. Again, these data suggest that UNG2 acts negatively on specific signaling pathways. Hence, promoters from different origins, either from the host or pathogens like HIV-1 can be affected.
Figure 6.
Assessment of UNG2 antiviral activity on cellular promoters. (A) 293T cells were cotransfected with 0.4 µg of a collagenase promoter-driven luciferase gene, pcDNA3.1-lacZ (β-galactosidase-containing reference plasmid; 0.1 µg), the c-Jun expression vector pcDNA-c-Jun (0.2 µg), and pUNG2-GFP (0.2 µg) or pGFP empty vector (0.2 µg). (B) 293T cells were co-tranfected with the p53 promoter-driven luciferase gene (pG13PY-Luc; 0.4 µg), pcDNA3.1-lacZ; 0.1 µg), the p53 expression vector pC53SN3 (0.2 µg), and pUNG2-GFP (0.2 µg) or pGFP empty vector (0.2 µg). Two days post-transfection, cell extracts were analyzed for their content in luciferase activity. Transfection efficiency was normalized by quantification of β-galactosidase activity in cells extracts. Experiment was carried out in duplicate and results are represented as the fold of activation observed in the pGFP control condition normalized to one.
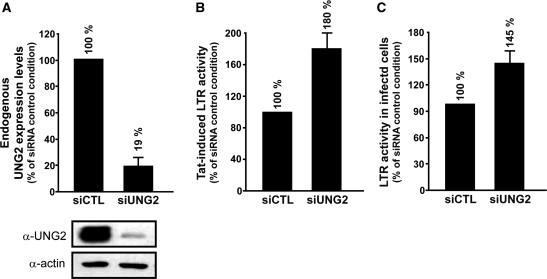
Depletion of endogenous UNG2 promotes Tat-mediated LTR transcription
The inhibitory effect of UNG2 on HIV-1 LTR activity has been observed in overexpressed conditions. Therefore, we decided to investigate the impact of endogenous UNG2 depletion on Tat-dependent transcription. Expression of endogenous UNG2 in the MAGIC-5B indicator cell line was reduced over 80% after transfection with a specific Stealth siRNA (siUNG2) in comparison with the control Stealth siRNA (siCTL) condition (Figure 7A). These cells, stably expressing the LTR-lacZ construct, were then used to monitor LTR activity after transfection with the Tat expression vector pCMV-Tat. As shown in Figure 4B, the analysis of β-galactosidase (β-gal) activity induced by Tat revealed that the LTR promoter was transactivated more efficiently (180%) in cells depleted for endogenous UNG2 by siUNG2 compared with cells transfected with the siCTL control. Similar experiments were finally designed to monitor LTR activity after infection with HIV-1 in MAGIC-5B cells depleted for endogenous UNG2. For that, MAGIC-5B cells were transfected with siUNG2 or siCTL stealth siRNA and were subsequently infected with NL4-3ΔVpr. As shown in Figure 7C, β-gal activity measured from cells transfected with siUNG2 was significantly enhanced (145%) when compared with cells expressing siCTL. Altogether, these observations suggest that endogenous UNG2 behaves as a negative regulatory factor that represses HIV-1 replication in human cells at the level of HIV-1 LTR promoter transactivation.
Figure 7.
Depletion of endogenous UNG2 promotes Tat-mediated LTR transcription. (A) MAGIC-5B cells (6 × 104) were cotransfected with 200 ng of pCMV-Tat and 5 pmol of Stealth RNAi directed against UNG2 (siUNG2) or Stealth RNAi control (siCTL). Two days after transfection, cells were analyzed in triplicate for endogenous UNG2 expression by immunoblot (see lower panel). Results are represented as the percentage of UNG2 expression observed in siCTL condition. (B) Cell lysates were analyzed in parallel for their content in β-gal activity used as an index of HIV-1 LTR activity. Data are represented as the percentage of LTR promoter activity observed in presence of siCTL with standard deviations. (C) MAGIC-5B cells (3 × 105) were transfected with 20 pmol of siUNG2 or siCTL. After 24 h, cells were diluted (6 × 104/well) and infected in triplicate with VSV-G pseudotyped NL4-3ΔVpr (RT of viral input 103 cpm). Cell lysates were analyzed two days post-infection for their content in β-gal activity as in B.
DISCUSSION
The function of UNG2 in the HIV-1 life cycle has been highly debated during the past few years. The DNA repair enzyme has been first found to be specifically incorporated into purified HIV-1 particles. This incorporation was successively proposed to be mediated through a direct interaction with the Vpr accessory protein (10) or with the viral integrase (8). More recently, the expression of Vpr was reported to induce a drastic proteasome-dependent depletion of the intracellular pool of UNG2 through the recruitment of DDB1 (15). Here, we do confirm that expression of Vpr in HIV-1 infected cells is sufficient to deplete endogenous UNG2. In light of repeated description of UNG2 packaging into HIV-1 particles, such depletion may however be incomplete, allowing either the incorporation of residual UNG2 or the packaging of degradation products that remain to be characterized. UNG2 depletion was less effective in HIV-2-infected cells where endogenous UNG2 persists at high levels despite an active replication of the virus. Interestingly, Priet et al. (11) have previously shown that depletion of endogenous UNG2 had no effect on HIV-2ROD infectivity. Moreover, they have shown that highly purified HIV-2ROD and SIVmac viral particles did not incorporate host UNG2, contrasting with the presence of UNG2 in HIV-1 viral particles (12). Such differences between HIV-1 and HIV-2 have been related to the capacity of UNG2 to interact preferentially with IN from HIV-1 and to a weaker extent with IN from HIV-2 or SIVmac (12). In our study, UNG2 was efficiently depleted following infection with HIV-1 INLK172-173AA mutant supporting no role for a direct IN-UNG2 interaction in UNG2 depletion. Accordingly, we hypothesized that HIV-2 Vpr may either bind to UNG2 with reduced efficiency or poorly redirect UNG2 to the DDB1-Cul4-Roc1 E3 Ubiquitin Ligase complex, leading to efficient expression of endogenous UNG2 in HIV-2 infected cells.
Next, we investigated the consequence of an HIV-1 Vpr mutation on the tryptophan residue 54 (VprW54G). Infection with HIV-1 VprW54G allows the expression of endogenous UNG2 at an intermediate level when compared with UNG2 expression in uninfected cells. The VprW54G mutant was reported previously to retain the capacity to recruit DBB1 (15), while it failed to interact with UNG2 (22). This mutant also retains the property to cause cell cycle G2 arrest (22). Interestingly, a recent investigation has shown that UNG2 expression is downregulated in the G2 phase of the cell cycle (32). Therefore, the partial recovery of UNG2 expression that we observed in cells infected with HIV-1 VprW54G may reflect the gain of UNG2 expression related to the absence of a direct interaction between Vpr and UNG2 while almost 70% of the intracellular pool of UNG2 may be depleted consecutively to the cell cycle G2 arrest. We have also observed that inhibition of the proteasome-dependent degradation pathway in Vpr-transfected cells revealed an incomplete recovery of endogenous UNG2 expression in cells treated with efficient amounts of MG132 proteasome inhibitor (Supplementary Figure 1). Hence, the existence of some other inhibitory mechanism of UNG2 expression in Vpr positive cells, like the cell cycle G2 arrest, needs to be considered. Nevertheless, such mechanism is unrelated to the capacity of UNG2 to interact with HIV-1 integrase. Indeed, we show that UNG2 expression was dramatically impaired in cells infected with HIV-1 INLK172-173AA, an IN mutant unable to interact with and to direct viral packaging of UNG2 (23). We also did not observe any effect of HIV-1 Vpr on UNG2 transcript levels using RT-QPCR experiments (not shown), excluding a potential effect of Vpr on UNG2 at the transcriptional level.
The main result reported herein is that HIV-1 replication is dramatically altered when UNG2 is overexpressed. The inhibition of viral particles release was found to correlate with a significant reduction in Tat-dependent LTR transactivation and decrease in full-length HIV-1 RNA synthesis. These data suggest that UNG2 harbors an inhibitory activity at the level of viral transcription. This model is reinforced by the observation that LTR-directed transcription mediated by Tat is increased in UNG2 depleted cells using an RNA interference approach. Vpr-mediated HIV-1 LTR activation has previously been attributed to the establishment of a cell cycle G2 arrest and also the direct interaction of Vpr with the glucocorticoid receptor and diverse transcription factors (Sp1, TFIIH) (33). Our data support the idea that depletion of UNG2 by Vpr might be a new way to increase HIV-1 replication at the level of LTR-directed transcription.
In the past literature, crosstalks have been identified between DNA repair pathways and gene transcription regulation. Of note, human DNA glycosylases have been implicated in transcription regulation, especially the thymine DNA glycosylase (TDG) and the Methyl-CpG binding domain protein 4 (MBD4) (34–38). The molecular mechanisms underlying UNG2-induced inhibition of HIV-1 transcription remain elusive at this time. However, results obtained in the present study indicate that the UNG2-induced transcriptional repression is independent of cell cycle progress. Moreover, LTR promoter inhibition by UNG2 depends on the type of stimuli or transactivators used to activate the LTR. Of note, PMA-induced LTR activity is barely affected by UNG2. Finally, UNG2-induced transcriptional repression is not only observed for markers under control of the HIV-1 LTR promoter but also under the control of some cellular promoters. Therefore, we hypothesize that such inhibition, may at least in part, rely on the sequestration of cellular factor(s) critical for transcriptional regulation of multiple cellular genes. In support of this hypothesis, human TDG and MBD4 were previously reported to interfere with gene transcription through specific interactions with diverse transcription factors, receptors and chromatin-remodeling complexes (34–37). Interestingly, UNG2 has recently been shown to be recruited to the promoter of Kaposi’s Sarcoma-associated Herpesvirus, leading to latent persistence of the virus (39). The ability of UNG2 to repress HIV-1-LTR transcription through a direct recruitment to the HIV-1 LTR promoter needs to be investigated.
Next, we found that transcriptional inhibition observed in presence of wild type UNG2 is abolished in cells expressing the UNG2QD153-154LE catalytic mutant. Given that upon transfection this mutant reached expression level comparable to that of WT protein without inhibiting LTR-driven transcription, this result may reflect the requirement for UNG2 enzymatic activity. However, UNG2 is a truly constrained protein whose structure has been previously defined (31). The insertion of mutations may drastically alter the overall structure of the protein and result in an unspecific alteration of the UNG2 antiviral property. Therefore, at this stage of the study, one should be cautious to draw any general conclusion regarding the role of this protein domain in the antiviral phenotype.
To our knowledge, this is the first time that UNG2 is linked to transcriptional regulation and more particularly described as a new class of viral transcription inhibitor. Despite the broad spectrum of action observed for UNG2 anti-transcriptional activity, HIV-1 has evolved and acquired a specific viral factor to counteract UNG2, identified as the Vpr regulatory protein. This observation is reminiscent of HIV-1 Vif described as a protective factor against the cytidine deaminase APOBEC3G (25). The antiviral activity of APOBE3G relies on its capacity to induce cytidine deamination and severe uracilation of the viral genome during the reverse transcription step, leading to G to A accumulation (26). Interestingly, the direct binding of Vif to APOBEC3G targets these latter to the proteasome (40). Thus, we propose that the specific interaction of HIV-1 Vpr with the DNA repair enzyme UNG2, an interaction described more than a decade ago, is likely to represent a new viral mechanism of protection against an innate antiviral activity exerted by UNG2 at the level of viral transcription. Our data support the idea that depletion of UNG2 by Vpr might be a new way to increase HIV-1 replication at the level of LTR-directed transcription.
In conclusion, our data together with other recent publications highlight the capacity of HIV-1 to counteract innate antiviral activity through the specific design of protective viral factors and support the idea that interactions between viruses and DNA repair components can result in inhibition of the infection rather than cooperation towards its establishment.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Institutional funding from the Centre National de la Recherche Scientifique (CNRS); the University of Montpellier (UM1 and UM2); and the French National Agency of Research Against AIDS (ANRS). Grant from ANRS and SIDACTION, respectively (to D.F. and L.H.); CNRS/Région Languedoc-Roussillon (to S.B.). Funding for open access charge: CNRS.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
The authors thank Joséphine Sire, Serge Bénichou, Geir Slupphaug and Warner C. Greene for providing us with plasmids and antibodies. They thank Masashi Tatsumi and Stéphane Emiliani for indicator cell lines.
REFERENCES
- 1.Van Maele B, Debyser Z. HIV-1 integration: an interplay between HIV-1 integrase, cellular and viral proteins. AIDS Rev. 2005;7:26–43. [PubMed] [Google Scholar]
- 2.Lloyd AG, Tateishi S, Bieniasz PD, Muesing MA, Yamaizumi M, Mulder LC. Effect of DNA repair protein Rad18 on viral infection. PLoS Pathog. 2006;2:e40. doi: 10.1371/journal.ppat.0020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lau A, Kanaar R, Jackson SP, O'C;onnor MJ. Suppression of retroviral infection by the RAD52 DNA repair protein. EMBO J. 2004;23:3421–3429. doi: 10.1038/sj.emboj.7600348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoder K, Sarasin A, Kraemer K, McIlhatton M, Bushman F, Fishel R. The DNA repair genes XPB and XPD defend cells from retroviral infection. Proc. Natl Acad. Sci. USA. 2006;103:4622–4627. doi: 10.1073/pnas.0509828103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kavli B, Sundheim O, Akbari M, Otterlei M, Nilsen H, Skorpen F, Aas PA, Hagen L, Krokan HE, Slupphaug G. hUNG2 is the major repair enzyme for removal of uracil from U:A matches, U:G mismatches, and U in single-stranded DNA, with hSMUG1 as a broad specificity backup. J. Biol. Chem. 2002;277:39926–39936. doi: 10.1074/jbc.M207107200. [DOI] [PubMed] [Google Scholar]
- 6.Nilsen H, Rosewell I, Robins P, Skjelbred CF, Andersen S, Slupphaug G, Daly G, Krokan HE, Lindahl T, Barnes DE. Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol. Cell. 2000;5:1059–1065. doi: 10.1016/s1097-2765(00)80271-3. [DOI] [PubMed] [Google Scholar]
- 7.Priet S, Navarro JM, Gros N, Querat G, Sire J. Functional role of HIV-1 virion-associated uracil DNA glycosylase 2 in the correction of G:U mispairs to G:C pairs. J. Biol. Chem. 2003;278:4566–4571. doi: 10.1074/jbc.M209311200. [DOI] [PubMed] [Google Scholar]
- 8.Willetts KE, Rey F, Agostini I, Navarro JM, Baudat Y, Vigne R, Sire J. DNA repair enzyme uracil DNA glycosylase is specifically incorporated into human immunodeficiency virus type 1 viral particles through a Vpr-independent mechanism. J. Virol. 1999;73:1682–1688. doi: 10.1128/jvi.73.2.1682-1688.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mansky LM, Preveral S, Selig L, Benarous R, Benichou S. The interaction of vpr with uracil DNA glycosylase modulates the human immunodeficiency virus type 1 In vivo mutation rate. J. Virol. 2000;74:7039–7047. doi: 10.1128/jvi.74.15.7039-7047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen R, Le Rouzic E, Kearney JA, Mansky LM, Benichou S. Vpr-mediated incorporation of UNG2 into HIV-1 particles is required to modulate the virus mutation rate and for replication in macrophages. J. Biol. Chem. 2004;279:28419–28425. doi: 10.1074/jbc.M403875200. [DOI] [PubMed] [Google Scholar]
- 11.Priet S, Gros N, Navarro JM, Boretto J, Canard B, Querat G, Sire J. HIV-1-associated uracil DNA glycosylase activity controls dUTP misincorporation in viral DNA and is essential to the HIV-1 life cycle. Mol. Cell. 2005;17:479–490. doi: 10.1016/j.molcel.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 12.Priet S, Navarro JM, Gros N, Querat G, Sire J. Differential incorporation of uracil DNA glycosylase UNG2 into HIV-1, HIV-2, and SIV(MAC) viral particles. Virology. 2003;307:283–289. doi: 10.1016/s0042-6822(02)00073-9. [DOI] [PubMed] [Google Scholar]
- 13.Kaiser SM, Emerman M. Uracil DNA Glycosylase Is Dispensable for Human Immunodeficiency Virus Type 1 Replication and Does Not Contribute to the Antiviral Effects of the Cytidine Deaminase Apobec3G. J. Virol. 2006;80:875–882. doi: 10.1128/JVI.80.2.875-882.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bouhamdan M, Benichou S, Rey F, Navarro JM, Agostini I, Spire B, Camonis J, Slupphaug G, Vigne R, Benarous R, et al. Human immunodeficiency virus type 1 Vpr protein binds to the uracil DNA glycosylase DNA repair enzyme. J. Virol. 1996;70:697–704. doi: 10.1128/jvi.70.2.697-704.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schrofelbauer B, Hakata Y, Landau NR. HIV-1 Vpr function is mediated by interaction with the damage-specific DNA-binding protein DDB1. Proc. Natl Acad. Sci. USA. 2007;104:4130–4135. doi: 10.1073/pnas.0610167104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schrofelbauer B, Yu Q, Zeitlin SG, Landau NR. Human immunodeficiency virus type 1 Vpr induces the degradation of the UNG and SMUG uracil-DNA glycosylases. J. Virol. 2005;79:10978–10987. doi: 10.1128/JVI.79.17.10978-10987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slupphaug G, Eftedal I, Kavli B, Bharati S, Helle NM, Haug T, Levine DW, Krokan HE. Properties of a recombinant human uracil-DNA glycosylase from the UNG gene and evidence that UNG encodes the major uracil-DNA glycosylase. Biochemistry. 1995;34:128–138. doi: 10.1021/bi00001a016. [DOI] [PubMed] [Google Scholar]
- 18.Brun S, Solignat M, Gay B, Bernard E, Chaloin L, Fenard D, Devaux C, Chazal N, Briant L. VSV-G pseudotyping rescues HIV-1 CA mutations that impair core assembly or stability. Retrovirology. 2008;5:57. doi: 10.1186/1742-4690-5-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cartier C, Hemonnot B, Gay B, Bardy M, Sanchiz C, Devaux C, Briant L. Active cAMP-dependent protein kinase incorporated within highly purified HIV-1 particles is required for viral infectivity and interacts with viral capsid protein. J. Biol. Chem. 2003;278:35211–35219. doi: 10.1074/jbc.M301257200. [DOI] [PubMed] [Google Scholar]
- 20.Houzet L, Paillart JC, Smagulova F, Maurel S, Morichaud Z, Marquet R, Mougel M. HIV controls the selective packaging of genomic, spliced viral and cellular RNAs into virions through different mechanisms. Nucleic Acids Res. 2007;35:2695–2704. doi: 10.1093/nar/gkm153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Houzet L, Morichaud Z, Mougel M. Fully-spliced HIV-1 RNAs are reverse transcribed with similar efficiencies as the genomic RNA in virions and cells, but more efficiently in AZT-treated cells. Retrovirology. 2007;4:30. doi: 10.1186/1742-4690-4-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Selig L, Benichou S, Rogel ME, Wu LI, Vodicka MA, Sire J, Benarous R, Emerman M. Uracil DNA glycosylase specifically interacts with Vpr of both human immunodeficiency virus type 1 and simian immunodeficiency virus of sooty mangabeys, but binding does not correlate with cell cycle arrest. J. Virol. 1997;71:4842–4846. doi: 10.1128/jvi.71.6.4842-4846.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Priet S, Navarro JM, Querat G, Sire J. Reversion of the lethal phenotype of an HIV-1 integrase mutant virus by overexpression of the same integrase mutant protein. J. Biol. Chem. 2003;278:20724–20730. doi: 10.1074/jbc.M301768200. [DOI] [PubMed] [Google Scholar]
- 24.Nilsen H, Otterlei M, Haug T, Solum K, Nagelhus TA, Skorpen F, Krokan HE. Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res. 1997;25:750–755. doi: 10.1093/nar/25.4.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sheehy AM, Gaddis NC, Choi JD, Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 26.Harris RS, Liddament MT. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 2004;4:868–877. doi: 10.1038/nri1489. [DOI] [PubMed] [Google Scholar]
- 27.Branzei D, Foiani M. Regulation of DNA repair throughout the cell cycle. Nat. Rev. Mol. Cell Biol. 2008;9:297–308. doi: 10.1038/nrm2351. [DOI] [PubMed] [Google Scholar]
- 28.Novina CD, Murray MF, Dykxhoorn DM, Beresford PJ, Riess J, Lee SK, Collman RG, Lieberman J, Shankar P, Sharp PA. siRNA-directed inhibition of HIV-1 infection. Nat. Med. 2002;8:681–686. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]
- 29.Blot G, Lopez-Verges S, Treand C, Kubat NJ, Delcroix-Genete D, Emiliani S, Benarous R, Berlioz-Torrent C. Luman, a new partner of HIV-1 TMgp41, interferes with Tat-mediated transcription of the HIV-1 LTR. J. Mol. Biol. 2006;364:1034–1047. doi: 10.1016/j.jmb.2006.09.080. [DOI] [PubMed] [Google Scholar]
- 30.Goutsias J, Kim S. A nonlinear discrete dynamical model for transcriptional regulation: construction and properties. Biophys. J. 2004;86:1922–1945. doi: 10.1016/S0006-3495(04)74257-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mol CD, Arvai AS, Slupphaug G, Kavli B, Alseth I, Krokan HE, Tainer JA. Crystal structure and mutational analysis of human uracil-DNA glycosylase: structural basis for specificity and catalysis. Cell. 1995;80:869–878. doi: 10.1016/0092-8674(95)90290-2. [DOI] [PubMed] [Google Scholar]
- 32.Hagen L, Kavli B, Sousa MM, Torseth K, Liabakk NB, Sundheim O, Pena-Diaz J, Otterlei M, Horning O, Jensen ON, et al. Cell cycle-specific UNG2 phosphorylations regulate protein turnover, activity and association with RPA. EMBO J. 2008;27:51–61. doi: 10.1038/sj.emboj.7601958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Rouzic E, Benichou S. The Vpr protein from HIV-1: distinct roles along the viral life cycle. Retrovirology. 2005;2:11. doi: 10.1186/1742-4690-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cortazar D, Kunz C, Saito Y, Steinacher R, Schar P. The enigmatic thymine DNA glycosylase. DNA Repair (Amst) 2007;6:489–504. doi: 10.1016/j.dnarep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 35.Kondo E, Gu Z, Horii A, Fukushige S. The thymine DNA glycosylase MBD4 represses transcription and is associated with methylated p16(INK4a) and hMLH1 genes. Mol. Cell Biol. 2005;25:4388–4396. doi: 10.1128/MCB.25.11.4388-4396.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kovtun IV, McMurray CT. Crosstalk of DNA glycosylases with pathways other than base excision repair. DNA Repair (Amst) 2007;6:517–529. doi: 10.1016/j.dnarep.2006.10.015. [DOI] [PubMed] [Google Scholar]
- 37.Missero C, Pirro MT, Simeone S, Pischetola M, Di Lauro R. The DNA glycosylase T:G mismatch-specific thymine DNA glycosylase represses thyroid transcription factor-1-activated transcription. J. Biol. Chem. 2001;276:33569–33575. doi: 10.1074/jbc.M104963200. [DOI] [PubMed] [Google Scholar]
- 38.Zhou J, Blue EK, Hu G, Herring BP. Thymine DNA glycosylase represses myocardin-induced smooth muscle cell differentiation by competing with serum response factor for myocardin binding. J. Biol. Chem. 2008;283:35383–35392. doi: 10.1074/jbc.M805489200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verma SC, Bajaj BG, Cai Q, Si H, Seelhammer T, Robertson ES. Latency-associated nuclear antigen of Kaposi's; sarcoma-associated herpesvirus recruits uracil DNA glycosylase 2 at the terminal repeats and is important for latent persistence of the virus. J. Virol. 2006;80:11178–11190. doi: 10.1128/JVI.01334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.