Abstract
High-throughput complementary DNA sequencing (RNA-Seq) is a powerful tool for whole-transcriptome analysis, supplying information about a transcript's expression level and structure. However, it is difficult to determine the polarity of transcripts, and therefore identify which strand is transcribed. Here, we present a simple cDNA sequencing protocol that preserves information about a transcript's direction. Using Saccharomyces cerevisiae and mouse brain transcriptomes as models, we demonstrate that knowing the transcript's orientation allows more accurate determination of the structure and expression of genes. It also helps to identify new genes and enables studying promoter-associated and antisense transcription. The transcriptional landscapes we obtained are available online.
INTRODUCTION
Recent studies have demonstrated an unexpected complexity of transcription in eukaryotes (1–5). In addition to classical mRNAs, which cover ∼1.5% of the genome in higher eukaryotes, numerous non-coding RNAs with widely varying expression levels have been identified. The biological function of these novel transcripts is largely unknown and represents a new research area, requiring high-throughput transcriptome studies.
Direct cDNA sequencing (RNA-Seq) is a new tool for whole-transcriptome analysis. Second generation sequencing machines have increased sequencing throughput by about two orders of magnitude compared to previous systems. They have also reduced the costs of sequencing by roughly two orders of magnitude, making global transcriptome sequencing feasible (4,6–9). Since sequencing costs are constantly decreasing (contrary to those of microarrays) it is likely that cDNA sequencing will capture a considerable portion of transcriptome analyses in the future.
The RNA-Seq procedure is simple, has a large dynamic range and high sensitivity, and can unequivocally identify splicing and RNA editing products as well as allele-specific transcripts. RNA-Seq provides a number of advantages over previous high throughput approaches: microarray hybridization, gene-specific and tiling arrays or SAGE-analyses (10,11). In contrast to SAGE, RNA-Seq does not depend on the presence of particular restriction sites within the cDNA. The depth of RNA-Seq analysis is flexible, providing a dynamic range typically an order of magnitude greater than one can achieve with hybridization arrays. The digital character of the RNA-Seq data permits to compare and pool results from different laboratories. No prior information about transcript sequences is required, allowing detection of novel transcripts. It is possible to estimate the absolute level of gene expression and to study structure of transcripts.
A weakness of RNA-Seq is the inability to determine the polarity of RNA transcripts without laborious modification of the protocol (12,13). The polarity of the transcript is important for correct annotation of novel genes, because it provides essential information about the possible function of a gene, both at the RNA (structure and hybridization to other nucleic acid molecules) and protein levels. In addition, many genomic regions give rise to transcripts from both strands. Antisense transcription is characteristic for eukaryotic genes and is thought to play an important regulatory role (2,12). Overlapping genes are common for compact genomes of prokaryotes and lower eukaryotes. Knowledge of a transcript's orientation helps to resolve colliding transcripts and to correctly determine gene expression levels in the presence of antisense transcripts.
Here, we describe a simple modification of RNA-Seq method that addresses this problem. Incorporation of deoxy-UTP during the second strand cDNA synthesis and subsequent destruction of the uridine-containing strand in the sequencing library allowed us to identify the orientation of transcripts. To illustrate the capabilities of the method we present our sequencing data for yeast and mouse transcriptomes.
MATERIALS AND METHODS
Detailed step-by-step protocols for polyA+ RNA purification and double stranded (ds) cDNA synthesis are presented in Supplementary Methods.
RNA isolation
Yeast strain BY4741 (MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0) was grown in rich medium (YPD; BD Company) at 30°C overnight, diluted to an OD600 of 0.15 and grown until reaching an OD600 of 0.87. The cells were harvested by centrifugation at room temperature, washed once with 1× PBS, and frozen in liquid nitrogen. Total RNA was extracted using the RiboPureTM-Yeast kit (Ambion) and analyzed by an Agilent 2100 bioanalyzer (Agilent Technologies).
Two 11-week-old female C57Bl/6J mice were dissected and whole brain was taken for RNA preparation. Total RNA was extracted using the Trizol method.
polyA+RNA purification
polyA+RNA was purified with the Dynabeads mRNA purification kit (Invitrogen) following the manufacturer's instructions and treated for 30 min at 37°C with 0.2 units of TURBO™ DNase (Ambion) per 1 μg of RNA.
First strand synthesis (FSS)
FSS reaction was prepared by mixing 0.5 μg of polyA+ RNA, 40 ng of (dN)6 primers (Invitrogen) and 25 pmol of oligo(dT) primer (Invitrogen) in 8.5 μl of 1× reverse transcription buffer (Invitrogen), 0.5 mM dNTPs, 5 mM MgCl2 and 10 mM DTT. The mixture was incubated at 98°C for 1 min to melt RNA secondary structures, then at 70°C for 5 min and was cooled to 15°C at 0.1°C/s. Slow temperature cooling was used to make annealing of secondary RNA structures and primers as reproducible as possible. At 15°C 0.5 μl of actinomycin D solution (120 ng/μl), 0.5 μl of RNase OUT (40 units/μl, Invitrogen) and 0.5 μl of SuperScript III polymerase (200 units/μl, Invitrogen) were added to the reaction. Temperature of reverse transcription reaction was increased gradually as a compromise between survival of the enzyme, stability of the primers and denaturation of RNA secondary structures: heating from 15 to 25°C at 0.1°C/s; incubation at 25°C for 10 min; heating from 25 to 42°C at 0.1°C/s; incubation at 42°C for 45 min; heating from 42 to 50°C at 0.1°C/s; incubation at 50°C for 25 min. SuperScript III polymerase was finally inactivated at 75°C for 15 min.
Removal of dNTPs
EB (20 μl) (10 mM Tris–Cl, pH 8.5, Qiagen) was added to the reaction. dNTPs were removed by purification of the first strand mixture on a self-made 200 μl G-50 gel filtration spin-column equilibrated with 1 mM Tris–Cl, pH 7.0.
Second strand synthesis (SSS)
Since the Invitrogen kit was used for the SSS, the FSS buffer had to be restored after gel filtration. Water was added to the purified FSS reaction to bring the final volume to 52.5 μl. The mixture was cooled on ice. Then, 22.5 μl of the ‘second strand mixture’ [1 μl of 10× reverse transcription buffer (Invitrogen); 0.5 μl of 100 mM MgCl2; 1 μl of 0.1 M DTT; 2 μl of 10 mM mixture of each: dATP, dGTP, dCTP, dUTP; 15 μl of 5× SSS buffer (Invitrogen); 0.5 μl of Escherichia coli ligase (10 units/μl, NEB); 2 μl of DNA polymerase I (10 units/μl, NEB); and 0.5 μl RNase H (2 units/μl, Invitrogen)] were added. SSS reactions were incubated at 16°C for 2 h. ds cDNA was purified on QIAquick columns (Qiagen) according to the manufacturer's instructions.
DNA fragmentation
About 250 ng of ds cDNA was fragmented by sonication with a UTR200 (Hielscher Ultrasonics GmbH, Germany) under the following conditions: 1 h, 50% pulse, 100% power and continuous cooling by 0°C water flow-through.
Preparation of libraries for Illumina sequencing platform
Libraries were prepared using the DNA sample kit (#FC-102-1002, Illumina), as described previously (4), but with the following modifications: just before library amplification uridine digestion was performed at 37°C for 15 min in 5 μl of 1× TE buffer, pH 7.5 with 1 units of Uracil-N-Glycosylase (UNG; Applied Biosystems).
The procedure of paired-end sequencing library preparation was the same as for single read libraries except that different ligation adapters and PCR primers were used (#PE-102-1002, Illumina).
Sequencing
Amplified material was loaded onto a flow-cell at a concentration of 4 pM. Sequencing was carried out on the Illumina 1G Genome Analyser by running 36 cycles according to the manufacturer's instructions.
Data analysis
A flowchart of the sequence analysis pipeline developed by us is shown on Supplementary Figure 7.
Image deconvolution, quality value calculation and the mapping of exon reads and exon junctions were performed as described previously (4). Sequencing reads were aligned to the Mus musculus (UCSC mm9) or Saccharomyces cerevisiae (UCSC sacCer1) genomes using a modification of the Eland software (Gerald module v.1.27, Illumina). The mapping criteria of Eland are the following: sequencing reads should be uniquely matched to the genome allowing up to two mismatches, without insertions or deletions. We applied the following recursive modification of the Eland procedure: the first 32 bp of reads (trimming the last 4 bp of 36 bp reads due to Eland limitations) were aligned, then reads that do not match according to Eland criteria were trimmed to 31 bp, and aligned again. This 3′-end trimming of unmatched reads was done recursively down to a length of 25 bp. This modified procedure typically increases the number of uniquely aligned fragments by 20–50%, because sequencing errors that prevent successful alignment by the Eland criteria are mostly located at the ends of reads, and these are gradually trimmed off. Under these conditions, ∼60% of the reads obtained here were matched to unique locations on the reference genome, whereas ∼25% of the reads map to more than one genomic position and ∼15% do not map to any location.
Mapping end tags
Unmapped sequencing reads with 1–11 nt long leading oligo(dT) stretches were used to map the 3′-gene boundaries. Leading oligo(dT) stretches were removed, and the remaining fragment was aligned on a reference genome.
Repetitive regions
The Eland program does not map reads with multiple hits on a genome. As a result, no sequencing reads were mapped to repetitive genomic regions. To visualize repeat-related gaps in the genome browser the following simulation was performed. The whole reference genome was sliced into 30-bp long fragments with a 10-bp overlap for mouse and a 1 bp overlap for yeast. These fragments were aligned back to the reference sequence using the standard Eland settings. About 80% of the reads for mouse and 90% for yeast were then aligned uniquely. The remaining reads producing multiple hits are shown in the genome browser by gray bars, representing repetitive genomic regions where in general expression levels cannot be resolved unambiguously.
Search for novel transcribed regions
The whole genome was split into 50 bp windows (non-overlapping). A ‘new transcribed region’ was defined as a joined group of more than two consecutive windows, with at least two sequence reads (in the same direction) mapped per window. The gap between ‘new transcribed regions’ should be at least 50 bp, and the gap between a ‘new transcribed region’ and an annotated gene (with the same transcription direction as the ‘new transcribed region’) at least 100 bp.
RESULTS
Our approach (strand-specific RNA-seq, ssRNA-Seq, Figure 1A) relies on the incorporation of deoxy-UTP during the SSS, allowing subsequent selective destruction of this strand by UNG. The detailed flowchart of the procedure at the nucleotide level is presented in Supplementary Figure 1. After the first strand cDNA synthesis non-incorporated nucleotides are removed and dTTP is substituted by dUTP during the synthesis of the second strand. After ligation with a Y-shaped adaptor, the deoxyuridin-containing strand is selectively removed with UNG, leaving the first cDNA strand intact. Due to the use of Y-shaped adaptors for library preparation, all molecules are sequenced in the same direction. Thus the sequencing library maintains the polarity information of the original RNA molecules.
Figure 1.
ssRNA-Seq method. (A) Flowchart of the ssRNA-Seq procedure. RNA is shown in red, DNA in green. Arrows are in the 5′ to 3′ direction. (B–E) Scatter plots comparing mouse mRNA expression data (number of reads in annotated genes). (B) The same mouse liver sample, strand-specific (ssRNA-Seq, X-axis) and strand-unspecific (RNA-Seq, Y-axis) protocols (Pearson correlation coefficient (cc) = 0.999). (C) ssRNA-Seq results for two biological replicas (mouse whole brain mRNA); cc = 0.990. (D) Our mouse whole brain expression data (X-axis) and data from (8) (Y-axis); cc = 0.817. (E) Sense (X-axis) and antisense (Y-axis) expression in mouse brain. (F and G) Overlap of the yeast YGR203W gene with a non-annotated gene in a head-to-head orientation. Transcriptional profile without orientation is shown in (F), with orientation in (G). Reads mapped in the forward direction are shown in blue; in the reverse direction in red. Vertical lines mark the boundaries of the YGR203W gene, as determined previously (6).
Reproducibility of the method
To demonstrate that SSS with deoxyuridine does not disturb the transcriptional landscape, we performed both strand-specific and non-strand-specific transcriptome sequencing with the same RNA sample. The resulting scatter plot (Figure 1B) shows that both RNA-Seq and ssRNA-Seq protocols produce identical transcription patterns.
The RNA-Seq method is highly reproducible both for biological replicas and results, obtained in different laboratories. Correlation coefficients for independent analyses of the whole mouse brain (Figure 1C) and S. cerevisiae (Supplementary Figure 2) transcriptomes are in the range of 0.98–0.99. Our data are also in good agreement (cc = 0.82) with previously published RNA-Seq results (8; Figure 1D).
The ssRNA-Seq protocol results in a high degree of certainty for identifying transcript polarity. In theory, one uridine base in a molecule of a sequencing library is enough to prevent a sequencing read from the false strand. Even if UNG occasionally does not remove the uridine base, the molecule would still not be amplified, since uridine-containing templates strongly suppress the Phusion DNA polymerase used for library amplification (14). Apparently, most second cDNA strands in the library contain more than one uridine base, since a 75%/25% dUTP/dTTP mixture in the SSS reaction gives essentially the same results as 100% dUTP (Supplementary Figure 3).
To prevent spurious second-strand cDNA synthesis, which was shown to be the major source of artifactual antisense transcripts (15), we included actinomycin D in the reverse transcription reaction. Actinomycin D specifically inhibits DNA-dependent, but not RNA-dependent, DNA synthesis (16). In our hands, the presence of actinomycin D in the reaction did not significantly influence the level of antisense transcription (Supplementary Figure 4). However, we used actinomycin D in our protocol to ensure that the reaction was more reproducible.
To illustrate the ssRNA-Seq method, we present here the transcriptome analyses of yeast and whole mouse brain. All sequencing data are submitted to the NCBI Short Read Archive (submission SRP000667). Results of the analyses are available online at http://genseq.molgen.mpg.de/ssRNA/. We have designed a special genome browser for visualization of sequencing data (Supplementary Figure 5). The browser shows annotated genes, mapped reads, repetitive genomic regions, mismatches between reads and reference sequence, end-tags and splice junctions. Mapped reads can be viewed ranging from whole-chromosome to single-nucleotide resolution levels. The sequencing statistics, reflecting total number of reads, number of unique and multiple-matching reads, end tags and splice junctions, is presented in Supplementary Table 1.
Gene expression level
Ignoring the orientation of transcription leads to an overestimation of expression level for genes where the level of reverse transcription is comparable with that of forward. Table 1 presents ssRNA-Seq data on antisense transription level for genes with at least 10 unique mapped reads. For ∼10% of mouse and ∼15% of yeast genes error related with ignoring of transcription orientation is higher than 30%.
Table 1.
Rough estimation of antisense transcription level in mouse and yeast genes
| Mouse | Yeast | |
|---|---|---|
| Total number of genes | 28 995 | 7527 |
| Genes with more than 10 sequence reads | 17 203 | 6325 |
| More than 30% of sequence reads are in antisense orientationa | 1769 | 922 |
| More than half of sequence reads are in antisense orientationa | 910 | 656 |
aOnly for genes with more than 10 sequence reads.
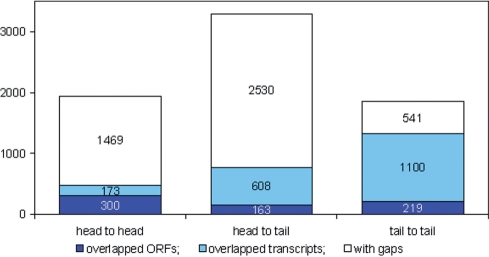
Information about transcription orientation helps to determine correctly expression levels of overlapped transcripts, both annotated (Supplementary Figure 6A, B, D and E) and novel Figure 1G, Supplementary Figure 6C. It is especially important for small genomes of prokaryotes and lower eukaryotes. The S. cerevisiae transcriptome is considerably more compact than that of the mouse, with four times fewer genes, packed into a 200 times smaller genome. Cumulative transcription plots in Figure 2 show that neighboring genes are located in close proximity. According to our results, in 36% of cases S. cerevisiae genes have no gaps between each other (Figure 3). About two-thirds of these overlaps are in opposite direction and therefore resolvable by ssRNA-seq.
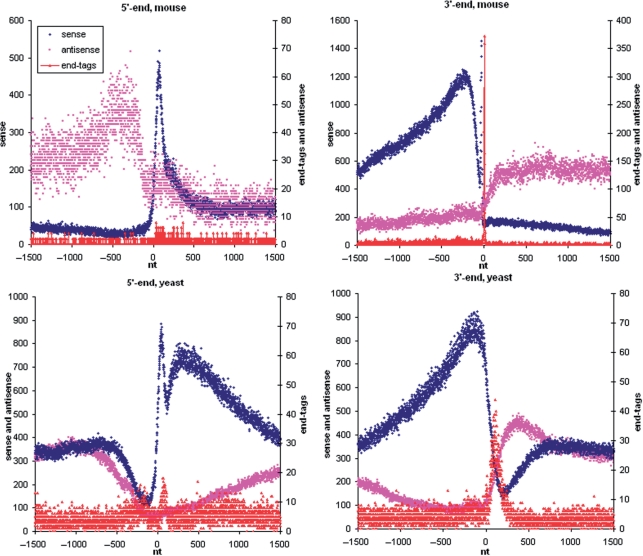
Figure 2.
Cumulative profiles of transcription (blue: sense, pink: antisense) and end tags for sense orientation (red) in 5′ and 3′ regions of mouse and yeast genes. X-axis: positions relative to the 5′ (left panels) or 3′ (right panels) end of the gene; Y-axis: total number of sequencing reads or end tags mapped in this position.
Figure 3.
Different types of intergenic regions in yeast. Stacked columns show the distribution of 7103 annotated intergenic regions according to orientation and relative position of neighboring genes. The neighboring genes were counted as overlapping if it was impossible to find a 30 nt ‘gap’ (the interval not covered by sequencing reads) between them. Transcription initiation requires more space than transcription termination: genes tend to be closer to each other in a tail-to-tail than in a head-to-head orientation. The mean distances between ORF's are 375 bp in tail-to-tail, 590 bp in head-to-tail and 703 bp in head-to-head orientations. About 49% of 3′-ends (tails) overlap with neighboring genes. This is about two times more than the fraction of overlapping 5′-ends (heads), which is 24%.
Gene structure
The ssRNA-Seq is indispensable whenever a lack of knowledge of transcript polarity can lead to misinterpretations. The current annotation of transcriptome is far from being complete. There are a lot of mistakes in determining gene and exon boundaries—Figure 1G, (Supplementary Figure 6A, B, D, E and F). Besides, there are still a number of unknown genes (Figure 1G, Supplementary Figure 6C and H) and exons (Supplementary Figure 6F and G).
About 400 novel yeast transcripts were identified in a previous study without taking into account the RNA orientation (6). Applying a simple algorithm for counting of gene-like transcribed regions for our data we found about the same number (377) of novel transcribed regions. To roughly estimate how this number might change due to available polarity information, the search was performed twice (for reads mapped in the forward direction and then for reads mapped in the reverse direction), yielding in about three times more novel gene candidates (549 forward and 512 reverse).
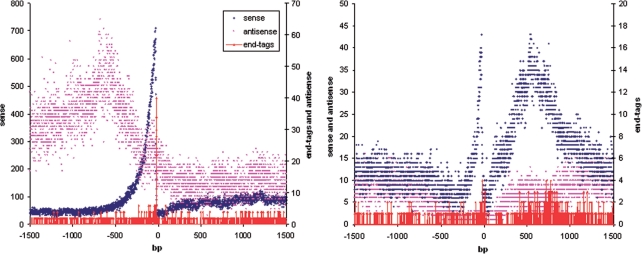
With the RNA-Seq procedure it is possible to determine the 3′-boundaries of genes using those sequencing reads, which overlap with the 3′-borders of genes (6). These reads may be mapped to the reference genome only after removal of the oligo(dT) tail. The ssRNA-Seq protocol has the advantage of reducing the noise compared to the RNA-Seq protocol because only one orientation of the homopolymeric stretch is allowed. Mapped end tags are shown in the online genome browser (Supplementary Figure 5).
It is interesting to analyze distribution of end tags within the transcriptome. Cumulative end tag profiles in the 5′- and 3′-regions of the annotated mouse and yeast genes are shown in Figure 2. Mouse end tags are grouped in a narrow peak at the 3′-region. Yeast end tags are grouped in two wide peaks close to 3′- and 5′-ends. These peaks are wider than those in the mouse, since genes are aligned using the borders of ORFs as opposed to the alignment by transcript borders for the mouse cumulative profile. The location of a surprisingly large fraction of end tags close to the promoter may reflect a new mechanism of transcription regulation. Additional analysis is required to prove this hypothesis. The similar (but not so distinguishable) end tag peak is present nearby the promoter on the mouse cumulative profile.
Mouse end tags are also strongly associated with 3′-edges of internal exons (Figure 4) indicating the dynamic interplay between polyadenylation and splicing reported previously (17).
Figure 4.
Cumulative profiles of transcription and sense end tags in 3′ regions of internal exons for mouse (left) and yeast (right).
Antisense and promoter-associated transcription
The ssRNA-Seq permits to study antisense and promoter-associated transcription both on a single gene—Supplementary Figure 6 (B, H)—and whole-transcriptome levels.
The level of antisense transcription weakly correlates with that of the sense transcription for highly expressed genes, with a ratio of about 1:100 (Figure 1E). However, it is not clear whether this weak correlation is due to a recently reported association of antisense transcription with highly expressed genes (18) or caused by a background of the ssRNA-Seq protocol. In the later case we expect at most 1% contamination for the incorrect strand.
Tiling array analysis of transcription in a human cell line demonstrated previously, that antisense transcription is enriched ∼1.3-fold in the 5′-region and ∼1.5-fold in the 3′-region of genes (2). Recent sequencing of short RNAs demonstrated that divergent transcription is associated with promoter regions of the majority of active genes in mammals (5,19,20). We have obtained similar results in our experiments. Figure 2 shows cumulative transcription profiles in the 5′ and 3′ regions of annotated mouse and yeast genes (sense transcription level in blue and antisense in pink). The mouse profiles demonstrate an about 2-fold increase of antisense transcription outside the boundaries of genes.
DISCUSSION
In the RNA-Seq approaches employed to date (4,6–9) RNA is first converted into ds cDNA, and then processed into a sequencing library. Two modifications of the ds cDNA synthesis have been suggested so far that allow one to preserve information about the direction of the transcripts (12,13). The first procedure is based on changing all cytidine residues in RNA to uridines by bisulfite treatment prior to cDNA synthesis (12). Another approach (13) involves first-strand cDNA synthesis from a tagged random hexamer primer, and SSS from a DNA–RNA template-switching primer. Both procedures are laborious. The bisulfite approach requires a non-standard sequencing data analysis scheme and also leads to the loss of ∼30% of uniquely matched sequencing reads because part of the genome complexity is lost during transformation of four bases into a three-base code. Combining a random primer with template switching may result in uneven coverage of the genes.
In other directional transcriptome profiling schemes adapters are ligated directly to single-stranded RNA molecules [21; DGE Small RNA Sample Prep Kit (Illumina); SOLiD Small RNA Expression Kit (Applied Biosystems)]. These schemes are laborious and time consuming, but they are the only choice for analysis of short RNAs. Adaptor-ligation methods are sensitive to ribosomal RNA contamination, so the RNA fraction of interest (mRNA, microRNA or short transcripts) must be pre-selected.
The suggested ssRNA-Seq approach is a modification of standard cDNA synthesis, and compatible with commercially available kits. The principle of the procedure—labeling of one of ds cDNA strands so that it can be removed—does not specifically require dUTP. For example, biotinylated nucleotides could be incorporated and the biotinylated strand then removed using streptavidin-coated magnetic particles. Strand labeling can also be performed during FSS (results not shown). The protocol can be easily adapted for other second generation sequencing platforms: SOLiD/ABI, 454/Roche.
We routinely use ssRNA-Seq for transcriptome analysis with the Illumina second-generation sequencing platform for both single read and paired end sequencing. After using ssRNA-Seq for more than a year for transcriptome analysis in different organisms (mammals, birds, fishes, plants, yeast), the procedure has proven to be convenient, reliable and highly reproducible.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Max Planck Society and the European Community's Seventh Framework Programme (FP7/2007–2013) under grant agreement number (HEALTH-F4-2008-201418) entitled READNA (REvolutionary Approaches and Devices for Nucleic Acid analysis). Funding for open access charge: Max Planck Society.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.The Encode Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 3.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 4.Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 5.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bähler J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- 8.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 9.Marioni J, Mason C, Mane S, Stephens M, Gilad Y. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stoughton RB. Applications of DNA microarrays in biology. Annu. Rev. Biochem. 2005;74:53–82. doi: 10.1146/annurev.biochem.74.082803.133212. [DOI] [PubMed] [Google Scholar]
- 11.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 12.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. The antisense transcriptomes of human cells. Science. 2008;322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, et al. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat. Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- 14.Hogrefe HH, Hansen CJ, Scott BR, Nielson KB. Archaeal dUTPase enhances PCR amplifications with archaeal DNA polymerases by preventing dUTP incorporation. Proc. Natl Acad. Sci. USA. 2002;99:596–601. doi: 10.1073/pnas.012372799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perocchi F, Xu Z, Clauder-Münster S, Steinmetz LM. Antisense artifacts in transcriptome microarray experiments are resolved by actinomycin D. Nucleic Acids Res. 2007;35:e128. doi: 10.1093/nar/gkm683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ruprecht RM, Goodman NC, Spiegelman S. Conditions for the selective synthesis of DNA complementary to template RNA. Biochim. Biophys. Acta. 1973;294:192–203. [PubMed] [Google Scholar]
- 17.Tian B, Pan Z, Lee JY. Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing. Genome Res. 2007;17:156–165. doi: 10.1101/gr.5532707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Györffy A, Surowiak P, Tulassay Z, Györffy B. Highly expressed genes are associated with inverse antisense transcription in mouse. J. Genet. 2007;86:103–109. doi: 10.1007/s12041-007-0015-x. [DOI] [PubMed] [Google Scholar]
- 19.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 21.Lister R, O'Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, Ecker JR. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.