Abstract
Short contiguous arrays of variant CTAGGG repeats in the human telomere are unstable in the male germline and somatic cells, suggesting formation of unusual structures by this repeat type. Here, we report on the structure of an intramolecular G-quadruplex formed by DNA sequences containing four human telomeric variant CTAGGG repeats in potassium solution. Our results reveal a new robust antiparallel G-quadruplex fold involving two G-tetrads sandwiched between a G·C base pair and a G·C·G·C tetrad, which could represent a new platform for drug design targeted to human telomeric DNA.
INTRODUCTION
Telomeres, the ends of linear eukaryotic chromosomes, consist of tandem repeats of G-rich sequences (1). Acting as the protective caps of the chromosomes, telomeres are critical for chromosomal stability, cell survival and proliferation (2). Human telomeres encompass thousands of canonical (wild-type) TTAGGG repeats (3), which can be interspersed with some sequence-variant repeats (4,5). It has been reported [see accompanying paper (6)] that a particular repeat type, CTAGGG (variation is underlined), when present as a short contiguous array within the telomere, causes an extraordinarily high level of localized telomere instability in the male germline and somatic cells. These variant repeats also bind to the telomeric factor POT1 more efficiently than TTAGGG repeats (6). Thus, it is important to understand if unusual structures are formed by these sequence-variant repeats, as they could lead to incongruities during telomere replication, contributing to the instability of the telomere.
Previous studies have established that human telomeric DNA sequences containing four canonical TTAGGG repeats can adopt at least five different G-quadruplex structures under different experimental conditions (7–29). These include three conformations observed in K+ solution (Supplementary Figure S1): the sequences d[TAGGG(TTAGGG)3] and d[TAGGG(TTAGGG)3TT] form predominantly intramolecular three-G-tetrad (3+1) G-quadruplexes Form 1 (21–26,28) and Form 2 (24,27,28), respectively, while the sequence d[GGG(TTAGGG)3T] forms mainly an intramolecular basket-type G-quadruplex involving only two G-tetrads, designated as Form 3 (29). Here, we report on the structure of an intramolecular G-quadruplex formed by DNA sequences containing four human telomeric variant CTAGGG repeats in K+ solution. Our results reveal a new robust antiparallel G-quadruplex fold involving two G-tetrads, a G·C base pair and a G·C·G·C tetrad, which could serve as a new platform for drug design targeted to human telomeric DNA.
METHODS
DNA sample preparation
Unlabeled and site-specific labeled DNA oligonucleotides (Table 1; Supplementary Tables S1 and S2) were chemically synthesized on an ABI 394 DNA/RNA synthesizer or purchased from Eurogentec (Belgium). DNA concentration was expressed in strand molarity using a nearest-neighbor approximation for the absorption coefficients of the unfolded species (30).
Table 1.
Representative DNA sequences used in this studya
| Name | Sequence | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 22wt | A | GGG | TTA | GGG | TTA | GGG | TTA | GGG | |
| 22CTA | A | GGG | CTA | GGG | CTA | GGG | CTA | GGG | |
| 23CTA | A | GGG | CTA | GGG | CTA | GGG | CTA | GGG | C |
| 22TCA | A | GGG | TCA | GGG | TCA | GGG | TCA | GGG | |
aVariations from the canonical repeat are underlined.
Gel electrophoresis
The molecular size of the structures formed by DNA oligonucleotides was probed by non-denaturing polyacrylamide gel electrophoresis (PAGE) as previously described (31). DNA samples (32P-labeled) of three different total concentrations (0.05, 4 and 80 μM) were incubated in a 10 mM Tris–HCl pH 7.5 buffer supplemented with 100 mM NaCl or KCl. They were heated at 90°C for 5 min and slowly cooled down (over 2 h) to 20°C. The samples were loaded on a 15% polyacrylamide gel supplemented with 20 mM of the corresponding salt and run at 26°C; 10% sucrose was added just before loading.
UV-melting experiments
The thermal stability of different oligonucleotides was characterized in heating/cooling experiments by recording the UV absorbance at 295 nm as a function of temperature (32) using a Kontron-Uvikon 940 UV/Vis spectrophotometer. UV-melting experiments were conducted as previously described (33) in a 10 mM lithium cacodylate pH 7.2 buffer containing 100 mM NaCl or KCl. The heating and cooling rates were 0.2°C per minute. Experiments were performed with 1-cm pathlength quartz cuvettes.
Thermal difference spectra
The thermal difference spectra (TDS) of a nucleic acid were obtained by recording the UV absorbance spectra over a range of temperatures (20–90°C) and subsequently taking the difference between each spectrum and that at 90°C. TDS provide specific signatures of different DNA and RNA structural conformations (34). Spectra were recorded between 220 and 320 nm on a Kontron-Uvikon 940 UV/Vis spectrophotometer using quartz cuvettes with an optical pathlength of 1 cm. DNA concentration was 4 µM.
Circular dichroism
Circular dichroism (CD) spectra were recorded on a JASCO-810 spectropolarimeter using a 1-cm pathlength quartz cuvette with a reaction volume of 600 µl. The DNA oligonucleotides (4 µM) were prepared in a 10 mM lithium cacodylate pH 7.2 buffer containing 100 mM NaCl or KCl. The samples were heated at 90°C for 5 min and cooled down to room temperature overnight. For each sample, an average of three scans was taken, the spectrum of the buffer was subtracted, and the data were zero-corrected at 320 nm.
Calorimetry
Microcalorimetry experiments were performed on a Nano DSC-II microcalorimeter as previously described (35). The oligonucleotides were prepared at concentrations ranging from 194 to 223 µM in a 10 mM lithium cacodylate pH 7.2 buffer containing 100 mM KCl. An average of six differential scanning calorimetric (DSC) heating and cooling profiles was taken.
NMR spectroscopy
Samples for NMR study were dialyzed successively against ∼50 mM KCl solution and against water. Unless otherwise stated, the strand concentration of the NMR samples was typically 0.5–2.0 mM; the solutions contained 70 mM KCl and 20 mM potassium phosphate (pH 7). NMR experiments were performed on 600 MHz and 700 MHz Bruker spectrometers at 25°C, unless otherwise specified. Resonances for guanine residues were assigned unambiguously by using site-specific low-enrichment 15N labeling (36), site-specific 2H labeling (37), and through-bond correlations at natural abundance (38,39). Resonances for thymine residues were assigned following systematic T-to-U substitutions. Spectral assignments were completed by NOESY, COSY, TOCSY, {13C-1H}-HMBC and {13C-1H}-HSQC, as previously described (29,39). Inter-proton distances were deduced from NOESY experiments at various mixing times. All spectral analyses were performed using the FELIX (Felix NMR, Inc.) program.
Structure calculation
Inter-proton distances for the d[AGGG(CTAGGG)3] quadruplex were deduced from NOESY experiments performed in H2O (mixing time, 200 ms) and D2O (mixing times, 100, 200 and 350 ms). Structure computations were performed using the XPLOR-NIH program (40) in three general steps essentially as previously described (29): (i) distance geometry simulated annealing, (ii) distance-restrained molecular dynamics refinement and (iii) relaxation matrix intensity refinement. Hydrogen bond restraints, inter-proton distance restraints, dihedral restraints, planarity restraints and repulsive restraints were imposed during structure calculations. Structures were displayed using the PyMOL program (41).
Data deposition
The coordinates for the d[AGGG(CTAGGG)3] quadruplex have been deposited in the Protein Data Bank (accession code 2KM3).
RESULTS AND DISCUSSION
Sequences containing four human telomeric variant CTAGGG repeats form a new G-quadruplex fold in K+ solution
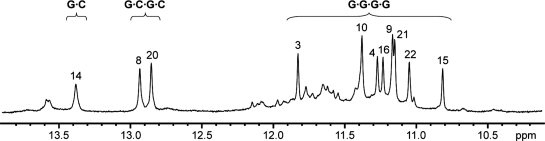
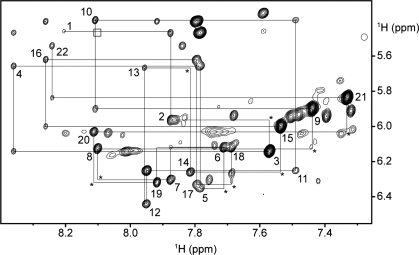
The imino proton spectrum of the 22-nt human telomeric variant d[AGGG(CTAGGG)3] sequence, henceforth designated as 22CTA (Table 1), in K+ solution (Figure 1) was distinct from those observed for the canonical four-repeat TTAGGG human telomeric sequences (22–29) and indicated the formation of a new G-quadruplex structure: eight major peaks at 10.8–11.9 p.p.m. corresponded to the formation of two G-tetrads and three major peaks at 12.8-13.4 p.p.m. corresponded to three Watson–Crick G·C base pairs (see below). The eight former peaks remained sharp at 45°C (Supplementary Figure S2), consistent with their G-tetrad origin (29). Minor G-quadruplex form(s) were also present in 22CTA, as shown by minor peaks at 10.6–12.2 p.p.m., whose relative population (as compared to the major form) augmented when the temperature was increased (Supplementary Figure S2). Imino proton spectra (Supplementary Figure S3) of many different sequences containing four human telomeric variant CTAGGG repeats with different flanking ends (Supplementary Table S1) showed eight major peaks at 10.8–11.9 p.p.m. along with major peaks at 12.8–13.6 p.p.m., suggesting that they also adopted predominantly a G-quadruplex containing Watson–Crick G·C base pairs in K+ solution. A preliminary analysis on a NOESY spectrum of the 23CTA sequence (Table 1) suggested that this sequence adopted in K+ solution the same overall G-quadruplex fold as 22CTA (see below).
Figure 1.
Imino proton spectrum of the 22-nt human telomeric variant d[AGGG(CTAGGG)3] sequence (22CTA) in K+ solution with assignments for the major form listed over the spectrum. The imino protons are classified into three categories, corresponding to their involvement in G·G·G·G tetrad, G·C·G·C tetrad, or G·C base pair formation.
The imino proton spectrum of 22CTA in Na+ solution (Supplementary Figure S4) indicated the presence of a major G-quadruplex form and minor conformation(s), but major sharp peaks around 13 p.p.m. (characteristic of stable Watson–Crick G·C base pairs) were not observed.
Native gel electrophoresis analysis
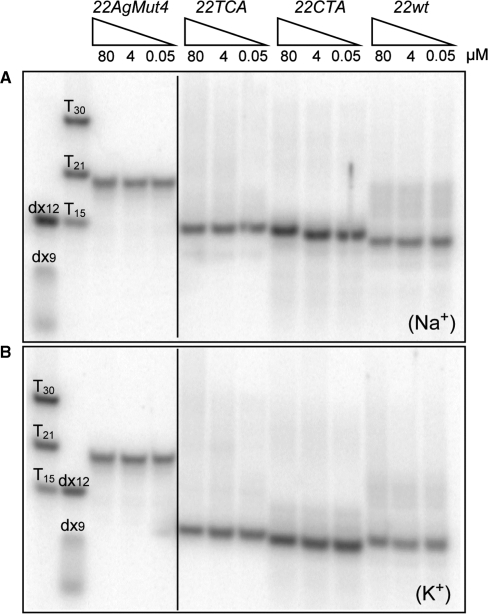
The molecular size of G-quadruplexes formed by variant human telomeric sequences (Table 1) was probed by native PAGE. In both Na+ and K+ solution, a single major band was observed for each of the 22-nt human telomeric sequences (Figure 2), 22wt (containing TTAGGG repeats), 22CTA (containing CTAGGG repeats) and 22TCA (containing TCAGGG repeats). The rate of migration of the band was independent of oligonucleotide concentrations (from 0.05 to 80 µM), arguing for an intramolecular structure for each of the three sequences. This was further substantiated by the fast migration of the band in relation to those of duplexes (9 and 12 bp), oligothymidylate sequences (15, 21 and 30 nt), and a 22-nt control sequence (22AgMut4) incapable of forming a quadruplex.
Figure 2.
Non-denaturing PAGE analysis of the 22wt, 22CTA and 22TCA human telomeric sequences (Table 1), which were pre-incubated in a 10 mM Tris–HCl pH 7.5 buffer supplemented with (A) 100 mM Na+ or (B) 100 mM K+, then loaded on a 15% polyacrylamide gel supplemented with 20 mM of the corresponding salt, and run at 26°C. Migration markers are provided on the left: oligothymidylate sequences (dT15, dT21 and dT30), duplexes (dx9: d[GCGATACGG] + d[CCGTATCGC] and dx12: d[GCGTGACTTCGG] + d[CCGAAGTCACGC]), and single-stranded 22AgMut4 control sequence d[ATGGTTAGTGTTAGGTTTAGTG] incapable of forming a quadruplex. Note that with respect to the markers, 22wt, 22CTA and 22TCA human telomeric sequences migrate faster in K+ solution than in Na+ solution.
Thermal stability: UV absorption and calorimetry studies
Melting experiments were conducted for variant human telomeric sequences by monitoring the UV absorbance at 295 nm (32). All transitions were reversible, indicating that the denaturation curves corresponded to a true equilibrium process (Supplementary Figure S5). The intramolecular nature of G-quadruplex formation was evaluated by varying the oligonucleotide concentration (Table 2). No significant difference (1°C or less) in melting temperature was observed upon about 40-fold increase in concentration (from 5 to ∼200 µM), indicating that the G-quadruplex folding was indeed intramolecular. The variant 22CTA sequence was found to be marginally less stable than the canonical 22wt sequence in 20 and 100 mM KCl (Table 2 and data not shown). The stability of both 22CTA and 22wt was lower in NaCl than in KCl (data not shown), as usually observed for the formation of G-quadruplexes. Note that the 23-nt human telomeric variant sequence 23CTA was slightly more stable than 22CTA. Thermodynamic parameters of the variant human telomeric sequences in K+ solution, extracted from the UV-melting experiments (32), are listed in Table 2.
Table 2.
Melting temperatures (°C) and thermodynamic parameters for quadruplex dissociation of various human telomeric sequences in 100 mM K+ solution, as measured by 295-nm UV absorbance and DSC
| Oligoa | UV-melting |
DSC: average excess enthalpy/entropyd |
DSC: deconvolution general model, 1 transitione |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Tmb (°C) | ΔH °VH (kcal mol−1) | ΔS °VH (cal K−1 mol−1) | ΔHcal (kcal mol−1) | ΔScal (cal K−1 mol−1) | Tt(°C) | ΔH (kcal mol−1) | ΔCp (kcal K−1 mol−1) | Tm (°C) | |
| 22wt | 64 | 54.8 ± 3.1 | 163 ± 9 | 39.6 ± 3.0 | 116 ± 9 | 66.9 ± 0.5 | 51.6 ± 0.9 | −0.54 ± 0.18 | 68.7 ± 0.5 |
| 22CTA | 62 | 53.7 ± 0.8 | 157 ± 4 | 41.7 ± 3.9 | 123 ± 12 | 65.4 ± 0.5 | 49.8 ± 0.6 | −0.45 ± 0.15 | 66.9 ± 0.4 |
| 23CTA | 64 | 53.0 ± 3.2 c | 157 ± 10c | 47.4 ± 4.8 | 139 ± 14 | 67.1 ± 0.5 | 55.1 ± 1.0 | −0.61 ± 0.21 | 68.3 ± 0.3 |
aConcentration of 22wt, 22CTA and 23CTA is 223, 194 and 205 μM, respectively.
bTm measured for 5 μM of 22wt, 22CTA and 23CTA is 63.5, 61 and 63°C, respectively.
cΔH °VH and ΔS °VH of 23CTA in KCl are provided for illustration only; in this case, the lnK versus 1/T graph significantly deviates from linearity.
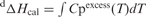 and
and 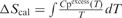 where Cpexcess is the excess heat capacity function.
where Cpexcess is the excess heat capacity function.
eThe general transition model directly fits the molar heat capacity Cp (and not the excess heat capacity Cpexcess). It is used for transitions with ΔCp≠0. In this model, ΔCp(T) is fitted with a second-order polynomial: ΔCp(T) = a + bT + cT2 = ΔCp(Tm) + b(T−Tm) + c(T 2−Tm2). ΔH and ΔCp are given at T = Tm.
Microcalorimetry experiments (35) were performed on the same set of oligonucleotides in a buffer containing 100 mM KCl (Supplementary Figure S6). Analysis of DSC scans confirmed that 22CTA was slightly less stable than 22wt. Both transitions gave similar enthalpy values (∼40 kcal/mol, determined using a calorimetry model-independent analysis, as compared to ∼50 kcal/mol, with a van t’ Hoff analysis; Table 2). A similar difference in enthalpy has been reported for the wild-type sequence (42).
TDS and CD signatures
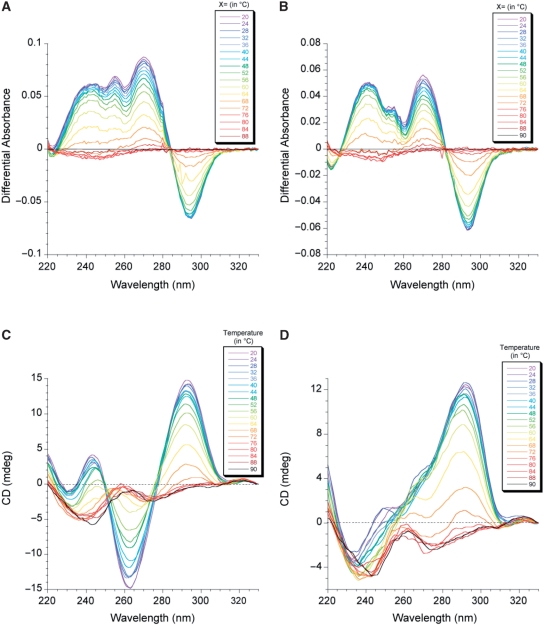
The TDS for both 22CTA and 22wt in K+ solution (Figure 3A and B; Supplementary Figure S7) exhibited typical patterns of a G-quadruplex structure, with two positive maxima at 240 and 275 nm and a negative minimum around 295 nm (34). There was an isosbestic point for both sequences around 285 nm, but the shapes of the two TDS differed around 255 nm.
Figure 3.
TDS (top panels) and CD spectra (bottom panels) recorded at different temperatures (color coded on the top right corner of each spectrum) for (A and C) 22CTA and (B and D) 22wt. TDS were obtained by substracting the UV absorbance at X°C from that at 90°C. DNA concentration was 4 µM; solution contained 100 mM KCl and 10 mM lithium cacodylate, pH 7.2.
CD spectra for both sequences in K+ solution (Figure 3C and D; Supplementary Figure S8) were in agreement with the formation of G-quadruplexes. The CD profile of 22CTA (with a very negative peak around 260 nm) differed significantly from that of the wild-type sequence 22wt, consistent with the formation of a different fold.
NMR spectral assignments
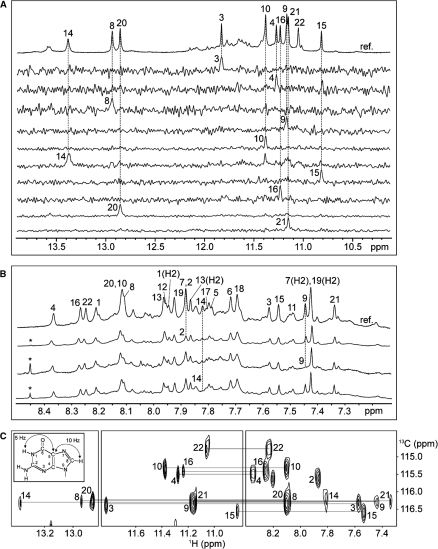
Guanine imino and H8 protons of 22CTA were unambiguously assigned using site-specific low-enrichment 15N labeling (36), site-specific 2H labeling (37) and through-bond correlations at natural abundance (38,39) (Figure 4; Supplementary Table S2). Resonances for thymine residues were unambiguously assigned by T-to-U substitutions (39) (Supplementary Figure S9 and Table S2). With the help of these unambiguous assignments and other through-bond correlation experiments (COSY, TOCSY, {13C-1H}-HMBC and {13C-1H}-HSQC) (data not shown), the classical H8/H6-H1’ NOE sequential connectivity could be traced from A1 through G22 (Figure 5). The intensity of intraresidue H8-H1’ NOE cross-peaks (Supplementary Figure S10) indicated syn glycosidic conformation for G3, G9, G15 and G21, in contrast to other residues, which adopted anti conformation. These results were supported by NMR spectra of modified sequences in which G3, G9, G15 and G21 were substituted by 8-bromoguanine (BrG) (Supplementary Figure S11 and data not shown).
Figure 4.
Guanine imino and H8 proton assignments of 22CTA in K+ solution. (A) Imino protons were assigned in 15N-filtered spectra of samples, 2% 15N-labeled at the indicated positions. The reference spectrum (ref.) is shown at the top. (B) Examples of H8 proton assignments by site-specific 2H labeling at the indicated positions. The reference spectrum (ref.) is shown at the top. The peak from a contamination is marked by an asterisk. (C) Through-bond correlations between guanine imino and H8 protons via 13C5 at natural abundance, using long-range J-couplings shown in the inset. Assignments are labeled with residue numbers.
Figure 5.
NOESY spectrum (mixing time, 350 ms) showing the H8/H6-H1’ connectivity of 22CTA in K+ solution. Intraresidue H8/H6-H1’ NOE cross-peaks are labeled with residue numbers. Weak or missing sequential connectivities are marked with asterisks. The box indicates the position of a very weak cross-peak (not seen at this threshold) corresponding to the NOE from the H8 proton of G8 to the H5 proton of C5, consistent with a slipped alignment for the G8·C17·G20·C5 tetrad (see text).
22CTA forms a chair-type G-quadruplex containing a G·C·G·C tetrad
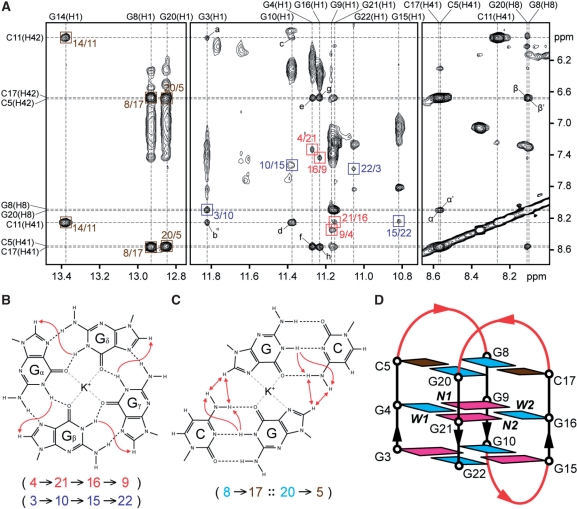
Analysis of imino-H8 connectivity patterns (Figure 6A) revealed the formation of an intramolecular chair-type G-quadruplex with two G-tetrad layers, G4·G21·G16·G9 and G3·G10·G15·G22 (Figure 6B and D): the hydrogen-bond directionalities of the G-tetrads alternate anticlockwise and clockwise; the glycosidic conformations of guanines around each tetrad are syn·anti·syn·anti; each strand is antiparallel to the two adjacent strands; there are two narrow and two wide grooves; the three connecting loops are edgewise; the first and third loops (on the top) span narrow grooves, while the second loop (at the bottom) spans a wide groove. The observation of the imino protons of G8, G20 and G14 at 12.8–13.4 p.p.m. (Figure 1) and their strong NOE cross-peaks to the amino protons of C17, C5 and C11, respectively, (Figure 6A) indicated the formation of three Watson–Crick G·C base pairs: G8·C17 and G20·C5 on the top and G14·C11 at the bottom of the G-tetrad core. The two former base pairs are further aligned to form a G·C·G·C tetrad (Figure 6C). Note that all three Watson–Crick G·C base pairs are formed across the wide grooves of the G-tetrad core. The central position of the G4·G21·G16·G9 tetrad (between the G3·G10·G15·G22 and the G8·C17·G20·C5 tetrads) is consistent with the observation of the imino protons of G4, G21, G16 and G9 being well protected from the exchange with solvent (Supplementary Figure S12).
Figure 6.
Formation of a chair-type G-quadruplex containing two G·G·G·G tetrads and a G·C·G·C tetrad by 22CTA in K+ solution. (A) NOESY spectrum (mixing time, 200 ms) showing the cross-peaks that identify the arrangement of two G-tetrads and three G·C base pairs. Cross-peaks arising from imino-H8 connectivity around the two G-tetrads are framed in red and blue, and labeled with the residue number of imino protons in the first position and that of H8 protons in the second position. Cross-peaks between guanine imino and cytosine amino protons from the three Watson–Crick G·C base pairs are framed in brown, and labeled with the residue number of guanine imino protons in the first position and that of cytosine amino protons in the second position. The NOE cross-peaks a to h are assigned as follows: a, G3(H1)-C11(H42); b, G3(H1)-C11(H41); c, G10(H1)-C11(H42); d, G10(H1)-C11(H41); e, G4(H1)-C5(H42); f, G4(H1)-C5(H41); g, G16(H1)-C17(H42); h, G16(H1)-C17(H41). The NOE cross-peaks α, α′, β and β′ correspond to G20(H8)-C17(H41), G8(H8)-C5(H41), G20(H8)-C17(H42) and G8(H8)-C5(H42), respectively. (B) Characteristic guanine imino-H8 NOE connectivity patterns around a Gα·Gβ·Gγ·Gδ tetrad as indicated with arrows, with the connectivities observed for the G4·G21·G16·G9 and G3·G10·G15·G22 tetrads shown below. (C) The slipped G·C·G·C tetrad arrangement of the two Watson–Crick G8·C17 and G20·C5 base pairs, as supported by NOE cross-peaks α, α′, β and β′ (represented by red double-headed arrows). NOEs from guanine imino protons to cytosine amino protons that establish the G8·C17 and G20·C5 base pairs are represented by single-headed arrows. (D) Schematic view of the 22CTA G-quadruplex. anti and syn guanines are colored cyan and magenta, respectively, while cytosines are colored brown. W1, W2, N1 and N2 represent wide 1, wide 2, narrow 1 and narrow 2 grooves, respectively. The backbone of the core and loops is colored black and red, respectively.
Solution structure of the 22CTA G-quadruplex
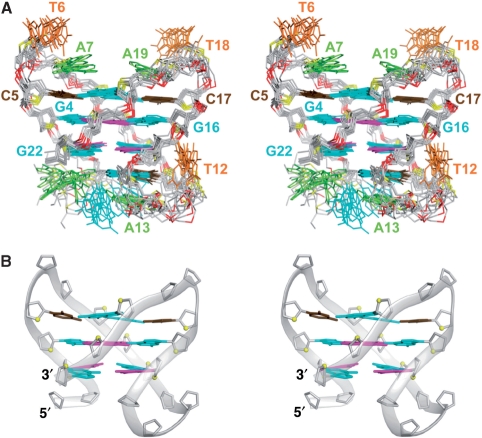
The structure of the 22CTA G-quadruplex in K+ solution (Figure 7) was calculated on the basis of NMR restraints (Table 3 and Supplementary Data). In the G-tetrad core, there is extensive stacking between five-membered rings of guanines (Supplementary Figure S13), which belong to the two G-tetrads with opposite hydrogen-bond directionalities (7,29).
Figure 7.
Stereo views of the 22CTA G-quadruplex structure in K+ solution. (A) Ten superimposed refined structures. (B) Ribbon view of a representative structure. anti and syn guanines are colored cyan and magenta, respectively; cytosines are colored brown, adenines, green; thymines, orange; backbone and sugar, gray; O4’ atoms, yellow; phosphorus atoms, red.
Table 3.
Statistics of the computed structures of the 22-nt human telomeric variant d[AGGG(CTAGGG)3] sequence
| NMR restraints | ||
| Distance restraints | D2O | H2O |
| Intraresidue distance restraints | 203 | 5 |
| Sequential (i, i + 1) distance restraints | 179 | 29 |
| Long-range (i, ≥ i + 2) distance restraints | 34 | 72 |
| Other restraints | ||
| Hydrogen bond restraints | 50 | |
| Dihedral restraints | 34 | |
| Repulsive restraintsa | 6 | |
| Intensity restraints | ||
| Non-exchangeable Protons (each of three mixing times) | 108 | |
| Structure statistics for 10 molecules following intensity refinement | ||
| NOE violations | ||
| Number (>0.2Å)b | 0.300 ± 0.458 | |
| Maximum violation (Å) | 0.168 ± 0.031 | |
| RMSD of violations (Å) | 0.019 ± 0.002 | |
| Deviations from the ideal covalent geometry | ||
| Bond lengths (Å) | 0.004 ± 0.000 | |
| Bond angles (deg) | 0.746 ± 0.010 | |
| Impropers (deg) | 0.413 ± 0.010 | |
| NMR R-factor (R1/6) | 0.016 ± 0.001 | |
| Pairwise all heavy atom RMSD values (Å) | ||
| All heavy atoms except A1 and G2 | 1.21 ± 0.14 | |
| All heavy atoms | 1.53 ± 0.25 | |
aDistance restraints between pairs of protons that do not exhibit NOE cross-peaks; these restraints are removed during relaxation matrix intensity refinement.
bThe total number of violations divided by the number of structures.
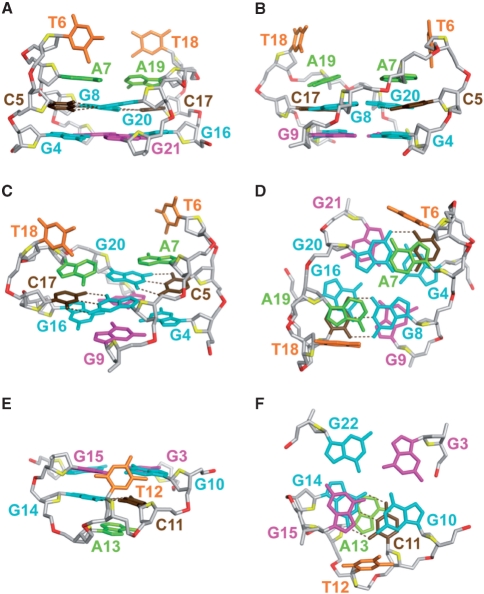
Previous studies have demonstrated two different G·C·G·C tetrads that are aligned via the major groove edges of the Watson–Crick G·C base pairs: the direct alignment (Supplementary Figure S14A and C) observed in Na+ solution (43–45) and the slipped alignment (Supplementary Figure S14B and D) observed in K+ solution (46). The latter was proposed to provide a K+ coordination site between the two G·C base pairs (Figure 6C; Supplementary Figure S14B) (46). In the direct alignment, the distances from the guanine H8 proton of one Watson–Crick G·C base pair to the cytosine H5 proton and the cytosine amino protons of the other Watson–Crick G·C base pair are ∼3 Å and ∼5 Å, respectively (Supplementary Figure S14C). Accordingly, the guanine H8 proton of one G·C base pair should exhibit a strong NOE to the cytosine H5 proton and weak NOEs to the cytosine amino protons of the other G·C base pair. The reverse scenario should be observed in the slipped alignment: the guanine H8 proton of each Watson–Crick G·C base pair is closer to the cytosine amino protons (∼3 Å) than to the cytosine H5 proton (∼5 Å) of the adjacent Watson–Crick G·C base pair (Supplementary Figure S14D). The G8·C17·G20·C5 tetrad at the top of the current structure adopted the slipped alignment (Figures 6–8). This alignment between the G8·C17 and G20·C5 base pairs was supported by the observations of strong NOEs from the H8 protons of G8 and G20 to the amino protons of C5 and C17 (Figure 6A, see figure legend), respectively, but very weak NOEs from the H8 protons of G8 and G20 to the H5 protons of C5 and C17 (Figure 5), respectively. NOEs from the imino protons of G8 and G20 to the imino protons of G16 and G4, respectively, define the position of the G8·C17·G20·C5 tetrad over the top G-tetrad.
Figure 8.
Detailed structure of the 22CTA G-quadruplex in K+ solution. (A–D) The top half: the segments G4-G9 and G16-G21, showing the G4·G21·G16·G9 tetrad, the slipped G8·C17·G20·C5 tetrad, and the two quasi-symmetric loops. (A) View from the side looking into the wide 1 (W1) groove. (B) View from the side looking into the narrow 1 (N1) groove. (C) Side view from a different angle. (D) Top-down view. (E and F) The bottom half: the G3·G10·G15·G22 tetrad, the G14·C11 base pair, and the bottom loop, looking from the side (E) and from the top down (F). Color coded as in Figure 7A. Hydrogen-bonds in the Watson–Crick G·C base pairs are shown by dotted lines.
Two adenine bases, A7 and A19, further stack over the G8·C17·G20·C5 tetrad (Figure 8A–D), supported by the observation of NOEs from the A7 and A19 protons to the C5 and C17 protons, respectively. The two thymine bases, T6 and T18, are projected outward (Figure 8A–D), consistent with the observation of only a few NOEs between the protons of these bases and their neighboring residues (data not shown). Quasi-symmetry (39,47) was observed for the top part of the structure (Figures 7 and 8), consistent with its two halves (segments G4–G9 and G16–G21) displaying many similar spectral characteristics including chemical shifts and NOE patterns (Figures 1, 4–6; Supplementary Figure S15).
The G14·C11 base pair is located at the bottom of the G-tetrad core, directly under G10 and G15 (Figure 8E and F). This placement was supported by the observations of a number of NOE cross-peaks [e.g. a NOE between the imino protons of G14 and G10 (data not shown) and NOEs between the protons of C11 and the bottom G-tetrad (Figure 6A)] and the results from solvent exchange experiment showing that the imino proton of G10 was better protected than that of the three other guanines (G3, G15 and G21) from the same G-tetrad (Supplementary Figure S12). At the bottom loop, the A13 base stacks underneath the G14·C11 base pair, while the T12 base folds into a hydrophobic groove (29).
The 22CTA G-quadruplex represents a robust fold
The structure of 22CTA in K+ solution consists of a two-G-tetrad antiparallel core sandwiched between a G·C base pair and a G·C·G·C tetrad. This robust fold appeared to be the preferred one for various four-repeat variant CTAGGG sequences with different flanking ends in K+ solution, contrasting the situation of sequences containing four canonical TTAGGG repeats, which were shown to adopt different G-quadruplex folds (7–29). Our modeling and preliminary NMR spectral analysis (including NOESY) suggested that sequences containing an additional C at the 3′-end (e.g. 23CTA) could form a similar G-quadruplex with a second G·C·G·C tetrad at the bottom of the structure. The formation of the G14·C11·G2·C23 tetrad for 23CTA would be consistent with its higher stability (Table 2) and considerable changes in the chemical shifts of G3 and G15 imino protons of the bottom G-tetrad (Supplementary Figure S3) as compared to 22CTA. Such a core structure with two G-tetrads sandwiched between two G·C·G·C tetrads, which has already been observed previously (46), would provide a well-aligned channel for K+ ion coordination. Our results indicate that G·C·G·C tetrads may positively contribute to G-quadruplex stability (43–46,48), in contrast to very detrimental effects observed for many tetrads of guanine derivatives (49).
Although the G-quadruplex fold of 22CTA contains only two G-tetrad layers, its stability is comparable to that of the structures containing three G-tetrads (29). This can be explained by molecular interactions seen in our structure including the formation of a G·C base pair and a G·C·G·C tetrad, and additional base stacking interactions in the loops. The principle of this folding and base arrangement is analogous to that observed for Form 3 human telomeric G-quadruplex in K+ solution (29), and reinforces the view that the overall folding topology of a G-quadruplex is defined not only by maximizing the number of G-tetrads, but also by maximizing all possible base pairing and stacking in the loops (29).
CONCLUSION
We have shown that sequences containing four human telomeric variant CTAGGG repeats adopt a novel antiparallel G-quadruplex involving two G-tetrads sandwiched between a G·C base pair and a G·C·G·C tetrad. This structure, with its robust and unique structural features, and its unusual binding properties to the telomeric factor POT1, could be used as a new scaffold for the design of specific G-quadruplex ligands directed against human telomeric DNA.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Singapore Biomedical Research Council (grant 07/1/22/19/542 to A.T.P.); Singapore Ministry of Education (ARC30/07 and RG62/07 to A.T.P.); Nanyang Technological University start-up (SUG5/06 and RG138/06 to A.T.P.); INSERM, CNRS, and the Muséum National d’Histoire Naturelle (to J.L.M.). Funding for open access charge: Singapore Biomedical Research Council grant 07/1/22/19/542.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Joefina Kim Cheow Lim and Jocelyn Mei Wen Lim from A.T.P's laboratory for DNA sample preparation.
REFERENCES
- 1.Wellinger RJ, Sen D. The DNA structures at the ends of eukaryotic chromosomes. Eur. J. Cancer. 1997;33:735–749. doi: 10.1016/S0959-8049(97)00067-1. [DOI] [PubMed] [Google Scholar]
- 2.De Cian A, Lacroix L, Douarre C, Temime-Smaali N, Trentesaux C, Riou JF, Mergny JL. Targeting telomeres and telomerase. Biochimie. 2008;90:131–155. doi: 10.1016/j.biochi.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 3.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl Acad. Sci. USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allshire RC, Dempster M, Hastie ND. Human telomeres contain at least 3 types of G-rich repeat distributed non-randomly. Nucleic Acids Res. 1989;17:4611–4627. doi: 10.1093/nar/17.12.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baird DM, Jeffreys AJ, Royle NJ. Mechanisms underlying telomere repeat turnover, revealed by hypervariable variant repeat distribution patterns in the human Xp/Yp telomere. EMBO J. 1995;14:5433–5443. doi: 10.1002/j.1460-2075.1995.tb00227.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mendez-Bermudez A, Hills M, Pickett HA, Phan AT, Mergny JL, Riou JF, Royle NJ. Human telomeres that contain (CTAGGG)n repeats show replication dependent instability in somatic cells and the male germline. Nucleic Acids Res. 2009;37:6225–6238. doi: 10.1093/nar/gkp629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Y, Patel DJ. Solution structure of the human telomeric repeat d[AG3(T2AG3)3] G-tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 8.Parkinson GN, Lee MPH, Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 9.Ying L, Green JJ, Li H, Klenerman D, Balasubramanian S. Studies on the structure and dynamics of the human telomeric G quadruplex by single-molecule fluorescence resonance energy transfer. Proc. Natl Acad. Sci. USA. 2003;100:14629–14634. doi: 10.1073/pnas.2433350100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Redon S, Bombard S, Elizondo-Riojas MA, Chottard JC. Platinum cross-linking of adenines and guanines on the quadruplex structures of the AG3(T2AG3)3 and (T2AG3)4 human telomere sequences in Na+ and K+ solutions. Nucleic Acids Res. 2003;31:1605–1613. doi: 10.1093/nar/gkg259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Risitano A, Fox KR. Inosine substitutions demonstrate that intramolecular DNA quadruplexes adopt different conformations in the presence of sodium and potassium. Bioorg. Med. Chem. Lett. 2005;15:2047–2050. doi: 10.1016/j.bmcl.2005.02.050. [DOI] [PubMed] [Google Scholar]
- 12.Rezler EM, Seenisamy J, Bashyam S, Kim MY, White E, Wilson WD, Hurley LH. Telomestatin and diseleno sapphyrin bind selectively to two different forms of the human telomeric G-quadruplex structure. J. Am. Chem. Soc. 2005;127:9439–9447. doi: 10.1021/ja0505088. [DOI] [PubMed] [Google Scholar]
- 13.Rujan IN, Meleney JC, Bolton PH. Vertebrate telomere repeat DNAs favor external loop propeller quadruplex structures in the presence of high concentrations of potassium. Nucleic Acids Res. 2005;33:2022–2031. doi: 10.1093/nar/gki345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qi J, Shafer RH. Covalent ligation studies on the human telomere quadruplex. Nucleic Acids Res. 2005;33:3185–3192. doi: 10.1093/nar/gki632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vorlickova M, Chladkova J, Kejnovska I, Fialova M, Kypr J. Guanine tetraplex topology of human telomere DNA is governed by the number of (TTAGGG) repeats. Nucleic Acids Res. 2005;33:5851–5860. doi: 10.1093/nar/gki898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Correia JJ, Wang L, Trent JO, Chaires JB. Not so crystal clear: the structure of the human telomere G-quadruplex in solution differs from that present in a crystal. Nucleic Acids Res. 2005;33:4649–4659. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee JY, Okumus B, Kim DS, Ha T. Extreme conformational diversity in human telomeric DNA. Proc. Natl Acad. Sci. USA. 2005;102:18938–18943. doi: 10.1073/pnas.0506144102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xue Y, Kan ZY, Wang Q, Yao Y, Liu J, Hao YH, Tan Z. Human telomeric DNA forms parallel-stranded intramolecular G-quadruplex in K+ solution under molecular crowding condition. J. Am. Chem. Soc. 2007;129:11185–11191. doi: 10.1021/ja0730462. [DOI] [PubMed] [Google Scholar]
- 19.Gaynutdinov TI, Neumann RD, Panyutin IG. Structural polymorphism of intramolecular quadruplex of human telomeric DNA: effect of cations, quadruplex-binding drugs and flanking sequences. Nucleic Acids Res. 2008;36:4079–4087. doi: 10.1093/nar/gkn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amrane S, Ang RW, Tan ZM, Li C, Lim JK, Lim JM, Lim KW, Phan AT. A novel chair-type G-quadruplex formed by a Bombyx mori telomeric sequence. Nucleic Acids Res. 2009;37:931–938. doi: 10.1093/nar/gkn990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Y, Noguchi Y, Sugiyama H. The new models of the human telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorg. Med. Chem. 2006;14:5584–5591. doi: 10.1016/j.bmc.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 22.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human telomeric sequence forms a hybrid-type intramolecular G-quadruplex structure with mixed parallel/antiparallel strands in potassium solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the human telomere in K+ solution: an intramolecular (3+1) G-quadruplex scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phan AT, Luu KN, Patel DJ. Different loop arrangements of intramolecular human telomeric (3+1) G-quadruplexes in K+ solution. Nucleic Acids Res. 2006;34:5715–5719. doi: 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai J, Punchihewa C, Ambrus A, Chen D, Jones RA, Yang D. Structure of the intramolecular human telomeric G-quadruplex in potassium solution: a novel adenine triple. Nucleic Acids Res. 2007;42:481–490. doi: 10.1093/nar/gkm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsugami A, Xu Y, Noguchi Y, Sugiyama H, Katahira M. Structure of a human telomeric DNA sequence stabilized by 8-bromoguanosine substitutions, as determined by NMR in a K+ solution. FEBS J. 2007;274:3545–3556. doi: 10.1111/j.1742-4658.2007.05881.x. [DOI] [PubMed] [Google Scholar]
- 27.Dai J, Carver M, Punchihewa C, Jones RA, Yang D. Structure of the Hybrid-2 type intramolecular human telomeric G-quadruplex in K+ solution: insights into structure polymorphism of the human telomeric sequence. Nucleic Acids Res. 2007;35:4927–4940. doi: 10.1093/nar/gkm522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phan AT, Kuryavyi V, Luu KN, Patel DJ. Structure of two intramolecular G-quadruplexes formed by natural human telomere sequences in K+ solution. Nucleic Acids Res. 2007;35:6517–6525. doi: 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim KW, Amrane S, Bouaziz S, Xu W, Mu Y, Patel DJ, Luu KN, Phan AT. Structure of the human telomere in K+ solution: a stable basket-type G-quadruplex with only two G-tetrad layers. J. Am. Chem. Soc. 2009;131:4301–4309. doi: 10.1021/ja807503g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cantor CR, Warshaw MM, Shapiro H. Oligonucleotide interactions. III. Circular dichroism studies of the conformation of deoxyoligonucleotides. Biopolymers. 1970;9:1059–1077. doi: 10.1002/bip.1970.360090909. [DOI] [PubMed] [Google Scholar]
- 31.Guédin A, De Cian A, Gros J, Lacroix L, Mergny JL. Sequence effects in single-base loops for quadruplexes. Biochimie. 2008;90:686–696. doi: 10.1016/j.biochi.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 32.Mergny JL, Phan AT, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 33.Mergny JL, Lacroix L. Analysis of thermal melting curves. Oligonucleotides. 2003;13:515–537. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]
- 34.Mergny JL, Li J, Lacroix L, Amrane S, Chaires JB. Thermal difference spectra: a specific signature for nucleic acid structures. Nucleic Acids Res. 2005;33:e138. doi: 10.1093/nar/gni134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Amrane S, Saccà B, Mills M, Chauhan M, Klump HH, Mergny JL. Length-dependent energetics of (CTG)n and (CAG)n trinucleotide repeats. Nucleic Acids Res. 2005;33:4065–4077. doi: 10.1093/nar/gki716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phan AT, Patel DJ. A site-specific low-enrichment 15N,13C isotope-labeling approach to unambiguous NMR spectral assignments in nucleic acids. J. Am. Chem. Soc. 2002;124:1160–1161. doi: 10.1021/ja011977m. [DOI] [PubMed] [Google Scholar]
- 37.Huang X, Yu P, LeProust E, Gao X. An efficient and economic site-specific deuteration strategy for NMR studies of homologous oligonucleotide repeat sequences. Nucleic Acids Res. 1997;25:4758–4763. doi: 10.1093/nar/25.23.4758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phan AT. Long-range imino proton-13C J-couplings and the through-bond correlation of imino and non-exchangeable protons in unlabeled DNA. J. Biomol. NMR. 2000;16:175–178. doi: 10.1023/a:1008355231085. [DOI] [PubMed] [Google Scholar]
- 39.Phan AT, Guéron M, Leroy JL. Investigation of unusual DNA motifs. Methods Enzymol. 2001;338:341–371. doi: 10.1016/s0076-6879(02)38228-4. [DOI] [PubMed] [Google Scholar]
- 40.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 41.DeLano WL. The PyMOL User's Manual. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
- 42.Lane AN, Chaires JB, Gray RD, Trent JO. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kettani A, Kumar RA, Patel DJ. Solution structure of a DNA quadruplex containing the fragile X syndrome triplet repeat. J. Mol. Biol. 1995;254:638–656. doi: 10.1006/jmbi.1995.0644. [DOI] [PubMed] [Google Scholar]
- 44.Kettani A, Bouaziz S, Gorin A, Zhao H, Jones RA, Patel DJ. Solution structure of a Na cation stabilized DNA quadruplex containing G·G·G·G and G·C·G·C tetrads formed by G-G-G-C repeats observed in adeno-associated viral DNA. J. Mol. Biol. 1998;282:619–636. doi: 10.1006/jmbi.1998.2030. [DOI] [PubMed] [Google Scholar]
- 45.Zhang N, Gorin A, Majumdar A, Kettani A, Chernichenko N, Skripkin E, Patel DJ. Dimeric DNA quadruplex containing major groove-aligned A·T·A·T and G·C·G·C tetrads stabilized by inter-subunit Watson-Crick A·T and G·C pairs. J. Mol. Biol. 2001;312:1073–1088. doi: 10.1006/jmbi.2001.5002. [DOI] [PubMed] [Google Scholar]
- 46.Bouaziz S, Kettani A, Patel DJ. A K cation-induced conformational switch within a loop spanning segment of a DNA quadruplex containing G-G-G-C repeats. J. Mol. Biol. 1998;282:637–652. doi: 10.1006/jmbi.1998.2031. [DOI] [PubMed] [Google Scholar]
- 47.Phan AT, Guéron M, Leroy JL. The solution structure and internal motions of a fragment of the cytidine-rich strand of the human telomere. J. Mol. Biol. 2000;299:123–144. doi: 10.1006/jmbi.2000.3613. [DOI] [PubMed] [Google Scholar]
- 48.Mergny JL, De Cian A, Amrane S, Webba da Silva M. Kinetics of double-chain reversals bridging contiguous quartets in tetramolecular quadruplexes. Nucleic Acids Res. 2006;34:2386–2397. doi: 10.1093/nar/gkl098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gros J, Rosu F, Amrane S, De Cian A, Gabelica V, Lacroix L, Mergny JL. Guanines are a quartet's best friend: impact of base substitutions on the kinetics and stability of tetramolecular quadruplexes. Nucleic Acids Res. 2007;35:3064–3075. doi: 10.1093/nar/gkm111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.