Abstract
A method is presented to assemble a gene of interest into a linear DNA template with all the components necessary for in vitro transcription and translation in ∼90 min. Assembly is achieved using a coupled uracil excision–ligation strategy based on USER Enzyme and T4 DNA ligase, which allows the simultaneous and seamless assembly of three different PCR products. The method is suitable for screening and selection systems of very high throughput as up to 1011 molecules can be efficiently assembled and purified in reaction volumes of 100 μl. The method is exemplified with the gene coding for a mutant version of O6-alkylguanine alkyltransferase, which is efficiently assembled with an N-terminal peptide tag and its 5′- and 3′-untranslated regions that include a T7 promoter, ribosome binding site and T7 terminator. The utility of the method is further corroborated by assembling error-prone PCR libraries and regenerating templates following model affinity selections. This fast and robust method should find widespread application in directed evolution for the assembly of gene libraries and the regeneration of linear DNA templates between successive screening and selection cycles.
INTRODUCTION
Directed evolution mimics evolution through natural selection in the test tube with the aim of creating genes that code for proteins with new or improved functional traits (1). For this purpose, a number of in vitro screening and selection systems have been developed over the past 10 years (2–4). Nucleic acid display systems are now routinely used for the directed evolution of protein binders (5–9) and water-in-oil emulsions provide a high-throughput platform for the screening and selection of catalytic activities (10–13) and binding interactions (14–16). Their main advantages over screening and selection systems that depend on steps in vivo, such as genetic screening and selection systems and surface display methods based on phage, yeast or Escherichia coli, are as follows: (i) greater sensitivity, (ii) controllable assay conditions with less interference from cellular functions, (iii) rapid successive selection cycles, (iv) subcloning-independent processing of DNA, (v) flexible in vitro expression systems that facilitate selections with non-natural amino acids and (vi) libraries that are neither limited by toxicity nor transformation efficiencies (2–4).
Between successive in vitro screening and selection cycles, the gene of interest (GOI) is propagated by PCR. Only the diversified region is usually amplified. This is to minimise the formation of non-specific by-products, regenerate sites for binding or conjugation and prevent the accumulation of mutations in constant parts, e.g. in regulatory regions, specialized linkers or fusion genes required for covalent or non-covalent conjugation (6–16). As a result, the GOI needs to be re-assembled into a functional template for in vitro transcription and translation (TS–TL). The preferred assembly methods are either based on overlap extension PCR (9,10) or sequential restriction–digestion and ligation reactions (7,8,12–15). Both strategies are, however, laborious, time-consuming and typically take an entire day to complete—in many cases longer than the selection itself. For instance, restriction–digestion–ligation protocols alone require several hours of sequential incubations. Furthermore, both strategies entail relatively long PCRs featuring 25–30 amplification cycles to amplify the desired template from an overlap extension or ligation reaction (7–10,12–15). While the purity of templates is critical for the success of a screening and selection process, obtaining pure templates after PCR amplification is not necessarily a trivial task in practice (8). Amplification from overlap extension and ligation reactions generates additional non-specific by-products. Therefore templates frequently need to be further purified by agarose gel electrophoresis (7–10,12). These additional steps significantly add to the work-up time and can also have unexpected detrimental effects on downstream applications caused by salt and residual agarose contaminants (7,17,18). Along with time, conventional methods are inherently complex and, therefore, susceptible to accidental errors. For instance, suboptimal template regeneration in one cycle of a multistep selection process may quickly diminish the size of a library by several orders of magnitude, thereby potentially cancelling out the advantage of screening large libraries. Considering the intrinsic amplification power of PCR, suboptimal assembly may not necessarily be obvious to the experimenter at the time, especially since it is generally not possible to monitor the integrity of an assembly process using simple means such as agarose gel electrophoresis, and instead requires more sophisticated methods such as competitive (13) or real-time PCR.
Here, a simple and efficient protocol is presented that allows the assembly of a gene with a peptide tag and its flanking untranslated regions (UTRs) in ∼90 min (Figure 1). Assembly is based on a coupled uracil excision–ligation strategy that is reminiscent of UDG (19–21) and USER enzyme cloning (22–24), but also includes T4 DNA ligase to generate DNA templates devoid of nicks. Pure templates that do not need to be purified further by agarose gel electrophoresis and that are functional for in vitro TS–TL are obtained following a short PCR over 10 amplification cycles. The procedure is exemplified for a mutant of O6-alkylguanine alkyltransferase (AGT) (25), which is assembled with an N-terminal peptide tag and its 5′- and 3′-UTRs that carry all the necessary components for in vitro TS–TL including a T7 promoter, a ribosome-binding site and a T7 terminator. The utility of the method is further corroborated by assembling error-prone PCR (epPCR) libraries and regenerating templates following model affinity selections. The method is suitable for screening and selection systems of very high throughput; up to 1011 molecules can be efficiently assembled and purified in reaction volumes of 100 μl.
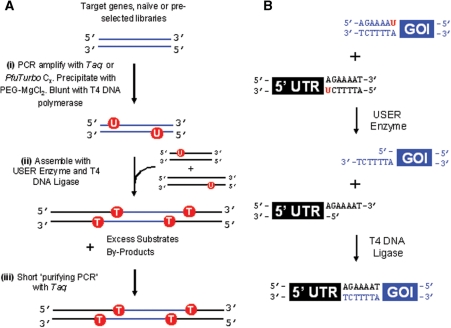
Figure 1.
(A) Assembly Scheme. (i) GOI or a derivative library is amplified with primers that specifically incorporate uracil nucleotides close to both 5′-ends. (ii) Assembly of the GOI with its 5′- and 3′-untranslated regions including any constant protein-coding regions based on a coupled uracil excision–ligation strategy. (iii) Pure templates are obtained following a short-purifying PCR, which effectively ‘removes’ excess substrates and partially assembled intermediates. (B) Mechanism of coupled uracil excision–ligation: first, USER enzyme catalyses the excision of uracil from DNA, thereby leaving a single base pair gap and a 3′-extension provided the 5′-portion can dissociate. Complementary overlapping 3′-extensions then direct the assembly of DNA fragments which are covalently sealed by T4 DNA ligase.
MATERIALS AND METHODS
General
General procedures, reagents, sequences of the oligonucleotides and the fully assembled Avi-AGT gene integrated into a pIVEX backbone are listed in the Supplementary Data.
Preparation of assembly substrates
The assembly substrates (5′-UTR-Avi, 3′-UTR and AGT) were prepared by PCR using BioTaq DNA polymerase. A typical PCR contained 1 × NH4-based reaction buffer [60 mM Tris–HCl, 6 mM (NH4)2SO4, 10 mM KCl, pH 8.3] supplemented with 2 mM MgCl2, 250 μM dNTPs, 2 μM oligonucleotides each (Supplementary Tables S1 and S2), 0.5 ng pIVEX-Avi-AGT and 5 U BioTaq in a volume of 100 μl. After an initial denaturation step for 2 min at 95°C, 30 cycles of PCR were run as follows: 30 s at 95°C, 1 min at 62°C and 1 min at 72°C. This was followed by a final extension step for 1 min at 72°C. To prevent repeated carryover contamination with wild-type DNA from the source plasmid, the PCRs of the flanking regions (5′-UTR-Avi and 3′-UTR) were digested with 40 U DpnI per 50 μl PCR for 20 min at 37°C. Alternatively, AGT was prepared with PfuTurbo Cx. In this case, a typical PCR contained 1 × PfuTurbo Cx reaction buffer, 250 μM dNTPs, 1 μM oligonucleotides each (Supplementary Tables S1 and S2), 0.5 ng pIVEX-Avi-AGT and 5 U PfuTurbo Cx in a volume of 100 μl. After an initial denaturation and heat activation step for 2 min at 95°C, 30 cycles of PCR were run as follows: 30 s at 95°C, 1 min at 62°C and 1.5 min at 72°C. This was followed by a final extension step for 10 min at 72°C.
PEG–MgCl2 precipitation of assembly substrates
To remove any primer dimers and other small non-specific by-products, all assembly substrates were purified by precipitation with PEG–MgCl2 (26). Briefly, a solution of DNA at a final concentration of 25–50 ng/μl in 10 mM MgCl2, 2–5 mM Tris–HC1, pH 8.5 and varying concentrations of PEG-8000 was spun at 16 000g for 10 min. The supernatant was then carefully removed by pipetting and the pellet resuspended in 10 mM Tris–HCl, pH 8.5. Yields were typically 60–70%. For optimal results, the concentration of PEG-8000 was titrated for each DNA fragment: 10% for 3′-UTR of 239 bp, 9.1% for 5′-UTR-Avi of 410 bp and 8.3% for AGT of 638 bp. See Supplementary Data for further details on titrating the optimal concentration of PEG-8000. Where applicable, assembly substrates were blunted with T4 DNA polymerase according to the manufacturer's instructions (NEB).
Coupled uracil excision–ligation
A typical assembly reaction contained 1×T4 DNA ligase buffer, 3 pmol AGT, 4.5 pmol 5′-UTR-Avi, 4.5 pmol 3′-UTR, 3 U USER enzyme (equivalent of 1 U/1 μg DNA) and 600 U T4 DNA ligase (equivalent of 200 U/1 μg DNA) in a volume of 100 μl. The reaction mixture was incubated for 10 min at 37°C. A formula is provided in the Supplementary Data to perform mass-to-molar conversions.
Purifying PCR
Template purification was performed with BioTaq DNA polymerase. A typical amplification reaction contained 1× NH4-based reaction buffer [60 mM Tris–HCl, 6 mM (NH4)2SO4, 10 mM KCl, pH 8.3] supplemented with 2 mM MgCl2, 250 μM dNTPs, 1 μM oligonucleotides LMB-2-6 and pIV-B1 (Supplementary Table S1), 50–500 ng DNA of the assembly reaction as indicated and 5 U BioTaq in a volume of 100 μl. After an initial denaturation step for 2 min at 95°C, 10 PCR cycles were run as follows: 30 s at 95°C, 1 min at 68°C and 2 min at 72°C. For sequencing, individual DNA templates were cloned into a pCR®2.1 vector using the TOPO TA cloning kit according to manufacturer's instructions (Invitrogen).
Template repair
DNA templates were repaired with the PreCR Repair Mix, which contains a cocktail of enzymes that can repair a range of different lesions in vitro. Repair reactions were prepared according to manufacturer's instructions (NEB) and contained 1.2 μg DNA of the assembly reaction in a volume of 150 μl.
Quantifying the assembly efficiency by real-time PCR
The assembly efficiency was quantified by real-time PCR. A typical amplification reaction contained 1× ImmoBuffer supplemented with 2 mM MgCl2, 250 μM dNTPs, 1 μM oligonucleotides LMB-2-6 and pIV-B1 (Supplementary Table S1), SYBR Green I diluted 30 000-fold, 1 pg DNA derived from the assembly reaction and 5 U IMMOLASE (Bioline) in a volume of 100 μl. After an initial heat activation and denaturation step for 10 min at 95°C, 30 PCR cycles were run as follows: 30 s at 95°C, 1 min at 68°C and 2 min at 72°C. The assembly efficiency was quantified using the ‘Comparative Quantification’ feature provided with the Rotor-Gene 6000 analysis software (Corbett). Errors are shown as standard deviations of four technical replicates.
In Vitro TS–TL from assembled DNA templates
DNA templates for in vitro TS–TL were either derived from a purifying PCR (Figure 2, L4 or Supplementary Figure S1, L12) or amplified from plasmid DNA. When templates were directly amplified from plasmid DNA, the PCR was run over 25 cycles with 0.5 ng pIVEX-Avi-AGT serving as template in a reaction volume of 100 μl. Extension times were 1 min 30 s and 3 min for Taq and PfuTurbo Cx respectively. A cell-free expression mix was subsequently prepared on ice according to the manufacturer's instructions (RTS100, E. coli HY kit, Roche Applied Science) with DNA templates at a final concentration of 5 nM. To label functional Avi-AGT, the in vitro TS–TL reaction was also supplemented with O6-benzylguanine-biotin (BG-biotin) at a final concentration of 5 μM. Following expression and conjugation for 4 h at 25°C, 4 μl of the TS–TL reaction were cleaned by acetone precipitation, separated by SDS–PAGE on 12% gels and blotted onto nitrocellulose membranes. Membranes were then transferred into TBS-T (20 mM Tris–HCl, 137 mM NaCl, 0.05% Tween 20, pH 7.5), blocked with 2.5% BSA, washed, primed with streptavidin-horseradish peroxidase (HRP; Sigma, 1:10 000 dilution) and washed. All steps were performed at room temperature under shaking conditions. HRP activity was developed by using ECL detection reagent. Detailed procedures are provided in the Supplementary Data.
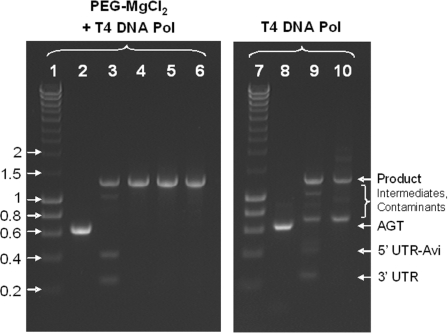
Figure 2.
Efficiency of template assembly by uracil excision–ligation and the purifying PCR for DNA templates prepared with Taq DNA polymerase. Precipitation with PEG–MgCl2 is necessary and sufficient for the efficient assembly. L1, hyperladder I in kilobase pairs; L2, substrate AGT amplified with Taq, precipitated with PEG–MgCl2 and blunted with T4 DNA polymerase; L3, assembly of L2 with its UTRs + Avi-tag; L4, purifying PCR of L3 with 50 ng DNA per 100 μl PCR; L5, purifying PCR of L3 with 250 ng DNA per 100 μl PCR; L6, purifying PCR of L3 with 500 ng DNA per 100 μl PCR; L7, hyperladder I in kb; L8, substrate AGT amplified with Taq and blunted with T4 DNA polymerase; L9, assembly of L8 with its UTRs + Avi-tag; L10, purifying PCR of L9 with 50 ng DNA per 100 μl PCR. Every sample lane contains ∼300 ng DNA.
Assembly of epPCR libraries
epPCR libraries of AGT were prepared with the GeneMorph II Random Mutagenesis Kit. Briefly, a typical PCR contained 1× Mutazyme II reaction buffer, 200 μM dNTPs each, 0.5 μM oligonucleotides Fw-AGT and Rw-AGT (Supplementary Table S1), 2.5 U Mutazyme II DNA polymerase and 20 ng pIVEX-Avi-AGT in a volume of 50 μl. After an initial denaturation and heat activation step for 2 min at 95°C, 30 cycles of PCR were run as follows: 30 s at 95°C, 1 min at 62°C and 1 min at 72°C. This was followed by a final extension step for 10 min at 72°C. The library was then purified with QIAquick PCR purification kit and subjected to a DpnI digest according to manufacturer's instructions (NEB). A portion of the library (50 ng or 7 × 1010 templates in a volume of 100 μl) was then re-amplified with Taq DNA polymerase under the same conditions described earlier to prepare suitable substrates for the assembly process except that amplification featured 25 cycles instead of 30. The re-amplified library was then precipitated with PEG–MgCl2, blunted with T4 DNA polymerase, assembled and purified by PCR. For sequencing, individual DNA templates were cloned into a pCR®2.1 vector using the TOPO TA cloning kit according to manufacturer's instructions (Invitrogen).
Model affinity selections
Model affinity selections were performed as previously described with slight modifications to accommodate for the selection of biotinylated protein–DNA conjugates (27). Detailed procedures are provided in the Supplementary Data.
RESULTS
Assembly strategy
A template regeneration strategy is presented that overcomes many of the practical shortcomings of the existing assembly strategies (Figure 1). The GOI is assembled into a functional template for in vitro TS–TL with USER Enzyme (22–24) and T4 DNA ligase. USER Enzyme is a commercially available enzyme mixture composed of uracil DNA glycolyase (UDG) and endonuclease VIII. In combination, the two enzymes excise uracil nucleotides from DNA thereby leaving a single base pair gap. If a uracil residue is incorporated close to the 5′-end of a linear DNA fragment by PCR and the resulting 5′-oligonucleotide can efficiently dissociate, single-stranded 3′-extensions are generated that subsequently direct the assembly of DNA fragments through complementary, overlapping ends. In the presence of T4 DNA ligase the assembled fragments are then covalently sealed which makes the desired product template amenable to amplification by PCR.
Assembly and PCR purification
Coupled uracil excision–ligation is efficient as the majority of substrate DNA fragments assembled into the desired product in ∼10 min (Figure 2, L3, Supplementary Figure S1). Suitable substrate DNA fragments with a uracil residue near the 5′-end were either prepared with Taq or PfuTurbo Cx (28). These constitute the only two commercially available thermostable DNA polymerases that can efficiently replicate uracil-containing DNA templates (24,28). For efficient assembly, it was crucial that templates were treated with T4 DNA polymerase in order to remove the 3′-adenine overhangs generated by the extendase activity of Taq. While PfuTurbo Cx generates blunt ends, we observe variable results in the assembly efficiency so that it may become necessary to blunt the assembly substrates with T4 DNA polymerase (Supplementary Data and Figure S1).
No further improvements could be achieved by lowering the temperature, higher quality oligonucleotides or increasing the reaction time and the amount of assembly substrates (data not shown). Similarly, it is unclear to what extent the assembled DNA templates are affected by gaps caused by singly recessed 3′-ends or non-specific uracil residues arising from oxidative damage such as heat induced conversion of cytidine to uridine (29). For instance, if templates are solely propagated by PCR in vitro, damaged DNA cannot be repaired by the bacterial repair machinery following transformation in vivo (22–24). Gaps and other types of DNA damage could thus significantly reduce the number of full-length DNA templates in vitro. This is not unlikely, especially if one considers that the assembly reactions performed here (Figure 2, L3 and Supplementary Figure S1) and elsewhere (22) were generally incomplete.
The desired product template was, therefore, purified through a short PCR run over 10 amplification cycles, which afforded clean templates in ∼30 min (Figure 2, L4-6). A typical PCR contained a total of 50–500 ng DNA in a reaction volume of 100 μl. Given that the desired product contains 25–33% of all DNA (Figure 2, L3), this is equivalent to 1010–1011 template molecules and constitutes a suitable library size for the majority of in vitro screening and selection systems. Amplification over more than 10 PCR cycles is not advisable as this may result in additional non-specific by-products, especially primer dimers (data not shown). To exclude that the assembled DNA templates were significantly affected by single base pair gaps caused by non-specific uracil residues, the assembly efficiency was additionally quantified by real-time PCR. For the purpose of quantification, the assembly reactions (Figure 2, L3 and Supplementary Figure S1, L3) were amplified in parallel with an equal amount of a full-length template that served as a comparative calibrator. The assembly efficiency was determined as 27 and 22% for substrates prepared with Taq and PfuTurbo Cx respectively. This is in good agreement with the assembly efficiency of 25–33% estimated from ethidium bromide stained agarose gels (Figure 2, L3 and Supplementary Figure S1, L3). Subjecting the entire assembly mix to repair with the PreCR kit prior to PCR amplification subsequently increased the portion of full-length templates in the assembly reaction to 34 and 29% for Taq and PfuTurbo Cx, respectively. This suggests that single base pair gaps generated after the excision of non-specific uracil residues and other lesions do occur, but ultimately do not impair the assembly process on a scale that would significantly affect the diversity of a library. The integrity of individual DNA templates was confirmed by sequencing six clones of each purifying PCR (Figure 2, L4–6). Assembly substrates prepared with Taq DNA polymerase contained on an average one mutation in the protein-coding region comprising 624 base pairs. No mutations were found if assembly substrates were prepared with PfuTurbo Cx (Supplementary Figure S1, L4).
In vitro TS–TL from assembled DNA templates
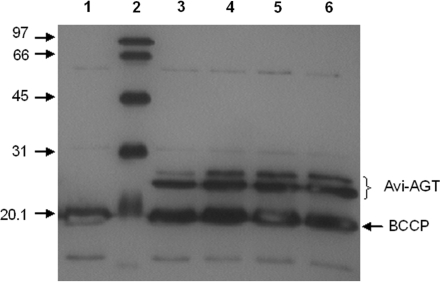
To test whether the newly assembled DNA templates were functional for in vitro TS–TL, Avi-AGT was either expressed from DNA templates that have been amplified from assembly reactions or plasmid DNA (Figure 3). To detect functional Avi-AGT, the in vitro TS–TL reaction was supplemented with BG-biotin, a suicide inhibitor of AGT which irreversibly reacts with the active site cysteine of AGT and by that covalently labels the protein with biotin (27,30). No difference in the functional expression of Avi-AGT could be detected between templates that have been amplified from assembly reactions or plasmid DNA (Figure 3). Similarly, the type of DNA polymerase made no difference in the functional expression levels either. Regarding the two bands that were observed in the functional expression test, we could exclude this to be a result of linker proteolysis since biotinylation of the Avi-tag with biotin ligase (as opposed to labelling of the active site with BG-biotin) similarly generated two distinct bands (Supplementary Data and Figure S2).
Figure 3.
In vitro expression tests measuring the activity of Avi-AGT following active site dependent labelling of AGT with BG-biotin. DNA templates were either amplified from assembly reactions or plasmid DNA. L1, no template control; L2, biotinylated protein marker; L3, amplified with Taq from plasmid DNA; L4, amplified with Taq from an assembly reaction; L5, amplified with PfuTurbo Cx from plasmid DNA; L6, amplified with PfuTurbo Cx from an assembly reaction. The band at ∼20 kDa corresponds to biotin carboxyl carrier protein. Linker proteolysis can be excluded as a cause for the two bands since biotinylation of the Avi-tag with biotin ligase similarly generated two distinct bands (Supplementary Data and Figure S2).
Assembly of epPCR libraries
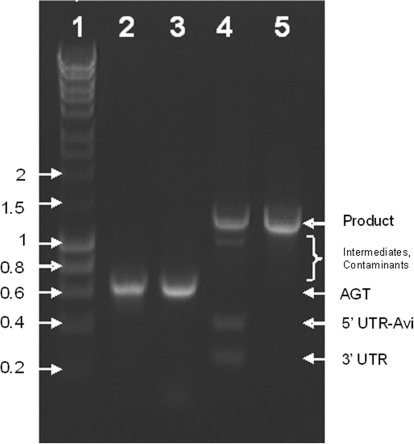
To test the method under more demanding conditions, the procedure was applied to assemble epPCR libraries of AGT (Figure 4). epPCR libraries were initially prepared with the Genemorph II kit (Figure 4, L2). A portion of the library (50 ng equivalent to a diversity of 7 × 1010) was then re-amplified with Taq DNA polymerase to generate suitable assembly substrates with uracil residues near the 5′-ends (Figure 4, L3). This additional step became necessary because amplification with uracil-containing primers yielded no PCR product in the first place (data not shown). Following precipitation with PEG–MgCl2 to remove residual primer dimers, the re-amplified library was assembled into the desired DNA template and purified over a short PCR run (Figure 4, L4-5). Sequencing of the library confirmed the integrity of the library with an average of five mutations over the 574 bp that were subject to mutagenesis, and none of the clones containing fewer than three mutations.
Figure 4.
Assembly of an epPCR library. L1, hyperladder I in kilobase pairs;. L2, epPCR library prepared with the Genemorph II kit; L3, epPCR library of L2 re-amplified with Taq DNA polymerase and uracil-containing primers; L4, assembly of L3 with its UTRs+Avi-tag after it has been precipitated with PEG–MgCl2 and blunted with T4 DNA polymerase; L5, purifying PCR of L4 with 50 ng DNA per 100 μl PCR. Every sample lane contains ∼300 ng DNA.
Assembly of DNA templates following model affinity selections
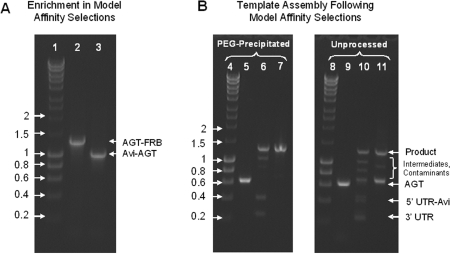
We also demonstrate that the method can be applied to regenerate DNA templates following model affinity selections (Figure 5). For this purpose, BG labelled DNA templates coding for Avi-AGT and AGT-FRB (FKBP12-rapamycin-binding domain) were mixed at a ratio of 1:250 and then compartmentalized along with an in vitro TS–TL reaction in a water-in-oil emulsion (13,27). Following expression and conjugation, the resulting protein–DNA conjugates were then extracted from the emulsion and the templates coding for Avi-AGT enriched on streptavidin-coated paramagnetic microbeads. This was followed by recovering the DNA in the supernatant and microbead fraction by PCR using primers that annealed to both templates (Figure 5, L2-3). DNA coding for Avi-AGT could only be detected in the microbead fraction (Figure 5, L3) and not in the supernatant (Figure 5, L2); this confirmed the fidelity of the selection process along with a >250-fold enrichment of Avi-AGT over AGT-FRB. In parallel, only the gene coding for AGT was recovered from the microbead fraction using primers that were specific to AGT. PCR products were obtained with both types of polymerases, PfuTurbo Cx and Taq (Figure 5, L5, Supplementary Figure S3, L2), and could subsequently be regenerated into their full-length DNA templates (Figure 5, L6-7, Supplementary Figure S3, L3-4). Once again, precipitation of the assembly substrate with PEG–MgCl2 proved necessary and sufficient for the specificity of the assembly process and subsequent purifying PCR (Figure 5, L9-11 and Supplementary Figure S3, L6-8). On this occasion, we also noticed that the protein–DNA conjugates did not have to be eluted from the microbeads as PCR amplification with PfuTurbo Cx remained highly functional in the presence of relatively high concentrations of microbeads (∼6.7 × 107 M-270 streptavidin coated paramagnetic microbeads per 1 ml PCR). This is surprising considering that paramagnetic microbeads are generally thought to interfere with the performance of PCR. Similarly, we found that the efficiency of PCR amplification with Taq DNA polymerase can be significantly improved by including β-mercaptoethanol at a final concentration of 20 mM in the amplification reaction.
Figure 5.
(A) The enrichment of Avi-AGT DNA in model affinity selections relative to a non-binding, control template coding for AGT-FRB was >250-fold. L1, hyperladder I in kilobase pairs; L2, PCR amplification of the supernatant; L3, PCR amplification of the bead fraction. (B) Assembly of recovered DNA fragments; L4, hyperladder I in kilobase pairs; L5, substrate AGT recovered from the bead fraction with PfuTurbo Cx, precipitated with PEG–MgCl2 and blunted with T4 DNA polymerase; L6, assembly of L5 with its UTRs + Avi-tag. L7: purifying PCR of L6 with 50 ng DNA per 100 μl PCR; L8, hyperladder I in kilobase pairs; L9, substrate AGT recovered from the bead fraction with PfuTurbo Cx and blunted with T4 DNA polymerase; L10, assembly of L9 with its UTRs + Avi-tag; L11, purifying PCR of L10 with 50 ng DNA per 100 μl PCR. Every sample lane contains ∼300 ng DNA.
DISCUSSION
A procedure was developed that allows the assembly of a GOI into a linear DNA template with all the components necessary for in vitro TS–TL. Assembly is achieved using a coupled uracil excision–ligation strategy based on USER enzyme and T4 DNA ligase, which allows the simultaneous and seamless assembly of three different PCR products with a minimal number of work-up steps by that conferring simplicity, robustness and speed to the method. For its success, several factors proved critical.
To achieve a high efficiency of assembly and efficient PCR purification, it was critical to purify all substrates by PEG–MgCl2 precipitation prior to assembly (24). Otherwise, primer dimers and other small non-specific by-products that could not readily be visualized with ethidium bromide interfered with the assembly process (Figure 2, L8-10). Precipitation with PEG–MgCl2 constitutes an inexpensive and fast method to purify PCR products. Size dependence can easily be exerted by titrating the optimal concentration of PEG-8000 for a given DNA fragment even though its resolution is less stringent compared to gel purification. Nonetheless, precipitation with PEG–MgCl2 proved necessary and sufficient in this case to generate clean substrates for efficient assembly through uracil excision–ligation.
Despite its widespread applicability to any assembly procedure that features PCR products including overlap extension PCR and restriction–digestion–ligation, precipitation with PEG–MgCl2 is virtually unused in practice. Our results emphasize that this step is critical and, if applied prior to the assembly step, overcomes the need to gel purify DNA templates after the assembly or purifying PCR steps: first of all, it saves time and labour as precipitation can be completed in ∼10–15 min as opposed to several hours needed to separate and extract DNA fragments from agarose gels. Furthermore, it prevents the GOI from becoming damaged following exposure to UV light during gel excision and pre-empts any inhibiting effects that contaminants such as residual agarose and salts may have on downstream reactions (7,17,18). For instance, the majority of commercial in vitro TS–TL systems explicitly advise not to purify DNA templates by agarose gel electrophoresis (17,18) Similarly, it has been reported in the literature that templates are not sufficiently clean for in vitro TS–TL following excision from agarose gels using commercial purification kits and generally need to be further purified by ethanol precipitation or include additional washing steps, e.g. with washing buffer during purification with the QIAquick kit (7). Similarly, we found that the efficiency of PCR is markedly reduced if the DNA was gel purified prior to amplification in the absence of additional purification steps (data not shown).
If a PCR is particularly prone to non-specific by-products that cannot easily be purified by precipitation with PEG–MgCl2, it may be necessary to perform two successive nested PCRs (12): a standard PCR to recover the desired DNA template followed by a short second run over 10 cycles to generate the assembly substrate with two uracil residues. This may particularly apply in real selections where very few DNA fragments serve as templates and the amplification efficiency of the target amplicon is compromised by greater competition from non-specific amplification reactions.
In terms of sequence requirements, template assembly through uracil excision–ligation only imposes minimal restrictions on the splice sites: only an adenine and a thymidine spaced apart by several nucleotides in the 5′→3′ direction to ensure efficient dissociation of the excised regions are required (22–24). In the context of directed evolution, the procedure allows rapid and flexible variation of the diversified regions saving laborious subcloning steps when different regions of a gene are targeted and no additional restriction sites need to be introduced. In practice it should be considered that all splice sites need to differ by at least two base pairs for a complementary overlap of 5–6 bp to prevent single-stranded extensions from cross-hybridizing (data not shown). Equally, it must be ensured that the single-stranded extensions cannot fold onto themselves to prevent the formation of covalently closed loops. Calculating the base pair probabilities of the single-stranded extensions with a suitable folding programme gives a good indication if secondary structures pose a problem, e.g. with the RNA-fold web server (http://rna.tbi.univie.ac.at/) which conveniently displays base pair probabilities as a dot plot (31,32).
In comparison, template assembly procedures based on overlap extension PCR or restriction–digestion and ligation take significantly longer—up to an entire day as opposed to ∼90 min. This can be attributed to sequential restriction–digestion and ligation reactions, and relatively long overlap extension and purifying PCRs featuring 25–30 cycles. Furthermore, the purifying PCR frequently generates non-specific fragments which require the desired DNA template to be purified by agarose gel electrophoresis (7–10,12). The reason for the purifying PCR frequently being ineffective is presumably a consequence of the reduced efficiency of the overlap extension and ligation reactions. For instance in ligation reactions, template circularization faces additional competition from template concatemerization. Furthermore, the palindromic nature of the single-stranded extensions generated by the majority of restriction enzymes enable DNA fragments to form homodimers which can be—just as the desired product—exponentially amplified by PCR when they are formed between the flanking regions or the plasmid backbone. In addition, the efficiency of restriction–digestion and ligation strategies is often limited by the idiosyncrasies of individual restriction enzymes, which can cause unexpected problems such as inefficient digestion at restriction sites located near the end of a linear DNA fragment (33). In overlap extension PCR, a qualitative analysis of DNA hybridization kinetics suggests that even under perfect conditions in the absence of any secondary structures only a fraction of templates recombines in a given thermal cycle. For instance, the hybridisation half lives for an overlap of 25 bp with both assembly substrates present at a concentration of 10 nM is on the order of 6–7 min (34,35). This compares to recombination times of ≤1 min that feature in most, if not all, overlap extension PCR protocols that have been published in textbooks (36,37), commercial product manuals (38) and scientific publications that either specifically deal with the subject of overlap extension PCR (39) or apply it in the context of directed evolution to assemble and regenerate libraries (9,10). In fact, it is unclear to what extent overlap extension PCR is suitable to assemble gene libraries. For instance, templates that by chance recombine in an earlier thermal cycle will also enter exponential amplification earlier, and thus increase in abundance so that templates are not uniformly amplified. If the size of a library then exceeds the recombination efficiency so that stochastic hybridization events cannot be averaged out for subpopulations of identical mutants, the library will become randomly biased. This makes the enrichment less dependent on a functional trait and may even reduce the diversity of a library as a portion of mutants can be eliminated during the assembly process.
In some cases, the entire template has been amplified after selection so that template reassembly is unnecessary in the first place (5,16). This strategy is, however, not broadly applicable; e.g. it has been reported that PCR amplification is inefficient if identical set of primers are used to prepare templates and recover genes after selection (7). This can be attributed to the partial, exonucleolytic degradation of the ends either by the 3′→5′ exonuclease activity of a proof-reading polymerase or due to any exonuclease activities in the cell extract (7,13). Successive nested PCRs of the whole template are equally impractical; e.g. if synthetic modifications are introduced (5,9,27), each set of nested oligonucleotides needs to be modified separately. Depending on the type of modification, this may significantly add to the costs. Furthermore, PCR amplification of the whole template is expected to accumulate mutations in constant parts, particularly in the coding regions of long and essential fusion genes, which can significantly compromise the performance of a screening and selection process (9). An exception relates to streptavidin that forms the basis of a non-covalent DNA display system (5) and has also been the subject of evolutionary optimization to improve binding towards a biotin analogue (16). In both cases, the entire streptavidin gene was amplified for at least five successive selection cycles with no detrimental effect on the selection process suggesting that streptavidin can tolerate this level of mutation. This may, however, be a protein-specific effect and does not necessarily apply to systems that rely on less robust proteins.
In summary, the assembly protocol presented here should be applicable to many different in vitro screening and selection systems, especially those that feature relatively long fusion genes that are susceptible to mutations and thus rely on nested PCRs. The protocol saves time, labour and enables many rapid successive selection cycles. This is highly practical when mimicking evolutionary processes such as genetic drifts.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
EU NEST project MiFem; an EPSRC Platform grant; financial support to Trinity College, Cambridge and the BBSRC (to V.S.). F.H. is an European Research Council (ERC) Starting Investigator. Funding for open access charge: EU NEST MiFem.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Yolanda Schärli for helpful advice with experimental methods and comments on the manuscript. We would also like to thank the reviewers for their helpful comments and suggestions.
REFERENCES
- 1.Jackel C, Kast P, Hilvert D. Protein design by directed evolution. Annu. Rev. Biophys. 2008;37:153–173. doi: 10.1146/annurev.biophys.37.032807.125832. [DOI] [PubMed] [Google Scholar]
- 2.Leemhuis H, Stein V, Griffiths AD, Hollfelder F. New genotype-phenotype linkages for directed evolution of functional proteins. Curr. Opin. Struct. Biol. 2005;15:472–478. doi: 10.1016/j.sbi.2005.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Aharoni A, Griffiths AD, Tawfik DS. High-throughput screens and selections of enzyme-encoding genes. Curr. Opin. Chem. Biol. 2005;9:210–216. doi: 10.1016/j.cbpa.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 4.Lipovsek D, Pluckthun A. In-vitro protein evolution by ribosome display and mRNA display. J. Immunol. Method. 2004;290:51–67. doi: 10.1016/j.jim.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 5.Yonezawa M, Doi N, Kawahashi Y, Higashinakagawa T, Yanagawa H. DNA display for in vitro selection of diverse peptide libraries. Nucleic Acids Res. 2003;31:e118. doi: 10.1093/nar/gng119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Odegrip R, Coomber D, Eldridge B, Hederer R, Kuhlman PA, Ullman C, FitzGerald K, McGregor D. CIS display: In vitro selection of peptides from libraries of protein-DNA complexes. Proc. Natl Acad. Sci. USA. 2004;101:2806–2810. doi: 10.1073/pnas.0400219101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reiersen H, Lobersli I, Loset GA, Hvattum E, Simonsen B, Stacy JE, McGregor D, Fitzgerald K, Welschof M, Brekke OH, et al. Covalent antibody display—an in vitro antibody-DNA library selection system. Nucleic Acids Res. 2005;33:e10. doi: 10.1093/nar/gni010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zahnd C, Amstutz P, Pluckthun A. Ribosome display: selecting and evolving proteins in vitro that specifically bind to a target. Nat. Method. 2007;4:269–279. doi: 10.1038/nmeth1003. [DOI] [PubMed] [Google Scholar]
- 9.Bertschinger J, Grabulovski D, Neri D. Selection of single domain binding proteins by covalent DNA display. Protein Eng. Des. Sel. 2007;20:57–68. doi: 10.1093/protein/gzl055. [DOI] [PubMed] [Google Scholar]
- 10.Doi N, Kumadaki S, Oishi Y, Matsumura N, Yanagawa H. In vitro selection of restriction endonucleases by in vitro compartmentalization. Nucleic Acids Res. 2004;32:e95. doi: 10.1093/nar/gnh096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen HM, Tawfik DS, Griffiths AD. Altering the sequence specificity of HaeIII methyltransferase by directed evolution using in vitro compartmentalization. Protein Eng. Des. Sel. 2004;17:3–11. doi: 10.1093/protein/gzh001. [DOI] [PubMed] [Google Scholar]
- 12.Griffiths AD, Tawfik DS. Directed evolution of an extremely fast phosphotriesterase by in vitro compartmentalization. EMBO J. 2003;22:24–35. doi: 10.1093/emboj/cdg014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller OJ, Bernath K, Agresti JJ, Amitai G, Kelly BT, Mastrobattista E, Taly V, Magdassi S, Tawfik DS, Griffiths AD. Directed evolution by in vitro compartmentalization. Nat. Method. 2006;3:561–570. doi: 10.1038/nmeth897. [DOI] [PubMed] [Google Scholar]
- 14.Fen CX, Coomber DW, Lane DP, Ghadessy FJ. Directed evolution of p53 variants with altered DNA-binding specificities by in vitro compartmentalization. J. Mol. Biol. 2007;371:1238–1248. doi: 10.1016/j.jmb.2007.05.099. [DOI] [PubMed] [Google Scholar]
- 15.Bernath K, Magdassi S, Tawfik DS. Directed evolution of protein inhibitors of DNA-nucleases by in vitro compartmentalization (IVC) and nano-droplet delivery. J. Mol. Biol. 2005;345:1015–1026. doi: 10.1016/j.jmb.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 16.Levy M, Ellington AD. Directed evolution of streptavidin variants using in vitro compartmentalization. Chem. Biol. 2008;15:979–989. doi: 10.1016/j.chembiol.2008.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roche-Applied-Science. Mannheim: 2007. Manual for RTS 100 E. coli, HY Kit. [Google Scholar]
- 18.Invitrogen. Carlsbad, CA: 2006. Manual for Expressway Cell-Free E. coli, Expression System. [Google Scholar]
- 19.Nisson PE, Rashtchian A, Watkins PC. Rapid and efficient cloning of Alu-PCR products using uracil DNA glycosylase. PCR Methods Appl. 1991;1:120–123. doi: 10.1101/gr.1.2.120. [DOI] [PubMed] [Google Scholar]
- 20.Smith C, Day PJ, Walker MR. Generation of cohesive ends on PCR products by UDG-mediated excision of dU, and application for cloning into restriction digest-linearized vectors. PCR Methods Appl. 1993;2:328–332. doi: 10.1101/gr.2.4.328. [DOI] [PubMed] [Google Scholar]
- 21.Rashtchian A. Novel methods for cloning and engineering genes using the polymerase chain reaction. Curr. Opin. Biotechnol. 1995;6:30–36. doi: 10.1016/0958-1669(95)80006-9. [DOI] [PubMed] [Google Scholar]
- 22.Bitinaite J, Rubino M, Varma KH, Schildkraut I, Vaisvila R, Vaiskunaite R. USER friendly DNA engineering and cloning method by uracil excision. Nucleic Acids Res. 2007;35:1992–2002. doi: 10.1093/nar/gkm041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Geu-Flores F, Nour-Eldin HH, Nielsen MT, Halkier BA. USER fusion: a rapid and efficient method for simultaneous fusion and cloning of multiple PCR products. Nucleic Acids Res. 2007;35:e55. doi: 10.1093/nar/gkm106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nour-Eldin HH, Hansen BG, Norholm MH, Jensen JK, Halkier BA. Advancing uracil-excision based cloning towards an ideal technique for cloning PCR fragments. Nucleic Acids Res. 2006;34:e122. doi: 10.1093/nar/gkl635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gronemeyer T, Chidley C, Juillerat A, Heinis C, Johnsson K. Directed evolution of O6-alkylguanine-DNA alkyltransferase for applications in protein labeling. Protein Eng. Des. Sel. 2006;19:309–316. doi: 10.1093/protein/gzl014. [DOI] [PubMed] [Google Scholar]
- 26.Paithankar KR, Prasad KS. Precipitation of DNA by polyethylene glycol and ethanol. Nucleic Acids Res. 1991;19:1346. doi: 10.1093/nar/19.6.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stein V, Sielaff I, Johnsson K, Hollfelder F. A covalent chemical genotype-phenotype linkage for in vitro protein evolution. Chembiochem. 2007;8:2191–2194. doi: 10.1002/cbic.200700459. [DOI] [PubMed] [Google Scholar]
- 28.Fogg MJ, Pearl LH, Connolly BA. Structural basis for uracil recognition by archaeal family B DNA polymerases. Nat. Struct. Biol. 2002;9:922–927. doi: 10.1038/nsb867. [DOI] [PubMed] [Google Scholar]
- 29.Hogrefe HH, Hansen CJ, Scott BR, Nielson KB. Archaeal dUTPase enhances PCR amplifications with archaeal DNA polymerases by preventing dUTP incorporation. Proc. Natl Acad. Sci. USA. 2002;99:596–601. doi: 10.1073/pnas.012372799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat. Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 31.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.SantaLucia J., Jr. A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl Acad. Sci. USA. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Russel DW. Molecular Cloning—A Laboratory Manual, Vol. 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. pp. 8.37–38.41. [Google Scholar]
- 34.Wetmur JG. DNA probes: applications of the principles of nucleic acid hybridization. Crit. Rev. Biochem. Mol. Biol. 1991;26:227–259. doi: 10.3109/10409239109114069. [DOI] [PubMed] [Google Scholar]
- 35.Young BD, Anderson MLM. In: Nucleic Acid Hybridization: A Practical Approach. Hames BD, Higgins SJ, editors. Oxford: IRL Press; 1985. pp. 47–71. [Google Scholar]
- 36.Georgescu R, Bandara G, Sun L. In: Directed Evolution Library Creation—Methods and Protocols, Arnold FH, Georgiou G, editors. Vol. 231. Totowa, NJ: Humana Press; 2003. pp. 75–83. [Google Scholar]
- 37.Sambrook J, Russel DW. Molecular Cloning—A Laboratory Manual, Vol. 2. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. pp. 13.36–13.39. [Google Scholar]
- 38.Roche-Applied-Science. Mannheim: 2001. Manual for RTS E. coli, Linear Template Generation Set. [Google Scholar]
- 39.Shevchuk NA, Bryksin AV, Nusinovich YA, Cabello FC, Sutherland M, Ladisch S. Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Res. 2004;32:e19. doi: 10.1093/nar/gnh014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.