Abstract
RNA aptamers against bovine prion protein (bPrP) were obtained, most of the obtained aptamers being found to contain the r(GGAGGAGGAGGA) (R12) sequence. Then, it was revealed that R12 binds to both bPrP and its β-isoform with high affinity. Here, we present the structure of R12. This is the first report on the structure of an RNA aptamer against prion protein. R12 forms an intramolecular parallel quadruplex. The quadruplex contains G:G:G:G tetrad and G(:A):G:G(:A):G hexad planes. Two quadruplexes form a dimer through intermolecular hexad–hexad stacking. Two lysine clusters of bPrP have been identified as binding sites for R12. The electrostatic interaction between the uniquely arranged phosphate groups of R12 and the lysine clusters is suggested to be responsible for the affinity of R12 to bPrP. The stacking interaction between the G:G:G:G tetrad planes and tryptophan residues may also contribute to the affinity. One R12 dimer molecule is supposed to simultaneously bind the two lysine clusters of one bPrP molecule, resulting in even higher affinity. The atomic coordinates of R12 would be useful for the development of R12 as a therapeutic agent against prion diseases and Alzheimer's disease.
INTRODUCTION
Prions are infectious particles and are composed exclusively of misfolded proteins, being devoid of nucleic acids. The prion protein (PrP) exhibits two forms; a normal cellular form (PrPC), which is a soluble α-helix-rich form, and an abnormal form (PrPSc), which is an insoluble β-sheet-rich form. PrPC is almost ubiquitously expressed and highly conserved in mammals, being anchored on the surface of cells. The conformational change from PrPC to PrPSc is thought to be crucial in prion pathogenesis, causing diseases such as a Creutzfeldt–Jacob disease (CJD) in humans, bovine spongiform encephalopathy (BSE) in cattle and scrapie in sheep (1–3). The detailed mechanism of the conformational conversion remains unknown.
Specific-anti-PrP probes, which exhibit high specificity and sensitivity, are required for the diagnosis of prion diseases. Study on the binding of these probes with PrP would facilitate elucidation of the mechanism of the conformational conversion. Furthermore, these probes may be utilized for therapeutic purposes. An RNA aptamer is an RNA molecule that can bind a target with high affinity and specificity like an antibody does. It was reported that some aptamer can significantly reduce PrPSc formation, which suggests the utility of an RNA aptamer as a therapeutic tool against prion diseases (4).
It was reported very recently that PrPC is a receptor of amyloid-β-oligomers (5). The accumulation of insoluble plaque containing the amyloid-β peptide is a pathological hallmark of Alzheimer's disease. Prefibrillar, soluble oligomers of amyloid-β have been recognized to be early and key intermediates in Alzheimer's-disease-related synaptic dysfunction (6–9). The amyloid-β-oligomers have been found to bind to PrPC (5). The interaction does not require the PrPSc conformation. The blockade of long-term potentiation by amyloid-β oligomers is absent in hippocampal slices from adult PrP null mice. Thus, PrPC is suggested to be a mediator of amyloid-β oligomer-induced synaptic dysfunction. It has been found that anti-PrP antibodies prevent amyloid-β oligomer binding to PrPC and rescue synaptic plasticity in hippocampal slices from oligomeric amyloid-β (5). This implies that PrPC-specific pharmaceutics may have therapeutic potential against Alzheimer's disease. Therefore, an RNA aptamer against PrP may also function as a therapeutic agent against Alzheimer's disease.
RNA aptamers against a bovine prion protein (bPrP) were obtained by means of an in vitro selection method (10,11) from RNA pools containing a 55-nt randomized region (12). The obtained RNA aptamers showed high affinity to both bPrP and its amyloidogenic β isoform (bPrP-β). It is well established that bPrP-β resembles PrPSc in terms of structural and biochemical properties (13). It was demonstrated that the RNA aptamer can specifically detect PrPC in a bovine brain homogenate on northwestern blotting assay (12). This detection has been conventionally accomplished with an immunoblotting assay involving an antibody. These RNA aptamers each comprise a four tandem repeat of the r(GGA) sequence, r(GGAGGAGGAGGA). Mutagenic studies indicated that the four tandem repeat is critical for specific binding of bPrP and bPrP-β. It was further demonstrated that the r(GGAGGAGGAGGA) molecule (R12) can bind bPrP and bPrP-β with high affinity, the dissociation constants being 8.5 and 280 nM, respectively (12). The DNA counterpart molecule, d(GGAGGAGGAGGA) (D12), can also bind bPrP, but the affinity is weaker by one order. The affinity of D12 to bPrP-β is also weaker than that of R12 (12).
It was shown that DNAs containing a repeat of the d(GGA) sequence form unique quadruplex structures. Four d(GGAGGA) strands form a quadruplex dimer composed of two G:G:G:G tetrad planes and two G(:A):G:G(:A):G hexad ones (14). Two d(GGAGGAGGAGGA) strands form a quadruplex dimer composed of two G:G:G:G tetrad planes and two G(:A):G(:A):G(:A):G heptad ones (15). One d(GGAGGAGGAGGAGGAGGAGGAGGA) strand forms a unimolecular quadruplex structure composed of two G:G:G:G tetrad planes and two G(:A):G(:A):G(:A):G heptad ones (16). It was also shown that two r(GGAGGUUUUGGAGG) strands form a quadruplex dimer composed of two G:G:G:G tetrad planes and two G(:A):G:G(:A):G hexad ones (17). These studies suggest that the four tandem repeat of the r(GGA) sequence present in RNA aptamers also forms a unique quadruplex structure. In fact, the formation of a quadruplex structure was suggested for R12 on the basis of its CD spectrum (12), although the detailed structure has not been elucidated. The unique quadruplex structure may be critically utilized for the recognition of bPrP and bPrP-β by these aptamers. Here, we have determined the structure of R12 by nuclear magnetic resonance (NMR). One R12 molecule forms a parallel quadruplex with a G:G:G:G tetrad plane and a G(:A):G:G(:A):G hexad one. Then, two R12 quadruplexes form a dimer in a tail-to-tail manner through hexad–hexad stacking. The R12 structure shares several common features with the structure formed by the DNA counterpart molecule, d(GGAGGAGGAGGA) (D12). Remarkable differences between the R12 and D12 structures have also been noted. The binding of R12 to a series of peptide fragments of bPrP was examined by means of microchip electrophoresis (ME), and two lysine clusters of bPrP were identified as binding sites. The combination of elucidation of the R12 structure and identification of binding sites has suggested that the affinity of R12 to bPrP is brought about by electrostatic and stacking interactions. One R12 dimer molecule may simultaneously bind the two lysine clusters of one bPrP molecule, which would increase the affinity further. The origin of the more preferable binding of R12 to bPrPC than to bPrP-β has been discussed. The potential of R12 as a starting molecule for therapeutic agents against prion diseases and Alzheimer's disease is addressed.
MATERIALS AND METHODS
Sample preparation and NMR spectroscopy
An RNA 12-mer (R12), r(GGAGGAGGAGGA), synthesized and purified by HPLC, was purchased (Nippon Seihun). R12 was dissolved in a solution comprising 100 mM KCl, 10 mM K-phosphate (pH 6.2) and 3 mM NaN3. The R12 concentration was 1.0 mM. DSS was used as an internal chemical shift reference. The sample was heated at 95°C for 5 min, followed by gradual cooling to room temperature prior to the measurements.
NMR spectra were recorded at 5 and 30°C with Bruker DRX600 and DRX800 spectrometers equipped with a cryoprobe with a Z-gradient. The following NMR experiments were performed to assign the resonances, and to obtain distance and dihedral angle constraints: NOESY, TOCSY, DQF-COSY, 1H-13C HSQC and JRHMBC (18). Spectra were processed and analyzed with XWIN-NMR/TopSpin (Bruker), NMRPipe (19), Capp/Pipp/Stapp (20) and Sparky (21).
Distance and dihedral angle constraints
Interproton distances were calculated from NOESY spectra with mixing times of 80 and 230 ms, as described previously (15–17,22), using the H2′–H3′ distance, which is almost constant for any conformation, 2.2–2.3 Å, as a reference. In total, 1054 distance constraints were obtained for a dimer.
Dihedral angle constraints for the δ, and endocyclic ν0, ν1, ν2, ν3 and ν4 torsion angles were derived from the 3JH1′–H2′ and 3JH3′–H4′ couplings (23,24), as described previously (15–17,22). The sugar pucker was determined as follows on the basis of the results of analysis of the intensities of the H1′–H2′ and H3′–H4′ cross-peaks in the DQF-COSY spectrum: the C2′-endo conformation for the G1, G2, G5, A6, G7 and G8 residues, the O4′-endo conformation for the A12 residue, the C4′-exo conformation for the G11 residue and the C3′-endo conformation for the A3, G4, A9 and G10 residues. The δ and endocyclic ν0–ν4 torsion angles of the residues taking on the C2′-endo conformation were moderately constrained, leaving the sugar free to take on any conformation without any energy penalty between C1′-exo and C3′-exo, including C2′-endo in the pseudorotation cycle. In the same way, those of the A12 residue taking on the O4′-endo conformation were constrained between C4′-exo and C1′-exo including O4′-endo, those of the G11 residue taking on the C4′-exo conformation between C3′-endo and O4′-endo, including C4′-exo, and those of the residues taking on the C3′-endo conformation between C2′-exo and C4′-exo, including C3′-endo.
Structure calculation
Structure calculations were carried out using distance and dihedral angle constraints with a simulated annealing protocol supplied with XPLOR-NIH v. 2.20 (25,26). Hydrogen-bonding constraints for G:G and G:A base pairs of the tetrad and hexad were included. Planarity constraints for the tetrad and hexad were also included. The initial structure for the simulated annealing protocol was generated by separating two chains, each 12 nt long, by 50 Å. After high temperature dynamics at 2000 K (80 000 steps with a time step of 0.001 ps), the system was gradually cooled to 100 K (240 000 steps with the same time step), and then energy minimized. Ten final structures were selected from 200 calculations on the basis of the criterion of the smallest residual energy values. None of them violated the distance constraints by >0.5 Å or the dihedral angle constraints by >5°.
The atomic coordinates have been deposited in the RCSB under Protein Data Bank accession code 2rqj.
Epitope mapping by ME
The Hitachi SV1210 microchip CE system (Hitachi Electronics Engineering Co. Ltd, Tokyo, Japan) was used for ME as described previously (27,28). The channel was filled with 0.6% hydroxypropyl methylcellulose (HPMC, Sigma, St Louis, MO, USA) in 1× TBE buffer (89 mM Tris, 89 mM borate, 2 mM EDTA) containing a 1 : 10 000 dilution of TO-PRO-3 (Molecular Probes, Inc. Carlsbad, CA, USA). The reaction mixture (10 µl) was applied to the sample well of the microchip device and the program was run at 600 V for 120 s (injection time), then at 1100 V for 150 s (separation time) under 350 V of return voltage at 20°C. During the electric separation, the separated RNA peaks were detected by laser induced fluorescence and analyzed.
The 12-mer peptide libraries covering bPrP 25–131 were overlapped with a +4 position shift between consecutive peptides (PEP screen, Sigma-Aldrich). At first R12 (100 nM) was incubated in binding buffer [20 mM Tris–HCl (pH 7.5), 10 mM KCl] under the refolding condition: 90°C for 2 min, 72°C for 5 min, 55°C for 5 min, 37°C for 2 min and kept at 4°C. Following the addition of Cy5-dUTP (10 nM, Amersham Biosciences), the internal standard, the RNA solution was incubated with peptides (0, 100 µM) for 15 min and analyzed using ME. The observed peak intensity of R12 was divided with the peak intensity of the internal standard, which gives the relative peak intensity. The value of the relative peak intensity of R12 was normalized by dividing the relative peak intensity of R12 in the absence of peptide, giving the normalized peak intensity. The relative binding of R12 was defined by the number [1–(the normalized peak intensity)] and plotted for each peptide. Experiments were performed at least twice.
RESULTS
Resonance assignments
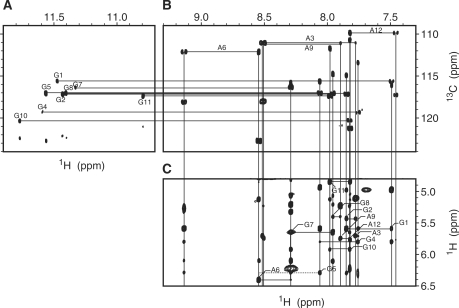
Figure 1B shows the base region of a JR-HMBC spectrum. An adenine base gives four correlation peaks, H2-C4, H8-C4, H2-C6 and H8-C5. It should be noted that the H2-C4, H8-C4 and H2-C6 correlation peaks are folded back by 40 p.p.m., which corresponds to the experimental spectral width, along the 13C dimension. Among them, the H2-C4 and H8-C4 correlation peaks for each adenine are aligned horizontally at the 13C chemical shift value of C4 of each adenine. Conversely, the H2-C4 and H2-C6 correlation peaks, and H8-C4 and H8-C5 ones are aligned vertically, respectively. Thus, a set of resonances for each adenine was identified. A guanine base gives two correlation peaks, H8-C4 and H8-C5, which are aligned vertically (Figure 1B). The H8-C4 correlation peak is also folded back along the 13C dimension. Thus, a set of resonances for each guanine was also identified. With the aid of the knowledge on discrimination between adenine and guanine resonances, the sequential assignment of the fingerprint region was accomplished (Figure 1C). The other nonexchangeable 1H resonances were assigned using standard methods (17), as described for other DNAs and RNAs (15–17,22,29), by means of NOESY, TOCSY and DQF-COSY spectra. Figure 1A shows the NH (imino proton)-C5 correlation peaks for each guanine base. The NH-C5 correlation peak is aligned horizontally with the H8-C5 one for each guanine base at the 13C chemical shift value of C5. On the basis of the assignment of H8, NH was assigned for eight guanines. NH2 (amino proton) resonances were assigned on the basis of the strong imino–amino NOESY cross-peaks.
Figure 1.
NMR spectra for resonance assignments at 30°C. The imino (A) and base (B) regions of the JR-HMBC spectrum, being recorded with the 13C spectral width of 40 p.p.m. The H2-C4, H8-C4, H2-C6 and H8-C5 correlation peaks for each adenine, and the H8-C4 and H8-C5 ones for each guanine are connected by lines, respectively, in (B). The NH (imino proton)-C5 correlation peak in (A) and the H8-C5 in (B) for each guanine are connected by a horizontal line. (C) The fingerprint region of the NOESY spectrum with the mixing time of 150 ms. The H1′ (i-1)-H6/H8(i)-H1′ (i) connectivities are traced, intraresidue cross-peaks being denoted by residue numbers. Dots indicate cross-peaks that are observable when the contour level is lowered.
Identification of the G:G:G:G tetrad and G(:A):G:G(:A):G hexad planes
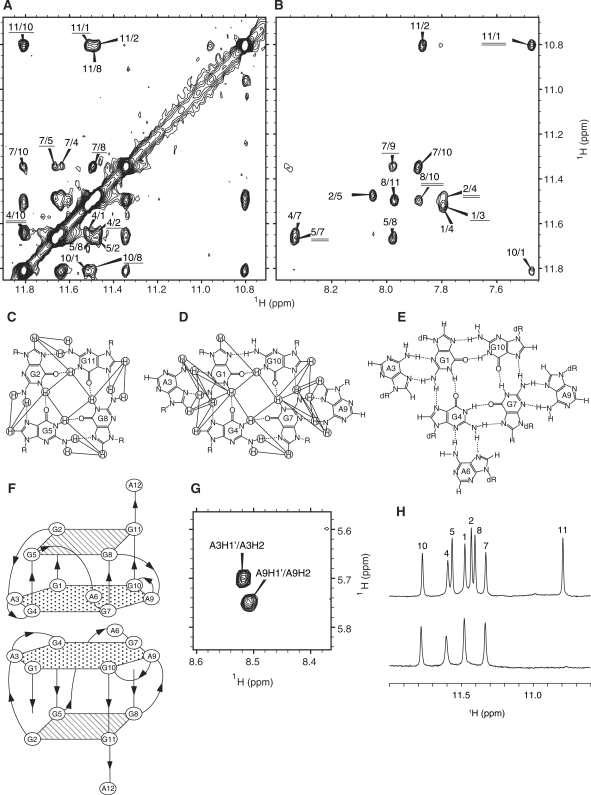
The observation of GNH (imino proton)/GNH2 (amino protons)-GH8 and GNH-GNH NOESY cross-peaks (Figure 2A and B) established the formation of G:G:G:G tetrad planes for G2:G5:G8:G11 and G1:G4:G7:G10 (Figure 2C and D). For the latter tetrad, the further involvement of two adenine bases, A3 and A9, and the resultant formation of a G(:A):G:G(:A):G hexad plane (Figure 2D) was concluded on the basis of the GNH/GNH2-AH8 (Figure 2B), GNH2-ANH2, GH8-AH8 and GH8-ANH2 NOEs. It was noted that the two amino protons both resonated in the low-field region for the G1 (8.87 and 10.56 p.p.m.) and G7 (8.99 and 10.15 p.p.m.) residues, while only one of them did so for the other G residues. This is consistent with the idea that both the amino protons are involved in hydrogen bonding for G1 and G7, while only one of them is involved for the other G residues (Figure 2C and D).
Figure 2.
The G:G:G:G tetrad and G(:A):G:G(:A):G hexad planes, and the dimeric architecture of R12. NH (imino)-NH (A) and NH-H8 (B) cross-peaks in the NOESY spectrum with the mixing time of 230 ms at 30°C. Cross-peaks are denoted by mutual residue numbers. In (A), the cross-peaks between the tetrad and hexad planes are underlined, and the intermolecular one between two hexad planes is double-underlined, respectively. In (B), the cross-peaks as to G:A base pairs are underlined, and those between the tetrad and hexad planes are double-underlined, respectively. The G:G:G:G tetrad (C) and G(:A):G:G(:A):G hexad (D) planes identified for R12. The observed NOESY cross-peaks are indicated by lines. Hydrogen bonds are indicated by dotted lines. (E) The G(:A):G(:A):G(:A):G heptad plane found in D12 (15) is shown for reference. (F) The dimeric architecture of two R12 parallel quadruplexes formed in a tail-to-tail manner through hexad-hexad stacking. (G) Intermolecular A3H2-A3H1′ and A9H2-A9H1′ NOESY cross-peaks confirming the dimeric architecture. (H) Imino proton spectra in H2O (upper) and D2O (lower) at 30°C with the assignments.
Dimer architecture
We reported that d(GGAGGAGGAGGA) (D12) forms a dimer in which the G(:A):G(:A):G(:A):G heptad plane of one monomer stacks on the heptad plane of the other. The H/D exchange experiment indicated that the imino protons of the G1, G4, G7 and G10 residues survive in D2O (Figure 2H). This strongly suggests that R12 also forms a dimer in which the G1(:A3):G4:G7(:A9):G10 hexad plane of one monomer stacks on the hexad plane of the other and that thus imino protons belonging to the hexad are protected from access to and exchange with solvent water molecules (Figure 2F). This dimeric architecture was confirmed by the observation of strong A3H2-A3H1′ and A9H2-A9H1′ NOESY cross-peaks (Figure 2G). The intraresidue AH2-AH1′ distance is >4.5 Å for any conformation (30). Therefore, the A3H2-A3H1′ and A9H2-A9H1′ NOESY cross-peaks cannot be an intraresidue ones, but they should be between the two monomers, which is consistent with the architecture described above. Then, the following NOESY cross-peaks were identified to support the architecture; A3H2-A3H2′, A3H2-A3H3′, A3H2-A3H8, A3NH2-A3H8, A9H2-A9H2′, A9H2-A9H3′, A9H2-A9H8 and A9NH2-A9H8. Moreover, eight NOESY cross-peaks between G1 and G4 residues and ten NOESY cross-peaks between G7 and G10 residues were also found to be consistent with the architecture.
Structure of R12
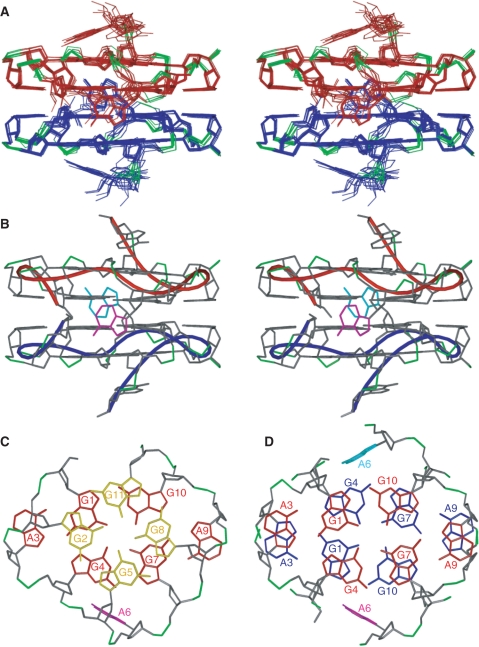
The structure of R12 was calculated on the basis of distance and dihedral angle constraints. The structure statistics are presented in Table 1. The root mean square deviations (RMSDs) of the 10 final structures versus the mean structure for all heavy atoms, the terminal A12 residues being excluded, were 0.48 ± 0.07 Å. The structure of R12 is shown in Figure 3. One tetrad and one hexad planes are present for each monomer, as already described qualitatively. The stacking between the tetrad and hexad planes is shown in Figure 3C. The five-membered ring of one guanine base is stacked on the six-membered ring of the other guanine base for each G-G segment. Additionally, the A3 and A9 bases are stacked on the G2 and G8 sugars, respectively (Figure 3C), which is consistent with the upfield shift of H4′ for G2 (3.13 p.p.m.) and G8 (3.07 p.p.m.). Furthermore, the A12 base stacks on the G11 base. The sugar-phosphate main-chain experiences successive turns, and four G-G segments, i.e. G1-G2, G4-G5, G7-G8 and G10-G11, are aligned parallel to each other. The two monomers form a dimer in a tail-to-tail manner through hexad–hexad stacking interaction. The stacking of the two hexad planes is shown in Figure 3D. The G1, A3, G4, G7, A9 and G10 bases of one hexad plane are stacked on the G4, A3, G1, G10, A9 and G7 bases of the other hexad plane, respectively.
Table 1.
NMR constraints and structural statistics for R12
| NMR constraints | |
| Distance constraints | 1054 |
| Intraresidue distance constraints | 440 |
| Sequential (i, i+1) distance constraints | 298 |
| Medium to long range ≥ (i, i+2) distance constraints | 212 |
| Interstrand distance constraints | 104 |
| Dihedral angle constraints | 144 |
| δ | 24 |
| ν0–ν4 | 120 |
| Planarity constraints for tetrad and hexad planes | 4 |
| Hydrogen-bonding constraints | 80 |
| Structural statistics for 10 final structures | |
| X-PLOR energies (kcal/mol) | |
| Etotal | 267 ± 1 |
| Ebond | 10 ± 0 |
| Eangle | 199 ± 1 |
| Eimproper | 16 ± 0 |
| Evdw | 6 ± 0 |
| Enoe | 21 ± 1 |
| Ecdih | 3 ± 0 |
| RMSD from idealized geometry | |
| Bond lengths (Å) | 0.003 ± 0.000 |
| Bond angles (°) | 0.91 ± 0.01 |
| Impropers (°) | 0.38 ± 0.00 |
| NOE violations | |
| Number of violations >0.5 Å | 0 ± 0 |
| RMSD of violations (Å) | 0.019 ± 0.000 |
| Dihedral angle violations | |
| Number of violations >5° | 0 ± 0 |
| RMSD of violations (°) | 0.62 ± 0.03 |
| RMSD of 10 final structures versus mean structure for all heavy atoms (Å) (terminal residues excluded) | 0.48 ± 0.07 |
Figure 3.
The structure of R12. (A) A stereoview of the superposition of 10 final structures of R12. The monomers are colored red and blue, respectively, phosphate groups being colored green. (B) A stereoview of the representative structure with the lowest energy. The A6 residues of each monomer are colored magenta and cyan, respectively. The chains connecting the C3′ and C4′ atoms of residues of each monomer are indicated by red and blue tubes, respectively. (C) Stacking between the tetrad (yellow) and hexad (red) planes of the upper monomer of the representative structure. (D) Stacking between the two hexad planes (red and blue) of each monomer of the representative structure.
Comparison of the structures of R12 and D12
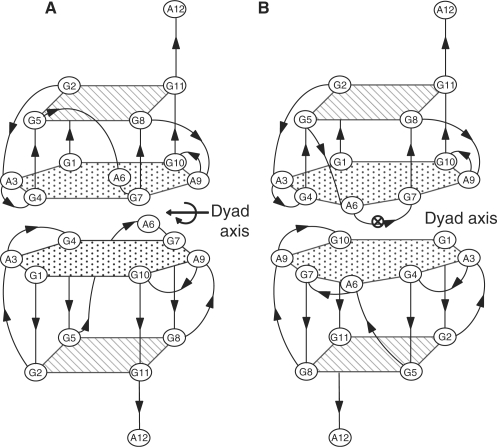
The structures of R12 and D12 are schematically shown in Figure 4. Similarities can be seen between them. One R12 strand forms a quadruplex with four G-G segments aligned parallel to each other, as one D12 strand does. The two R12 quadruplexes form a dimer in a tail-to-tail manner, as the two D12 quadruplexes do. Differences can also be noted between the two structures. The R12 quadruplex comprises a G(:A):G:G(:A):G hexad plane in addition to a G:G:G:G tetrad plane, while the D12 quadruplex comprises a G(:A):G(:A):G(:A):G heptad plane in addition to a G:G:G:G tetrad plane. This difference shows that A6 of R12 is not involved in the plane, while that of D12 is. Moreover, the dyad axis correlating two monomers is along the A3-A9 direction for R12, but is perpendicular to the A3-A9 direction for D12. The origins of these differences are discussed later.
Figure 4.
Comparison of the structures of R12 and D12. Schematic representation of the R12 structure (this study) (A) and the D12 structure (15) (B). A dyad axis correlating the two monomers is indicated for each structure.
Identification of the binding sites of bPrP as to R12
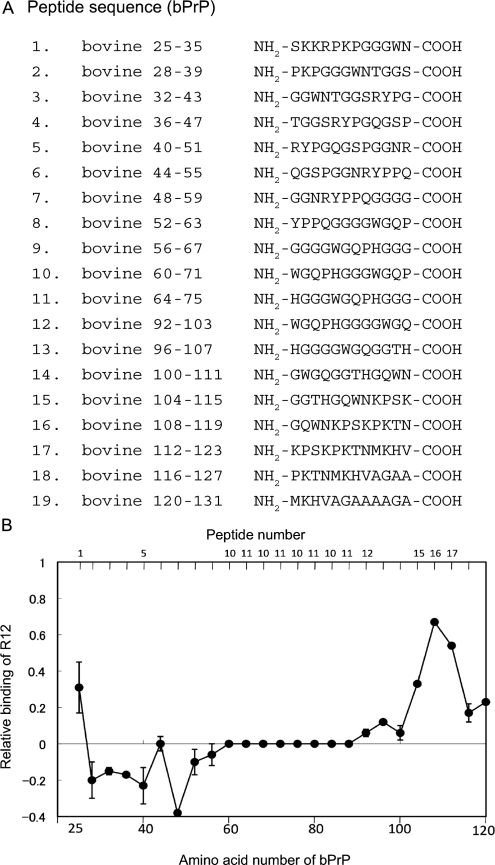
It was found that the RNA aptamer comprising the R12 sequence binds to the 25–131 region of bPrP (12). In order to exactly identify the sites of bPrP involved in the binding to R12, the affinity of a series of 12-mer peptide fragments of bPrP for R12 was examined by means of ME (Figure 5). Four peptide fragments, peptides 1, 15, 16 and 17, exhibited high affinity to R12. Peptide 1 corresponds to the first lysine cluster of bPrP, and three consecutive peptides 15–17 to the second lysine cluster, respectively. Peptide 16 exhibited the highest affinity. Thus, two lysine clusters of bPrP were identified as the binding sites for R12.
Figure 5.
Identification of the interaction sites on bPrP for R12. (A) The amino acid sequences of a series of peptide fragments of bPrP. (B) The plot of the relative binding of R12 with a series of 12-mer peptides covering bPrP 25–131. Amino acid positions 60–88 of bPrP is composed of the four repeats of peptides 10 and 11 (octapeptide repeat region).
DISCUSSION
Several RNA aptamers against PrP have been isolated. No 3D structure has been reported for these aptamers. This study revealed the 3D structure of an RNA aptamer against PrP for the first time. It has been proposed for some of these RNA aptamers that the G-quadruplex structure plays a critical role in the binding of PrP (4,31). For these aptamers, antiparallel quadruplexes have been postulated. The quadruplex structure of R12, the minimized RNA aptamer, turns out to be quite different from the postulated structures. One R12 molecule folds into an intramolecular parallel quadruplex with a G:G:G:G tetrad plane and a G(:A):G:G(:A):G hexad plane. Then, two R12 molecules dimerize in a tail-to-tail manner through intermolecular hexad–hexad stacking. This unique structure of R12 may be utilized to bind PrP, as discussed later.
Two major differences are noted between the R12 and D12 structures. First, the position of the A6 residue is different between them. The A6 residue of R12 is located close to the G(:A):G:G(:A):G hexad plane, but is not linked to the hydrogen bonding network of the hexad plane (Figures 3B and 4A). On the contrary, the A6 residue of D12 is linked to the hexad plane through the G4NH2-A6N7 and G4N3-A6NH2 hydrogen bonds, resulting in the formation of a G(:A):G(:A):G(:A):G heptad plane (15) (Figures 2E and 4B). A6 of R12 takes on the C2′-endo sugar conformation, while that of D12 is C3′-endo. This difference in the sugar conformation may be related to the different positioning of A6. Another remarkable difference is that the χ angle of the A6 residue around the glycoside bond is approximately from +50 to +90° for R12, while it is nearly –90° for D12 (15). Thus, the A6 base of R12 is rotated around the glycoside bond by ∼180° compared to that of D12. This rotation of the A6 base is also partly responsible for the different positioning of A6 and the nonformation of the hexad plane for R12. The direct driving force that causes the differences in local structure, particularly for A6 between R12 and D12, is not clear, although it must be related to the ribose/deoxyribose difference. A6 may preferably form the hydrogen bond involving 2′OH of either A6 or other residues, although this is not established at this moment.
Second, the position of the dyad correlating two monomers is different, the two dyad axes of each structure being perpendicular to each other. This difference can be rationalized from the viewpoint of the maximum stacking interaction between the two monomers. In order to achieve the maximum stacking between two heptad planes, two monomers should be arranged as shown in Figure 4B. All seven bases have their stacking partners in Figure 4B; G1 of the upper monomer stacks on G10 of the lower monomer, A3 on A9, and so on. In the case of the other arrangement, the stacking would be less. For example, if the lower monomer is rotated by 90° in an anticlockwise direction around a vertical axis passing through the center of the heptad plane, then the A3 bases of both the upper and lower monomers would lose their stacking partners. Similarly, if the lower monomer is rotated by 180°, then the A6 bases of both the upper and lower monomers would lose their stacking partners. On the contrary, the maximum stacking between two hexad planes can be achieved in two ways. One way is as shown in Figure 4A. All six bases have their stacking partners in Figure 4A; G1 of the upper monomer stacks on G4 of the lower monomer, A3 of the upper monomers on A3 of the lower monomer, and so on (Figures 3D and 4A). The maximum stacking is also achieved when the lower monomer is rotated by 180° around a vertical axis passing through the center of the hexad. All six bases have their stacking partners also in the resulting arrangement; G1 of the upper monomer stacks on G10 of the lower monomer, A3 on A9, and so on. For this arrangement, the position of the dyad axis is the same as that for D12 in Figure 4B. Thus, for R12, two positions are equally allowed for the dyad axis in terms of achieving the maximum stacking interaction between the two hexad planes; one is as actually found for R12 and the other is as found for D12. Then, why is the former favored for R12? In the case of the latter position of the dyad, the A6 residues of both the upper and lower monomers are located on the same side, which may result in unfavorable steric hindrance between them. In fact, such steric hindrance is predicted from inspection of the determined monomer structure of R12. Conversely, in the case of the former position of the dyad, the two A6 residues are located away on the opposite side, resulting in no steric hindrance between them. This could be why the former position of the dyad is favored for R12 over the latter one. In this way, the difference in the position of the dyad between R12 and D12 can be rationally explained on the basis of the fact that the heptad is formed for D12, while the hexad is formed instead for R12. It is interesting to note that r(GGAGGUUUUGGAGG) adopts the same fold as R12 does (17). Two strands of this RNA form a quadruplex dimer composed of two G:G:G:G tetrad planes and two G(:A):G:G(:A):G hexad ones (17). This RNA cannot form the heptad, because it has only two A residues per a strand. The fold of R12 may be generally found for the quadruplex composed of the tetrad and hexad planes.
It was found that the binding sites of bPrP for R12 are two lysine clusters (Figure 5). The first lysine cluster comprises three lysine residues, K25, K26 and K29, and the second one comprises four lysine residues, K112, K115, K117 and K121. The electrostatic interaction between the positive charges of the lysine clusters and the negative charges of the phosphate groups of R12 must be the main factor for the high affinity between bPrP and R12. The affinity of the second lysine cluster to R12 is higher than that of the first lysine cluster (Figure 5). It was reported that two lysine clusters of ovine PrP (residues 25–34 and 102–110, respectively) are primary binding sites of the anti-ovine RNA aptamer and that the first lysine cluster is the major binding site (32). These lysine clusters correspond to residues 25–34 and 110–118 of bPrP. Thus, although the two lysine clusters are the RNA aptamer binding sites for both bPrP and ovine PrP, the major binding sites are reversed for the two PrPs. Because of the formation of the remarkable quadruplex structure, the arrangement of the negatively charged phosphate groups of R12 is unique and quite different from those of canonical structures such as an A-form duplex (Figure 3B). Moreover, the density of the negative charges is high for the R12 structure. The unique arrangement and condensation of negative charges exhibited by the R12 structure are supposed to be responsible for high affinity of R12 to bPrP through an electrostatic interaction.
It was suggested for the ovine PrP-aptamer system that the stacking interaction of the aptamer with a tryptophan residue may also contribute to the binding (32). The first and second lysine clusters of bPrP also comprise tryptophan residues, W34 and W110, respectively (Figure 5). R12 has two G:G:G:G tetrad planes at the top and bottom of its dimer structure. These planes are suitable for the stacking interaction with the tryptophan residues. It is notable that the affinity to R12 of peptide 16 comprising three lysine residues and one tryptophan residue is higher than that of peptide 17 comprising four lysine residues (Figure 5). This suggests that W110 of peptide 16 contributes to binding more than K121 of peptide 17. When the W110 residue of peptide 16 was replaced by an alanine residue, the affinity to R12 decreased to 39% of that of the original peptide 16. This confirms the importance of the contribution of W110 to binding. When the W34 of the peptide 1 was replaced by an alanine residue, the affinity decreased to 78% of that of the original peptide 1. This suggests that W34 also contributes to binding to certain extent. In the case of the antiparallel quadruplex, either a diagonal or lateral loop passes over or below the tetrad plane and covers a part of the tetrad plane. The remarkable point of the parallel quadruplex structure of R12 is that neither a diagonal nor lateral loop passes over or below the G:G:G:G tetrad plane. Therefore, it is supposed to be easy for a tryptophan residue to approach and stack on the tetrad plane of R12, although A12 stacks on G11 and partly covers the tetrad plane. This may be related to the high affinity of R12 for bPrP. The detailed mode of electrostatic and stacking interactions in the ovine PrP–aptamer system has not been addressed, because the structure of an aptamer against ovine PrP has not been determined. The structural determination of R12 has enabled us to rationally interpret the contribution of electrostatic and stacking interactions to the high affinity for the bPrP–R12 aptamer system.
It was revealed that the 25–241 region of bPrP comprising both the first and second lysine clusters binds to an aptamer much more strongly than the 102–241 region comprising only the second lysine cluster (12). This indicates that the addition of the first lysine cluster dramatically increases the affinity, although the affinity of the first lysine cluster is lower than that of the second lysine cluster. These results strongly suggest that the two lysine clusters of bPrP are simultaneously bound by one R12 dimer molecule, resulting in the much higher affinity. From this point of view, dimer formation of R12 may contribute to the high affinity by presenting two binding sites to bPrP.
So far, the structural study of R12 in complex with a whole prion protein is not feasible due to low solubility of the complex. The affinity of either peptide 1 or peptide 16 to R12 seems to be lower than that of a whole prion protein by two orders. This may be due to the reason that a whole protein would be bound at two different regions by the R12 dimer, as discussed above. Although it is necessary to further examine whether the peptides are an adequate model for PrP association, the structural study of R12 in complex with peptides is expected to give important information.
Judging from available structures of PrPC (33–37), the second lysine cluster as well as the first one is supposed to be exposed to the solvent for bPrPC. Conversely, the second lysine cluster may partly be involved in the β-sheet structure of bPrP-β, because this cluster locates close to the short β-sheet structure of bPrPC and the extension of the β-sheet structure is assumed for bPrP-β. In this case, the access and binding of the aptamer to this cluster would be hindered to some extent. This may explain why R12 binds 30 times more strongly to bPrPC than to bPrP-β (12).
The binding of D12 to bPrP is weaker than that of R12 (12). The differences in the structure between R12 and D12, which were described above, may be responsible for the difference in the affinity, although the difference in the chemical structure of the sugar, ribose versus deoxyribose, may also be responsible.
It was reported that an RNA aptamer obtained against PrPC can reduce the de novo formation of PrPSc probably through the stabilization of PrPC, suggesting the therapeutic potential of the aptamer against prion diseases (4). R12 may also reduce the de novo formation of PrPSc and thus may be utilized as a therapeutic agent against prion diseases, because it binds to PrPC more preferably than to PrPSc (12), as described and rationalized above. Very recently, it was found that PrPC mediates impairment of synaptic plasticity by amyloid-β oligomers. Then, it was demonstrated that anti-PrP antibodies prevent the binding of amyloid-β oligomers to PrPC and rescue synaptic plasticity in hippocampal slices from oligomeric amyloid-β (5). Thus, a molecule that binds to PrPC with high affinity and specificity has great therapeutic potential as to Alzheimer's disease. Therefore, R12 may be utilized as a therapeutic agent for Alzheimer's disease due to its high affinity and specificity to PrPC. The atomic coordinates of R12 provided by this work would be useful as a starting point for designing modification or mutation for the further development of R12 as a therapeutic agent.
FUNDING
Ministry of Education, Science, Sports and Culture of Japan (19036026 and 21370047 to M.K.); Japan Science and Technology (04-183 to M.K.); Yokohama City University (K2102 to M.K.); and Advanced Medical Research Center of Yokohama City University. Funding for open access charge: Ministry of Education, Science, Sports and Culture of Japan, Japan Science and Technology, and Yokohama City University.
Conflict of interest statement. None declared.
REFERENCES
- 1.Prusiner SD. Novel proteinaceous infectious particles cause scrapie. Science. 2004;216:136–144. doi: 10.1126/science.6801762. [DOI] [PubMed] [Google Scholar]
- 2.Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick RJ, Cohen FE. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc. Natl Acad. Sci. USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huang Z, Gabriel JM, Baldwin MA, Fletterick RJ, Prusiner SB, Cohen FE. Proposed three-dimensional structure for the cellular prion protein. Proc. Natl Acad. Sci. USA. 1994;91:7139–7143. doi: 10.1073/pnas.91.15.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Proske D, Gilch S, Wopfner F, Schatzl HM, Winnacker EL, Famulok M. Prion-protein-specific aptamer reduces PrPSc formation. Chembiochem. 2002;3:717–725. doi: 10.1002/1439-7633(20020802)3:8<717::AID-CBIC717>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 5.Lauren J, Gimbel DA, Nygaard HB, Gilbert JW, Strittmatter SM. Cellular prion protein mediates impairment of synaptic plasticity by amyloid-β-oligomers. Nature. 2009;457:1128–1132. doi: 10.1038/nature07761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Walsh DM, Klyubin I, Fadeeva JV, Vullen WK, Anwyl R, Wolfe MS, Rowan MJ, Selkoe DJ. Naturally secreted oligomers of amyloid β protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416:535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 7.Cleary JP, Walsh DM, Hofmeister JJ, Shankar GM, Kuskowski MA, Selkoe DJ, Ashe KH. Natural oligomers of the amyloid-β protein specifically disrupt cognitive function. Nat. Neurosci. 2005;8:79–84. doi: 10.1038/nn1372. [DOI] [PubMed] [Google Scholar]
- 8.Lesne S, Koh MT, Kotilinek L, Kayed R, Glabe CG, Yang A, Gallaqher M, Ashe KH. A specific amyloid-β protein assembly in the brain impairs memory. Nature. 2006;440:352–357. doi: 10.1038/nature04533. [DOI] [PubMed] [Google Scholar]
- 9.Shankar GM, Li S, Mehta TH, Garcia-Munoz A, Shepardson NE, Smith I, Brett FM, Farrell MA, Rowan MJ, Lemere CA, et al. Amyloid-β protein dimers isolated directly from Alzheimer's brains impair synaptic plasticity and memory. Nat. Med. 2008;14:837–842. doi: 10.1038/nm1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 11.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 12.Murakami K, Nishikawa F, Noda K, Yokoyama T, Nishikawa S. Anti-bovine prion protein RNA aptamer containing tandem GGA repeat interacts both with recombinant prion protein and its β isoform with high affinity. Prion. 2008;2:73–78. doi: 10.4161/pri.2.2.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lührs T, Zahn R, Wüthrich K. Amyloid formation by recombinant full-length prion proteins in phospholipid bicelle sokutions. J. Mol. Biol. 2006;357:833–841. doi: 10.1016/j.jmb.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 14.Kettani A, Gorin A, Majumdar A, Hermann T, Skripkin E, Zhao H, Jones R, Patel DJ. A dimeric DNA interface stabilized by stacked A:(G:G:G:G):A hexads and coordinated monovalent cations. J. Mol. Biol. 2000;297:627–644. doi: 10.1006/jmbi.2000.3524. [DOI] [PubMed] [Google Scholar]
- 15.Matsugami A, Ouhashi K, Kanagawa M, Liu H, Kanagawa S, Uesugi S, Katahira M. An intramolecular quadruplex of (GGA)(4) triplet repeat DNA with a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad, and its dimeric interaction. J. Mol. Biol. 2001;313:255–269. doi: 10.1006/jmbi.2001.5047. [DOI] [PubMed] [Google Scholar]
- 16.Matsugami A, Okuizumi T, Uesugi S, Katahira M. Intramolecular higher order packing of parallel quadruplexes comprising a G:G:G:G tetrad and a G(:A):G(:A):G(:A):G heptad of GGA triplet repeat DNA. J. Biol. Chem. 2003;278:28147–28153. doi: 10.1074/jbc.M303694200. [DOI] [PubMed] [Google Scholar]
- 17.Liu H, Matsugami A, Katahira M, Uesugi S. A dimeric RNA quadruplex architecture comprised of two G:G(:A):G:G(:A) hexads, G:G:G:G tetrads and UUUU loops. J. Mol. Biol. 2002;322:955–970. doi: 10.1016/s0022-2836(02)00876-8. [DOI] [PubMed] [Google Scholar]
- 18.Phan AT. Long-range imino proton-13C J-couplings and the through-bond correlation of imino and non-exchangeable protons in unlabeled DNA. J. Biomol. NMR. 2000;16:175–178. doi: 10.1023/a:1008355231085. [DOI] [PubMed] [Google Scholar]
- 19.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 20.Garrett DS, Powers R, Gronenborn AM, Clore GM. A common sense approach to peak picking in two-, three-, and four-dimensional spectra using automatic computer analysis of contour diagrams. J. Magn. Reson. 1991;95:214–220. doi: 10.1016/j.jmr.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 21.Goddard TD, Kneller DG. SPARKY 3. San Francisco: University of California; 2006. [Google Scholar]
- 22.Matsugami A, Ohyama T, Inada M, Inoue N, Minakawa N, Matsuda A, Katahira M. Unexpected A-form formation of 4′-thioDNA in solution, revealed by NMR and the implications as to the mechanism of nuclease resistance. Nucleic Acids Res. 2008;36:1805–1812. doi: 10.1093/nar/gkn011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith JS, Nikonowicz EP. Phosphorothioate substitution can substantially alter RNA conformation. Biochemistry. 2000;39:5642–5652. doi: 10.1021/bi992712b. [DOI] [PubMed] [Google Scholar]
- 24.Varani G, Aboul-ela F, Allain FHT. NMR investigation of RNA structure. Prog. Nucl. Magn. Reson. Spect. 1996;29:51–127. [Google Scholar]
- 25.Brünger AT. New Haven: Yale University Press; 1993. X-PLOR Version 3.1: A System for X-ray Crystallography and NMR. [Google Scholar]
- 26.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J. Magn. Reson. 2003;160:65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 27.Nishikawa F, Arakawa H, Nishikawa S. Application of microchip electrophoresis in the analysis of RNA aptamer-protein interactions. Nucleosides Nucleotides Nucleic Acids. 2006;25:369–382. doi: 10.1080/15257770600683953. [DOI] [PubMed] [Google Scholar]
- 28.Nishikawa F, Murakami K, Matsugami A, Katahira M, Nishikawa S. Structural studies of an RNA aptamer containing GGA repeats under ionic conditions using microchip electrophoresis, circular dichroism, and 1D-NMR. Oligonucleotides. 2009;19:179–190. doi: 10.1089/oli.2008.0167. [DOI] [PubMed] [Google Scholar]
- 29.Matsugami A, Xu Y, Noguchi Y, Sugiyama H, Katahira M. Structure of a human telomeric DNA sequence stabilized by 8-bromoguanosine substitutions, as determined by NMR in a K+ solution. FEBS J. 2007;274:3545–3556. doi: 10.1111/j.1742-4658.2007.05881.x. [DOI] [PubMed] [Google Scholar]
- 30.Wüthrich K. NMR of Proteins and Nucleic Acids. New York, NY: John Wiley & Sons Inc.; 1986. [Google Scholar]
- 31.Weiss S, Proske D, Neumann M, Groschup MH, Kretzschmar HA, Famulok M, Winnacker EL. RNA aptamers specifically interact with the prion protein PrP. J. Viol. 1997;71:8790–8797. doi: 10.1128/jvi.71.11.8790-8797.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mercey R, Lantier I, Maurel MC, Grosclaude J, Lantier F, Marc D. Fast, reversible interaction of prion protein with RNA aptamers containing specific sequence patterns. Arch. Virol. 2006;151:2197–2214. doi: 10.1007/s00705-006-0790-3. [DOI] [PubMed] [Google Scholar]
- 33.Riek R, Hornemann S, Wider G, Billeter M, Glockshuber R, Wüthrich K. NMR structure of the mouse prion protein domain PrP (121-321) Nature. 1996;382:180–182. doi: 10.1038/382180a0. [DOI] [PubMed] [Google Scholar]
- 34.James TL, Liu H, Ulyanov NB, Farr-Jones S, Zhang H, Donne DG, Kaneko K, Groth D, Mehlhorn I, Prusiner SB, Cohen FE. Solution structure of a 142-residue recombinant prion protein corresponding to the infectious fragment of the scrapie isoform. Proc. Natl Acad. Sci. USA. 1997;94:10086–10091. doi: 10.1073/pnas.94.19.10086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riek R, Hornemann S, Wider G, Glockshuber R, Wüthrich K. NMR characterization of the full-length recombinant murine prion protein mPrP (23-231) FEBS Lett. 1997;413:282–288. doi: 10.1016/s0014-5793(97)00920-4. [DOI] [PubMed] [Google Scholar]
- 36.Donne DG, Viles JH, Groth D, Mehlhorn I, James TL, Cohen FE, Prusiner SB, Wright PE, Dyson HJ. Structure of the recombinant full-length hamster prion protein PrP (29-231): the N terminus is highly flexible. Proc. Natl Acad. Sci. USA. 1997;94:13452–13457. doi: 10.1073/pnas.94.25.13452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zahn R, Liu A, Lührs T, Riek R, von Schroetter C, Garcia FL, Billeter M, Calzolai L, Wider G, Wüthrich K. NMR solution structure of the human prion protein. Proc. Natl Acad. Sci. USA. 2000;97:145–150. doi: 10.1073/pnas.97.1.145. [DOI] [PMC free article] [PubMed] [Google Scholar]