Abstract
The formation of G-quadruplexes in G-rich regions of DNA is believed to affect DNA transcription and replication. However, it is currently unclear how this formation occurs in the presence of a complementary strand. We have used atomic force microscopy (AFM) to image stable RNA/DNA hybrid loops generated by transcription of the plasmid pPH600, which contains a 604-bp fragment of the murine immunoglobulin Sγ3 switch region. We show that the non-RNA-containing portion folds into G-quadruplexes, consistent with computational predictions. We also show that hybrid formation prevents further transcription from occurring, implying a regulatory role. After in vitro transcription, almost all (93%) of the plasmids had an asymmetric loop, a large asymmetric blob or a spur-like projection at the appropriate position on the DNA contour. The loops disappeared following treatment of the transcribed plasmid with RNase H, which removes mRNA hybridized with the template strand. Replacement of K+ in the transcription buffer with either Na+ or Li+ caused a reduction in the percentage of plasmids containing loops, blobs or spurs, consistent with the known effects of monovalent cations on G-quadruplex stability. The minimal sample preparation required for AFM imaging has permitted direct observation of the structural changes resulting from G-quadruplex formation.
INTRODUCTION
In addition to the normal Watson–Crick duplex, DNA has the potential to form a number of alternative structures. One of these, found in guanine-rich regions, is the G-quadruplex, which is a four-stranded structure stabilized by the formation of planar arrays of four hydrogen-bonded guanines (1–3). Once formed, G-quadruplex DNA is very stable because of the hydrogen bonding within the G-quartets, the stacking of the G-quartets upon one another and the coordination of charge by monovalent cations in the central cavity of the G-quartets. Certain G-rich regions of chromosomes are especially prone to formation of G-quadruplex DNA (4). Much effort has been focused on telomeres, where G-quadruplexes have been shown to form in vivo, and there is considerable effort to develop G-quadruplex binding drugs with anti-cancer activity (5). G-quadruplexes have been implicated in the regulation of processes such as transcription and translation in a wide variety of genes (6–9), and G-rich regions are also found in immunoglobulin heavy chain switch regions, ribosomal DNA and G-rich minisatellites (2). In addition, there is evidence that the expansion of the CGG repeat associated with the human neurodegenerative disease, fragile X syndrome, involves the ability of this repeat to form a G-quadruplex structure (10–12). Furthermore, a large number of natural proteins have been found to interact specifically with G-quadruplexes (13). Given its potential involvement in both normal and abnormal cellular function, it is important to understand the mechanism of G-quadruplex formation and the perturbations in DNA structure involved in its generation.
One challenge to the G-quadruplex hypothesis is that DNA G-quadruplex formation in vivo would have to compete with the complementary strand, which would favour the formation of the standard Watson–Crick duplex. It is therefore interesting to note that some of these G-rich structures, such as the immunoglobulin switch regions, form an unusual post-transcriptional structure, where the RNA produced remains bound to the template DNA strand, leaving the coding strand free and forming a loop (14). These structures are known as RNA/DNA hybrids, and the loops are known as G-loops, and have been found to be involved in a range of processes, such as the regulation of E. coli plasmid replication (15). In these structures, the G-rich sequence, with potential to form a G-quadruplex, is in the free single strand and hence is readily able to fold into a G-quadruplex without competition from double strand formation. We therefore considered this an interesting model for in vivo G-quadruplex formation.
Recently, electron microscopy (EM) has been used to investigate G-quadruplex formation in a G-loop (14). The assay involved transcription of a plasmid containing a G-rich coding strand, taken from the mouse immunoglobulin switch region. The structures observed contained a loop in which one strand was interpreted as an RNA/DNA hybrid and the other a region of G-quadruplex DNA, although the extended nature of the strand implied minimal G-quadruplex formation in the final structure. Evidence for G-quadruplex formation was 2-fold: cleavage of one arm of the loop by the G-quadruplex-specific endonuclease GQN1 and recognition by Nucleolin-428. Both of these treatments were carried out before surface deposition.
Of course, EM analysis requires extensive sample preparation and the imaging of the sample in vacuo, processes that might result in the loss of some structural information. In contrast, atomic force microscopy (AFM) is able to image DNA at the single molecule level with only minimal sample preparation (16–18). In the present study, we have used AFM to examine the structures produced when a plasmid containing a G-rich coding strand was transcribed in vitro. In this way we have observed additional structural features that were not seen previously using EM imaging. We have also used recent theoretical results to investigate the formation of the RNA/DNA hybrid itself and have experimentally studied the effects of G-loop formation on future rounds of transcription.
MATERIALS AND METHODS
The DNA substrate
The plasmid used in these experiments—pPH600, kindly provided by N. Maizels—is a derivative of pBluescript KS(+) (2958 bp). The pPH600 plasmid (3562 bp) has a 604 bp PvuII-HindIII fragment of the murine Sγ3 switch region, which contains the repeat d(CTGGGCAGCTCTGGGGGAGCTGGGGTAGGTTGGGAGTGTGGGGACCAGG), inserted just downstream of the T7 promoter. pBluescript SK(–) containing a 660 bp insert, consisting of the syncollin open reading frame plus flanking sequences, was used as a control plasmid (19).
In vitro transcription
pPH600 (or control syncollin plasmid) was incubated for 30 min at 37°C in a buffer solution containing 200 μg/ml of DNA, 40 mM Tris–HCl, pH 7.9, 6 mM MgCl2, 10 mM dithiothreitol, 2 mM spermidine, 40 mM KCl, 1 mM each of ATP, UTP, GTP, CTP and 1 U/μl of T7 polymerase. The sample was then subjected to a 15-min RNaseA treatment to digest free mRNA. Both plasmids were linearized with AflIII before being purified by phenol/chloroform extraction and ethanol precipitation. When appropriate, hybridized mRNA was removed by treatment of the samples with RNaseH for 30 min at 37°C after the RNaseA treatment and before AflIII digestion. In some experiments, KCl in the transcription buffer was replaced by either 40 mM NaCl or 40 mM LiCl.
AFM imaging
Disks of ruby muscovite mica (Goodfellow, PA, USA) were attached to 13-mm steel pucks using Aron Alpha high-strength rapid bonding adhesive, alpha cyanoacrylate (Agar Scientific Limited, Essex, UK). Immediately prior to DNA deposition, the top layer of the mica was cleaved using Scotch tape to reveal an atomically flat surface. Purified DNA was diluted to a concentration of ∼0.5 × 10–14 mol/µl in imaging buffer (10 mM MgCl2, 10 mM KCl, 10 mM Tris–HCl, pH 7.9). Droplets (20 μl) were deposited onto freshly cleaved mica disks and incubated at room temperature for 2 min. Excess DNA was rinsed off with MilliQ water (Millipore System, MA, USA) and the water was wicked from the surface using tissue paper. The disks were then dried under a stream of nitrogen gas.
Imaging was performed using a Multimode Nanoscope IIIa atomic force microscope (Veeco Digital Instruments, Santa Barbara, CA, USA). All samples were imaged in air using tapping mode with a root-mean-square amplitude of ∼2 V and a drive frequency of ∼350 kHz. Commercially available silicon probes with a specified spring constant of 42 N/m were used (Olympus atomic force OMCL-AC160TS-E). Data were gathered by capturing 2 × 2 µm2 scans.
Data analysis
DNA lengths were measured by drawing a series of very short lines along the DNA contour and summating the lengths. All errors quoted are standard errors of the mean (SE).
Small raised structures were occasionally seen on untranscribed DNA (due to variations in the imaging surface and/or kinks in the DNA), so a criterion for distinguishing new structures (blobs) from features found on untranscribed DNA was required. The normal height and the peak height were determined for 20 DNA molecules. The mean normal height was 0.53 ± 0.01 nm, and the mean peak height was 0.72 ± 0.02 nm, with a highest absolute value of 0.97 nm. Consequently, any blob <1 nm in height was excluded from the dataset and any blob >1 nm was included. In the case of loops and spurs, the observed shape was significantly different from anything seen on pure DNA; hence, all of these structures were included in the dataset. The height measurements were taken across the middle of each blob, across the highest point on the short arm of each loop and across the base of each spur.
Computational prediction
The thermodynamic stabilities of the Sγ3 or syncollin RNA/DNA hybrid form in comparison to the duplex DNA form were calculated using the online tool at http://www.bss.phy.cam.ac.uk/interactive/hybrid/, using the methodology in (20). A window size of 10 bases was used for the short Sγ3 and 100 bases for the longer syncollin sequence. G-quadruplex prediction was performed using the default parameters of quadparser (21), which was accessed using the online interface at http://www.quadruplex.org.
RESULTS
We chose as a model system the murine Sγ3 switch region (14), which has a G-rich consensus repeat sequence of d(CTGGGCAGCTCTGGGGGAGCTGGGGTAGGTTGGGAGTGTGGGGACCAGG). An extension of nearest-neighbour analysis methods, together with folding models (20), allowed us to calculate whether an RNA/DNA hybrid and G-loop are likely to be thermodynamically stable and whether the hybrid-forming strand would be the G-rich strand or the complementary C-rich strand. In general, randomly chosen sequences are very unlikely to be predicted to form G-loops, as we found for the control syncollin sequence, which had a predicted ΔG for loop formation of over 2.5 kcal/mol, implying that G-loop formation would be extremely unlikely. However, we found that for the G-rich strand of the murine Sγ3 switch region we did predict stable G-loop formation, with a predicted ΔG of –1.07 kcal/mol over a 10-base window. In contrast, the C-rich strand would not be predicted to form such loops if it were transcribed (ΔG of +1.10 kcal/mol). Similarly, we used the quadparser algorithm (21; online at http://www.quadruplex.org) to predict whether the Sγ3 switch region would be predicted to form G-quadruplexes. This algorithm confirmed that the G-rich strand would be expected to form G-quadruplexes, whereas the complementary strand would not. The control syncollin sequence had no region predicted to form a G-quadruplex.
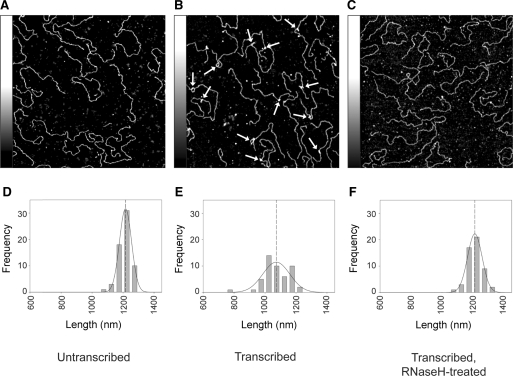
We then looked experimentally for G-loop and G-quadruplex formation by AFM imaging of a plasmid (pPH600) that carries a G-rich 604 bp PvuII-HindIII fragment of the murine Sγ3 switch region (14). Images of untranscribed, linearized plasmids showed homogenous spreads of DNA molecules, as expected (Figure 1A). The mean length of the molecules (±SE) was 1190 ± 7 nm (n = 63). Given that the expected size of the plasmid is 3562 bp, the nm-to-bp conversion factor will be 0.334 (nm/bp). This factor corresponds to the expected value for a ∼3 kb DNA molecule under dry AFM imaging conditions (22) and is used from here on to convert between bp and nm values, where appropriate. After in vitro transcription of the plasmids, followed by RNase A treatment (to remove free mRNA) and linearization, 93% (n = 56) of the DNA molecules contained structures that were not present on untranscribed DNA (arrows in Figure 1B). The structures observed could be categorized into three groups—asymmetric loops, blobs and spurs, which occurred with approximately equal frequency. The lengths of a number of DNA molecules were measured, taking the shorter route where there was a loop. The mean length of the DNA molecules was 1053 ± 12 nm. This is 137-nm (equivalent to 410 bp) shorter than the length of untreated DNA and indicates that the formation of the observed structures is accompanied by DNA condensation. (When the longer route was taken on the looped molecules, the average length was 1177 ± 17 nm (n = 15), comparable to that of the control DNA.) When transcribed plasmids were subjected to RNaseH treatment, which removes hybridized mRNA, the number of structures observed and the level of DNA condensation fell dramatically (Figure 1C). Now only 8% (n = 53) of the molecules showed structures (all were small blobs) and the mean DNA length was 1190 ± 6 nm, identical to that of untreated plasmid. Frequency distributions of plasmid lengths for each of the three conditions described above are shown in Figure 1D–F, below the corresponding AFM images. These distributions clearly show the shortening of the plasmid after transcription and the return to the original plasmid length after RNaseH treatment. Note also that both the control and RNaseH distributions are relatively narrow, whereas the distribution for the transcribed plasmid is much broader, consistent with a greater variation in plasmid length.
Figure 1.
Low-magnification AFM images of G-quadruplex structures on pPH600 plasmids. Scans (2 × 2 μm2) of samples of untranscribed plasmid (A), transcribed plasmid with positions of structures indicated by arrows (B), and transcribed and RNaseH-treated plasmid (C). Shaded bars on the left of each panel show the height scale from 0–1 nm. (D–F) Frequency distributions of the lengths (in nm) of untranscribed plasmids (D), transcribed plasmids (E) and transcribed and RNaseH-treated plasmids (F). The curves indicate the fitted Gaussian functions. The peaks of the distributions are indicated by the dashed lines.
In control experiments, we found that transcription of a plasmid containing a 660 bp syncollin insert, which was not predicted to form G-quadruplexes, caused structure formation in only 7% of cases, similar to the 8% of pPH600 plasmids showing structures after RNaseH treatment (Supplementary Data 1). Note also that for both transcribed control plasmids and RNaseH-treated pPH600 plasmids, over half of the structures observed were not at the expected position on the DNA, and the mean height of these structures was only 1.07 ± 0.08 nm (n = 7), significantly smaller than the height of the structures on the transcribed G-rich plasmid (1.57 ± 0.06 nm, see Table 1). It is likely, therefore, that these structures do not represent G-quadruplex formation. Significantly, when the pPH600 plasmid was linearized before transcription, the percentage of plasmids showing structures fell to 38% (n = 97; Supplementary data 2), consistent with the known ability of supercoiling to support G-quadruplex formation (23).
Table 1.
Quantitation of structure formation on DNA
| Condition | Plasmids with structures (%) | Structure height (nm) | DNA length (nm) | N |
|---|---|---|---|---|
| Untranscribed | 0 | N/A | 1190 ± 7 | 63 |
| Transcribed (K+) | 93 | 1.57 ± 0.06 | 1053 ± 12 | 56 |
| Transcribed/RNaseH | 8 | 1.10 ± 0.09 | 1190 ± 6 | 53 |
| Transcribed (Na+) | 77 | 1.34 ± 0.01 | 1082 ± 13 | 59 |
| Transcribed (Li+) | 50 | 1.16 ± 0.04 | 1098 ± 11 | 50 |
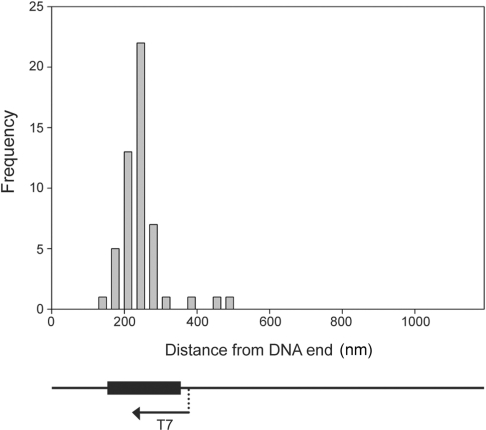
A frequency distribution of the positions of the structures seen on transcribed plasmids is shown in Figure 2, along with a map showing the position of the 604 bp (equivalent to 202 nm) G-rich insert in the plasmid, and the direction of transcription. The position of each structure was taken as the distance of the beginning of the structure from the nearer end of the DNA. Given the direction of transcription, this position represents the point at which transcription terminates. It is clear that nearly all of the structures fall within the region of the insert. Note that there is no way of determining the orientation of the molecule from the AFM images (i.e. which end of the DNA is which).
Figure 2.
Frequency distribution of the positions of the structures on the DNA molecules. The line at the bottom represents the DNA molecule with the position of the T7 promoter shown. The black box represents the G-rich repeat. Both the line and the box correspond to the x-axis of the graph. Since the orientation of the molecule cannot be determined from the AFM images the distribution gives the distances of the structures (the start of the structure in the case of loops) from the nearer end of the DNA.
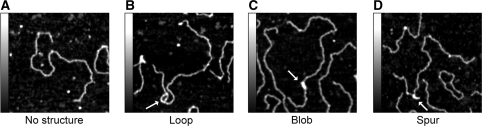
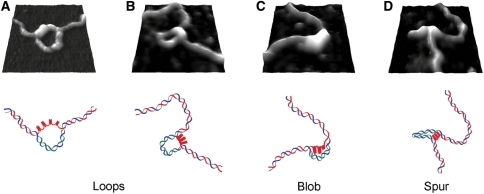
Figure 3 shows images of each type of structure (i.e. loop, blob and spur; arrows) in the context of the entire DNA molecule. Zoomed images, along with interpretive sketches of the DNA molecule together with hybridized mRNA, are shown in Figure 4. We propose that the various structures observed represent different manifestations of the same basic DNA/RNA arrangement, with the different appearances being caused by variations in how the plasmids have attached to the mica substrate. In the asymmetric loops (Figure 4A), for instance, the short strand likely represents the G-quadruplex DNA and the longer strand the RNA/DNA hybrid. Consistent with this idea, small blobs and other sub-structures were often observed on the short strand, which had an average maximum height of 1.3 ± 0.07 nm (n = 15). This is substantially larger than the height of double-stranded DNA (around 0.5 nm, as measured by AFM). In contrast, the long strands had a height comparable to that of double-stranded DNA, as would be expected for a RNA/DNA hybrid. Blobs (Figure 4B) are probably caused by a small number of closely spaced G-quartets. Consistent with this idea, the mean height of the blobs was 1.9 ± 0.1 nm (n = 18). The displaced RNA/DNA could be lying on the mica substrate in such an orientation as to be indistinguishable from the G-quartet DNA. Spurs (Figure 4C) are likely similar to the blob/loop structures, except that both sides of the loop are simply lying on the surface in an orientation where the loophole is not visible. As expected, the spurs appeared thicker than the double-stranded DNA and the mean height at the base of the spurs (the region likely to contain G-quadruplex DNA) was 1.5 ± 0.7 nm (n = 19).
Figure 3.
Zoomed AFM images of G-quadruplex structures on pPH600 plasmids. 600 × 600 nm2 areas of samples of transcribed plasmid showing no structure (A), a loop (B), a blob (C) and a spur (D). Structures are indicated by arrows. Shaded bars on the left of each panel show the height scale from 0–1 nm.
Figure 4.
Zoomed AFM images of G-quadruplex structures with corresponding diagrammatic representations of the DNA arrangment. Areas (140 × 140 nm2) showing regions of transcribed plasmids containing loops (A, B), a blob (C) and a spur (D). The blue lines in the diagrams represent the non-coding strand of the DNA; the red lines represent the G-rich coding DNA strand and the green lines represent hybridized mRNA.
It has been shown previously that the stability of G-quadruplexes falls progressively when the monovalent cation in the buffer is changed from K+ to Na+ and from Na+ to Li+ (24). Table 1 shows a comparison of the relative prevalence of structures and the mean DNA lengths when transcription was carried out in buffer solutions containing KCl, NaCl or LiCl. It is evident that from K+ to Na+ to Li+ the number of structures falls and the measured lengths of the DNA rise. However, all conditions yield a greater number of structures and shorter mean DNA lengths than the untreated plasmid control (and the RNaseH-treated plasmid). Table 1 also shows the values for the mean heights of all the structures (i.e. loops, blobs and spurs) for the various conditions. It is clear that the mean height falls from K+ to Na+ to Li+, indicating a reduction in the mean numbers of G-quadruplexes present in the structures.
The next question we wished to tackle was whether G-loop formation had physiological consequences. G-loops are clearly thermally stable once formed; otherwise we would not be able to visualize them. But do they affect further rounds of transcription? Are they always formed cotranscriptionally or is the process stochastic? We envisaged three possible scenarios: (i) they have no effect on transcriptional activity; (ii) they form after only a single round of transcription and prevent further transcription from occurring or (iii) they form stochastically, but once formed prevent further transcription from occurring.
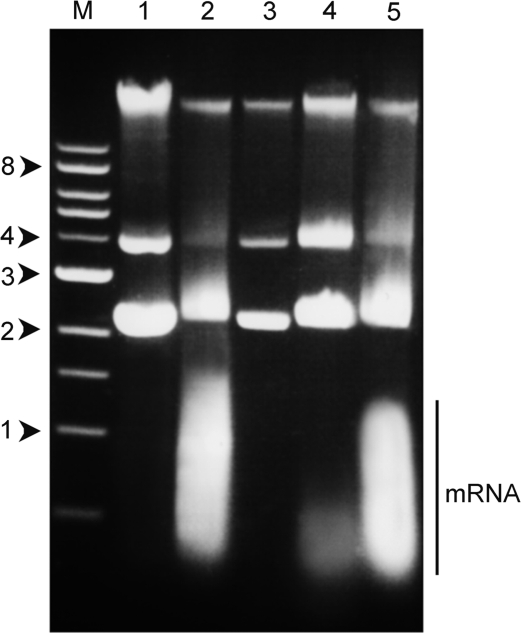
In order to investigate this question, we used agarose gel electrophoresis to estimate the amount of free mRNA formed during transcription of the plasmid. Figure 5 shows a representative gel from this analysis. Lane 1 shows the plasmid alone, with no transcription. The three bands result from different supercoiled states. Lane 2 shows the results of transcription as described earlier, but without the addition of RNase A. A very large smear corresponding to the transcribed mRNA is seen. This result confirms that transcription has occurred successfully and the large ratio of mRNA: plasmid DNA indicates that multiple rounds of transcription have occurred on each plasmid, eliminating scenario (ii). When the transcribed plasmid was treated with RNase A and then purified, the mRNA smear disappeared (lane 3). Lane 4 shows the results of performing a second cycle of transcription on the purified pre-transcribed DNA from lane 2, using freshly added components. There is very little mRNA, indicating that transcription has been curtailed. In contrast, lane 5 shows a plasmid that was treated identically, except that the first cycle was a mock transcription, in which no polymerase was added, although all other components were the same. mRNA production is now clearly visible, indicating that the formation of the G-loop structures seen using AFM imaging prevents further transcription from occurring, as in scenario (iii), above. The small amount of mRNA produced in the second cycle of transcription can be attributed to the 7% of plasmids that did not form a G-loop in the first cycle.
Figure 5.
Effect of prior transcription on mRNA production. Agarose gel electrophoresis was used to analyse the products of transcription of the pPH600 plasmid under various conditions. M: markers (kbp). Lane 1: plasmid alone, with no transcription. Lane 2: plasmid after transcription, showing a large smear corresponding to transcribed mRNA. Lane 3: plasmid after RNase A treatment and purification, showing that all free mRNA has been digested. Lane 4: plasmid after digestion of free mRNA from the first transcription, followed by a second transcription using freshly added components. Note that there is virtually no mRNA, indicating that transcription has been curtailed. Lane 5: plasmid after a mock first transcription, followed by a second real transcription. Note that mRNA production is clearly visible. The position of the mRNA smear is indicated.
DISCUSSION
EM imaging has been used previously to show that transcription of G-rich DNA produces stable loops, indicative of the production of G-quadruplexes (14). We have imaged the same transcribed DNA by AFM. Because of the decreased sample preparation involved, AFM permits visualization of these systems in a more physiologically relevant context, allowing us to observe additional structural features. For instance, we found that the loops are asymmetric and we showed directly that the G-rich strand forms a condensed structure, with the height appropriate to a four-stranded structure. This is in stark contrast to the extended single-stranded regions seen by EM, which are not consistent with the presence of G-quadruplexes in the form visualized. We also detected a greater percentage of structures than the value reported previously (93% of DNA molecules, compared with 63%) and a greater effect of prior linearization of the plasmid on the extent of G-quadruplex formation (14).
It is clear that the observed structures are forming specifically in the G-rich insert region of the DNA. We presume that the structures are formed upon unwinding of the DNA duplex, and that the coding strand contains an mRNA/DNA hybrid, whereas the G-rich non-coding strand contains G-quadruplexes. In comparison with pure native DNA, the transcribed DNA was significantly (12%) shorter in contour length, suggesting that the structures observed are associated with DNA condensation, and strongly supporting the idea that they contain G-quadruplexes. After treatment with RNaseH, the structures disappeared almost completely, and the length of the DNA returned to its native value. This observation indicates that the loop region does indeed contain an mRNA/DNA hybrid and that this region is crucial for the stability of the observed structures. This conclusion is supported by computational predictions, which are entirely consistent with the results observed. The 8% of plasmids still showing structures after RNase H treatment might indicate incomplete digestion of hybridized mRNA. However, these structures were not located at the expected position on the plasmid and they were relatively small, suggesting that transcription might occasionally have created secondary structures other than G-quadruplexes on the non-template strand.
Manipulating the ionic composition of the transcription buffer had a significant effect on the stability of the G-quadruplexes. Replacement of K+ with Na+ and then with Li+ caused a progressive reduction in the percentage of plasmids containing structures and an increase in the contour length of the DNA. These trends indicate that the stability of the G-quadruplexes falls from K+ to Na+ to Li+, consistent with the known effects of monovalent cations on G-quadruplex stability (24). This result also demonstrated that the formation of G-quadruplexes in the coding strand directly affects the formation of the G-loops rather than being an incidental side-effect of their formation.
We have also elucidated for the first time the physiological consequences of G-loop formation on future rounds of transcription. We find that once these structures have been formed and purified, they abolish transcriptional activity, in contrast to direct purification, which allows transcription to continue. Such switching could therefore be used biologically as an off-switch for transcriptional regulation. This behaviour is consistent with the known regulatory role of RNA/DNA hybrids in E. coli plasmid replication (15). We have not investigated the consequences of the presence of a G-loop distal to the promoter, but hypothesize that it may lead to premature termination of the mRNA, possibly leading to different protein products. While the murine Sγ3 switch region appears not to form G-loops on every transcriptional cycle, other sequences may lead to even more stable G-loops, resulting in single-read transcripts, a mechanism that might permit precise regulation of transcript numbers. Further work is required to understand how many transcriptional rounds are typically required to lead to G-loop silencing and to investigate whether other proteins play a role in adjusting this number.
AFM imaging has been used extensively to study nucleic acid structure and the interactions of various proteins with DNA (e.g. 16–18). Significantly, AFM analysis has previously been applied to the structure of immunoglobulin switch region RNA/DNA complexes (25). As in our study, mRNA hybridization to the transcribed DNA was detected, in this case through relaxation of the supercoiling of intact, circular plasmids. However, in contrast to our findings, and for reasons that are not clear to us, no loop formation was observed. In addition, it was reported that omission of RNaseA treatment resulted in the formation of large RNA/DNA aggregates. RNaseH-sensitive loops, similar to the structures seen in our study, were also seen in AFM images of mouse mitochondrial DNA to which RNA had been annealed (26). In contrast, mitochondrial DNA–RNA isolated from a mouse cell line showed no loops, although changes in supercoiling of the mitochondrial DNA on RNaseA treatment did indicate the presence of bound RNA, likely in the form of triplexes.
The preparation of a plasmid for AFM imaging requires only a brief (2 min) incubation with the mica substrate followed by gentle washing. In our study, this simple procedure has provided images of G-quadruplexes that have not previously been seen and revealed how the structure of a DNA molecule is affected by G-loop and G-quadruplex formation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Biotechnology and Biological Sciences Research Council Studentship (to K.J.N.); Research Councils UK Academic Fellowship (to J.L.H.). Funding for open access charge: Department of Pharmacology, and the Isaac Newton Trust, University of Cambridge.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENT
The authors thank N. Maizels for providing the pPH600 plasmid and for helpful discussions.
REFERENCES
- 1.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neidle S, Balasubramanian S. Quadruplex Nucleic Acids. Cambridge: RSC; 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patel DJ, Phan AT, Kuryavyi V. Human telomere, oncogenic promoter and 5′-UTR G-quadruplexes: diverse higher order DNA and RNA targets for cancer therapeutics. Nucleic Acids Res. 2007;35:7429–7455. doi: 10.1093/nar/gkm711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huppert JL. Four-stranded nucleic acids: structure, function and targeting of G-quadruplexes. Chem. Soc. Rev. 2008;37:1375–1384. doi: 10.1039/b702491f. [DOI] [PubMed] [Google Scholar]
- 5.Oganesian L, Bryan TM. Physiological relevance of telomeric G-quadruplex formation: a potential drug target. Bioessays. 2007;29:155–165. doi: 10.1002/bies.20523. [DOI] [PubMed] [Google Scholar]
- 6.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huppert JL, Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35:406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huppert JL, Bugaut A, Kumari S, Balasubramanian S. G-quadruplexes: the beginning and end of UTRs. Nucleic Acids Res. 2008;36:6260–6268. doi: 10.1093/nar/gkn511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin Y, Hurley LH. Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie. 2008;90:1149–1171. doi: 10.1016/j.biochi.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinden RR. Biological implications of the DNA structures associated with disease-causing triplet repeats. Am. J. Hum. Genet. 1999;64:346–353. doi: 10.1086/302271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fojtik P, Vorlickova M. The fragile X chromosome (GCC) repeat folds into a DNA tetraplex at neutral pH. Nucleic Acids Res. 2001;29:4684–4690. doi: 10.1093/nar/29.22.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fry M, Loeb LA. The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl Acad. Sci. USA. 1994;91:4950–4954. doi: 10.1073/pnas.91.11.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fry M. Tetraplex DNA and its interacting proteins. Front. Biosci. 2007;12:4336–4351. doi: 10.2741/2391. [DOI] [PubMed] [Google Scholar]
- 14.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2007;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cesareni G, Helmer-Citterich M, Castagnoli L. Control of ColE1 plasmid replication by antisense RNA. Trends Genet. 1991;7:230–235. doi: 10.1016/0168-9525(91)90370-6. [DOI] [PubMed] [Google Scholar]
- 16.Crampton N, Yokokawa M, Dryden DTF, Edwardson JM, Rao DN, Takeyasu K, Yoshimura SH, Henderson RM. Fast-scan atomic force microscopy reveals that the type III restriction enzyme EcoP15I is capable of DNA translocation and looping. Proc. Natl Acad. Sci. USA. 2007;104:12755–12760. doi: 10.1073/pnas.0700483104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crampton N, Roes S, Dryden D.TF, Rao DN, Edwardson JM, Henderson RM. DNA looping and translocation provide an optimal cleavage mechanism for the type III restriction enzymes. EMBO J. 2007;26:3815–3825. doi: 10.1038/sj.emboj.7601807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Neaves KJ, Cooper LP, White JH, Carnally SM, Dryden D.TF, Edwardson JM, Henderson RM. Atomic force microscopy of the EcoKI type I DNA restriction enzyme bound to DNA shows enzyme dimerisation and DNA looping. Nucleic Acids Res. 2009;37:2053–2063. doi: 10.1093/nar/gkp042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Edwardson JM, An S, Jahn R. The secretory granule protein syncollin binds to syntaxin in a Ca2+-sensitive manner. Cell. 1997;90:325–333. doi: 10.1016/s0092-8674(00)80340-2. [DOI] [PubMed] [Google Scholar]
- 20.Huppert JL. Thermodynamic prediction of RNA-DNA duplex-forming regions in the human genome. Mol. Biosyst. 2008;4:686–691. doi: 10.1039/b800354h. [DOI] [PubMed] [Google Scholar]
- 21.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hori K, Takahashi T, Okada T. The measurement of exonuclease activities by atomic force microscopy. Eur. Biophys. J. 1998;27:63–68. [Google Scholar]
- 23.Sun D, Hurley LH. The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: implications for drug targeting and control of gene expression. J. Med. Chem. 2009;52:2863–2874. doi: 10.1021/jm900055s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bardin C, Leroy JL. The formation pathway of tetramolecular G-quadruplexes. Nucleic Acids Res. 2008;36:477–488. doi: 10.1093/nar/gkm1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mizuta R, Iwai K, Shigeno M, Mizuta M, Uemura T, Ushiki T, Kitamura D. Molecular visualization of immunoglobulin switch region RNA/DNA complex by atomic force microscope. J. Biol. Chem. 2003;278:4431–4434. doi: 10.1074/jbc.M209262200. [DOI] [PubMed] [Google Scholar]
- 26.Brown TA, Tkachuk AN, Clayton DA. Native R-loops persist throughout the mouse mitochondrial DNA genome. J. Biol. Chem. 2008;283:36743–36751. doi: 10.1074/jbc.M806174200. [DOI] [PMC free article] [PubMed] [Google Scholar]