Abstract
Three-way junction RNAs adopt a recurrent Y shape when two of the helices form a coaxial stack and the third helix establishes one or more tertiary contacts several base pairs away from the junction. In this review, the structure, distribution, and functional relevance of these motifs are examined. Structurally, the folds exhibit conserved junction topologies, and the distal tertiary interactions play a crucial role in determining the final shape of the structures. The junctions and remote tertiary contacts behave as flexible hinge motifs that respond to changes in the other region, providing these folds with switching mechanisms that have been shown to be functionally useful in a variety of contexts. In addition, the juxtaposition of RNA domains at the junction and at the distal tertiary complexes enables the RNA helices to adopt unusual conformations that are frequently used by proteins, RNA molecules, and antibiotics as platforms for specific binding. As a consequence of these properties, Y-shaped junctions are widely distributed in all kingdoms of life, having been observed in small naked RNAs such as riboswitches and ribozymes or embedded in complex ribonucleoprotein systems like ribosomal RNAs, RNase P, or the signal recognition particle. In all cases, the folds were found to play an essential role for the functioning or assembly of the RNA or ribonucleoprotein systems that contain them.
Keywords: folding, hammerhead ribozyme, loop, ribonucleoprotein, ribosome, riboswitch, RNase P, signal recognition particle, structure, tertiary interaction, three-way junction
INTRODUCTION
In addition to the classical mRNA, tRNA, and rRNA molecules charged with transferring the genetic information from DNA to proteins, in recent years a plethora of new RNA molecules playing very diverse biological roles, alone or in association with proteins, have been described (Sharp 2009). In many cases, these RNAs fold into well-defined structures ranging in complexity from the simple hairpin or three-way junction folds found in some small ribozymes and riboswitches to the intricate architecture of ribosomal RNA or group II introns (Serganov and Patel 2007).
In the same way as protein architecture can be viewed as the assembly of two main types of structural units (α-helices and β-sheets) connected by loops and packaged by tertiary interactions in three-dimensional space, RNA architecture is essentially built through the assembly of double-stranded helices that pack in space side by side, orthogonally or on top of each other as continuous stacks. Key for organizing the three-dimensional packing of RNA helices are the junctions where three or more helices meet with each other, and tertiary contacts between different helical domains. Whereas the junctions impose topological constraints on three-dimensional helix organization (Lescoute and Westhof 2006; Laing and Schlick 2009), tertiary contacts are thought to modulate the final arrangement of stem–loops in space (Chauhan and Woodson 2008).
In this review we analyze simple RNA systems that are evolutionary and functionally very different, yet similar in terms of three-dimensional structure organization. These RNAs consist of three double helices that intersect at a three-way junction, adopting a recurrent-Y shape stabilized by coaxial stacking and remote tertiary contacts between the helices (Fig. 1). These folds are ubiquitously found in naked RNAs such as riboswitches or ribozymes, and ribonucleoprotein systems like ribosomal RNAs, RNase P, or the signal recognition particle (SRP). The juxtaposition of RNA domains at the junction and at the remote tertiary complexes enables the RNA helices of these motifs to adopt unusual shapes that are used as binding platforms by proteins and other RNA molecules. Moreover, the helical junction and the remote tertiary contact behave as flexible hinges whose structure is capable of responding to changes in the other region. Due to these properties, these motifs play essential functions in a variety of biological processes, representing a key building block in RNA architecture.
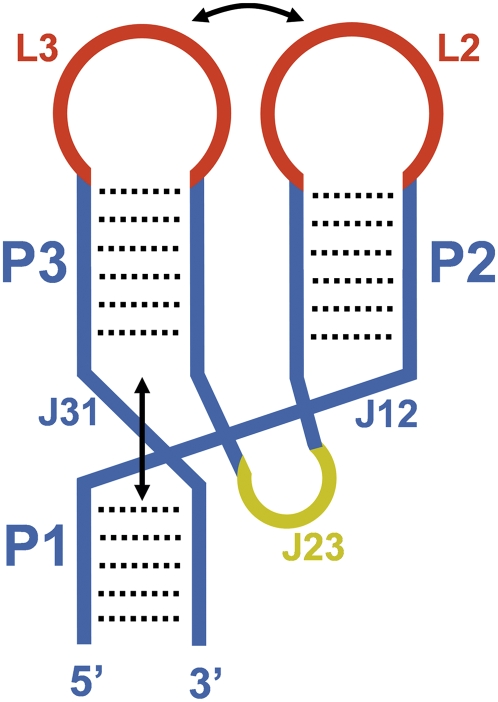
FIGURE 1.
Schematic representation of a three-way junction RNA adopting an archetypical parallel-Y fold. The numbering of the helical domains (P) and loops (L) follows the 5′-to-3′ direction of the RNA molecule. Junctions are indicated with the letter “J” followed by the number of the helices they connect. Base-paired regions and junctions are colored blue except for the J23 junction, which is colored yellow. The loops establishing the remote tertiary interactions are depicted in red.
DESCRIPTION OF THE FOLD, AND FUNCTIONAL AND ARCHITECTURAL CONTEXTS
The RNA folds analyzed in this review comprise three double helices (domains) that, starting from the 5′ end of the RNA, will be identified as P1, P2, and P3 (Fig. 1). The three helices intersect at a three-way junction and adopt a “Y” shape, where the helix occupying the lower part of the “Y” coaxially stacks with one of the helices forming the two upper branches. Tertiary contacts between the two upper domains keep the remaining helix in parallel with the coaxial stack. These tertiary contacts are several base pairs away from the junction and usually involve loops, which will be named L or IL depending on whether they are apical or internal. In order to allow the parallel juxtaposition of the two upper domains, the junction strand (J) connecting these two helices is usually longer than the other two junction strands. In many cases, this strand forms a structured loop that establishes tertiary contacts with the lower helix. As a result of the sum of these tertiary interactions, all of the branches of the “Y” are parallel to each other, and for this reason these motifs will be hereafter identified as parallel-Y folds. These folds would fall within the C family of three-way junctions previously defined by Lescoute and Westhof (2006).
In many parallel-Y folds P1 and P3 are coaxially stacked, P2 and P3 occupy the upper branches of the “Y” and establish the remote tertiary contact, and J23 is the longest of the three junctions; an archetypical system of this type is shown in Figure 1. However, the coaxial stack can also be established between P1 and P2 or P2 and P3, and P1 may occupy one of the two upper branches rather than the lower part of the “Y” (Table 1).
TABLE 1.
Structural features of parallel-Y RNAs
The structural context in which these motifs have been observed is very diverse: they can be stable as isolated motifs (like hammerhead ribozymes) or embedded within larger RNA structures like RNase P or rRNA and can function either as naked RNA structures or forming complexes with proteins or metabolites (Table 1). Despite their global structural similarity (Figs. 2, 3), parallel-Y RNAs play very different functions across all kingdoms of life. This review first follows a functional criterion to group and to describe the structure and biological context of the motifs, and then compares all parallel-Y structures and discusses their mechanistic implications in the final sections of the article.
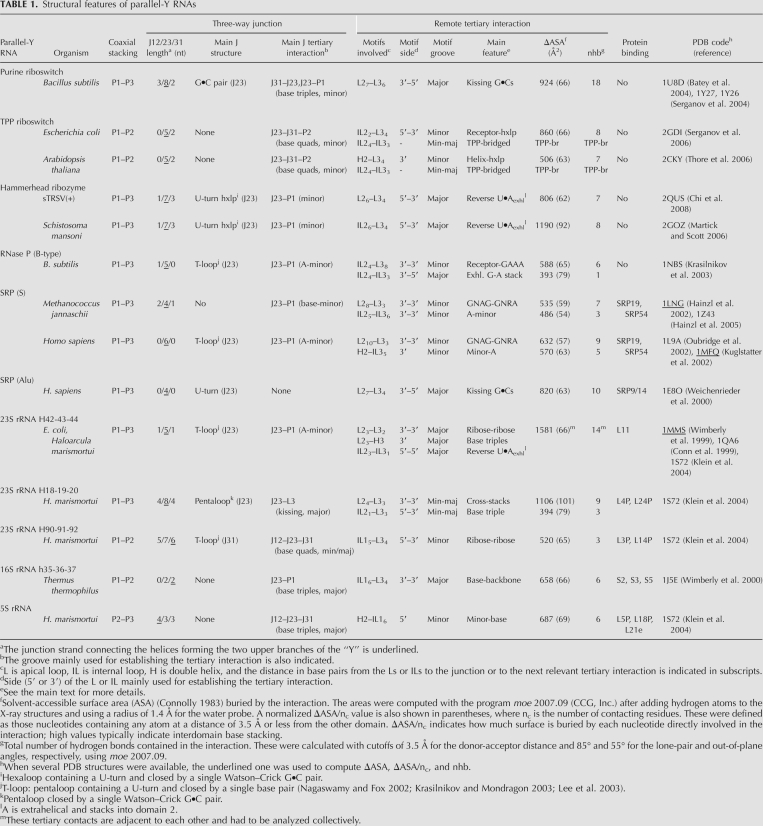
FIGURE 2.
Three-dimensional structures and schematic representations of naked RNAs adopting parallel-Y folds. (A) Aptamer domain of the guanine-responsive riboswitch complexed with the metabolite hypoxanthine (PDB code 1U8D) (Batey et al. 2004); (B) sensing domain of the TPP riboswitch complexed with its ligand (2CKY) (Thore et al. 2006); (C) sTRSV(+) hammerhead ribozyme (2QUS) (Chi et al. 2008); (D) S-domain of B-type RNase P (1NBS) (Krasilnikov et al. 2003). (A–D) Three-dimensional structures are on the left and schematic representations on the right, with the main remote tertiary interactions indicated with symbols (Leontis and Westhof 2001). The color codes are as in Figure 1, and the metabolites in A and B are depicted as spheres.
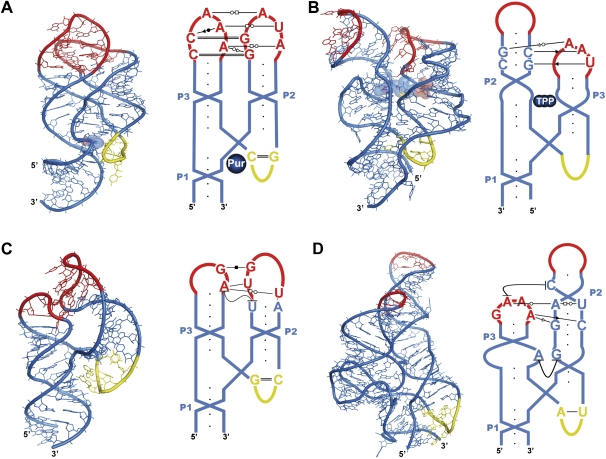
FIGURE 3.
Three-dimensional structures and schematic representations of parallel-Y ribonucleoprotein RNAs and their protein complexes. (A) S-domain SRP RNA complexed with the SRP19 and SRP54 proteins (PDB code 1MFQ) (Kuglstatter et al. 2002); (B) H42–H43–H44 23S rRNA junction complexed with ribosomal protein L11 (1MMS) (Wimberly et al. 1999); (C) Alu-domain SRP RNA complexed with the SRP9/14 heterodimer (1E8O) (Weichenrieder et al. 2000); (D) H18–H19–H20 23S rRNA junction complexed with L4P and L24P (1S72) (Klein et al. 2004); (E) H90–H91–H92 23S rRNA junction complexed with L3P and L14P (1S72) (Klein et al. 2004); (F) 5S rRNA complexed with L5P, L18P, and L21e (1S72) (Klein et al. 2004); (G) h35–h36–h37 16S rRNA junction complexed with ribosomal proteins S2, S3, and S5 (1J5E) (Wimberly et al. 2000). (A–G) Three-dimensional structures are on the left and schematic representations on the right, with the most important remote tertiary interactions indicated with symbols (Leontis and Westhof 2001). The color codes are as in Figure 1 and proteins are represented in green cartoon mode. For clarity, only protein domains with direct interactions with the parallel-Y motifs are shown.
RIBOSWITCHES
Riboswitches are protein-free regulatory systems typically found embedded in the untranslated regions of bacterial metabolic genes. They generally consist of a 5′-sensing (aptamer) domain, which specifically binds metabolites when concentrations exceed a threshold, and a 3′-domain (expression platform) that controls gene expression. Depending on metabolite binding, the sensing domain can adopt alternative structures that influence the folding of the expression platform (Montange and Batey 2008; Batey 2009).
The adenine- and guanine-responsive riboswitches control the transcription of genes associated with purine metabolism in numerous bacterial species. The sensing domains of these riboswitches form similar 70-nucleotide (nt) parallel-Y folds (Fig. 2A; Batey et al. 2004; Serganov et al. 2004) and have strong sequence similarities, containing highly conserved nucleotides at the junction and at the L2 and L3 loops. These loops establish an extensive tertiary interaction across the major groove (Table 1), primarily comprising two Gs in the 3′ side of L2 and two Cs in the 5′ side of L3, which open toward the major groove to form two consecutive antiparallel Watson–Crick G•C pairs, a kissing loop-type interaction. Two noncanonical A•U and A•A pairs buttress the minor groove flanks of the G•C pairs, effectively forming with them two stacked tetrads (Fig. 2A). The ligand binding site is located at the three-way junction, completely encapsulated by the P1 helix and by J12, J31, and J23 nucleotides (Fig. 2A): it stacks above a P1 A•U pair forming a triple with a J23 C nucleotide, and below another triple formed by junction J31, J12, and J23 nucleotides; G versus A selectivity is provided by Watson–Crick pairing with a J31 C or U nucleotide, respectively, the only functionally significant sequence variation between the two types of purine riboswitches (Batey et al. 2004; Serganov et al. 2004). The L2–L3 interaction is essential for ligand binding by these riboswitches (Batey et al. 2004); replacement of the wild-type loops with UUCG tetraloops abolishes ligand binding (Batey et al. 2004), and single mutations of the L2–L3 G•C kissing nucleotides cause strong reductions in ligand affinity (Lemay et al. 2006; Gilbert et al. 2007). These results, together with NMR (Buck et al. 2007; Noeske et al. 2007; Ottink et al. 2007), chemical probing (Stoddard et al. 2008), and single-molecule force microscopy experiments (Greenleaf et al. 2008), indicate that the primary role of the loop–loop interaction is a preorganization of the parallel-Y fold that precedes ligand binding, where the junction remains in a locally disordered state (necessary for ligand entry). Purine binding then leads to a local structuring of the junction via the network of interactions with P1 and junction nucleotides (Batey et al. 2004; Serganov et al. 2004), which also results in a further tightening of the remote L2–L3 tertiary structure (Buck et al. 2007).
TPP-sensing riboswitches also adopt parallel-Y folds. They respond to the coenzyme thiamine pyrophosphate and have been identified in thiamine-biosynthetic genes of bacteria and also fungi and plants, where they regulate mRNA splicing and stability (Cheah et al. 2007). The topology of TPP riboswitches is different from all of the three-way junctions studied in this review, because while P2 and P3 occupy the two upper branches of the “Y,” the coaxial stacking is between helices P1 and P2 instead of P1 and P3 (Fig. 2B; Table 1; Serganov et al. 2006; Thore et al. 2006). There are two groups of remote tertiary contacts between helices P2 and P3: a first set of contacts takes place between a L3 apical hexaloop (poorly conserved in sequence) and the upper part of domain P2. This interaction takes place across the minor groove and mainly involves nucleotides of the 3′ side of L3 (Fig. 2B), resembling the interaction between GNRA tetraloops and their receptors (Cate et al. 1996a). The second group of interactions takes place between two highly conserved internal loops of helices P2 and P3 (IL2 and IL3) and is bridged by the TPP ligand, which is positioned perpendicular to the two helical domains in an extended conformation. The minor groove side of IL2 forms a cavity that binds the pyrimidine ring of TPP via hydrogen bonding and stacking interactions, whereas the pyrophosphate group is lodged in the major groove of IL3 with the help of several magnesium ions (Fig. 2B; Serganov et al. 2006; Thore et al. 2006). In the ligand-bound structures, the conformation of the three-way junction is quite compact, because J23 and J31 form two consecutive, highly conserved A•G and A•A pairs whose A's buttress two adjacent G•C pairs within P2 by forming A-minor (Doherty et al. 2001; Nissen et al. 2001) interactions. Whereas the upper L3–P2 tertiary interaction probably contributes to lock into register IL2 and IL3 for ligand binding, the bridging by the TPP molecule of these two loops is expected to play a decisive role in the stabilization of the three-way junction.
Either by direct binding to the junction or by an allosteric mechanism involving P2 and P3 bridging, ligand binding to the purine and TPP riboswitches always results in the tightening of the junction structure and the stabilization of the nearby P1 helix containing the switching sequence (Fig. 2A,B); the sequestering of this sequence by P1 impedes alternative foldings of the expression platform (Montange and Batey 2008; Serganov 2009).
HAMMERHEAD RIBOZYMES
Hammerhead ribozymes are small catalytic RNAs initially discovered in viroids and virus satellite RNAs (Flores et al. 2001) and more recently identified in mRNAs of plant (Przybilski et al. 2005) and animal (Martick et al. 2008) species. They catalyze the specific cleavage of their own RNA backbone through a transesterification reaction, a step required for the replication of infectious circular RNA molecules.
Hammerhead ribozymes fold into typical parallel-Y structures (Fig. 2C). The catalytic center is located at the three-way junction and self-cleavage takes place at a J12 nucleotide, releasing 2′,3′-cyclic phosphodiester and 5′-OH termini. Remote loop–loop interactions between domains P2 (domain I in the literature) and P3 (II) have been shown to greatly increase (from 100- to 1000-fold) the catalytic activity of these ribozymes (De la Peña et al. 2003; Khvorova et al. 2003) by promoting the adoption by the junction of a catalytically active conformation (Martick and Scott 2006). The L2–L3 interactions are established across the major groove of the helices and are quite extensive (Table 1), comprising a complex network of noncanonical base pairs as well as base interdigitations (Martick and Scott 2006; Chi et al. 2008) (Fig. 2C).
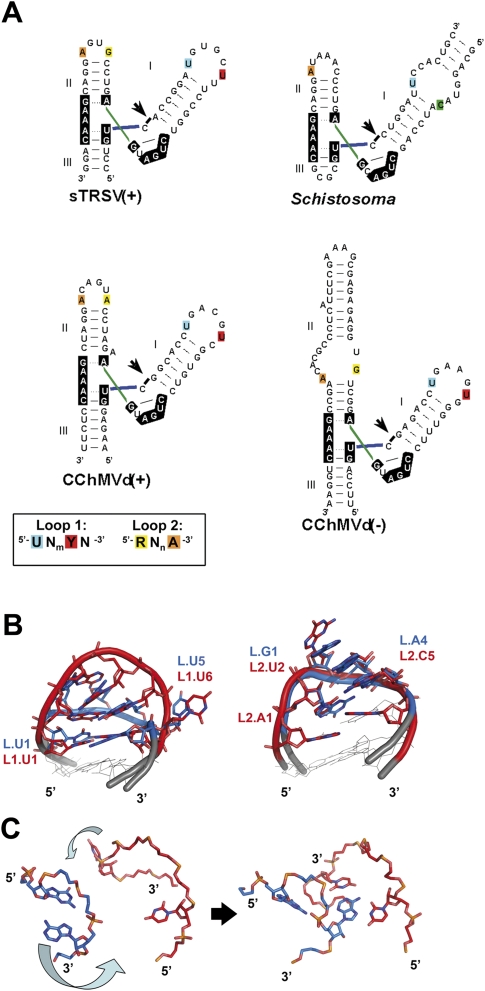
In contrast to the highly conserved sequence of the catalytic site nucleotides, there are many sequence and topology differences within the L2 loops and L3 loops of natural hammerheads (Fig. 4A; Dufour et al. 2009). Despite these differences, a conserved tertiary interaction motif embedded within the loop–loop complexes has been detected in at least four different hammerheads (Fig. 4; Dufour et al. 2009). The 5′ U of L2 and the 3′ A of L3 form a reverse Hoogsteen pair across the major groove in the X-ray structures of the Schistosoma mansoni (Martick and Scott 2006) and tobacco ring spot virus satellite RNA (Chi et al. 2008) (sTRSV) hammerheads, a base pair also supported by site-directed mutagenesis experiments in the hammerheads of the (+) and (−) polarity RNAs of chrysanthemum chlorotic mottle viroid (CChMVd) (Dufour et al. 2009). In addition, the penultimate U of L2 in the sTRSV hammerhead forms a Watson–Crick pair with the 3′ A of L3, so that a base triple is formed by these two nucleotides and the L2 5′ U (Chi et al. 2008); this base-triple interaction is also consistent with the mutagenesis analyses of CchMVd(+) and CchMVd(−) hammerheads (Dufour et al. 2009). In the Schistosoma hammerhead this U:A:U base triple is not observed, but the last pyrimidine of L2 is extrahelical and protrudes into L3, facilitating the displacement of the 3′ A of L3 to form the reverse Hoogsteen pair with the 5′ U of L2 (Martick and Scott 2006). An NMR spectroscopy study of the undocked loops of the CChMVd(+) and CChMVd(−) hammerheads, combined with a comparative analysis of L2s and L3s in 27 natural hammerheads, revealed that the 5′ nucleotide of L2s is usually a U that is exposed in the major groove, whereas the penultimate nucleotide is an extrahelical pyrimidine. On the other hand, the last two nucleotides of L3s are usually dynamic and/or opened toward the minor groove (Fig. 4; Dufour et al. 2009). The presence of the available pyrimidines in the major groove of L2 would trigger a further turning of the last two nucleotides of L3 toward the minor groove, so that the 3′ A of L3 forms a reverse Hoogsteen pair with the 5′ U of L2 across the major groove and, in some cases [i.e., in sTRSV(+) and CChMVd(+) and CChMVd(−) hammerheads], a base triple with the extrahelical L2 pyrimidine (Fig. 4C). The L2 and L3 nucleotides involved in this tertiary interaction motif are conserved among natural hammerheads (Fig. 4A), and mutations affecting these residues produce large reductions of catalytic activity (Khvorova et al. 2003; Dufour et al. 2009).
FIGURE 4.
Three-dimensional structure conservation of the undocked L2 and L3 loops of natural hammerhead ribozymes and proposed triggering mechanism of the conserved tertiary interaction motif. (A) Sequence and secondary structure of the sTRSV(+), Schistosoma, CChMVd(+), and CChMVd(−) hammerhead ribozymes. Nucleotides matching the UNmYN L2- and RNnA L3-conserved sequence motifs (Dufour et al. 2009) are color coded as indicated in the scheme. (B, left) Superposition of the UGAAGUG CChMVd(−) L2 heptaloop (red) (PDB code 2RO2) (Dufour et al. 2009) with a UCAAUG hexaloop (blue) from 23S rRNA (1NJP) (Harms et al. 2001). UCAAUG-related hexaloops form the most frequently found class of L2’s in natural hammerheads (Dufour et al. 2009). (Right) Superposition of the AUGACA CChMVd(+) L3 hexaloop (red) (2RPK) (Dufour et al. 2009) with a GAAA tetraloop (blue) (1KH6) (Kieft et al. 2002). GNRA tetraloops (Jucker et al. 1996) are the most frequently found class of L3’s in natural hammerheads (Dufour et al. 2009). (C) Proposed triggering mechanism of the tertiary interaction motif likely conserved in most natural hammerheads. L2 and L3 are colored red and blue, respectively, and only the bases of the nucleotides involved in the conserved motif are shown. The undocked L2 (UGAAGUG; 2RO2) (Dufour et al. 2009) and L3 (GAAA; 1KH6) (Kieft et al. 2002) are on the left, and the sTRSV(+) loop–loop complex (PDB 2QUS) (Chi et al. 2008) on the right.
The substantial opening toward the minor groove (about 180°) of the last two nucleotides of L3 to interact with L2 across the major groove is highly unusual: L3s include, for example, GNRA loops (Fig. 4B), which commonly establish tertiary interactions across the minor groove (Cate et al. 1996a). This unusual L2–L3 interaction likely contributes to the observed overwinding of helix P3 and to the severe bending and unwinding of helix P2, which in turn produce crucial conformational changes at the catalytic junction located 5–6 base pairs (bp) away from the interaction (Fig. 5A,B). Namely, the J23 junction becomes more structured, gaining one Watson–Crick base pair and moving away from the P1 helix, and the J12 cleavage nucleotide shifts to the J31 side of the junction, becoming properly oriented for in-line attack by J31 residues (Martick and Scott 2006).
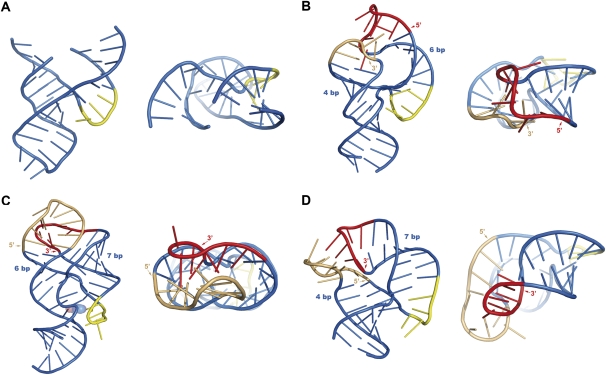
FIGURE 5.
Effects of distal tertiary interactions on the shape of parallel-Y RNAs. (A) Minimal hammerhead ribozyme construct without L2 and L3 loops (PDB code 1MME) (Scott et al. 1996); (B) sTRSV(+) full-length hammerhead ribozyme (2QUS) (Chi et al. 2008); (C) sensing domain of the guanine-responsive riboswitch (1U8D) (Batey et al. 2004); (D) SRP Alu domain RNA (1E8O) (Weichenrieder et al. 2000). (B–D) The side (5′ or 3′) used by the loops for establishing the remote tertiary interaction is indicated together with the number of base pairs separating the loops from the three-way junction. Two views of each structure are provided, from the major groove of L2 (left) and down the long axis of the “Y” (right). The tertiary interaction is established across the major groove of the loops in all three structures, but depending on loop sides and stem lengths, P2 and P3 have different degrees of bending and winding, and the effect on the global shape of the parallel-Y and on the structure of the three-way junction can be easily observed.
SIGNAL RECOGNITION PARTICLE (S DOMAIN)
The SRP is a universally conserved ribonucleoprotein that mediates the cotranslational targeting of membrane proteins to the endoplasmic reticulum or the bacterial plasma membrane (Egea et al. 2005; Halic and Beckmann 2005). In eukaryotic and archaeal cells, the SRP RNA is composed of ∼300 nt arranged in two domains, named S and Alu. In mammals, six different proteins bind to the SRP RNA, whereas in archaea only SRP19 and SRP54 homologs have been identified so far. In eubacteria the SRP comprises a smaller RNA bound to an SRP54 homolog, the only SRP protein component present in all organisms. SRP54 plays a major role in SRP function, since it binds the signal sequence of the nascent peptides and directly interacts with the SRP receptor at the membrane.
The S domain of eukaryotic and archaeal SRPs is ∼100-nt long and adopts a typical parallel-Y fold (Fig. 3A). The loops capping helices P2 (helix 6 in the literature) and P3 (helix 8) are conserved GNAG and GNRA tetraloops, respectively, and establish tertiary interactions across the minor groove that mainly consist of side-by-side base–base and base–backbone hydrogen-bonding contacts, with no base interdigitations (Table 1; Hainzl et al. 2002; Kuglstatter et al. 2002; Oubridge et al. 2002; Hainzl et al. 2005). Both tetraloops adopt similar conformations in their undocked form, with the last three nucleotides stacked and opened toward the minor groove (Jucker et al. 1996; Sakamoto et al. 2002). In fact, GNRA loops usually establish this type of tertiary interactions across the minor groove (Cate et al. 1996a), which is quite different from the one observed in hammerhead ribozymes (see above). The SRP19 protein binds to this L2–L3 complex from the major groove side (Fig. 3A), contributing to clamp together helices P2 and P3 (Hainzl et al. 2002; Kuglstatter et al. 2002; Oubridge et al. 2002). SRP19 needs a preformed L2–L3 complex to bind, since mutations affecting nucleotides involved in important RNA tertiary contacts destabilize the protein–RNA complex (Zwieb 1992).
Further down the parallel-Y, helix P2 consists of an almost regular A-form stem, but helix P3 contains two highly conserved internal loops that comprise the binding site of the SRP54 protein (Fig. 3A). The internal loop closest to the tip of P3 is symmetric and likely to be rigid because of the mostly continuous stacking of base pairs (Hainzl et al. 2002; Oubridge et al. 2002), whereas the other one (IL3) is asymmetric and provides a further flexible hinge region (Schmitz et al. 1999) in addition to the three-way junction (Fig. 3A). IL3 undergoes a dramatic structural rearrangement upon SRP54 binding, exposing some bases outside of the RNA helix for interaction with the protein (Batey et al. 2000; Kuglstatter et al. 2002; Oubridge et al. 2002). The main function of helix P2 and of the apical L2–L3 tertiary interaction is the stabilization of this protein–RNA interaction, which is essential for SRP function. In the archaeal SRP, two consecutive A's loop out of helix P2 and form A-minor motif interactions (Doherty et al. 2001; Nissen et al. 2001) with base pairs flanking IL3, providing another point of attachment between the two helices—in addition to the apical L2–L3 interaction—which promotes a compact side-by-side association of the two helices (Yin et al. 2004). Binding of SRP19 stabilizes the L2–L3 interaction and causes a substantial repositioning of helix P2 relative to helix P3 that, transmitted through the A-minor P2–P3 interaction, results in the splaying out of the bases of IL3 (Hainzl et al. 2002, 2005), which can now interact with SRP54. However, spontaneous fluctuations of the two helices in the absence of SRP19 likely generate similar favorable conformations for SRP54 binding (Hainzl et al. 2005) and this probably accounts for the observed intrinsic affinity of free archaeal SRP RNA for SRP54 (Bhuiyan et al. 2000; Diener and Wilson 2000). In mammalian SRP RNA the additional A-minor P2–P3 interaction does not exist in the absence of SRP54 binding, so that due to the flexibility provided by the three-way junction and asymmetric IL3 (Schmitz et al. 1999) hinge regions, the relatively weak L2–L3 apical interaction does not bring the two helices side-by-side as efficiently. The clamping of the apical loops by SRP19 induces a closer side-by-side interaction of P2 and P3, so that when SRP54 binds to P3 and collapses the asymmetric loop, two A's in the short strand of IL3 flip out of the helix to form A-minor motif interactions with helix P2, stabilizing the SRP54-P3–P2 ternary complex. The structure of the SRP54 bound-P3 would not be stable in the absence of a firm juxtaposition of helices P2 and P3 (Kuglstatter et al. 2002), explaining the greater requirement of mammalian SRPs for SRP19 (Walter and Blobel 1983).
RIBOSOMAL RNA (23S rRNA COMPLEX WITH L11)
The highly conserved 58-nt parallel-Y motif formed by helices 42, 43, and 44 of 23S rRNA (Fig. 3B) was one of the first structures of this type to be described (Conn et al. 1999; Wimberly et al. 1999). The two upper domains of the “Y” (H43 and H44) bind to ribosomal protein L11, and the complex forms a knob protruding above the peptidyl transferase cleft of the large ribosomal subunit (Ban et al. 2000). This complex interacts with the EF-G and EF-Tu elongation factors during protein synthesis. The parallel-Y fold is characterized by severely distorted and unwound P2 (H43) and P3 (H44) domains. P2 comprises an internal loop (IL2) and a large and bent 9-nt apical loop (L2), whereas P3 has an internal loop (IL3) just below two G•C pairs leading to its apical loop (Fig. 3B). The fold is quite compact because the two unwound helices establish extensive tertiary interactions along their major grooves (Table 1; Conn et al. 1999; Wimberly et al. 1999). Two G and C nucleotides protruding out of the 3′ side of L2 bind to the major groove of the P3 G•C pairs, forming two consecutive triples that rest above a third triple formed by IL3 nucleotides, which acts as a stacking platform reminiscent of the A platforms observed in group I introns (Cate et al. 1996b). Just before this triple, the essential 5′ A of IL3 (A1088 in E. coli) protrudes out of the P3 domain in a syn conformation and stacks within the P2 helix, forming a reverse Hoogsteen pair with the locally inverted 5′ U nucleotide of IL2, which is part of an S-turn motif (Szewczak et al. 1993; Wimberly et al. 1993). At the junction, the J23 nucleotides form a T-type pentaloop (Nagaswamy and Fox 2002; Krasilnikov and Mondragon 2003; Lee et al. 2003), and two A's protruding from this loop buttress the minor groove of the P1 helix via ribose–ribose (Cate et al. 1996a) and A-minor (Doherty et al. 2001; Nissen et al. 2001) interactions. The nucleotides involved in the P2–P3 and J23–P1 tertiary interactions described above are conserved across all phylogenetic domains, and mutations that destabilize the Y fold above a certain threshold (like A1088U) abolish L11 binding and are lethal in vivo (Lu and Draper 1994; Conn et al. 1998; Maeder et al. 2006). The Ct domain of L11 recognizes the flat surface provided by the minor groove side of the bent and unwound P2 domain. Although the interaction mainly involves contacts with the RNA backbone, L11 also establishes direct hydrogen bonds with the 5′ U of IL2 and the 5′A of IL3 (A1088) forming the reverse Hoogsteen pair (Conn et al. 1999; Wimberly et al. 1999). Thus, L11 indirectly and directly promotes the P2–P3 tertiary interaction by stabilizing the P2 conformation and by locking into place the P3–P2 A•U base pair.
OTHER PARALLEL-Y RNAS
RNase P
A parallel-Y fold can also be found embedded in the specificity (S) domain of B-type bacterial RNase P (Krasilnikov et al. 2003). RNase P is a universal ribonucleoprotein that processes the 5′ end of pre-tRNA molecules (Kazantsev and Pace 2006; Torres-Larios et al. 2006). The main tertiary interaction between domains P2 and P3 (P10.1 and P12 in the literature) is a GAAA tetraloop–receptor interaction (Cate et al. 1996b) established across the minor groove of the helices (Fig. 2D). Further down the “Y,” the P3 helix contains a large and highly structured internal loop that provides additional tertiary contacts with an internal loop in helix P2 via stacking of bulged-out A and G nucleotides. The nucleotides of the J23 junction form a T-type pentaloop (Nagaswamy and Fox 2002; Krasilnikov and Mondragon 2003; Lee et al. 2003), from which two A's buttress the minor groove of the P1 helix at the bottom of the “Y” via A-minor interactions (Doherty et al. 2001; Nissen et al. 2001). The structure of the P1 domain is quite complex, since immediately below the Y-junction it forms an X (four-way)-junction, where P1 coaxially stacks with another helix (P7) leading to the catalytic domain, and packs side by side with helix P9 via tertiary stacks between bulged-out A's (Krasilnikov et al. 2003). The presence of these consecutive parallel-Y and parallel-X motifs within the S-domain of RNase P is particularly interesting, because they likely play a role in pre-tRNA recognition: footprinting experiments (LaGrandeur et al. 1994; Odell et al. 1998) indicate that the TΨC arm of the pre-tRNA docks into a clamp-like opening located between helix P9 and the complex internal loop of helix P3, which contains universally conserved nucleotides (Fig. 2D; Krasilnikov et al. 2003). Thus, multiple tertiary interactions within the S domain may modulate tRNA recognition in addition to RNase P folding.
Alu domain of the SRP
In mammals, this 100-nt domain contains the 5′ and 3′ ends of the SRP RNA and, together with the SRP9/14 protein heterodimer, delays the elongation of the nascent protein chain targeted by the SRP (Egea et al. 2005; Halic and Beckmann 2005). The 5′ region of the domain forms a parallel-Y fold stabilized by a L2–L3 tertiary interaction involving three antiparallel G•C Watson–Crick pairs (Weichenrieder et al. 2000). The junction J23 nucleotides are highly conserved and form a U-turn, to which the SRP14 component of SRP9/14 binds from the side opposite P1 (Fig. 3C). It has been suggested that the P1 helix, which links the SRP Alu and S domains, folds back on itself and binds the SRP9 side of the heterodimer (Weichenrieder et al. 2000). This involves a sharp turn that would nevertheless be facilitated by the presence of the SRP RNA 5′- and 3′-ends within the P1 helix, 3 bp away from the junction. The function of the parallel-Y motif formed by the Alu RNA appears to be, in this case, the formation of an appropriate platform for SRP9/14 binding: mutations disrupting the loop–loop interaction abolish SRP9/14-RNA binding and SRP assembly (Huck et al. 2004).
Additional ribosomal motifs
In addition to the H42–H43–H44 complex with L11, the 23S rRNA H18–H19–H20 and H90–H91–H92 junctions adopt parallel-Y folds (Ban et al. 2000). The first is located within 23S rRNA domain I, relatively close to the rRNA 5′ end, is contacted from opposite sides by ribosomal proteins L4P and L24P (Klein et al. 2004) (Fig. 3D), and plays a critical role in the assembly of the large subunit (Skinner et al. 1985). The second junction belongs to domain V of 23S rRNA. In this motif, the coaxial stacking is established between P1 and P2 rather than P1 and P3; P1 and P3 are the interacting domains occupying the two upper branches and P2 forms the lower part of the “Y” (Fig. 3E; Table 1). The solvent-exposed side of this parallel-Y (called loop A in the literature) forms the A-site of the peptidyl transferase cleft, whereas the opposite side is bound by proteins L3P and L14P (Fig. 3E). In contrast to the H42–H43–H44 junction, this parallel-Y is inverted relative to the surface of the ribosome. The P1 (H90)–P3 (H92) complex is at the bottom of the subunit cleft. Two G and U solvent-exposed bases at the tip of L3 have been shown to form antiparallel base pairs with the 3′ CCA tail of aminoacyl tRNA analogs (Schmeing et al. 2005). The P1 internal loop that establishes the remote contact with L3 accommodates the tRNA aminoacyl moiety (Schmeing et al. 2005) and is close to the catalytic nucleotides of the ribosome at the base of helix 89. At the other side of the “Y,” the third domain (P2 or H91) protrudes out of the subunit cleft and is packed between the upper portions of H89 and H95. In particular, at the top of helix 95 is the sarcin-ricin loop (SRL), which rests against the tip of P2 (Ban et al. 2000) and is essential for EF-G and EF-Tu binding (Moazed et al. 1988; Wool et al. 1992). The 120-nt 5S rRNA of the 50S subunit adopts a parallel-Y conformation, where the coaxial stacking is established between domains P2 and P3 (Fig. 3F; Table 1). This Y fold is bound by ribosomal proteins L5P, L18P, and L21e, which contribute to glue the 5S rRNA molecule to the rest of the subunit (Ban et al. 2000). At the head of the small ribosomal subunit, helix 35 (P1) also forms a parallel-Y fold with h36 (P2) and h37 (P3), which is bound by ribosomal proteins S2, S3, and S5 (Fig. 3G; Wimberly et al. 2000).
STRUCTURE–FUNCTION RELATIONSHIPS
Three-way junction
A number of structural features are shared by most parallel-Y folds studied in this review (Table 1). In order to allow for the parallel juxtaposition of the coaxial stack with the third helix, the junction strand connecting the two upper branches of the “Y” usually contains more nucleotides than the other two junction strands (Table 1; Lescoute and Westhof 2006) and makes a sharp turn when traveling from one helix to the next helix (Figs. 1, 2, 3). In addition, the topology of the three-way junction is remarkably conserved: one of the strands of the helix occupying the lower branch of the “Y” always travels in a 5′-to-3′ direction across the coaxial stack and toward the third helix (Figs. 2, 3), leaving the minor groove of the lower helix available for interactions with nucleotides of the long junction (Table 1; Lescoute and Westhof 2006). Most junctions maintain this topology regardless of the P1–P3 distribution among the three branches of the “Y.” In the H90–H91–H92 and h35–h36–h37 ribosomal junctions, for example, the coaxial stack is established between P1 and P2, P1 and P3 occupy the upper branches of the “Y,” and the junction strand making the sharp turn is J31 (Fig. 3E,G); in 5S rRNA there is P2–P3 coaxial stacking and P1 and P2 occupy the upper branches of the “Y” connected by the J12 junction strand (Fig. 3F). The topology of these junctions, however, does not vary relative to that of classical folds with P1–P3 stacking (Figs. 1–3). The fold contained in the TPP riboswitch represents the only example of a parallel-Y with an unusual junction topology. In this system P2 and P3 occupy the two upper branches of the “Y,” but the coaxial stack is between P1 and P2 instead of P1 and P3, and the direction of the strand connecting the third helix with the lower domain of the coaxial stack is inverted (Fig. 2B). The global shape of this fold, however, is remarkably similar to that of parallel-Y's with standard topologies (Figs. 2, 3).
Regardless of junction topology, the long and bent junction strand connecting the two upper helices of the “Y” often forms a U-turn structure or a more complex loop structure containing a U-turn, like T-type pentaloops (Nagaswamy and Fox 2002; Krasilnikov and Mondragon 2003; Lee et al. 2003) or U-turn-containing hexaloops closed by a single base pair (Table 1). From these T- or T-like loops, one or two bases often contact the minor groove of the lower helix through A-minor (Doherty et al. 2001; Nissen et al. 2001) or equivalent interactions. In several cases, however, the long junction does not form a structured loop, but participates in a network of interactions (usually involving base triples or quadruples) with other junction strands or helical domains. These unusually compact junctions are observed in riboswitches and ribosomal junctions (Table 1) and are connected to the switching and folding roles played by these systems.
Distal tertiary contacts
The remote tertiary interactions between the domains occupying the upper branches of the Y are more variable, since each system has evolved in a different way by varying the nature of the contacts, the number of base pairs separating them from the junction or other relevant regions of the motif, and the presence or absence of internal loops distorting the stems (Table 1). On average, these interactions take place at about half a helix turn (5 bp) from the junction (Table 1). A quantification of the surface areas and hydrogen bonds involved in the contacts (Table 1) indicates that, in general, tertiary interactions established across the major groove tend to be more extensive and contain more interdomain base stacks than those established across the minor groove, which often consist of side-by-side contacts without extensive base cross-stacking (Table 1). This is a logical consequence of the groove shapes: whereas the bases and sugars are more accessible in the shallow (minor) groove, greater distortions are required to make the nucleoside groups available for tertiary interactions across the deep (major) groove. For example, the apical regions of the parallel-Y motifs contained in the TPP riboswitch, the S domain of the SRP, and RNase P interact across the minor groove, but these interactions are always reinforced by additional tertiary interactions further down the fold, or by protein binding to the RNA tertiary complex (Table 1). In contrast, interactions across the major groove usually comprise extensive antiparallel kissing interactions with multiple hydrogen bonds, as observed in purine riboswitches, or involve the intercalation of key nucleotides within the base stack of the other domain, as observed in hammerhead ribozymes and the H42–H43–H44 23S rRNA junction (Table 1).
A comparison of the parallel-Y folds formed by a minimal hammerhead construct lacking the distal tertiary interaction (Fig. 5A; Scott et al. 1996), the sTRSV full-length hammerhead (Fig. 5B; Chi et al. 2008), and the aptamer domain of purine riboswitches (Fig. 5C; Batey et al. 2004; Serganov et al. 2004) provides clues about the effects of the distal tertiary contact and the length of the stems separating it from the junction. Both the sTRSV hammerhead and the purine riboswitch aptamer form naked Y-folds containing a single P2–P3 tertiary interaction. This interaction is established across the major groove of two apical loops separated from the junction by mostly standard base-paired stems (Table 1). Mainly due to a shorter P3 stem and to the unusual opening and interdomain stacking of the conserved 3′ A of L3 to interact with the 5′ U of L2 in the tertiary complex (Fig. 4C), P2 is bent and P3 is partially overwound in the full-length hammerhead (Fig. 5B) compared with the minimal hammerhead (Fig. 5A) and the purine riboswitch (Fig. 5C) folds. In the purine riboswitch fold, the P2 and P3 stems have similar lengths, and the main tertiary interaction involves different loop sides (the 3′ side of L2 and 5′ side of L3) relative to the hammerhead fold. As a consequence, P2 and P3 are not overwound or bent, and a tight binding site can be formed for the purine ligand at the junction. The bending of P2 and overwinding of P3 in full-length hammerheads, on the other hand, allows the junction to adopt a catalytically competent conformation (Martick and Scott 2006). In fact, different natural hammerhead ribozymes may have tuned their self-cleaving activities by varying the length and irregularities of their P2 and P3 stems (Fig. 4A).
The parallel-Y fold formed by the Alu domain of SRP RNA (Weichenrieder et al. 2000) also contains a single tertiary interaction established across the major groove of two apical loops and provides another example of how the distal tertiary contact determines the shape of the Y structure. As in hammerhead ribozymes, P2 is longer than P3, but in the Alu fold, the P3 domain is bent and unwound instead of overwound (Fig. 5, cf. B and D). This is because the main tertiary interactions take place between the 3′ side of L2 and the 5′ side of L3; in this case, P3 needs to unwind to bring into plane the bases forming the two L2–L3 kissing G•C pairs (Fig. 5D).
Distortions in the P2 and P3 domains relative to regular A-form geometry also have an important impact in the conformation of the motif as illustrated by the 23S rRNA–L11 complex (Fig. 3B; Conn et al. 1999; Wimberly et al. 1999). In this case the motif is unusually compact because the P2 and P3 helices are distorted by IL2 and IL3 internal loops and a large and twisted L2 apical loop. These deformations unwind the helices and allow both domains to contact each other across the major groove along their entire length (Table 1).
Folding
The folding mechanism of these RNA motifs is relevant because some of these structures exert their function in a dynamic context, as they are transcribed. From the observations gathered in this review, the most important factor governing the shape of these motifs appears to be the presence of a junction strand that has a sufficient number of nucleotides to allow the parallel arrangement of the helices occupying the two upper branches of the “Y.” In fact, the most highly conserved structural motif in parallel-Y's is a U-turn within this long junction strand (Table 1), allowing it to make a sharp turn when traveling from the first to the second upper domains. The position of this junction in the nucleotide sequence governs the distribution of the P1, P2, and P3 domains among the three branches of the Y (Table 1). However, once this condition is fulfilled, the remote tertiary contacts established between the two upper domains play a major role in fixing the global shape of the parallel-Y (Fig. 5), which is a likely requirement for function. This distal interaction usually involves apical or internal loops that have either conserved sequences or conserved three-dimensional structures (Fig. 4). As long as these contacts are sufficiently stable, parallel-Y's can keep a globally similar three-dimensional shape while exhibiting an unusual junction topology, as observed in the TPP riboswitch (Fig. 2B). In this regard, tertiary interactions between helices have been recently shown to determine the accuracy of group I intron RNA folding, biasing the ensemble of secondary structures toward native-like conformations (Chauhan and Woodson 2008), and the conserved undocked conformations of the interacting loops have been proposed to play a key role in triggering the specific loop–loop contact leading to catalytically active hammerhead ribozymes (Fig. 4; Dufour et al. 2009).
The side-by-side packing of double-helices observed in parallel-Y RNAs is surprisingly common among RNA structures (Murthy and Rose 2000), showing that tertiary interactions and ion-binding effects can offset the electrostatic repulsion between the negatively charged phosphate backbones (Bassi et al. 1997; Murthy and Rose 2000; Penedo et al. 2004).
Dynamic effects
An important property of a number of parallel-Y folds is that both the junction and the remote tertiary interaction have been shown to behave as flexible hinges that can respond to distal conformational changes in the other region. These changes may be caused by the binding of metabolites, auxiliary proteins, or even by the folding of the 3′ domain (P3) during transcription. The changes are transmitted through the regular base-paired stems and provide these motifs with the possibility of behaving as molecular switches, a property often connected with their biological function. The clearest examples in this respect are provided by riboswitches and hammerhead ribozymes together with the SRP S RNA. In the sensing domain of TPP riboswitches, the bridging by the TPP ligand of the P2 and P3 internal loops likely induces an allosteric stabilization of the three-way junction via tertiary interactions that sequesters the switching strand contained within the P1 helix (Serganov et al. 2006; Thore et al. 2006). In purine riboswitches, the L2–L3 interaction is necessary to preorganize the junction for purine binding (Batey et al. 2004; Lemay et al. 2006; Buck et al. 2007; Gilbert et al. 2007; Noeske et al. 2007; Ottink et al. 2007; Greenleaf et al. 2008; Stoddard et al. 2008), and ligand binding at the junction also tightens the remote L2–L3 complex (Buck et al. 2007). In hammerhead ribozymes, the L2–L3 interactions have been shown to reorganize the helical junction (Fig. 5A,B; Martick and Scott 2006), which becomes catalytically competent only upon the establishment of the remote loop–loop contacts (De la Peña et al. 2003; Khvorova et al. 2003; Martick and Scott 2006). These allosteric mechanisms are also observed in parallel-Y motifs contained in ribonucleoprotein systems. In the S domain of the mammalian SRP, the L2–L3 contact (stabilized by the SRP19 protein) brings the P2 and P3 helices together and enables a conformational change in a P3 internal loop located 10-bp away from the interaction that is necessary for SRP54 binding, since the protein–RNA interaction is stable in the context of the SRP54–P3–P2 ternary complex only (Batey et al. 2000; Kuglstatter et al. 2002; Oubridge et al. 2002). In addition, X-ray experiments have revealed that the J23 junction becomes ordered in the presence of SRP54 (Kuglstatter et al. 2002; Oubridge et al. 2002).
Protein and RNA recognition
In addition to their dynamic properties, the Y folds contained in ribonucleoprotein systems provide platforms for specific protein binding. This is because the juxtaposition of RNA domains at the junction and at the remote tertiary complexes enables the RNA helices to adopt unusual shapes that would not be stable in the absence of the RNA partner. For example, in the large ribosomal subunit, L11 recognizes the unusual shape of the P2 (H43) minor groove (Fig. 3B), whose conformation would not be possible in the absence of the H43–H44 tertiary interactions taking place in the parallel-Y fold (Conn et al. 1999; Wimberly et al. 1999). In the SRP S domain, the side-by-side arrangement of helices P2 and P3 induced by tertiary L2–L3 interactions (stabilized by SRP19) creates a binding platform for the essential SRP54 protein (Fig. 3A; Kuglstatter et al. 2002). In other cases, the proteins bind to the three-way junction: the SRP9/14 complex with the Alu junction (Weichenrieder et al. 2000) or ribosomal junction H18–H19–H20 bound to proteins L4P and L24P (Ban et al. 2000) are two representative examples of this type of complex (Fig. 3C,D). In most instances, protein binding typically has a stabilization role, because the RNA tertiary complexes are usually preformed in the absence of the protein partner (Zwieb 1992; Lu and Draper 1994; Conn et al. 1998; Huck et al. 2004; Maeder et al. 2006). Ribosomal three- and four-way RNA junctions and their protein complexes are believed to play an important role in rRNA folding and ribosome assembly (Ban et al. 2000; Wimberly et al. 2000). In this respect, the H18–H19–H20 parallel-Y, which is relatively close to the 5′ end of 23S rRNA, has been shown to play a critical role in the assembly of the large subunit (Skinner et al. 1985). Remarkably, the H18–H19–H20 and H90–H91–H92 parallel-Y's are contacted by L24P and L3P, respectively, the only proteins capable of initiating 50S assembly in vitro (Nowotny and Nierhaus 1982). Likewise, the complex between the Alu SRP junction and the SRP9/14 protein heterodimer contributes to pack the P1 helix containing the SRP RNA 5′ and 3′ ends and is critical for SRP folding and assembly (Weichenrieder et al. 2000; Huck et al. 2004).
At least two of these motifs are also involved in the recognition of other RNA molecules. In the parallel-Y embedded within the S-domain of B-type RNase P (Fig. 2D), the large internal loop of domain P3 (P12) contributes to the recognition of the TΨC arm of the pre-tRNA molecule (LaGrandeur et al. 1994; Odell et al. 1998; Krasilnikov et al. 2003). In the H90–H91–H92 junction forming the A-site of the large ribosomal subunit, exposed bases within the tertiary complex formed by the apical loop of domain P3 (H92) and the internal loop of domain P1 (H90) participate in the recognition of the 3′ CCA tail of aminoacyl tRNA molecules (Schmeing et al. 2005).
PARALLEL-Y JUNCTIONS AS TARGETS FOR DRUG ACTION
Several parallel-Y RNAs have been shown to be the target of antibiotic action, a fact that adds further support to their functional relevance. The phosphorylated form of the antibiotic pyrithiamine exerts its activity by directly interacting with TPP riboswitches (Sudarsan et al. 2005). The thiopeptide antibiotics thiostrepton and micrococcin interact with the H42–H43–H44 junction of the large ribosomal subunit. They bind to a cavity formed by the tip of the L2 (H43)–L3 (H44) tertiary complex and the Nt domain of L11, interfering with the interaction of elongation factor EF-G with the ribosome (Harms et al. 2008). Junction H90–H91–H92 forms the A-cavity of the peptidyl transferase center and is also the binding site of numerous antibiotics: puromycin (Nissen et al. 2000), chloramphenicol (Schlunzen et al. 2001), clindamycin (Schlunzen et al. 2001), anisomycin (Hansen et al. 2003), the pleuromutilin tiamulin (Schlunzen et al. 2004), and the recently developed oxazolidinone linezolid (Wilson et al. 2008) bind to the internal loop at the base of H90 and exert their action by interfering with the placement of the tRNA aminoacyl moiety in the A-site.
CONCLUDING REMARKS
Three-way junction RNAs adopt a recurrent parallel-Y shape when two of the helices form a coaxial stack and the third helix establishes one or more tertiary contacts several base pairs away from the junction. Structurally, these motifs are characterized by a conserved junction topology, where one of the strands of the lower helix crosses the coaxial stack in a 5′-to-3′ direction toward the third helix. In addition, the junction strand connecting the two upper helices is usually longer than the other two junction strands and often forms a U-turn structure. The remote tertiary contacts have a strong influence on the three-dimensional shape of the parallel-Y folds and usually involve apical or internal loops with conserved sequence or conserved three-dimensional structure. In fact, the helical junctions and the distal tertiary complexes have been shown to behave as flexible hinges that respond to conformational changes in the other region, a property that is often connected with the biological roles played by these motifs. In addition, the juxtaposition of RNA domains at the junction and at the remote tertiary complexes enables the RNA helices to adopt unusual shapes that are frequently used by proteins or other RNA molecules for specific binding. Not surprisingly, these motifs are targets for antibiotic action and play essential functions in a variety of biological processes including regulation of gene expression, translation, and ribonucleoprotein assembly.
ACKNOWLEDGMENTS
This work was supported by the Ministerio de Educación y Ciencia of Spain (grant SAF2007-60243 to J.G. and Ramón y Cajal contracts to M.d.l.P. and J.G.), Generalitat Valenciana (GV06/206 to M.d.l.P. and ACOMP2009/075 to J.G.), and the CIPF.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1889509.
REFERENCES
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Bassi GS, Murchie AIH, Walter F, Clegg RM, Lilley DMJ. Ion-induced folding of the hammerhead ribozyme: A fluorescence resonance energy transfer study. EMBO J. 1997;16:7481–7489. doi: 10.1093/emboj/16.24.7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey RT, Rambo RP, Lucast L, Rha B, Doudna JA. Crystal structure of the ribonucleoprotein core of the signal recognition particle. Science. 2000;287:1232–1239. doi: 10.1126/science.287.5456.1232. [DOI] [PubMed] [Google Scholar]
- Batey RT, Gilbert SD, Montange RK. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- Bhuiyan SH, Gowda K, Hotokezaka H, Zwieb C. Assembly of archaeal signal recognition particle from recombinant components. Nucleic Acids Res. 2000;28:1365–1373. doi: 10.1093/nar/28.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck J, Furtig B, Noeske J, Wohnert J, Schwalbe H. Time-resolved NMR methods resolving ligand-induced RNA folding at atomic resolution. Proc Natl Acad Sci. 2007;104:15699–15704. doi: 10.1073/pnas.0703182104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Kundrot CE, Cech TR, Doudna JA. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science. 1996a;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- Cate JH, Gooding AR, Podell E, Zhou K, Golden BL, Szewczak AA, Kundrot CE, Cech TR, Doudna JA. RNA tertiary structure mediation by adenosine platforms. Science. 1996b;273:1696–1699. doi: 10.1126/science.273.5282.1696. [DOI] [PubMed] [Google Scholar]
- Chauhan S, Woodson SA. Tertiary interactions determine the accuracy of RNA folding. J Am Chem Soc. 2008;130:1296–1303. doi: 10.1021/ja076166i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah MT, Wachter A, Sudarsan N, Breaker RR. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature. 2007;447:497–500. doi: 10.1038/nature05769. [DOI] [PubMed] [Google Scholar]
- Chi YI, Martick M, Lares M, Kim R, Scott WG, Kim SH. Capturing hammerhead ribozyme structures in action by modulating general base catalysis. PLoS Biol. 2008;6:e234. doi: 10.1371/journal.pbio.0060234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn GL, Gutell RR, Draper DE. A functional ribosomal RNA tertiary structure involves a base triple interaction. Biochemistry. 1998;37:11980–11988. doi: 10.1021/bi980825+. [DOI] [PubMed] [Google Scholar]
- Conn GL, Draper DE, Lattman EE, Gittis AG. Crystal structure of a conserved ribosomal protein–RNA complex. Science. 1999;284:1171–1174. doi: 10.1126/science.284.5417.1171. [DOI] [PubMed] [Google Scholar]
- Connolly ML. Solvent-accessible surfaces of proteins and nucleic acids. Science. 1983;221:709–713. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- De la Peña M, Gago S, Flores R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 2003;22:5561–5570. doi: 10.1093/emboj/cdg530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener JL, Wilson C. Role of SRP19 in assembly of the Archaeoglobus fulgidus signal recognition particle. Biochemistry. 2000;39:12862–12874. doi: 10.1021/bi001180s. [DOI] [PubMed] [Google Scholar]
- Doherty EA, Batey RT, Masquida B, Doudna JA. A universal mode of helix packing in RNA. Nat Struct Biol. 2001;8:339–343. doi: 10.1038/86221. [DOI] [PubMed] [Google Scholar]
- Dufour D, de la Peña M, Gago S, Flores R, Gallego J. Structure–function analysis of the ribozymes of chrysanthemum chlorotic mottle viroid: A loop–loop interaction motif conserved in most natural hammerheads. Nucleic Acids Res. 2009;37:368–381. doi: 10.1093/nar/gkn918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea PF, Stroud RM, Walter P. Targeting proteins to membranes: Structure of the signal recognition particle. Curr Opin Struct Biol. 2005;15:213–220. doi: 10.1016/j.sbi.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Flores R, Hernandez C, de la Peña M, Vera A, Daros JA. Hammerhead ribozyme structure and function in plant RNA replication. Methods Enzymol. 2001;341:540–552. doi: 10.1016/s0076-6879(01)41175-x. [DOI] [PubMed] [Google Scholar]
- Gilbert SD, Love CE, Edwards AL, Batey RT. Mutational analysis of the purine riboswitch aptamer domain. Biochemistry. 2007;46:13297–13309. doi: 10.1021/bi700410g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenleaf WJ, Frieda KL, Foster DA, Woodside MT, Block SM. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hainzl T, Huang S, Sauer-Eriksson AE. Structure of the SRP19 RNA complex and implications for signal recognition particle assembly. Nature. 2002;417:767–771. doi: 10.1038/nature00768. [DOI] [PubMed] [Google Scholar]
- Hainzl T, Huang S, Sauer-Eriksson AE. Structural insights into SRP RNA: An induced fit mechanism for SRP assembly. RNA. 2005;11:1043–1050. doi: 10.1261/rna.2080205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halic M, Beckmann R. The signal recognition particle and its interactions during protein targeting. Curr Opin Struct Biol. 2005;15:116–125. doi: 10.1016/j.sbi.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Hansen JL, Moore PB, Steitz TA. Structures of five antibiotics bound at the peptidyl transferase center of the large ribosomal subunit. J Mol Biol. 2003;330:1061–1075. doi: 10.1016/s0022-2836(03)00668-5. [DOI] [PubMed] [Google Scholar]
- Harms J, Schluenzen F, Zarivach R, Bashan A, Gat S, Agmon I, Bartels H, Franceschi F, Yonath A. High-resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- Harms JM, Wilson DN, Schluenzen F, Connell SR, Stachelhaus T, Zaborowska Z, Spahn CM, Fucini P. Translational regulation via L11: Molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol Cell. 2008;30:26–38. doi: 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Huck L, Scherrer A, Terzi L, Johnson AE, Bernstein HD, Cusack S, Weichenrieder O, Strub K. Conserved tertiary base pairing ensures proper RNA folding and efficient assembly of the signal recognition particle Alu domain. Nucleic Acids Res. 2004;32:4915–4924. doi: 10.1093/nar/gkh837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jucker FM, Heus HA, Yip PF, Moors EHM, Pardi A. A network of hetergeneous hydrogen bonds in GNRA tetraloops. J Mol Biol. 1996;264:968–980. doi: 10.1006/jmbi.1996.0690. [DOI] [PubMed] [Google Scholar]
- Kazantsev AV, Pace NR. Bacterial RNase P: A new view of an ancient enzyme. Nat Rev Microbiol. 2006;4:729–740. doi: 10.1038/nrmicro1491. [DOI] [PubMed] [Google Scholar]
- Khvorova A, Lescoute A, Westhof E, Jayasena SD. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- Kieft JS, Zhou K, Grech A, Jubin R, Doudna JA. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated initiation of translation. Nat Struct Biol. 2002;9:370–374. doi: 10.1038/nsb781. [DOI] [PubMed] [Google Scholar]
- Klein DJ, Moore PB, Steitz TA. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J Mol Biol. 2004;340:141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- Krasilnikov AS, Mondragon A. On the occurrence of the T-loop RNA folding motif in large RNA molecules. RNA. 2003;9:640–643. doi: 10.1261/rna.2202703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasilnikov AS, Yang X, Pan T, Mondragon A. Crystal structure of the specificity domain of ribonuclease P. Nature. 2003;421:760–764. doi: 10.1038/nature01386. [DOI] [PubMed] [Google Scholar]
- Kuglstatter A, Oubridge C, Nagai K. Induced structural changes of 7SL RNA during the assembly of human signal recognition particle. Nat Struct Biol. 2002;9:740–744. doi: 10.1038/nsb843. [DOI] [PubMed] [Google Scholar]
- LaGrandeur TE, Huttenhofer A, Noller HF, Pace NR. Phylogenetic comparative chemical footprint analysis of the interaction between ribonuclease P RNA and tRNA. EMBO J. 1994;13:3945–3952. doi: 10.1002/j.1460-2075.1994.tb06710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laing C, Schlick T. Analysis of four-way junctions in RNA structures. J Mol Biol. 2009;390:547–559. doi: 10.1016/j.jmb.2009.04.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JC, Cannone JJ, Gutell RR. The lone pair triloop: A new motif in RNA structure. J Mol Biol. 2003;325:65–83. doi: 10.1016/s0022-2836(02)01106-3. [DOI] [PubMed] [Google Scholar]
- Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA. Folding of the adenine riboswitch. Chem Biol. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA. 2006;12:83–93. doi: 10.1261/rna.2208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Draper DE. Bases defining an ammonium and magnesium ion-dependent tertiary structure within the large subunit ribosomal RNA. J Mol Biol. 1994;244:572–585. doi: 10.1006/jmbi.1994.1753. [DOI] [PubMed] [Google Scholar]
- Maeder C, Conn GL, Draper DE. Optimization of a ribosomal structural domain by natural selection. Biochemistry. 2006;45:6635–6643. doi: 10.1021/bi052544p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martick M, Scott WG. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martick M, Horan LH, Noller HF, Scott WG. A discontinuous hammerhead ribozyme embedded in a mammalian messenger RNA. Nature. 2008;454:899–902. doi: 10.1038/nature07117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D, Robertson JM, Noller HF. Interaction of elongation factors EF-G and EF-Tu with a conserved loop in 23S RNA. Nature. 1988;334:362–364. doi: 10.1038/334362a0. [DOI] [PubMed] [Google Scholar]
- Montange RK, Batey RT. Riboswitches: Emerging themes in RNA structure and function. Annu Rev Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- Murthy VL, Rose GD. Is counterion delocalization responsible for collapse in RNA folding? Biochemistry. 2000;39:14365–14370. doi: 10.1021/bi001820r. [DOI] [PubMed] [Google Scholar]
- Nagaswamy U, Fox GE. Frequent occurrence of the T-loop RNA folding motif in ribosomal RNAs. RNA. 2002;8:1112–1119. doi: 10.1017/s135583820202006x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc Natl Acad Sci. 2001;98:4899–4503. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noeske J, Buck J, Furtig B, Nasiri HR, Schwalbe H, Wohnert J. Interplay of ‘induced fit’ and preorganization in the ligand induced folding of the aptamer domain of the guanine binding riboswitch. Nucleic Acids Res. 2007;35:572–583. doi: 10.1093/nar/gkl1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotny V, Nierhaus KH. Initiator proteins for the assembly of the 50S subunit from Escherichia coli ribosomes. Proc Natl Acad Sci. 1982;79:7238–7242. doi: 10.1073/pnas.79.23.7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odell L, Huang V, Jakacka M, Pan T. Interaction of structural modules in substrate binding by the ribozyme from Bacillus subtilis RNase P. Nucleic Acids Res. 1998;26:3717–3723. doi: 10.1093/nar/26.16.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottink OM, Rampersad SM, Tessari M, Zaman GJ, Heus HA, Wijmenga SS. Ligand-induced folding of the guanine-sensing riboswitch is controlled by a combined predetermined induced fit mechanism. RNA. 2007;13:2202–2212. doi: 10.1261/rna.635307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oubridge C, Kuglstatter A, Jovine L, Nagai K. Crystal structure of SRP19 in complex with the S domain of SRP RNA and its implication for the assembly of the signal recognition particle. Mol Cell. 2002;9:1251–1261. doi: 10.1016/s1097-2765(02)00530-0. [DOI] [PubMed] [Google Scholar]
- Penedo JC, Wilson TJ, Jayasena SD, Khvorova A, Lilley DM. Folding of the natural hammerhead ribozyme is enhanced by interaction of auxiliary elements. RNA. 2004;10:880–888. doi: 10.1261/rna.5268404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Przybilski R, Graf S, Lescoute A, Nellen W, Westhof E, Steger G, Hammann C. Functional hammerhead ribozymes naturally encoded in the genome of Arabidopsis thaliana. Plant Cell. 2005;17:1877–1885. doi: 10.1105/tpc.105.032730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T, Morita S, Tabata K, Nakamura K, Kawai G. Solution structure of a SRP19 binding domain in human SRP RNA. J Biochem. 2002;132:177–182. doi: 10.1093/oxfordjournals.jbchem.a003207. [DOI] [PubMed] [Google Scholar]
- Schlunzen F, Zarivach R, Harms J, Bashan A, Tocilj A, Albrecht R, Yonath A, Franceschi F. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature. 2001;413:814–821. doi: 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- Schlunzen F, Pyetan E, Fucini P, Yonath A, Harms JM. Inhibition of peptide bond formation by pleuromutilins: The structure of the 50S ribosomal subunit from Deinococcus radiodurans in complex with tiamulin. Mol Microbiol. 2004;54:1287–1294. doi: 10.1111/j.1365-2958.2004.04346.x. [DOI] [PubMed] [Google Scholar]
- Schmeing TM, Huang KS, Strobel SA, Steitz TA. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature. 2005;438:520–524. doi: 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- Schmitz U, Behrens S, Freymann DM, Keenan RJ, Lukavsky P, Walter P, James TL. Structure of the phylogenetically most conserved domain of SRP RNA. RNA. 1999;5:1419–1429. doi: 10.1017/s1355838299991458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott WG, Murray JB, Arnold JRP, Stoddard BL, Klug A. Capturing the structure of a catalytic RNA intermediate: The hammerhead ribozyme. Science. 1996;274:2065–2069. doi: 10.1126/science.274.5295.2065. [DOI] [PubMed] [Google Scholar]
- Serganov A. The long and the short of riboswitches. Curr Opin Struct Biol. 2009;19:251–259. doi: 10.1016/j.sbi.2009.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Patel DJ. Ribozymes, riboswitches, and beyond: Regulation of gene expression without proteins. Nat Rev Genet. 2007;8:776–790. doi: 10.1038/nrg2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PA. The centrality of RNA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- Skinner RH, Stark MJ, Dahlberg AE. Mutations within the 23S rRNA coding sequence of E. coli which block ribosome assembly. EMBO J. 1985;4:1605–1608. doi: 10.1002/j.1460-2075.1985.tb03824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoddard CD, Gilbert SD, Batey RT. Ligand-dependent folding of the three-way junction in the purine riboswitch. RNA. 2008;14:675–684. doi: 10.1261/rna.736908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Cohen-Chalamish S, Nakamura S, Emilsson GM, Breaker RR. Thiamine pyrophosphate riboswitches are targets for the antimicrobial compound pyrithiamine. Chem Biol. 2005;12:1325–1335. doi: 10.1016/j.chembiol.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Szewczak AA, Moore PB, Chan Y-L, Wool IG. The conformation of the sarcin/ricin loop from 28S ribosomal RNA. Proc Natl Acad Sci. 1993;90:9581–9585. doi: 10.1073/pnas.90.20.9581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thore S, Leibundgut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- Torres-Larios A, Swinger KK, Pan T, Mondragon A. Structure of ribonuclease P–a universal ribozyme. Curr Opin Struct Biol. 2006;16:327–335. doi: 10.1016/j.sbi.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Walter P, Blobel G. Disassembly and reconstitution of signal recognition particle. Cell. 1983;34:525–533. doi: 10.1016/0092-8674(83)90385-9. [DOI] [PubMed] [Google Scholar]
- Weichenrieder O, Wild K, Strub K, Cusack S. Structure and assembly of the Alu domain of the mammalian signal recognition particle. Nature. 2000;408:167–173. doi: 10.1038/35041507. [DOI] [PubMed] [Google Scholar]
- Wilson DN, Schluenzen F, Harms JM, Starosta AL, Connell SR, Fucini P. The oxazolidinone antibiotics perturb the ribosomal peptidyl-transferase center and effect tRNA positioning. Proc Natl Acad Sci. 2008;105:13339–13344. doi: 10.1073/pnas.0804276105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly B, Varani G, Tinoco I., Jr The conformation of loop E of eukaryotic 5S ribosomal RNA. Biochemistry. 1993;32:1078–1087. doi: 10.1021/bi00055a013. [DOI] [PubMed] [Google Scholar]
- Wimberly BT, Guymon R, McCutcheon JP, White SW, Ramakrishnan V. A detailed view of a ribosomal active site: The structure of the L11-RNA complex. Cell. 1999;97:491–502. doi: 10.1016/s0092-8674(00)80759-x. [DOI] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WMJ, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- Wool IG, Glück A, Endo Y. Ribotoxin recognition of ribosomal RNA and a proposal for the mechanism of translocation. Trends Biochem Sci. 1992;17:266–269. doi: 10.1016/0968-0004(92)90407-z. [DOI] [PubMed] [Google Scholar]
- Yin J, Huang Q, Pakhomova ON, Hinck AP, Zwieb C. The conserved adenosine in helix 6 of Archaeoglobus fulgidus signal recognition particle RNA initiates SRP assembly. Archaea. 2004;1:269–275. doi: 10.1155/2004/134861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C. Recognition of a tetranucleotide loop of signal recognition particle RNA by protein SRP19. J Biol Chem. 1992;267:15650–15656. [PubMed] [Google Scholar]