Abstract
In eukaryotic cells, dozens to hundreds of different mRNAs are localized by specialized motor-dependent transport complexes. One of the best-studied examples for directional mRNA transport is the localization of ASH1 mRNA in Saccharomyces cerevisiae. For transport, ASH1 mRNA is bound by the unusual RNA-binding protein She2p. Although previous results indicated that She2p forms dimers required for RNA binding and transcript localization, it remained unclear if the dimer constitutes the minimal RNA-binding unit assembling in vivo. By using analytical ultracentrifugation we found that She2p forms larger oligomeric complexes in solution. We also identified a point mutant that shows impaired oligomer formation. Size-exclusion chromatography suggests that She2p forms defined tetramers at physiological concentrations. Subsequent structural studies by small-angle X-ray scattering confirmed this finding and demonstrated that the previously observed She2p dimers interact in a head-to-head conformation to form an elongated tetrameric complex. This She2p tetramer suggests the generation of large continuous RNA-binding surfaces at both sides of the complex. Biochemical studies and immunostaining of cells confirmed that She2p tetramer formation is required for RNA binding, efficient mRNP assembly, and mRNA localization in vivo. Our finding on She2p tetramerization resolves previously raised questions on complex formation and mRNP function.
Keywords: She2p, She3p, mRNP, RNA localization, RNA binding, SAXS
INTRODUCTION
Eukaryotic cells maintain an elaborate network of microtubules and actin filaments that are used by motor proteins for the asymmetric localization of cellular cargo (Vale 2003). Directional transport and localization of mRNA is widely used to regulate gene expression and to generate cellular asymmetry (Shav-Tal and Singer 2005; St Johnston 2005; Müller et al. 2007). For this process, motor-containing messenger ribonucleoprotein particles (mRNPs) translocate transcripts that are translationally silent from perinuclear areas to their peripheral destination. After anchoring, translation of mRNAs is activated and the encoded proteins are produced (Shav-Tal and Singer 2005; Dahm et al. 2007; Müller et al. 2007).
One of the best-studied examples of localizing mRNPs is the ASH1 mRNA-translocation process during mitosis of the yeast Saccharomyces cerevisiae (Müller et al. 2007). ASH1 mRNA is transported as part of a large mRNP from the mother cell to the tip of the daughter cell (Takizawa et al. 2000; Shepard et al. 2003). ASH1 mRNA contains four cis-acting regions, termed zip-code elements, that mediate mRNA incorporation into the mRNPs and subsequent localization. After anchoring and ASH1 mRNA translation, the protein product Ash1p acts as a repressor of mating-type switching exclusively in the daughter cell (Bobola et al. 1996; Sil and Herskowitz 1996). In addition to ASH1 mRNA, more than 30 transcripts are selectively transported by this mRNP (Takizawa et al. 2000; Shepard et al. 2003; Oeffinger et al. 2007; Hogan et al. 2008).
The only core RNA-binding protein of this transport complex, termed She2p (Kruse et al. 2002), binds ASH1 mRNA already in the nucleus at the site of transcription (Du et al. 2008) and escorts it through the nucleolus into the cytoplasm (Kruse et al. 2002). She2p is an unusual type of nucleic-acid binding protein (Niessing et al. 2004) that binds to the four zip-code elements of ASH1 mRNA (Chartrand et al. 1999; Gonzalez et al. 1999; Böhl et al. 2000; Chartrand et al. 2002). After nuclear export, the She2p-ASH1 mRNA complex binds to the adapter protein She3p, which itself forms a co-complex with the type V myosin motor Myo4p (Münchow et al. 1999; Böhl et al. 2000; Long et al. 2000; Takizawa and Vale 2000; Heuck et al. 2007; Hodges et al. 2008). Binding of cytoplasmic She3p to the co-complex of She2p and ASH1 mRNA is thought to stabilize the nucleic acid–protein interaction (Böhl et al. 2000).
Since She2p is an unusual type of RNA-binding protein, it is still unclear how this protein interacts with its mRNA cargo and what its requirements are for mRNP assembly. Because in a recently published crystal structure, two She2p monomers were shown to interact with a large buried surface interface of more than 2000 Å2 / monomer (Table 1), it was assumed that She2p forms dimers in solution (Niessing et al. 2004). The crystal structure further helped to identify a positively charged surface required for RNA binding. Interaction studies with different zip-code elements revealed that one She2p dimer binds to either one or two zip-code elements (Gonsalvez et al. 2003; Niessing et al. 2004). Assuming She2p dimers, it has been suggested that for a one-to-one stoichiometry, zip-code elements would have to wrap around the entire She2p structure. In addition, mutations at the upper uncharged surface of She2p (Niessing et al. 2004) were shown to impair RNA binding in vitro (positions T47 and L130) (Gonsalvez et al. 2003; Niessing et al. 2004). Based on these results, it was speculated that the upper surface of She2p is an integral part of the RNA-binding motif and that zip-code elements would arch over this upper surface. However, the upper uncharged surface of She2p shows classical features of a hydrophobic protein interaction surface rather than of a nucleic acids binding interface (Dasgupta et al. 1997; Conte et al. 1999). In addition, a mutation in position L130 at the upper She2p surface was suggested to affect the interaction of She2p with She3p (see Supplemental Table 1 in Gonsalvez et al. 2003; Niessing et al. 2004). For these reasons, it has also been speculated that the main function of the upper surface is the interaction with the adapter She3p. To date, it is unclear how well either of these contradicting hypotheses matches the function of She2p.
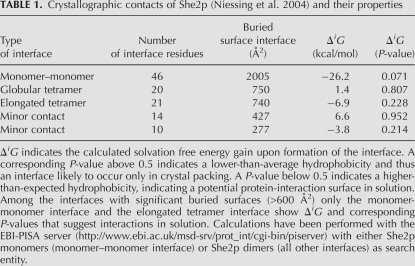
TABLE 1.
Crystallographic contacts of She2p (Niessing et al. 2004) and their properties
In this study, we show that none of these two hypotheses are likely. Instead, we find that RNA binding and RNA localization require the formation of higher She2p oligomers, involving its upper, uncharged surface. At high protein concentrations, She2p forms oligomers in the range of 8–12mers. At physiological concentrations, however, we rather observe stable tetramers. Structural analyses of She2p in solution revealed an extended conformation of two She2p dimers in a head-to-head conformation. This elongated tetramer creates a continuous RNA-binding surface that is large enough to accommodate RNA zip-code elements on She2p in the reported stoichiometries. We also found that mutation of residue L130 disrupts the formation of She2p tetramers, resulting in impaired RNA binding, mRNP assembly, and abolished She2p localization to the bud tip. Together, these results demonstrate the functional importance of She2p tetramer formation. The requirement of She2p tetramers also indicates that She2p might interconnect different zip-code-containing mRNAs to form larger mRNPs of variable size.
RESULTS
She2p forms large oligomeric complexes
During protein purification of recombinantly expressed She2p we noticed that wild-type She2p elutes in size-exclusion chromatography at a calculated molecular weight of about 340 kDa (data not shown), which is far above the theoretical molecular weight of a She2p dimer (57.2 kDa). We also found that the previously described mutant She2p (L130→Y) elutes at a much lower calculated molecular weight (data not shown). These observations provided a first indication that wild-type She2p forms large oligomeric complexes and indicated that the mutation L130→Y disrupts this higher oligomeric state.
In order to determine the molecular weight of She2p in solution more precisely, we performed analytical-ultracentrifugation experiments. To avoid RNA-mediated oligomerization of She2p during ultracentrifugation experiments, we purified She2p under high-salt conditions, treated the sample with RNase A, and monitored protein purity by the ratio of OD280/254.
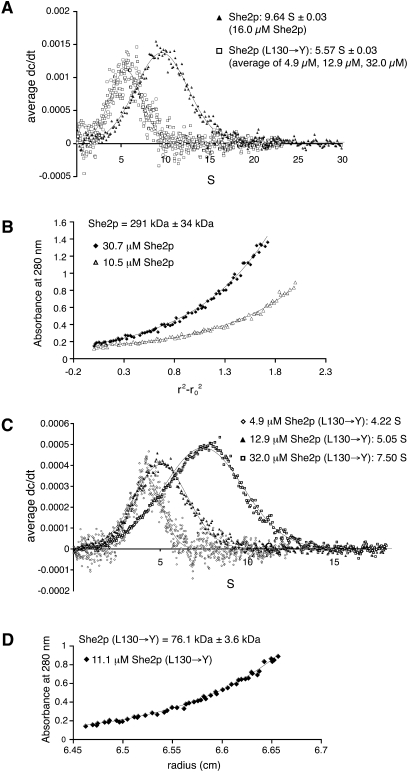
Sedimentation–velocity experiments revealed that wild-type She2p sediments in a single species at 9.6 S (Fig. 1A). Subsequent sedimentation–equilibrium experiments revealed a concentration-independent molecular weight for this complex of about 290 kDa (Fig. 1B). Since the She2p dimer has a calculated molecular weight of 57.2 kDa, we concluded that She2p forms an oligomer of about 10 monomers, corresponding to five dimers in solution. However, because of experimental error and symmetry considerations, She2p might instead form an octa- or dodecameric complex.
FIGURE 1.
Analytical-ultracentrifugation experiments with She2p. (A) In sedimentation–velocity experiments wild-type She2p sediments at 9.6 S. In contrast, She2p (L130→Y) sedimentation is concentration dependent with an average of 5.6 S (individual experiments are shown in C). The reduced sedimentation coefficient for She2p (L130→Y) is consistent with a reduced molecular weight. (B) Sedimentation–equilibrium experiments with wild-type She2p at different protein concentrations indicate a concentration-independent oligomer of 290 kDa. Plot shows two representative curves. (C) Sedimentation–velocity experiments with She2p (L130→Y) at different protein concentrations show a concentration-dependent change in sedimentation. (D) Sedimentation–equilibrium experiments with She2p (L130→Y) at an intermediate concentration of 11.1 μM revealed a molecular weight of 76 kDa, confirming that higher molecular weight oligomers are disrupted by this mutation.
Analytical-ultracentrifugation experiments with She2p (L130→Y) showed that this mutant protein sediments in a concentration-dependent manner between 4.2 and 7.5 S (Fig. 1C), average: 5.6 S (Fig. 1A). Thus, even at high concentrations the oligomeric state of wild-type She2p (9.6 S) is not achieved. Subsequent sedimentation–equilibrium experiments with an intermediate protein concentration (11.1 μM) revealed a molecular weight of 76 kDa (Fig. 1D), corresponding to an oligomerization state between a dimer and a tetramer. From this result, we conclude that the L130→Y mutation interferes with She2p oligomerization through the upper, uncharged surface region of the dimer.
She2p forms tetramers at physiological concentrations
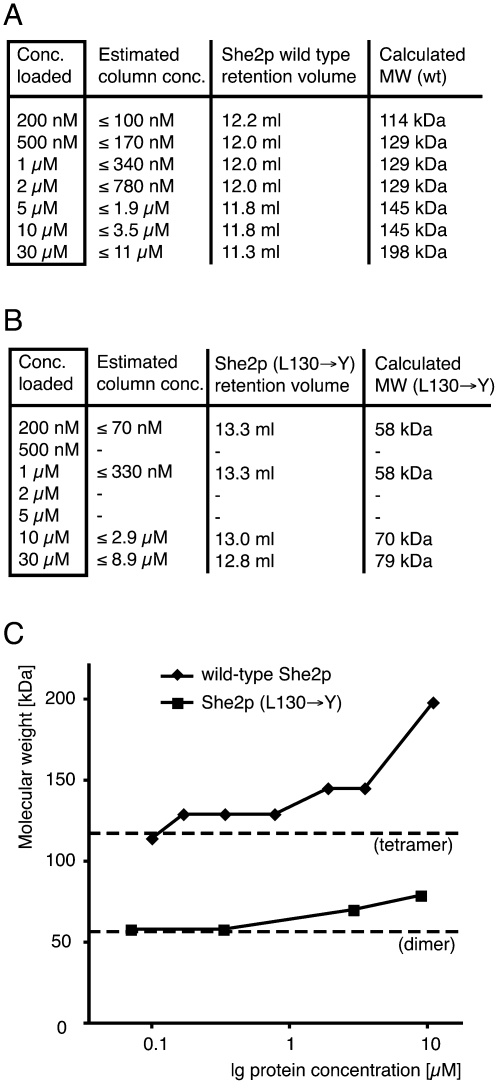
Our results from size-exclusion chromatography and analytical-ultracentrifugation experiments put into question whether the crystallographic dimers with N- and C-terminally truncated, cysteine-mutated She2p (Niessing et al. 2004) indeed represent the oligomeric state in solution. Because in our chromatography and centrifugation experiments high protein concentrations were used, we also performed analytical size-exclusion chromatography at a range of more physiological concentrations (Fig. 2A,B). The lowest concentrations applied were below the estimated physiological She2p concentration of 230 nM (Niessing et al. 2004). To avoid RNA-mediated oligomerization, we ensured that no RNA was present in these experiments (see Materials and Methods). When considering sample dilution (for details, see the Fig. 2 legend), wild-type She2p forms a distinct oligomeric complex at experimental concentrations up to 3.5 μM (Fig. 2A,C) with an average molecular weight of 132 kDa. Although molecular weight calculation by size-exclusion chromatography is usually not very accurate, this result suggests the presence of a She2p tetramer (28.6 kDa × 4 = 114.4 kDa) or even a pentamer (143 kDa). Thus, also at physiological concentrations She2p forms higher oligomers than the dimers observed in the crystal structure.
FIGURE 2.
Characterization of the oligomeric state of She2p at a physiological concentration range. Size-exclusion chromatography was performed to assess the oligomeric states of wild-type She2p (A) and She2p (L130→Y) (B). Whereas wild-type She2p forms a distinct oligomeric complex corresponding to a tetramer at concentrations up to 3.5 μM, the mutant She2p (L130→Y) shows impaired oligomerization over the entire concentration range. In addition to the protein concentration loaded, the estimated protein concentration after sample dilution on the column is listed. The real average concentrations might be even lower. Molecular weights were calculated from peak elution volumes. (C) Graph summarizing results from (A) and (B) using the estimated sample concentration on the column. Dashed lines indicate either the molecular weight of a She2p dimer (57 kDa) or of a tetramer (114 kDa).
In contrast, She2p (L130→Y) failed to form oligomers corresponding to a putative tetramer (Fig. 2B,C). At experimental concentrations up to 330 nM, the She2p (L130→Y) mutant forms complexes with a calculated molecular weight of 58 kDa, corresponding to a She2p dimer (57.2 kDa). Thus, a mutation at amino acid position 130 impairs She2p oligomerization at physiological concentrations. At higher concentrations of She2p (L130→Y) an increase of molecular weight is observed.
Small-angle X-ray scattering confirms She2p-tetramer formation in solution
In order to determine the oligomeric state of She2p observed at physiological concentrations (Fig. 2) more accurately and to obtain information on the shape of She2p oligomers, we performed small-angle X-ray scattering (SAXS) experiments. SAXS is a reliable method for the calculation of molecular masses of proteins with an average error below 10% (Gherardi et al. 2006; Mylonas and Svergun 2007).
Like analytical ultracentrifugation, SAXS experiments require high protein concentrations. Because at such high protein concentrations She2p forms complexes significantly larger than at physiological concentrations (cf. Fig. 1A,B and Fig. 2A,C), we screened different buffer conditions for their ability to prevent formation of these large oligomers at high concentration. Using dynamic light scattering (DLS), She2p was characterized in various buffers for its tendency to form large complexes. Although absolute hydrodynamic radii (RH) derived by DLS have to be considered with caution, this technique is well suited to rapidly screen buffer conditions for their effect on the size of protein complexes. A buffer that prevents the formation of large oligomers at high She2p concentrations would yield significantly smaller RH values than observed in standard buffer. The use of HEPES buffer, as well as the addition of detergents, resulted in large She2p complexes with an average RH of about 10 nm (Supplemental Table S1). In contrast, Tris-buffer with increased salt concentration (300 mM NaCl) and phosphate buffer with standard salt concentration (150 mM NaCl) yielded RH values in the range of 6 nm (Supplemental Table S1). Both conditions were tested in SAXS experiments, but only measurements with She2p in phosphate buffer were suitable for high-quality data collection.
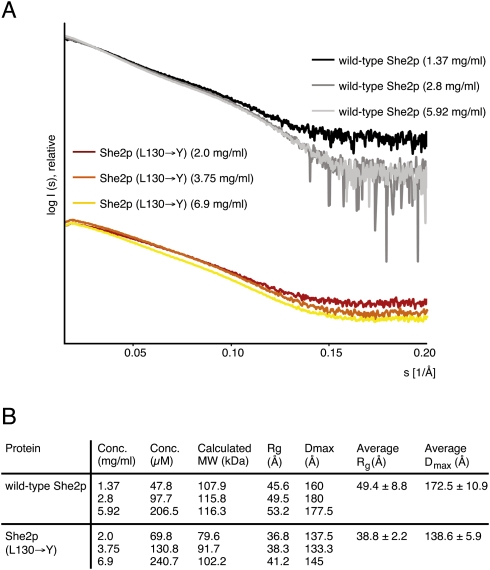
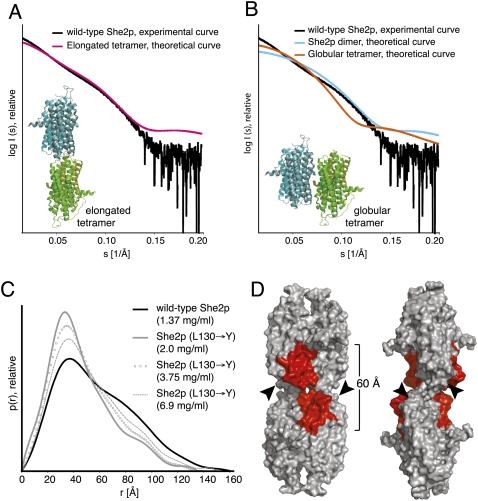
The obtained scattering curves (Fig. 3A) of wild-type She2p and She2p (L130→Y) revealed no signs of aggregation, intermolecular forces between particles, or radiation damage. The molecular mass of She2p was calculated from SAXS curves to range between 108 and 116 kDa (Fig. 3B). This corresponds to a She2p tetramer (114.4 kDa) and thus confirms the molecular weight calculated by size-exclusion chromatography (Fig. 2A,C). Corresponding calculations for She2p (L130→Y) revealed that this mutant associates concentration-dependently into complexes with average molecular masses between 80 and 102 kDa (Fig. 3B). Thus, also in SAXS measurements She2p (L130→Y) fails to associate into tetramers. Instead, oligomers were formed in equilibrium between dimers and tetramers. The observation that the L130→Y mutation affects tetramer formation in size-exclusion chromatography and in SAXS measurements in a similar way indicates that these tetramers are indeed identical.
FIGURE 3.
Small-angle X-ray scattering studies with She2p. (A) Scattering curves obtained from wild-type She2p and She2p (L130→Y) at three different concentrations after buffer subtraction. The scattering intensity I is plotted logarithmically against the scattering angle s. With increasing scattering angles (s > 0.15/Å) the signal-to-noise ratio decreases. These data were excluded from calculation and are shown for completeness only. (Table in B) Summary of structural parameters determined from scattering curves.
Small-angle X-ray scattering indicates that She2p forms an elongated tetramer
In order to obtain information about the overall size of oligomers, we calculated the radii of gyration (Rg) from the scattering curves. We found the average Rg values of She2p (L130→Y) to be substantially smaller than that of wild-type She2p (38.8 and 49.4 Å, respectively; Fig. 3B). In addition, the maximum dimension of the particle (Dmax) was determined to be between 160 and 180 Å for wild-type She2p and between 133.3 and 145 Å for She2p (L130→Y) (Fig. 3B).
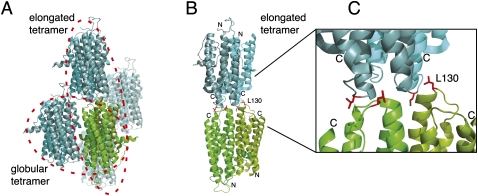
Since leucine in position 130 is located on the conserved upper surface of She2p (Niessing et al. 2004), tetramers might assemble through this upper surface of the She2p dimer. Because in the published She2p crystal structure contacts between She2p dimers were already observed (Niessing et al. 2004), we decided to re-examine the potential role of these crystal contacts for tetramer formation in solution. Among the different crystallographic contacts (Fig. 4A) only two dimer interactions had buried surface areas large enough to be potentially relevant in solution (Table 1). One dimer interaction involves the basic helical hairpin-containing surface at the front of She2p (Fig. 4A). It buries a surface area of 750 Å2/dimer and was termed globular tetramer. The other dimer interaction involves the upper, uncharged surface of She2p (Fig. 4A–C) and buries a surface area of 740 Å2/dimer. It was termed elongated tetramer. Leucine in amino acid position 130 is located at the dimer interface of the elongated tetramer (Fig. 4B,C).
FIGURE 4.
Crystallographic contacts of She2p. (A) In the published crystal structure (PDB ID: 1XLY) a stable She2p dimer (subunits in light and dark green) was observed with a large buried surface interface of over 2000 Å2/monomer (Table 1; Niessing et al. 2004). Among the several crystallographic contacts (subunits shown in light and dark blue, respectively) only two dimer contacts have significant buried surface interfaces (Table 1). In one case, the She2p dimer binds on its flat, basic-helical hairpin-containing side to another dimer in an upside-down orientation. We termed this complex globular tetramer. In the other case, She2p interacts with another dimer through its uncharged upper surface in a head-to-head conformation. This complex was termed elongated tetramer. As defined in Niessing et al. (2004), the upper surface refers to the uncharged area on top of the She2p dimer when oriented with the N-termini to the bottom and the C-termini to the top. (B) The elongated tetramer with leucine 130 shown with its side chains in red and rotated slightly when compared to (A). Leucine 130 is located at the dimer interface of the elongated tetramer. Formation of tetramers is disrupted by mutation of leucine 130 into tyrosine. The positions of N- and C-termini are marked. (C) Close-up of the tetramer–interaction surface shown in (B). Images were generated using PyMOL (DeLano Scientific).
In order to find out if one of these crystallographic tetramers is consistent with experimental SAXS curves, we calculated the theoretical scattering curves of both potential She2p tetrameric assemblies as well as of the dimeric She2p from the crystal structure. The theoretical scattering curve of She2p forming the elongated tetramer fits well to the experimental scattering curve of wild-type She2p (Fig. 5A). In contrast, the theoretical scattering curve of dimeric She2p fits less exactly and the theoretical scattering curve of the globular She2p tetramer differs substantially from the experimental She2p curve (Fig. 5B). The latter finding is also consistent with the previous observation that mutations in this crystallographic dimer interface do not change the oligomeric state of She2p in solution (Niessing et al. 2004). Thus, it is unlikely that She2p preferentially exists as a dimer or as globular tetramer, whereas the elongated tetramer might indeed be the preferred conformation of She2p in solution.
FIGURE 5.
Evaluation of crystallographic tetramers for their presence in solution. (A) The theoretical scattering curve of She2p forming the elongated tetramer via the upper uncharged surface (see inset, and also Fig. 4A–C) shows a good fit with the experimental scattering curve of wild-type She2p (shown is the curve measured with 2.8 mg/mL She2p). (B) In contrast, neither the theoretical scattering curve of the She2p globular tetramer (inset, and also Fig. 4A) nor the theoretical curve of a She2p dimer matches the experimental curve of She2p. Both tetramer models were directly generated from the published crystal structure (PDB-ID: 1XLY; Niessing et al. 2004) and show, therefore, the original difference in completeness. (C) The pair-distribution functions of wild-type She2p suggest an elongated particle, whereas for She2p (L130→Y) a rather globular molecule is assumed. With increasing protein concentration, the pair-distribution functions for She2p (L130→Y) approach the shape of the wild-type She2p p(r)-distribution, suggesting a concentration-dependent oligomerization of the mutant protein. (D) Surface representation of the elongated tetramer depicted from the front (left) and rotated by about 90° (right). The basic-helical hairpin as RNA-binding motif is highlighted in red. The positions of the C-terminal tails, which were absent in the crystal structure, are indicated by arrowheads. This structural model has been derived by substituting the less complete monomer from the crystal structure by its more complete counterpart. For further details, see Niessing et al. (2004). The indicated distance shows the dimension of the continuous RNA-binding surface on both sides of the tetramer.
In the crystal structure, She2p forms the elongated tetramer in a head-to-head conformation (Fig. 4A–C; Niessing et al. 2004). However, the effect of mutation L130→Y could also be explained by a head-to-tail interaction. We assessed this possibility by generating a She2p mutant, in which amino acids 81–89 in the main loop region at the bottom of She2p is deleted. Since this mutant is indistinguishable from wild-type She2p, as judged by size-exclusion chromatography and by RNA-filter binding assays (data not shown), we concluded that the elongated She2p tetramer is indeed likely to associate in a head-to-head conformation.
Calculation of the pair-distribution function allows us to obtain additional information on the overall shape of molecules in solution. The p(r)-distribution of wild-type She2p shows the typical skewed shape of an elongated molecule having a peak at short distances and an extended tail (Fig. 5C), which is fully consistent with an elongated She2p tetramer in solution. In contrast, the p(r) distribution of She2p (L130→Y) is rather symmetric with a single peak and shows the shape of a more globular molecule (Fig. 5C). We previously observed for She2p (L130→Y) a concentration-dependent increase of molecular weight (Figs. 1C, 2B,C), which never reached the molecular weight of a tetramer. This behavior is also observable in the pair-distribution functions of She2p (L130→Y). With increasing concentrations of She2p (L130→Y), its p(r) curves approached the shape of the wild-type She2p curve without ever merging with it (Fig. 5C).
In summary, the SAXS analyses indicate that wild-type She2p forms an elongated tetramer via the upper surface (Fig. 5D). We note that in certain buffer conditions, She2p tends to form oligomers even larger than tetramers (Fig. 1A,B), with She2p (L130→Y) still failing to reach a tetrameric state (Fig. 1A,C,D). Although unlikely, we cannot rule out that oligomers larger than tetramers may also form at physiological concentrations. She2p tetramers, however, are the likely building blocks for such potentially larger oligomers.
She2p binds to zip-code elements with Kd's in the nanomolar range
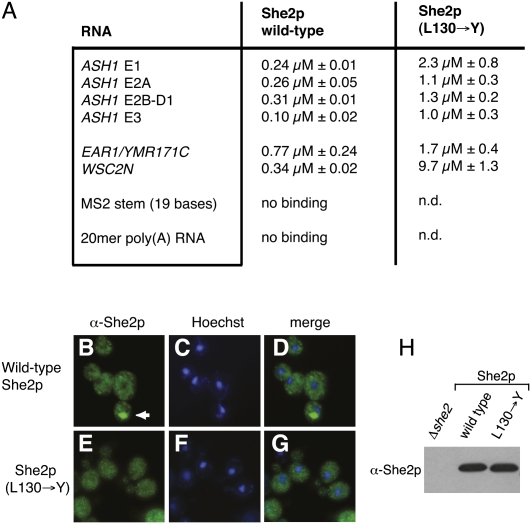
It has been previously shown that She3p (L130→Y) has a reduced affinity for the E3 zip-code element, suggesting that She2p tetramer formation is important for RNA binding. In order to find out if this effect also extends to other zip-code elements, we performed RNA-filter binding studies. Since, to date, only the binding of She2p to the ASH1 E3 zip-code element has been quantified (Niessing et al. 2004), we measured the interaction of She2p with all four zip-code elements of the ASH1 mRNA (E1, E2A, E2B, and E3), with the zip-code element of EAR1 mRNA, and with the N-terminal zip-code element of WSC2 mRNA (WSC2N) (Supplemental Fig. S1A,B; Jambhekar et al. 2005; Olivier et al. 2005). Secondary-structure predictions for these RNA elements indicate that they all fold into stem–loop structures (Gonzalez et al. 1999; Chartrand et al. 2002; Jambhekar et al. 2005; Olivier et al. 2005). Our binding studies showed that She2p interacts with all zip-code elements with an equilibrium-dissociation constant (Kd) in the nanomolar range (Fig. 6A). The strongest binding with a Kd of 0.10 μM was observed for the most 3′-located E3 element of the ASH1 mRNA and the weakest binding for the EAR1 zip-code element (Kd = 0.77 μM).
FIGURE 6.
Functional analyses with wild-type and oligomerization-mutant She2p. (A) RNA filter-binding assays with She2p. All Kd values shown are derived from at least three independent binding experiments. The label “no binding” indicates that at the highest protein concentration measured, no significant RNA binding was observed (see Materials and Methods). “n.d.” indicates that an experiment has not been performed. (B–G) Bud–tip localization in response to mutation of amino acid 130 in She2p. (B–D) Antibody staining reveals that wild-type She2p localizes to the bud tip in 69.2% of otherwise ΔSHE2 cells. (E–G) In contrast, no localization above background level (i.e., 3%) was observed in ΔSHE2 cells expressing She2p (L130→Y). (B,E) show anti-She2p antibody staining, (C,F) show nuclear Hoechst staining, and (D,G) display merged images of the respective antibody and nuclear stainings. Arrow indicates the position of She2p localization at the bud tip. In all experiments, at least 100 budding cells were inspected for She2p localization. (H) Western-blot analysis with anti-She2p antibody shows that She2p (L130→Y) is expressed at wild-type levels.
She2p tetramer formation is required for the recognition of zip-code RNAs
Mutation of leucine 130 in the conserved upper surface area of She2p was previously shown to reduce She2p's affinity for the ASH1-E3 element (Niessing et al. 2004) and was mentioned to affect She3p interaction (Supplemental Table 1 in Gonsalvez et al. 2003). Here, we found that mutation of leucine 130 to tyrosine resulted in reduced affinity to all zip-code elements, with the WSC2N element showing the strongest (29-fold) and EAR1 the weakest (2.2-fold) reduction (Fig. 6A). This observation indicates that She2p tetramer formation is required for the binding to zip-code elements in general.
Impaired tetramer formation abolishes She2p localization
In principle, She2p mutants with impaired RNA binding should also be unable to mediate mRNA localization in vivo. If She2p localization fails, all She2p-dependent mRNAs should be mislocalized. In contrast to Δshe2 cells that express wild-type She2p (Fig. 6B–D), She2p (L130→Y)-expressing cells showed no She2p localization to the bud tip above background levels (3%, n > 100; Fig. 6E–G). Western-blot analyses demonstrated that the She2p (L130→Y) mutant was expressed at wild-type levels (Fig. 6H). Thus, tetramer formation of She2p is required for She2p-containing mRNP transport in vivo.
She2p with impaired tetramer formation shows defects in translocation-complex assembly
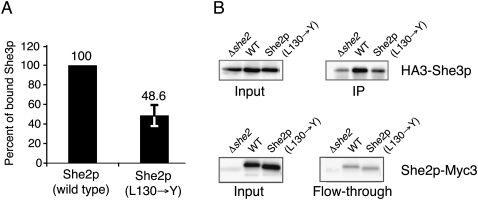
The inability of the She2p mutant to localize to the bud tip raises the question whether assembly of stable translocation complexes still occurs with this mutant. To find out if She2p (L130→Y) is still able to mediate complex assembly, we performed co-immunoprecipitation experiments of Myc-tagged She2p followed by Western-blot analyses against HA-tagged She3p. Experiments were performed in a Δshe2 background. Wild-type She2p-Myc efficiently co-precipitated She3p (Fig. 7A,B IP lanes), indicating that assembled translocation complexes can be detected. In contrast, She2p-Myc (L130→Y) immunoprecipitated only half the amount of She3p compared to wild-type She2p (Fig. 7A,B), suggesting that transport-complex assembly is impaired. Since in this mutant no bud localization is observed (Fig. 6E–G), we conclude that translocation complexes assembling with She2p (L130→Y) are insufficient for localization and may be inactive.
FIGURE 7.
Assessment of mRNP assembly with She2p (L130→Y) mutant. Co-immunoprecipitation experiments were performed with anti-Myc antibodies against Myc-tagged She2p, translocation-complex assembly was monitored by Western-blot analyses with anti-HA antibodies against HA-tagged She3p. (A) Compared to wild-type She2p, complex assembly was impaired in She2p (L130→Y). Relative amount of co-immunoprecipitated She3p was quantified from two independent blots after normalization. For further details, see Materials and Methods. (B) Input and IP fractions, as well as input and flow-through fractions, are shown from the same blot. She2p mutant was expressed at levels comparable to wild-type She2p and She3p expression was unchanged in She2p-mutant expressing cells.
DISCUSSION
Previously, She2p dimers were reported from crystal-structure analyses and analytical ultracentrifugation (Niessing et al. 2004). However, these experiments were performed with mutated versions of She2p designed to facilitate crystallization. This mutated She2p lacked the N-terminal five and C-terminal seven residues, which are predicted to be unstructured, and had all four cysteines mutated into serines (Niessing et al. 2004). In light of the present results with wild-type She2p, it has to be assumed that these alternations of She2p impaired the oligomeric state of She2p. In the present study, size-exclusion chromatography with wild-type She2p showed that the protein forms tetrameric complexes at physiological concentrations. Because the previously described dimer buries an extensive surface interface of more than 2000 Å2/monomer (Table 1; Niessing et al. 2004) and since mutations disrupting dimer formation also inactivate She2p function in vivo (Niessing et al. 2004), these dimers are likely subunits of the She2p tetramer identified in our study. Analysis of the oligomerization mutant She2p (L130→Y) revealed that the upper, uncharged surface region of She2p might be involved in tetramer formation.
The SAXS analyses show that She2p forms an elongated tetramer, most likely through an interaction of the upper surface area. A calculated scattering curve of an elongated tetramer that was assembled via crystallographic dimer contacts (Niessing et al. 2004) matches well with the experimental scattering curve of She2p (Fig. 5A). In addition, mutation of leucine 130 into tyrosine at the dimer interface of this elongated tetramer (Fig. 4B,C) results in a steric clash (data not shown), potentially disrupting tetramer formation. Thus, She2p is likely to form an elongated tetramer through an asymmetric head-to head interaction, for which an atomic model can be derived (Fig. 5A,D). During SAXS analyses we also attempted to calculate surface envelopes. Unfortunately, results were too ambiguous to produce trustworthy models. Although calculated surface envelopes roughly matched the elongated tetramer, we cannot rule out the possibility that crystal packing and the lack of the C-terminal tail in the crystal structure modified the exact conformation of the elongated tetramer.
A recent study showed that She2p contains a nuclear localization signal (NLS) that is required for its function (Shen et al. 2009). In the same study, mutants were characterized that have disrupted the previously described, extensive dimer interface of She2p (>2000 Å2/monomer) (Table 1; Niessing et al. 2004). Based on observations with these mutants, the authors discuss the potential existence of monomeric She2p, which is actively imported into the nucleus. In our study, we do not find support for the existence of She2p dimers or even monomers at physiological concentrations (Fig. 2A,C). However, we cannot exclude that post-translational modifications affect the oligomeric state, for instance, of a minor fraction of She2p in vivo, as suggested by Shen et al. (2009). Calculations of buried surface interfaces yield a considerably smaller buried surface for the elongated tetramer than for the monomer–monomer interface (Table 1). Thus, structural considerations suggest that She2p tetramers potentially disassemble more easily into dimers than She2p dimers into monomers.
Mutant She2p (L130→Y) with impaired tetramer formation also showed impaired binding to all RNA zip-code elements tested in this study (Fig. 6A). From this we conclude that specific mRNAs may interact with the She2p tetramer by binding to both dimer subunits simultaneously. A second mutation (T47→Y) in the upper surface was reported to impair RNA binding (Niessing et al. 2004). Since this mutation is also located at the dimer interface of the elongated tetramer (not shown), it provides further support for our conclusions derived from She2p (L130→Y). Previous in vitro studies indicated an interaction of assumed She2p dimers and ASH1 mRNA zip-code elements with molar ratios of 1:1 and 1:2 (Niessing et al. 2004; Olivier et al. 2005). Whereas a ratio of one She2p dimer per two zip-code elements could be explained by RNA binding at the two opposing sides of the dimer (Niessing et al. 2004; Olivier et al. 2005), the ratio of one dimer per one zip-code element was structurally difficult to comprehend. For the 1:1 ratio it was suggested that the RNA might wrap around the core of She2p and thus interacts with both known binding surfaces at opposing sides of the dimer (Niessing et al. 2004). It was also speculated that the conserved upper surface of the She2p dimer would either be involved in RNA binding or participates in the interaction with the adapter She3p.
Our finding that the upper surface of She2p is a dimer–dimer interaction surface clarifies this issue. The upper surface is directly required for the formation of functionally important She2p tetramers, rendering the above-mentioned possibilities unlikely. When applying the stoichiometries of RNA binding to the She2p tetramer, RNA zip-code elements may bind with ratios of either two or four zip-code elements per tetramer. Two zip-code elements can be easily accommodated at both opposing surface interfaces (Fig. 5D). Four zip-code elements binding to one tetramer requires two elements binding at the same surface interface of She2p (Fig. 5D). The two continuous RNA-binding surfaces at the opposing sides of a She2p tetramer each have a length of about 60 Å. Considering average diameters of RNA stem loops of less than 25 Å, two RNAs could also be accommodated by this interface. Thus, binding of two zip-code elements at each side of She2p tetramers should also be possible without steric clashes.
These considerations on RNA binding also suggest a simultaneous interaction of different mRNA molecules to a She2p tetramer. This interpretation is consistent with the observation that different localizing mRNAs are transported together in one particle (Lange et al. 2008). An additional level of cross-linking between She2p tetramers into larger mRNPs may be achieved by the binding of She2p to mRNAs with multiple zip-code elements, like the ASH1 or WSC2 mRNAs. This reasoning is supported by the observation that overexpression of ASH1 mRNA produces fewer, but larger, ASH1 mRNPs in vivo (Bertrand et al. 1998; Lange et al. 2008). Thus, it is unlikely that She2p-dependent mRNPs have a defined, limited size. They may be rather interconnected in a dynamic way, resulting in mRNPs with variable size. It has recently been reported that translational repression of oskar mRNA in Drosophila is mediated by Bruno-dependent RNA oligomerization (Chekulaeva et al. 2006). Thus, the formation of large She2p mRNPs might support the previously described translational silencing during ASH1-mRNA transport (Paquin et al. 2007; Deng et al. 2008; Du et al. 2008).
MATERIALS AND METHODS
Plasmids and yeast strains
Detailed information on plasmids is found in Supplemental Table S2, on oligonucleotides in Supplemental Table S3, and on yeast strains in Supplemental Table S4. All SHE2 fragments for recombinant protein expression were amplified by PCR using plasmid p01 as the template (Niessing et al. 2004) and cloned into the vector pGEX-6P-1 (GE Healthcare). Artificially introduced BamHI sites in the respective 5′ primer and XhoI sites in the 3′ primer were used to clone the resulting fragments into the vector. Amino acid exchange in plasmid p08 (pGEX-SHE2-L130Y) was introduced by site-directed mutagenesis using oligonucleotides S2-3 and S2-24, and S2-23 and S2-1. To create plasmid p15, full-length SHE2 was amplified by PCR using primer pairs S2-17 and S2-19 and subcloned via NsiI and XbaI into RJP1565. RJP1565 was created by cutting out SHE2-myc3 of RJP428 with HindIII, ligation into RJP138, and following site-directed mutagenesis of an additional 5′ located XbaI site. p16 was created by using plasmid p08 as the template for a PCR reaction with primers S2-17 and S2-19, followed by subcloning into RJP1565. Construction of plasmid RJP916 has been reported previously (Du et al. 2008). Plasmid p19 was created by PCR amplification of the SHE2 L130Y fragment from plasmid p08 with oligonucleotides S2-17/18, followed by digestion with NsiI and StuI and cloning into NsiI/StuI digested RJP916.
General methods to culture and manipulate yeast strains were used as described (Gietz and Schiestl 1991; Adams et al. 1997). Yeast strains used in this study are listed in Supplemental Table S4. Yeast strain RJY2053 has been obtained from EUROSCARF, Frankfurt (accession no. Y14980). Strains y01, y02, and y05 were derived from RJY2053 by transformation of plasmid RJP132 and corresponding plasmids RJP145, RJP916, and p19, respectively. SHE2 deletion in strain RJY570 was generated by a PCR-based knockout method (Knop et al. 1999), resulting in strain RJY3364. Strains y06, y07, and y10 were derived from RJY3364 by transformation of corresponding plasmids RJP138, p15, and p16, respectively.
Protein purification
Wild-type She2p and She2p (L130→Y) were recombinantly expressed as GST-fusion proteins in E. coli BL21 Star (DE3) (Invitrogen) and purified to >95% homogeneity using standard chromatography techniques (Niessing et al. 2004). GST tags were removed using PreScission Protease (GE Healthcare).
Analytical ultracentrifugation
Analytical ultracentrifugation has been performed using a Beckman XL-A instrument (Beckman Coulter) equipped with UV/VIS optical systems. For all experiments, proteins had been pretreated with RNaseA (20 μg/mL) and were measured in a buffer containing 20 mM Tris (pH 7.5), 150 mM NaCl, 5 mM DTT, and 1 mM EDTA. In order to investigate concentration-dependent oligomerization events, the following protein concentrations were used in sedimentation–equilibrium experiments: wild-type-She2p: 1.7, 2.6, 3.5, 5.2, 7.3, 10.5, 15.0, 21.6, and 30.7 μM, and She2p (L130→ Y): 1.9, 2.6, 3.7, 5.2, 7.7, 10.8, 15.7, 22.3, and 32.0 μM. Six-channel centerpieces were filled with 105 μL of sample volume and 15 μL heptacosafluorotributylamine. After 48 h at 4°C, equilibrium conditions were obtained for wild-type-She2p (9000 rpm) and She2p (L130→Y) (12,500, 18,000, and 23,000 rpm), as judged from stable sedimentation profiles. Data were analyzed with the ULTRASCAN 9.8 software package (Demeler 2005). Sedimentation–velocity experiments were performed at 50,000 rpm and 20°C with two channel centerpieces (Ti-60 rotor), and data acquired at 280 nm wavelength. The following protein concentrations were measured: wild-type-She2p: 16.0 μM; She2p (L130→Y): 4.9, 12.9, and 32.0 μM. For analysis of sedimentation traces the dc/dt approach in the ULTRASCAN software package was used. In order to accurately determine S-values, resulting dc/dt curves were fitted in SigmaPlot (Systat) to a Gaussian function. Despite the potentially complex association model, this approach still resulted in consistent values with little nonstochastic residuals remaining after the data fitting.
Size-exclusion chromatography
Chromatography was performed with a Superose 12 10/300 GL column. One-half milliliter of She2p was loaded onto the column in buffer containing 20 mM Tris (pH 7.5), 150 mM NaCl, and 2 mM DTT with a flow rate of 0.5 mL/min. Peak fractions at 280 nm absorbance were analyzed by SDS PAGE for the presence of She2p. Potential DNA and RNA contaminations were removed by extensive washing steps with 1 M NaCL-containing buffer during protein purification. Sample dilution was calculated by dividing the sample peak volume through the sample volume loaded. Because we limited the included peak volumes to absorption areas above 10% of the peak maximum, leading and trailing portions of the peaks were ignored. In addition, sample loss and void peaks were not included in the calculations. Thus, the real average concentrations might be even lower.
Small-angle X-ray scattering
Synchrotron SAXS data were collected at the X33 beamline (EMBL/DESY) and at the ID14-3 beamline (ESRF). Potential nucleic acid contaminations in the protein samples were removed by washing with 1 M NaCl-containing buffer during protein purification. Protein purity was monitored by the ratio of OD280/254. Scattering curves were measured in 50 mM sodium phosphate (pH 7.4), 100 mM NaCl, and 2 mM DTT with exposures of one time 2 min (wild-type She2p at DESY, X-33) and ten times 30 sec (She2p [L130→Y] at ESRF, ID14-3), respectively. Data evaluation and processing was performed using the ATSAS program package (Konarev et al. 2006). For molecular-mass determination, scattering intensities were extrapolated to zero angle (I0), using bovine serum albumine and lysozyme as references. The radius of gyration Rg was calculated using the Guinier approximation with the constraint s*Rg < 1.3. The pair-distribution function p(r) and the maximum dimension of the particle Dmax were computed using the indirect transform package GNOM. The correct Dmax was iteratively determined by evaluating the resulting Rg value and the shape of the p(r) distribution. CRYSOL was used to determine the theoretical scattering curves based on the She2p crystal structure (PDB ID: 1XLY).
RNA preparation and RNA filter-binding assay
RNAs (see Supplemental Table S5) were either in vitro transcribed using the MEGAshortscript T7 Kit (Ambion) and subsequently purified via native polyacrylamide gel electrophoresis or produced by total chemical synthesis (EAR1 RNA, WSC2N RNA, and MS2 stem–loop RNA). RNA sequences are listed in Supplemental Table S5. RNA filter-binding assays were essentially performed as previously described (Du et al. 2008). Serial protein dilutions and constant amount of radiolabeled RNA (0.5 nM) were incubated in 20 mM HEPES (pH 7.5), 100 mM NaCl, 2 mM DTT, 2 mM MgCl2, and 30 μg/mL yeast tRNA at 22°C for 20 min. Reaction mixture (80 μL) was applied on a nitrocellulose membrane in a Bio-Dot microfiltration apparatus. Radioactivity retained was measured by phosphoimaging. Equilibrium-dissociation constants were calculated from a plot of the fraction of bound RNA versus protein concentration using the Langmuir isotherm. For quantification, in each experiment eight serial dilutions were used: wild-type-She2p binding to ASH1 zip-code elements: 0–4 μM; wild-type-She2p binding to EAR1 and WSC2N RNAs: 0–12 μM; wild-type-She2p binding to MS2 stem–loop RNA: 0–6 μM; She2p-mutant binding to bud-localizing RNAs: 0–16 μM; wild-type-She2p binding to poly(A) RNA: 0–32 μM. Indicated standard deviations were calculated from at least three independent experiments.
Monoclonal antibody
Monoclonal antibodies from rat against She2p were generated using standard procedures. Clones She2p-1C3, She2p-4G8, and She2p-5A1 (rat IgG2a) recognize wild-type and mutant She2p specifically in Western blotting. In this study, She2p-1C3 was used.
Fluorescence microscopy
For indirect immunofluorescence staining of She2p, yeast spheroplasts were incubated with polyclonal anti-She2p antibody (Du et al. 2008) and goat anti-rabbit antibody coupled to AlexaFluor488. Nuclei were stained with Hoechst Stain Solution (SIGMA). Cells were inspected with an Olympus BX60 fluorescence microscope and images were acquired on a Hamamatsu OrcaER CCD camera controlled by using Openlab 4.0 software (Improvision). She2p-expression levels were tested by Western-blot analysis using the monoclonal anti-She2p antibody She2p-1C3.
Co-immunoprecipitation
Co-immunoprecipitation of Myc-tagged She2p and HA-tagged She3p was performed by using monoclonal anti-Myc antibody (9E11, Acris Antibodies) coupled to magnetic Protein G beads (Invitrogen) in reaction buffer (50 mM Hepes [pH 7.5], 20 mM potassium acetate, 2 mM EDTA, 0.1% [v/v] Triton X-100, 5% [v/v] glycerol, 1X Complete Protease Inhibitor mix [Roche]). After washing, proteins were eluted from the beads by boiling at 95°C for 10 min in 1X Laemmli buffer. One-fiftieth (v/v) of the input fraction and 1/3 (v/v) of the IP elution fraction were subjected to Western-blot analysis, using anti-Myc antibody 9E10 (Roche) and anti-HA antibody 3F10 (Roche). For quantification, Western blots of two independent experiments were analyzed using the LAS-3000 mini system and Multi Gauge software (FUJIFILM). In all experiments, background signals from Δshe2 lanes were subtracted from respective Western blot signals. Subsequently, signals from the flow-through fraction were subtracted from the input fractions and used to normalize co-immunoprecipitated She3p against the bead-bound fraction of She2p-Myc3.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We are grateful to Olivier Gires, Sophia Hartung, Vigo Heissmeyer, Sigrun Jaklin, Susanne Lange, Maria Schmid, and Gregor Witte for their support. We acknowledge EMBL/DESY and the European Synchrotron Radiation Facility for provision of synchrotron radiation facilities. We thank Dmitry Svergun at DESY-X33 for support, and Adam Round and Petra Pernot for assistance in using beamline ID14-3. This work was supported by the Helmholtz Association (VG-NH 142 to D.N.), Deutsche Forschungsgemeinschaft (FOR855 to M.M. and D.N; SFB646 to M.M., R.-P.J., and D.N.; RI1873-1/1 to K.R.), and the Boehringer Ingelheim Fonds (to M.M.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1753309.
REFERENCES
- Adams A, Gottschling DE, Kaiser CA, Stearns T. Methods in yeast genetics. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1997. [Google Scholar]
- Bertrand E, Chartrand P, Schaefer M, Shenoy SM, Singer RH, Long RM. Localization of ASH1 mRNA particles in living yeast. Mol Cell. 1998;2:437–445. doi: 10.1016/s1097-2765(00)80143-4. [DOI] [PubMed] [Google Scholar]
- Bobola N, Jansen RP, Shin TH, Nasmyth K. Asymmetric accumulation of Ash1p in postanaphase nuclei depends on a myosin and restricts yeast mating-type switching to mother cells. Cell. 1996;84:699–709. doi: 10.1016/s0092-8674(00)81048-x. [DOI] [PubMed] [Google Scholar]
- Böhl F, Kruse C, Frank A, Ferring D, Jansen RP. She2p, a novel RNA-binding protein tethers ASH1 mRNA to the Myo4p myosin motor via She3p. EMBO J. 2000;19:5514–5524. doi: 10.1093/emboj/19.20.5514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartrand P, Meng XH, Singer RH, Long RM. Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr Biol. 1999;9:333–336. doi: 10.1016/s0960-9822(99)80144-4. [DOI] [PubMed] [Google Scholar]
- Chartrand P, Meng XH, Hüttelmaier S, Donato D, Singer RH. Asymmetric sorting of ash1p in yeast results from inhibition of translation by localization elements in the mRNA. Mol Cell. 2002;10:1319–1330. doi: 10.1016/s1097-2765(02)00694-9. [DOI] [PubMed] [Google Scholar]
- Chekulaeva M, Hentze MW, Ephrussi A. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell. 2006;124:521–533. doi: 10.1016/j.cell.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Conte LL, Chothia C, Janin J. The atomic structure of protein–protein recognition sites. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- Dahm R, Kiebler M, Macchi P. RNA localization in the nervous system. Semin Cell Dev Biol. 2007;18:216–223. doi: 10.1016/j.semcdb.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Dasgupta S, Iyer GH, Bryant SH, Lawrence CE, Bell JA. Extent and nature of contacts between protein molecules in crystal lattices and between subunits of protein oligomers. Proteins. 1997;28:494–514. doi: 10.1002/(sici)1097-0134(199708)28:4<494::aid-prot4>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Demeler B. UltraScan: A comprehensive data analysis software package for analytical ultracentrifugation experiments. In: Scott DJ, et al., editors. Modern analytical ultracentrifugation: Techniques and methods. Royal Society of Chemistry; London: 2005. pp. 210–229. [Google Scholar]
- Deng Y, Singer RH, Gu W. Translation of ASH1 mRNA is repressed by Puf6p-Fun12p/eIF5B interaction and released by CK2 phosphorylation. Genes & Dev. 2008;22:1037–1050. doi: 10.1101/gad.1611308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du TG, Jellbauer S, Müller M, Schmid M, Niessing D, Jansen RP. Nuclear transit of the RNA-binding protein She2p is required for translational control of localized ASH1 mRNA. EMBO Rep. 2008;9:781–787. doi: 10.1038/embor.2008.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gherardi E, Sandin S, Petoukhov MV, Finch J, Youles ME, Ofverstedt LG, Miguel RN, Blundell TL, Vande Woude GF, Skoglund U, et al. Structural basis of hepatocyte growth factor/scatter factor and MET signalling. Proc Natl Acad Sci. 2006;103:4046–4051. doi: 10.1073/pnas.0509040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH. Applications of high efficiency lithium acetate transformation of intact yeast cells using single-stranded nucleic acids as carrier. Yeast. 1991;7:253–263. doi: 10.1002/yea.320070307. [DOI] [PubMed] [Google Scholar]
- Gonsalvez GB, Lehmann KA, Ho DK, Stanitsa ES, Williamson JR, Long RM. RNA–protein interactions promote asymmetric sorting of the ASH1 mRNA ribonucleoprotein complex. RNA. 2003;9:1383–1399. doi: 10.1261/rna.5120803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez I, Buonomo SB, Nasmyth K, von Ahsen U. ASH1 mRNA localization in yeast involves multiple secondary structural elements and Ash1 protein translation. Curr Biol. 1999;9:337–340. doi: 10.1016/s0960-9822(99)80145-6. [DOI] [PubMed] [Google Scholar]
- Heuck A, Du TG, Jellbauer S, Richter K, Kruse C, Jaklin S, Muller M, Buchner J, Jansen RP, Niessing D. Monomeric myosin V uses two binding regions for the assembly of stable translocation complexes. Proc Natl Acad Sci. 2007;104:19778–19783. doi: 10.1073/pnas.0706780104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges AR, Krementsova EB, Trybus KM. She3p binds to the rod of yeast myosin V and prevents it from dimerizing, forming a single-headed motor complex. J Biol Chem. 2008;283:6906–6914. doi: 10.1074/jbc.M708865200. [DOI] [PubMed] [Google Scholar]
- Hogan DJ, Riordan DP, Gerber AP, Herschlag D, Brown PO. Diverse RNA-binding proteins interact with functionally related sets of RNAs, suggesting an extensive regulatory system. PLoS Biol. 2008;6:e255. doi: 10.1371/journal.pbio.0060255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jambhekar A, McDermott K, Sorber K, Shepard KA, Vale RD, Takizawa PA, DeRisi JL. Unbiased selection of localization elements reveals cis-acting determinants of mRNA bud localization in Saccharomyces cerevisiae. Proc Natl Acad Sci. 2005;102:18005–18010. doi: 10.1073/pnas.0509229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: More tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Konarev PV, Petoukhov MV, Volkov VV, Svergun DI. ATSAS2.1, a program package for small-angle scattering data analysis. J Appl Crystallogr. 2006;39:277–286. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse C, Jaedicke A, Beaudouin J, Böhl F, Ferring D, Güttler T, Ellenberg J, Jansen RP. Ribonucleoprotein-dependent localization of the yeast class V myosin Myo4p. J Cell Biol. 2002;159:971–982. doi: 10.1083/jcb.200207101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange S, Katayama Y, Schmid M, Burkacky O, Brauchle C, Lamb DC, Jansen RP. Simultaneous transport of different localized mRNA species revealed by live-cell imaging. Traffic. 2008;9:1256–1267. doi: 10.1111/j.1600-0854.2008.00763.x. [DOI] [PubMed] [Google Scholar]
- Long RM, Gu W, Lorimer E, Singer RH, Chartrand P. She2p is a novel RNA-binding protein that recruits the Myo4p–She3p complex to ASH1 mRNA. EMBO J. 2000;19:6592–6601. doi: 10.1093/emboj/19.23.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, Heuck A, Niessing D. Directional mRNA transport in eukaryotes: Lessons from yeast. Cell Mol Life Sci. 2007;64:171–180. doi: 10.1007/s00018-006-6286-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münchow S, Sauter C, Jansen RP. Association of the class V myosin Myo4p with a localized messenger RNA in budding yeast depends on She proteins. J Cell Sci. 1999;112:1511–1518. doi: 10.1242/jcs.112.10.1511. [DOI] [PubMed] [Google Scholar]
- Mylonas E, Svergun DI. Accuracy of molecular mass determination of proteins in solution by small-angle X-ray scattering. J Appl Crystallogr. 2007;40:245–249. [Google Scholar]
- Niessing D, Hüttelmaier S, Zenklusen D, Singer RH, Burley SK. She2p is a novel RNA-binding protein with a basic helical hairpin motif. Cell. 2004;119:491–502. doi: 10.1016/j.cell.2004.10.018. [DOI] [PubMed] [Google Scholar]
- Oeffinger M, Wei KE, Rogers R, Degrasse JA, Chait BT, Aitchison JD, Rout MP. Comprehensive analysis of diverse ribonucleoprotein complexes. Nat Methods. 2007;4:951–956. doi: 10.1038/nmeth1101. [DOI] [PubMed] [Google Scholar]
- Olivier C, Poirier G, Gendron P, Boisgontier A, Major F, Chartrand P. Identification of a conserved RNA motif essential for She2p recognition and mRNA localization to the yeast bud. Mol Cell Biol. 2005;25:4752–4766. doi: 10.1128/MCB.25.11.4752-4766.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquin N, Menade M, Poirier G, Donato D, Drouet E, Chartrand P. Local activation of yeast ASH1 mRNA translation through phosphorylation of Khd1p by the casein kinase Yck1p. Mol Cell. 2007;26:795–809. doi: 10.1016/j.molcel.2007.05.016. [DOI] [PubMed] [Google Scholar]
- Shav-Tal Y, Singer RH. RNA localization. J Cell Sci. 2005;118:4077–4081. doi: 10.1242/jcs.02543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, Paquin N, Forget A, Chartrand P. Nuclear shuttling of She2p couples ASH1 mRNA localization to its translational repression by recruiting Loc1p and Puf6p. Mol Biol Cell. 2009;20:2265–2275. doi: 10.1091/mbc.E08-11-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard KA, Gerber AP, Jambhekar A, Takizawa PA, Brown PO, Herschlag D, DeRisi JL, Vale RD. Widespread cytoplasmic mRNA transport in yeast: Identification of 22 bud-localized transcripts using DNA microarray analysis. Proc Natl Acad Sci. 2003;100:11429–11434. doi: 10.1073/pnas.2033246100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sil A, Herskowitz I. Identification of asymmetrically localized determinant, Ash1p, required for lineage-specific transcription of the yeast HO gene. Cell. 1996;84:711–722. doi: 10.1016/s0092-8674(00)81049-1. [DOI] [PubMed] [Google Scholar]
- St Johnston D. Moving messages: The intracellular localization of mRNAs. Nat Rev Mol Cell Biol. 2005;6:363–375. doi: 10.1038/nrm1643. [DOI] [PubMed] [Google Scholar]
- Takizawa PA, Vale RD. The myosin motor, Myo4p, binds ASH1 mRNA via the adapter protein, She3p. Proc Natl Acad Sci. 2000;97:5273–5278. doi: 10.1073/pnas.080585897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa PA, DeRisi JL, Wilhelm JE, Vale RD. Plasma membrane compartmentalization in yeast by messenger RNA transport and a septin diffusion barrier. Science. 2000;290:341–344. doi: 10.1126/science.290.5490.341. [DOI] [PubMed] [Google Scholar]
- Vale RD. The molecular motor toolbox for intracellular transport. Cell. 2003;112:467–480. doi: 10.1016/s0092-8674(03)00111-9. [DOI] [PubMed] [Google Scholar]