Abstract
Cosuppression is a classical form of eukaryotic post-transcriptional gene silencing. It was first reported in transgenic petunia, where a sense transgene meant to overexpress the host Chalcone Synthase-A (CHS-A) gene caused the degradation of the homologous transcripts and the loss of flower pigmentation. In this work, we used deep sequencing technology to characterize in detail the small RNA population generated from the CHS-A sequence in cosuppressed transgenic petunia. Unexpectedly, two distinct small interfering RNAs (siRNAs) were found to vastly predominate. Our demonstration that they guide prominent cleavage events in CHS-A mRNA provides compelling and previously lacking evidence of a causative association between induction of individual siRNAs and an example of cosuppression. The preferential accumulation of these siRNAs provides new insights about sense cosuppression that may apply to other natural and engineered RNA silencing events.
Keywords: cosuppression, petunia, small RNA, post-transcriptional silencing, gene silencing, RNA silencing
INTRODUCTION
Sense cosuppression was the first form of eukaryotic post-transcriptional gene silencing reported (Jorgensen 1995). It arises when transgenes designed to overexpress a host protein instead cause suppression of homologous host transcripts by inducing mRNA degradation. This phenomenon was originally described in transgenic petunia in which expression of the chalcone synthase-A (CHS-A) gene, required for the biosynthesis of anthocyanin pigments in the flower, was inhibited by the overexpression of a homologous sense transgene (Napoli et al. 1990). Later, CHS-A expression loss was associated with mRNA degradation by endonucleolytic cleavage (Metzlaff et al. 1997), which identified cosuppression as an RNA silencing mechanism (Baulcombe 2004). Nevertheless, as for most forms of gene silencing in transgenic organisms, the production of small interfering RNAs (siRNAs), assumed to be determinants of CHS-A silencing, has yet to be characterized. Here we provide evidence from deep sequencing of a causative relationship between the process of CHS-A sense cosuppression and the elevation of two specific siRNAs that vastly predominate and guide prominent mRNA cleavage events.
RESULTS AND DISCUSSION
Our analysis compared the small RNA populations of petals from wild-type (WT) (Petunia hybrida, strain V26, purple petals) or transgenic petunia that expressed a single copy of CHS-A coding sequence under the control of CaMV 35S promoter and exhibited only fully white (silenced) flowers. We used Illumina's SBS technology for sequencing of the small RNA libraries, producing a total of 2,549,729 trimmed sequences from transgenic petal and 7,757,806 from the WT. When distinct (unique) small RNAs were taken into account, WT and transgenic petals exhibited 3,384,212 and 516,906 sequences, respectively. In both libraries, sequences ranged substantially from 18 to 25 nucleotides (nt) in size (Supplemental Fig. S1); longer sequences, some of which represented nonproperly trimmed inserts, were also found at a negligible level. We observed three prominent peaks at 19, 21, and 24 nt in both libraries (Supplemental Fig. S1). The 21-nt and 24-nt peaks were recognized as the major subgroups of plant small RNAs, mostly represented by miRNAs/endo-siRNAs and heterochromatic siRNAs, respectively (Ghildiyal and Zamore 2009), while the additional 19-nt peak was found to be largely represented by a unique highly abundant tRNA-related small RNA.
The small RNAs sequenced from both transgenic and WT petals were mapped on the sequence of the CHS-A gene as well as those of additional CHS sequences available from petunia (B, D, F, J, and H genes). Among these genes, only CHS-A, and to a lower extent CHS-J, matched to a significant number of small RNAs. The other CHS genes, which are not expressed in flower tissues (Koes et al. 1989), showed negligible association with small RNAs in either genetic background and were not further investigated.
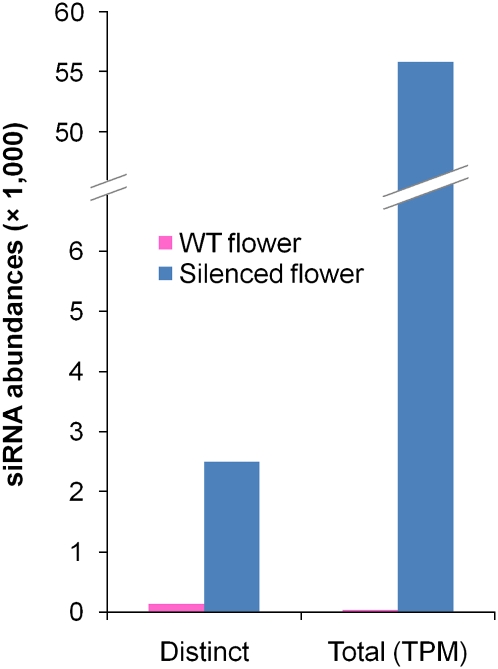
In transgenic petals, the entire population of siRNAs matching the CHS-A gene was dramatically larger than in WT (Fig. 1). The small RNAs from WT matching CHS-A corresponded to 128 distinct sequences accounting for 28 transcripts per million (TPM), which is in the range observed for other petunia genes. In contrast, the CHS-A small RNAs in silenced flowers corresponded to 2495 distinct sequences with a total abundance of ∼56,000 TPM. Nearly all of the latter (98%) matched to the coding region, and were also present in the CHS-A transgene. The siRNAs were distributed unevenly across the CHS-A sequence in silenced flowers with a prominent hot spot in the second exon (Fig. 2A). Surprisingly, two antisense, partially overlapping 21-mers (phy-siR1 and phy-siR2) were exceedingly abundant, contributing more than half of the total siRNA abundance across the gene. Interestingly, neither the hot spot nor the two outstanding small RNAs were observed in the WT.
FIGURE 1.
Number of CHS-A siRNAs in WT versus silenced petals. TPM indicates transcripts per million.
FIGURE 2.
Position and abundance of CHS-A gene-matching siRNAs in silenced flowers (A) compared with WT flowers (B). Upper chart: sense strand; lower chart: antisense strand. Nucleotide positions and annotations are relative to the CHS-A gene sequence (GenBank X14591.1).
The dramatic elevation of the two 21-mers associated with silencing was reproducible in gel assays and with an independent biological replicate (Supplemental Fig. S2), suggesting a direct role for these siRNAs in targeting CHS-A degradation. Indeed, RLM 5′ RACE (RNA ligase mediated 5′ rapid amplification of cDNA ends) showed that, within the 130-bp window of CHS-A mRNA examined, the most frequently detected cleavage occurred between the sites complementary to the 10th and 11th position of one siRNA and was prominent for the other as well (Fig. 3A). This result demonstrated the ability of the two siRNAs to cause the expected cleavage of the CHS-A transcript and prompted us to further investigate the small RNA population matching the CHS-A mRNA sequence in the search of additional characteristics that have been reported to accompany target RNA cleavage in some circumstances.
FIGURE 3.
(A) Cleavage site mapping around the extremely abundant siRNAs in CHS-A mRNA in silenced flowers. Numbers of RLM 5′ RACE clones are reported for each nucleotide position (genomic coordinates are reported for convenience). Although nonquantitative, one of the predicted cleavage sites predominated and the other was prominent. (B) Abundance distribution and phasing of small RNAs relative to the CHS-A coding sequence in silenced flowers. Top: total number of small RNA reads matching to CHS-A coding sequence in each position (genomic coordinates are reported for convenience). The most abundant siRNAs were not included in the analysis to avoid a spurious phasing signal. The approximate position of the major cleavage site is shown as a vertical gray line. TPM, transcripts per million. Bottom: phasing signals from small RNAs. Phasing scores are computed according to Howell et al. (2007) with modifications as described in Materials and Methods. Scores are assigned to the intermediate interval position in the phasing plot. Gridlines correspond to 21-nt intervals. Phase drift is illustrated by two regions where the best phase is shifted with respect to the major cleavage site by 1 and 2 nt, respectively. Phasing was not observed in WT flowers (data not shows).
In some well-studied examples, as in the case of TAS loci, small RNA-directed cleavage is known to trigger the generation of 21-nt, phased siRNAs beginning from a small RNA-directed cleavage site (Vaucheret 2005). Indeed, a small RNA phasing pattern, possibly triggered by the major cleavage, was also detected for CHS-A in silenced flowers (Fig. 3B). Sharp peaks were observed that represent a robust 21-nt phasing pattern extending from the target site to ∼400 nt upstream, while no clear phase was found downstream. The upper section of the best phase (nucleotides 2823–2907) showed a 2-nt backward shift with respect to the major cleavage site, whereas in the lower section (nucleotides 2928–3033), the shift was just 1 nt because of phase drift. A similar deviation was previously observed for a family of trans-acting siRNAs (Howell et al. 2007), although in that case no shift was observed with respect to the initiating cleavage. We investigated the possibility that the generation of phased siRNAs could be initiated by a cleavage in a different region of the transcript. However, no small RNAs likely to target CHS-A mRNA and trigger the phasing pattern were detected other than phy-siR1 and phy-siR2, even for comparisons allowing up to five mismatches.
In conclusion, our results implicate the two extremely abundant siRNAs as antisense guides for determining CHS-A sense cosuppression via mRNA cleavage. Moreover, the observed phasing pattern is reminiscent of other mechanisms that produce phased siRNAs from flanking sequences after a small RNA generated by an unlinked locus causes a nucleating cleavage, as in the case of trans-acting siRNA loci (Allen et al. 2005). However, in contrast to those systems, the biogenesis of the cleavage-guiding siRNAs in CHS-A sense cosuppression seems to be directly related to the primary target, and therefore a different primary mechanism may be operating.
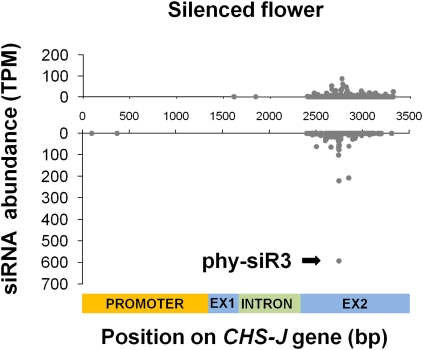
A likely model for CHS-A is that the two major antisense siRNAs along with the lower abundance sense and antisense siRNAs originate from double-stranded RNA generated by RDR6; the latter is an RNA-dependent RNA polymerase (RdRP) shown to be required for sense cosuppression in Arabidopsis (Beclin et al. 2002; Mallory and Vaucheret 2009). The origin of extremely abundant siRNAs from such a limited region strongly indicates that the processing of CHS-A transcripts relies on intrinsic properties. In support of this, we also observed elevation of siRNA levels from the homologous region of CHS-J (Fig. 4); the homology of its prominent 22-mer (phy-siR3) to the two CHS-A siRNAs as well as a predicted secondary structure conserved between CHS-A and CHS-J (Supplemental Fig. S3) suggest that local sequence determinants may play a critical role in recruiting the silencing machinery. Other members of the CHS family (CHS-B, CHS-D, and CHS-H) are also predicted to form similar secondary structures (data not shown), but the lack of their expression in floral tissues does not enable us to evaluate their availability to generate homologous siRNAs similar to those of CHS-J.
FIGURE 4.
Position and abundance of CHS-J gene-matching siRNAs in silenced flowers. Upper chart: sense strand; lower chart: antisense strand. Nucleotide positions and annotation are relative to the CHS-J genomic sequence (GenBank X14597.1). Eighty-two percent of the siRNAs, including the most abundant one (phy-siR3), were unique to CHS-J among all CHS sequences available from petunia. In the WT, matching of siRNAs to CHS-J was negligible (data not shown).
In light of our results, the major contribution of the transgene to the silencing mechanism would be to provide the necessary level of sense RNA transcription to trigger the amplification process catalyzed by RDR6. Whether the generated small RNAs may have an active role in enhancing this process, as in siRNA transitivity (see, for instance, Moissiard et al. 2007), is an issue yet to be clarified. However, the moderate elevation of CHS-J-specific siRNAs upon CHS-A overexpression suggests the attractive hypothesis that CHS-A-specific siRNAs could direct the machinery for siRNA biogenesis against the homologous CHS-J transcripts; CHS-J exhibits the highest degree of sequence identity with CHS-A (86%) compared with the other CHS genes. An alternative but unlikely explanation may evoke increased transcription of the CHS-J gene, in response to CHS-A depletion, and triggering of a mechanism similar to the one we have proposed for CHS-A.
The reasons why cosuppression efficiency varies greatly from gene to gene are not fully understood. Besides the strength of the transgene promoter (Vaucheret et al. 1995; Que et al. 1997) and the length of homology between the transgene and the endogenous gene (Vaucheret et al. 1997; Crete and Vaucheret 1999), features unique to the target genes are thought to promote the triggering of cosuppression. Our observations suggest the intriguing hypothesis that, at least in some cases, the silencing mechanism causing cosuppression may ultimately rely on the availability of a specific transcript region to be processed into stable and abundant small RNAs when particular conditions are met. We expect that further insights into the preferential generation of siRNAs with respect to local sequence elements or structural features may come from the deep sequencing of small RNAs from other well-studied examples of post-transcriptional silencing (e.g., Palauqui et al. 1996; Ruiz et al. 1998). Most importantly, our work provides definitive evidence for the role of antisense siRNAs as crucial players in CHS-A sense cosuppression. As such, our study suggests that the understanding of cosuppression would strongly benefit from the investigation of the threshold mechanism that activates and amplifies siRNA biogenesis. Moreover, it demonstrates that this classic example of post-transcriptional silencing is still capable of providing new insights that may extend to other natural and engineered RNA silencing events.
MATERIALS AND METHODS
Production of transgenic petunia and plant growth conditions
Young leaves of petunia line V26 were cocultivated with disarmed Agrobacterium tumefaciens LBA4404 harboring the complete coding region of CHS-A gene under the control of a 2x35S promoter and the nopaline synthase (NOS) terminator (pGSA2644). Transformants were selected and regenerated as described in Jorgensen et al. (1996) except that rooting of adventitious shoots was performed with 100 μg/mL kanamycin sulfate. After rooting, plantlets were transplanted to soil and grown in a greenhouse as described in Jorgensen et al. (1996).
RNA isolation
Total RNA was extracted from pre-anthesis petals of both WT (Petunia hybrida V26) and silenced petunia plants (two biological replicates per type, respectively) with the Plant RNA Purification Reagent (Invitrogen). Quality of total RNA was evaluated by agarose gel electrophoresis, micro-electrophoresis (Agilent 2100 Bioanalyzer, Ambion) and O.D. measurements (Nanodrop ND-1000).
Small RNA library construction
RNA from transgenic petals was submitted to Illumina (http://www.illumina.com) for small RNA library construction using approaches described in Lu et al. (2007) with minor modifications. This small RNA library was sequenced with the Sequencing-by-synthesis (SBS) technology by Illumina. The small RNA library from WT petals was constructed in-house according to Lu et al. (2007) and sequenced on the Illumina Genome Analyzer II at the Delaware Biotechnology Institute according to the manufacturer's instructions. Raw data were analyzed using Pipeline v1.3.2 by Illumina. Sequence data have been deposited in NCBI's Gene Expression Omnibus and are accessible through GEO Series accession number GSE13764 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13764).
Small RNA data analysis
Small RNA reads were trimmed and mapped on CHS-A and CHS-J genomic sequences available in GenBank (X14591.1 and X14597.1, respectively), allowing only perfect matches. Up to five mismatches were allowed to identify additional cleavage-directing small RNAs.
For the phasing analysis, we adjusted the algorithm developed by Howell et al. (2007) to make it suitable to the analysis of SBS data, which, because of its depth, has increased levels of background noise compared with the 454 technology (Margulies et al. 2005). Phasing scores were obtained with the following equation:
 |
where n = number of phase cycle positions occupied by at least one small RNA read within a 10-cycle window (phase cycle length was set at 21 nt); P = total number of reads for all small RNAs with start coordinates in a given phase within a 10-cycle window; and U = total number of reads for all small RNAs with start coordinates out of the given phase. Pairs of small RNAs, from the sense and antisense strand, respectively, that were offset by 2 nt with the overhang at the 3′ ends, were combined. The most abundant siRNAs were not included in the analysis to avoid a spurious phasing signal.
RNA secondary structures of CHS-A and CHS-J transcripts with the lowest ΔG were determined by Mfold (Zuker 2003) with default settings.
Small RNA hybridization
Low-molecular-weight (LMW) RNA isolation from total RNA, gel blots, radiolabeling, and hybridization/wash conditions were described previously (Lu et al. 2005). The radiolabeled probes for phy-siR1 and phy-siR2 were 5′-TCCTTGTTGTTTGTTCAGAAA and 5′-CGAGTCCTTGTTGTTTGTTCA, respectively.
Splinted ligation was performed by using the miRtect-IT miRNA labeling and detection kit (USB) as described in Maroney et al. (2007). Specific bridge oligonucleotides (5′-GAATGTCATAAGCGTCCTTGTTGTTTGTTCAGAAA-3′ and 5′-GAATGTCATAAGCGCGAGTCCTTGTTGTTTGTTCA-3′) were designed according to the manufacturer's directions. Using 1 μg of total RNA, the surveyed small RNAs were captured by a specific bridge oligonucleotide and ligated to the P32-labeled detection oligonucleotide with T4 DNA ligase. Ligated products were separated on 15% urea-polyacrylamide gel and visualized with a phosphorimaging system.
RNA ligase-mediated (RLM) 5′ RACE
To map the cleavage site of the CHS-A target transcript, a modified procedure for RLM 5′ RACE was performed by using the FirstChoice RLM-RACE Kit (Ambion). Total RNAs (5 μg) from WT and transgenic petals were ligated to the adapter for 5′ RACE without treatment with calf intestine alkaline phosphatase and tobacco acid pyrophosphatase. Initial PCR for cDNA synthesis was carried out by using the 5′ RACE outer primer from the manufacturer and a gene-specific outer primer (5′-GAAACGAGCTCGAACAAAGG-3′). Nested PCR was carried out by using 1/50 of the initial PCR, the 5′ RACE inner primer from the manufacturer, and a gene-specific inner primer (5′-CTCTCGACTCCTGGAATTGG-3′). RACE fragments were reamplified, gel-purified, and cloned with the TOPO TA cloning kit (Invitrogen), and 65 clones per sample were sequenced.
SUPPLEMENTAL MATERIAL
Supplemental material can be found at http://www.rnajournal.org.
ACKNOWLEDGMENTS
We thank Marcelo German, Scott Poethig, and anonymous reviewers for providing helpful comments. This work was supported by National Science Foundation Grant 0638525 (to P.J.G. and B.C.M.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.1706109.
REFERENCES
- Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Baulcombe D. RNA silencing in plants. Nature. 2004;431:356–363. doi: 10.1038/nature02874. [DOI] [PubMed] [Google Scholar]
- Beclin C, Boutet S, Waterhouse P, Vaucheret H. A branched pathway for transgene-induced RNA silencing in plants. Curr Biol. 2002;12:684–688. doi: 10.1016/s0960-9822(02)00792-3. [DOI] [PubMed] [Google Scholar]
- Crete P, Vaucheret H. Expression and sequence requirements for nitrite reductase cosuppression. Plant Mol Biol. 1999;41:105–114. doi: 10.1023/a:1006364323494. [DOI] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: An expanding universe. Nat Rev Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell MD, Fahlgren N, Chapman EJ, Cumbie JS, Sullivan CM, Givan SA, Kasschau KD, Carrington JC. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell. 2007;19:926–942. doi: 10.1105/tpc.107.050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen RA. Cosuppression, flower color patterns, and metastable gene expression states. Science. 1995;268:686–691. doi: 10.1126/science.268.5211.686. [DOI] [PubMed] [Google Scholar]
- Jorgensen RA, Cluster PD, English J, Que Q, Napoli CA. Chalcone synthase cosuppression phenotypes in petunia flowers: Comparison of sense versus antisense constructs and single-copy versus complex T-DNA sequences. Plant Mol Biol. 1996;31:957–973. doi: 10.1007/BF00040715. [DOI] [PubMed] [Google Scholar]
- Koes RE, Spelt CE, van den Elzen PJ, Mol JN. Cloning and molecular characterization of the chalcone synthase multigene family of Petunia hybrida. Gene. 1989;81:245–257. doi: 10.1016/0378-1119(89)90185-6. [DOI] [PubMed] [Google Scholar]
- Lu C, Tej SS, Luo S, Haudenschild CD, Meyers BC, Green PJ. Elucidation of the small RNA component of the transcriptome. Science. 2005;309:1567–1569. doi: 10.1126/science.1114112. [DOI] [PubMed] [Google Scholar]
- Lu C, Meyers BC, Green PJ. Construction of small RNA cDNA libraries for deep sequencing. Methods. 2007;43:110–117. doi: 10.1016/j.ymeth.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Mallory AC, Vaucheret H. ARGONAUTE 1 homeostasis invokes the coordinate action of the microRNA and siRNA pathways. EMBO Rep. 2009;10:521–526. doi: 10.1038/embor.2009.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroney PA, Chamnongpol S, Souret F, Nilsen TW. A rapid, quantitative assay for direct detection of microRNAs and other small RNAs using splinted ligation. RNA. 2007;13:930–936. doi: 10.1261/rna.518107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzlaff M, O'Dell M, Cluster PD, Flavell RB. RNA-mediated RNA degradation and chalcone synthase A silencing in petunia. Cell. 1997;88:845–854. doi: 10.1016/s0092-8674(00)81930-3. [DOI] [PubMed] [Google Scholar]
- Moissiard G, Parizotto EA, Himber C, Voinnet O. Transitivity in Arabidopsis can be primed, requires the redundant action of the antiviral Dicer-like 4 and Dicer-like 2, and is compromised by viral-encoded suppressor proteins. RNA. 2007;13:1268–1278. doi: 10.1261/rna.541307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli C, Lemieux C, Jorgensen R. Introduction of a chimeric chalcone synthase gene into petunia results in reversible cosuppression of homologous genes in trans. Plant Cell. 1990;2:279–289. doi: 10.1105/tpc.2.4.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palauqui JC, Elmayan T, De Borne FD, Crete P, Charles C, Vaucheret H. Frequencies, timing, and spatial patterns of cosuppression of nitrate reductase and nitrite reductase in transgenic tobacco plants. Plant Physiol. 1996;112:1447–1456. doi: 10.1104/pp.112.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que Q, Wang HY, English JJ, Jorgensen RA. The frequency and degree of cosuppression by sense chalcone synthase transgenes are dependent on transgene promoter strength and are reduced by premature nonsense codons in the transgene coding sequence. Plant Cell. 1997;9:1357–1368. doi: 10.1105/tpc.9.8.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz MT, Voinnet O, Baulcombe DC. Initiation and maintenance of virus-induced gene silencing. Plant Cell. 1998;10:937–946. doi: 10.1105/tpc.10.6.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H. MicroRNA-dependent trans-acting siRNA production. Sci STKE. 2005;2005:pe43. doi: 10.1126/stke.3002005pe42. [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Palauqui JC, Elmayan T, Moffatt B. Molecular and genetic analysis of nitrite reductase cosuppression in transgenic tobacco plants. Mol Gen Genet. 1995;248:311–317. doi: 10.1007/BF02191598. [DOI] [PubMed] [Google Scholar]
- Vaucheret H, Nussaume L, Palauqui JC, Quillere I, Elmayan T. A transcriptionally active state is required for post-transcriptional silencing (cosuppression) of nitrate reductase host genes and transgenes. Plant Cell. 1997;9:1495–1504. doi: 10.1105/tpc.9.8.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]