Abstract
Human cells contain several hundred ribosomal genes (rDNA) that are clustered into nucleolar organizer regions (NORs) on the short arms of five different acrocentric chromosomes. Only ∼50% of the gene copies are actually expressed in somatic cells. Here, we used a new cytological technique to demonstrate that rDNA is regulated allelically in a regional manner, with one parental copy of each NOR being repressed in any individual cell. This process is similar to that of X-chromosome inactivation in females. Early in development, one copy of each NOR becomes late-replicating, thus probably marking it for inactivation and subsequent targeted de novo methylation at rDNA promoter regions. Once established, this multichromosomal allelic pattern is then maintained clonally in somatic cells. This pathway may serve as an epigenetic mechanism for controlling the number of available rDNA copies during development.
Keywords: Ribosomal DNA, replication timing, monoallelic expression, early development, DNA methylation
Despite the clear-cut organization of rDNA copies into specific loci on the human genome, little is known about the logic of ribosomal gene expression. All ribosomal RNA synthesis is carried out by RNA polymerase I (polI) within a small number of distinct nucleoli that appear to be formed for this purpose (Boisvert et al. 2007; McStay and Grummt 2008). Even though all of the rDNA copies are probably associated with these nucleoli, only ∼50% are actually transcribed (Grummt 2007), with the remaining genes being inactivated through a combination of different epigenetic mechanisms that take advantage of specific repression factors (Grummt et al. 1986; Santoro et al. 2002), noncoding RNAi (Mayer et al. 2006, 2008), DNA methylation (Santoro and Grummt 2001; Ghoshal et al. 2004), and late replication timing (Li et al. 2005). Despite this general understanding of silencing at the individual gene level, little is known about how genes are chosen for inactivation in each cell. In addition, it is not yet clear how the ribosomal genes are regulated during normal development.
DNA replication timing is a regional epigenetic marking mechanism that is correlated with gene expression. The entire genome is divided into replication time zones that are organized in a distinct banding pattern on each chromosome. Active gene regions usually replicate early during S phase, while inactive genes replicate late, with only a relatively small number of genes not adhering to this correlation (Hiratani and Gilbert 2009). In keeping with this correlation, regions of monoallelic expression have been found to replicate asynchronously, with one allele copied earlier than the other (Goldmit and Bergman 2004). In the case of imprinted gene regions, this differential replication pattern is actually established in the gametes with the paternal allele preferentially replicating early in S (Simon et al. 1999). In contrast, for many other monoallelic regions, differential replication timing is set up randomly in the early embryo, with one allele, either the maternal or paternal, being early and the other allele being late. Once established, this binary pattern is then clonally maintained in somatic cells (Mostoslavsky et al. 2001). This early marking system apparently plays a role in allele usage during development. The establishment of late replication timing, for example, represents one of the earliest events of X-chromosome inactivation in female embryos (Keohane et al. 1998), and for many autosomal regions, asynchronous replication is instrumental in determining the preferred expression of one allele in each cell (Gimelbrant et al. 2005). This early differential mark also plays a role in the preselection of a single receptor allele for recombination in the immune system (Goldmit et al. 2005). In light of the regional nature of replication timing, we reasoned that this mechanism may also be involved in the silencing of rDNA.
Results
Replication timing-specific hybridization (ReTiSH)
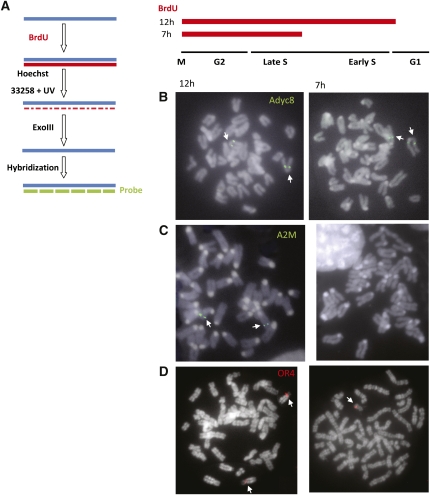
In order to determine how rDNA clusters replicate during S phase, we developed a new approach for visualizing BrdU uptake specifically at these loci. In this method, cells are labeled with BrdU for various times and then blocked in metaphase. Incorporation into mitotic chromosomes is visualized by a modification of CO-FISH (Cornforth and Eberle 2001), a technique in which replicated regions (BrdU-labeled) are converted to ssDNA and then hybridized directly with specific probes (Fig. 1A). To test this new methodology, we analyzed the hybridization patterns of control genes that replicate at different times during S phase in mouse embryonic fibroblasts (MEFs). Hybridization using a probe for the Adyc8 gene region (Fig. 1B), for example, revealed a signal on both chromosome 11 homologs when BrdU was initiated in either early S (12 h) or late S (7 h), consistent with the fact that this region replicates in late S phase. In contrast, labeling of the early-replicating A2M gene is only observed when BrdU is present from the beginning of S phase, and no signal was obtained if BrdU was added for only 7 h (Fig. 1C). Analysis of an olfactory receptor locus in human lymphoblasts revealed that both alleles are labeled if BrdU is initiated during early S phase, but only one can be observed when BrdU is added during late S (Fig. 1D), consistent with the fact that this gene undergoes asynchronous replication (Chess et al. 1994). In all ReTiSH experiments, the labeling and detection procedures were calibrated to ensure that all signals are indeed BrdU-dependent.
Figure 1.
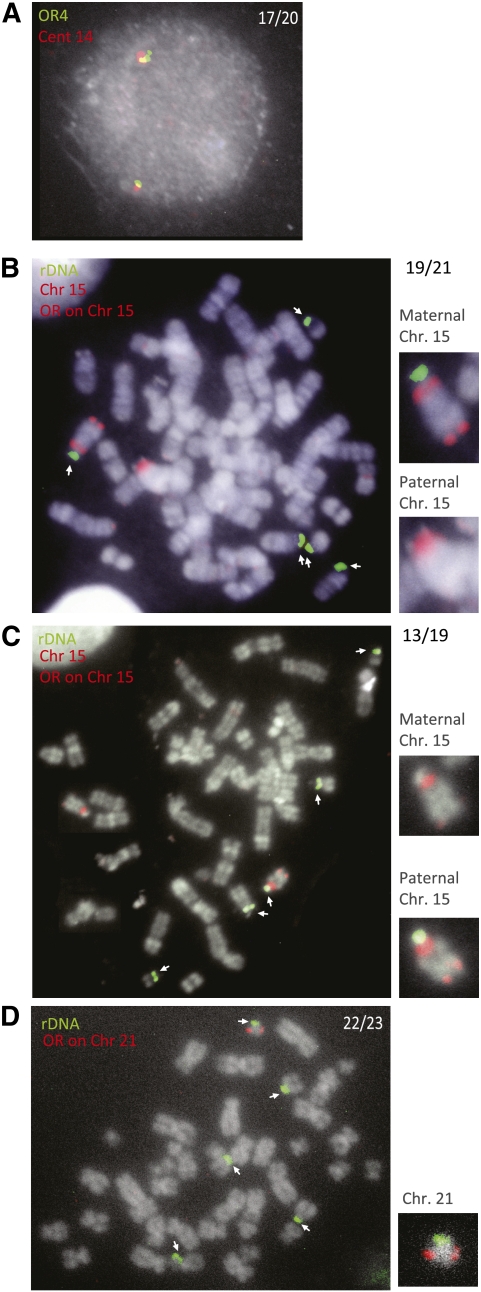
ReTiSH. (A) Scheme of ReTiSH method showing how BrdU-labeled regions are converted to ssDNA so that they can be identified by hybridization with specific probes. MEFs were labeled with BrdU for 7 or 12 h, arrested in metaphase, and subjected to ReTiSH using BAC probes for the late-replicating gene Adyc8 (RP23-352F10) (B) or the early-replicating gene A2M (RP23-191C6) (C). (D) Human lymphoblasts were labeled with BrdU for 7 or 12 h, arrested in metaphase, and subjected to ReTiSH using a BAC probe (RP11-55G7) for the asynchronously replicating olfactory receptor (family 4) gene cluster on chromosome 14 (OR4). Arrows indicate relevant gene signals.
Ribosomal genes replicate bimodally
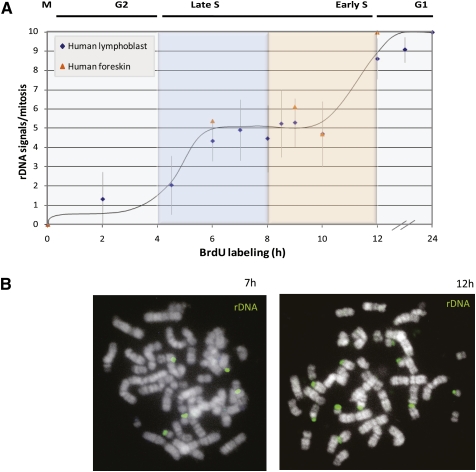
The ReTiSH technique was then applied for analyzing replication timing at the five pairs of rDNA loci in the nucleus by labeling human lymphoblast cells with BrdU for various times. Strikingly, replication at these sites is seen to occur in a bimodal pattern, with approximately five loci replicating early in S phase and the remaining five replicating late (Fig. 2). Similar results were observed in primary human foreskin fibroblasts, as well. While this experiment confirms that ∼50% of the rDNA copies replicate late in S phase (Li et al. 2005), it also indicates for the first time that the control of rDNA replication timing is carried out in a regional manner with each individual locus undergoing uniform replication either early or late in the cell cycle. These results are consistent with previous FISH experiments showing the presence of both replicated and unreplicated rDNA loci in the same interphase nucleus (Haaf 1997).
Figure 2.
Replication timing of rDNA. (A) Human lymphoblasts or foreskin fibroblasts were labeled with BrdU for various times, arrested in metaphase, and subjected to ReTiSH using a probe for rDNA. The number of rDNA signals in each metaphase spread (n = 461) was counted, and standard deviation was calculated for each data point (see error bars). (B) Examples at 7 and 12 h are shown.
rDNA replication is allelic
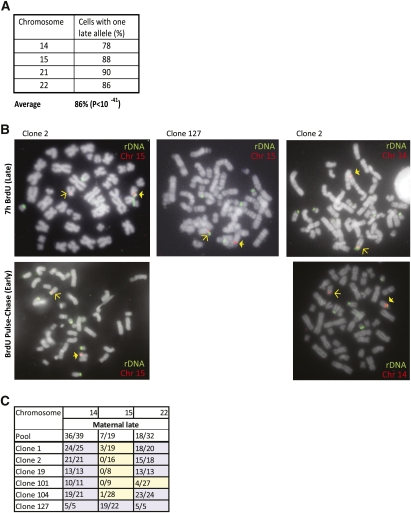
In order to test whether replication timing is regulated in an allelic manner, we labeled cells for 7 h with BrdU and carried out ReTiSH using one probe for detecting rDNA and the second specific for the subcentromeric region on chromosome 14. Under these conditions, we observed five labeled rDNA loci in each cell. We then counted the percentage of cells that have only a single copy of rDNA replicated on chromosome 14 (Fig. 3A). If replication timing occurs in a completely random manner, one would expect to find a single copy of rDNA from chromosome 14 among the five late-replicating loci in only 55% of the metaphase spreads. In contrast, our analysis showed that this occurred in ∼80% of the cases. A similar pattern was observed for rDNA replication on other chromosomes (15, 21, and 22), as well. Altogether, we analyzed 430 metaphase spreads, with 86% of them showing a single replicated rDNA locus for each chromosome. This indicates (P < 10−41) that replication of rDNA is controlled allelically, with one allele replicating early and one replicating late in almost every cell. This pattern clearly accounts for the observation that ∼50% of rDNA copies are late-replicating and inactive in any given cell (Li et al. 2005).
Figure 3.
Asynchronous replication timing is allelic. Human lymphoblasts were labeled with BrdU for 7 h, arrested in metaphase, and subjected to ReTiSH using probes for rDNA (green) and the subcentromeric regions on chromosome 14, 15, 21, or 22 (red). (A) The percentage of metaphase spreads (n = 431) showing rDNA hybridization on a single copy of the labeled chromosome was recorded. (B) Examples of metaphase spreads. Early-replicating alleles were observed using a pulse-chase BrdU-labeling protocol (Materials and Methods). Arrows indicate locations of polymorphic subcentromeric regions. (C) Pools and single-cell subclones of human lymphoblasts with polymorphic centromeric or subcentromeric regions on chromosomes 14, 15, and 22 were analyzed by ReTiSH using probes for ribosomal and centromeric DNA. The number of metaphases (n = 430) with maternal late replication of the rDNA is recorded. The probability of having a single copy of any particular chromosome labeled by ReTiSH in middle S is five out of nine (0.555). The P-value was calculated by the hypergeometric method, (0.555)370 × (0.445)60 × 430!/(370!60!) = 0.5 × 10−42.
rDNA asynchrony is clonally maintained
In all of the other examples of asynchronous replication in the genome, once set up in the early embryo, the allelic pattern is clonally inherited, with the maternal allele replicating early in some clones and the paternal allele replicating early in others (Mostoslavsky et al. 2001; Singh et al. 2003; Ensminger and Chess 2004). In order to determine whether this is also true for the rDNA loci, we carried out ReTiSH on single-cell-derived clones from human lymphoblasts containing a polymorphically enlarged centromeric region (Kitsberg et al. 1993b) on one homolog of chromosomes 14, 15, and 22. This enabled us to distinguish between the two parental rDNA alleles on these chromosomes (Fig. 3B). When this technique was applied to a pool of human lymphoblasts using a 7-h BrdU incubation, we found that rDNA on chromosome 15 or 22 replicates late on the large centromeric allele in some cells while the other copy replicates late in others (Fig. 3C), and this is consistent with observations on other asynchronous replicating regions in the genome. For some unknown reason, the rDNA locus on chromosome 14 appears to have a fixed allelic pattern in this particular individual.
We then analyzed single-cell subclones by ReTiSH using a 7-h incubation with BrdU. In contrast to the pattern seen in the overall cell population, metaphase spreads showed that one particular allele (the maternal or paternal) in each clone is preferentially replicated in late S phase. In order to confirm this result, we carried out a BrdU pulse-chase experiment that specifically labels DNA undergoing replication during early S phase. In this way, we were able to demonstrate that it is indeed the opposite centromeric allele that is early-replicating in these same cells (Fig. 3B). As expected, for the nucleloar organizer regions (NORs) located on chromosomes 15 and 22, we identified clones having either the paternal or maternal allele late-replicating, and these loci appear to segregate independently. Overall, this epigenetic clonality is consistent with previous experiments showing that the same rDNA molecules are late-replicating and methylated in succeeding cell generations (Li et al. 2005).
Histone modification is monoallelic at each rDNA locus
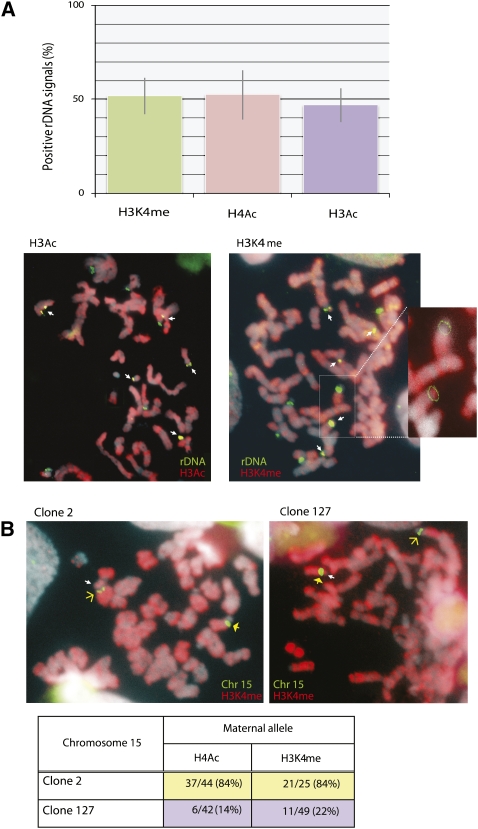
In light of the general correlation between replication timing and gene expression profiles, our results showing rDNA allelic replication strongly suggest that these genes may be selectively transcribed in a regional manner from a single allele at each locus. Preliminary experiments indicated that both primary RNA transcripts as well as the RNA polI machinery are not retained at rDNA loci during metaphase, making it difficult to assay expression in a direct manner. Thus, in order to identify active and inactive rDNA regions, we took advantage of the fact that actively transcribed gene regions are packaged with acetylated histones, while repressed genes are not. This turns out to be an excellent indicator of gene expression, since these structural features are known to be maintained on metaphase chromosomes (Jeppesen and Turner 1993). As a first step, we prepared metaphase nuclei from human lymphoblast cell lines, using FISH to label all of the rDNA loci and immunohistochemistry with anti-H4Ac or anti-H3Ac to detect regions of histone acetylation (Fig. 4A). In these initial assays, we were able to demonstrate that approximately half of the rDNA loci are packaged with acetylated histones and methylated H3K4 (H3K4me), another structural marker of active gene expression (Fig. 4A). This regional pattern is consistent with previous experiments showing that ∼50% of individual rDNA copies are packaged in an open chromatin conformation (Grummt 2007).
Figure 4.
Visualization of histone modification on metaphase chromosomes. (A) Metaphase spreads from YC lymphoblasts were visualized by immunohistochemistry with fluorescent antibodies (red) either to H3Ac (left) or H3K4me (right). The magnified area presents the histochemistry signal underlying two rDNA loci, one positive and one negative for H3K4me. The number of rDNA sites colocalized (yellow) with H4Ac, H3Ac, or H3K4me were counted (n = 78) and presented in graphic form as the percent of the rDNA loci labeled with modified histones. Arrows indicate colocalization (yellow) of rDNA and H3Ac or H3K4me. (B) Metaphase spreads from human YC lymphoblast clones (2 or 127) were used for immunohistochemistry (red) with anti-H4Ac or anti-H3K4me and then hybridized with a probe for the centromere on chromosome 15. In this experiment, the centromeric region is small on the maternal allele (open arrow) and large on the paternal allele (closed arrow). In each nucleus, the H3K4me-positive allele (red) is labeled with a white arrow. These results, including the number of nuclei examined, are summarized in the table. The maternal allele is early-replicating in clone 127 and late-replicating in clone 2.
In order to confirm the relationship between early replication and histone acetylation at rDNA loci, we then analyzed individual lymphoblast clones carrying a centromeric polymorphism on both chromosomes 14 and 15, thereby allowing us to distinguish between the two parental alleles. Our results already showed that in clone 2 the maternal (small centromere) rDNA allele on chromosome 15 replicates early (Fig. 3). Using anti-H4Ac, we were able to demonstrate that this same allele is preferentially packaged with acetylated histones, suggesting that this rDNA gene cluster is in an active chromatin conformation, as opposed to the unlabeled paternal allele (Fig. 4B), which replicates late. In contrast, in clone 127, it is the paternal allele (large centromere) that is marked by H4Ac, and this is in keeping with it being early-replicating in these cells (Fig. 3). Similar results were obtained using an antibody to H3K4me. These markers are also correlated with early replication at the rDNA locus on chromosome 14 (legend for Fig. 4). Taken together, these observations show that early replication at rDNA loci is associated with an open chromatin structure, strongly suggesting that these regions are expressed monoallelically.
rDNA replication is coordinated with other monoallelic markers
Previous studies have shown that asynchronous replication of many monoallelic gene regions is coordinated at the chromosomal level (Singh et al. 2003; Ensminger and Chess 2004). Thus, for example, both the Igκ and Tcrβ loci on chromosome 6 replicate asynchronously, but the early allele of one is always on the same parental chromosome that has the early allele of the other. This coordination has also been seen for many olfactory receptor loci as well as other monoallelic markers. These results suggest that random asynchronous replication may be controlled by independent master regulators associated with each individual chromosome. In order to test whether rDNA loci also replicate in coordination with other markers on the same chromosome, we performed several double-label experiments (Fig. 5A). As a first step, we carried out standard FISH analysis on interphase nuclei from a single clone containing a pericentromeric polymorphism on chromosome 14, using a probe for an olfactory receptor gene located nearby on the same chromosome. Analysis of nuclei with a single/double hybridization pattern for the olfactory receptor locus revealed that the early allele (double dot) is almost always associated with the enlarged pericentric heterochromatin region (paternal allele), and this is the same chromosome that was found to have the early-replicating rDNA locus in these same cells (Fig. 3C).
Figure 5.
Chromosomal coordination of rDNA asynchronous replication. (A) FISH analysis of human lymphoblast clone 2 BrdU-labeled (gray) nuclei using one probe (red) for the chromosome 14 subcentromeric region and another for an olfactory receptor cluster (green) located very close on this same chromosome (14q11.2). The early-replicating (double) olfactory receptor gene was associated with the larger paternally derived subcentromeric region on 17 out of 20 asynchronous nuclei, and this is the same parental chromosome that has an early-replicating NOR in these same cells (Fig. 3C). ReTiSH analysis of human lymphoblast clones 127 (B) and 2 (C) labeled with BrdU for 7 h and arrested in metaphase using a rDNA probe (green), a chromosome 15 olfactory receptor (15q26) probe (red), and a centromeric probe (red). In B, the late-replicating olfactory receptor locus and late-replicating rDNA locus are both associated with the smaller maternal centromeric region (19 out of 21), while in C, both late-replicating loci are associated with the larger paternal subcentromeric region (13 out of 19), and this is consistent with the data in Figure 3C. The parental alleles are also shown in the accompanying magnification. (D) ReTiSH analysis of human lymphoblast cells labeled with BrdU for 7 h and arrested in metaphase using a rDNA probe (green) and a chromosome 21 olfactory receptor probe (red). The labeled late-replicating olfactory receptor locus is almost always (22out of 23) associated with the same parental chromosome carrying a late-replicating rDNA locus (see magnification). Arrows indicate rDNA loci.
Similar results were also obtained using the ReTiSH technique. In this case, a single clone was labeled with BrdU exclusively in late S (7 h), and resulting metaphase chromosomes were analyzed by hybridization with probes for the centromeric region of chromosome 15, rDNA, and an olfactory receptor region located on this same chromosome. These studies showed that whenever the rDNA is labeled, the olfactory receptor locus is also labeled on the same chromosome (Fig. 5B,C), indicating that both regions are late-replicating in a coordinate manner. A similar pattern was observed for an olfactory receptor gene region on chromosome 21 (Fig. 5D).
Developmental control of rDNA replication timing
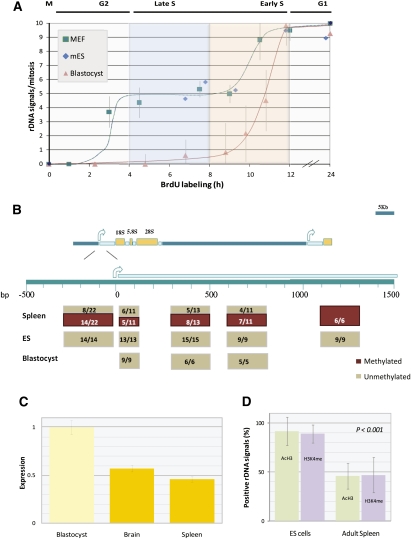
In other examples of monoallelic expression, asynchronous replication is first established in the early embryo at about the time of implantation (Mostoslavsky et al. 2001). In order to test if this is also true for rDNA, we carried out ReTiSH kinetics on early blastocysts grown from fertilized mouse oocytes in vitro. This analysis revealed that, unlike the asynchronous pattern seen in somatic cells, in blastocysts, replication of all 10 rDNA loci is highly synchronized and takes place early in the cell cycle (Fig. 6A). In contrast to these very early embryos, rDNA replication was already found to be asynchronous in mouse embryonic stem (ES) cells. Even though these experiments were carried out in culture, the results strongly suggest that, in vivo, the switch to differential replication actually occurs at about the time of implantation (Tesar et al. 2007).
Figure 6.
Regulation of rDNA loci during embryogenesis. (A) MEFs, mouse ES cells, and mouse blastocyst cells were labeled with BrdU for various times; arrested in metaphase; and subjected to ReTiSH using a probe for rDNA. The number of rDNA signals in each metaphase spread (n = 193) was counted, and the standard deviation was calculated for each data point (error bars). The data were adjusted to account for the shorter G2 (2 h) in blastocysts, as determined by the time at which 50% of metaphases are labeled with BrdU. (B) Diagram showing the stereotypic rDNA repeat indicating the transcription start site and the internal (ITS) and external (ETS) spacer regions. Methylation of the 5′ rDNA region was determined by bisulfite analysis on individual molecules (Supplemental Fig. 1), and the results are recorded as a function of position. The numbers of unmethylated (tan) and fully methylated (brown) molecules are shown. (C) Levels (±SD) of rRNA from mouse blastocysts and adult brain or spleen were measured by quantitative RT–PCR and normalized against a set of three housekeeping genes. (D) Metaphase spreads from mouse cells were used for immunohistochemistry with anti-H3Ac or anti-H3K4me and then hybridized. Results show the percent of rDNA loci in each nucleus (n = 127) that are labeled with H3Ac or H3K4me.
Methylation of rDNA
Although it is well known that ∼50% of the rDNA copies are methylated at selected sites within the promoter region (Santoro and Grummt 2001; Ghoshal et al. 2004), the exact developmental pattern of methylation at these loci has never been determined. To this end, we carried out bisulfite analysis on rDNA from early embryos, ES cells, and somatic tissues (Fig. 6B; Supplemental Fig. 1). The results show that while some regions of the rDNA repeat domain are completely methylated in spleen cells, a small window covering the gene promoter and extending ∼700 base pairs (bp) downstream appears to have a differential pattern, with ∼50% of the molecules being completely unmethylated. In contrast, this same region is unmethylated on all alleles both in the early embryo (blastocysts) and in ES cells, and may still remain unmodified in MEFs (Schmmitz et al. 2009). This strongly suggests that the differential pattern seen in somatic tissues is established by de novo methylation of one allele in each cell, in a process that apparently takes place post-implantation. Previous studies have already demonstrated that this DNA modification is always associated with the late-replicating copies of rDNA (Li et al. 2005).
rRNA expression during early development
The fact that rDNA is early-replicating and completely unmethylated in the blastocyst as opposed to somatic tissues raises the possibility that the expression pattern of these genes may be developmentally regulated. To determine if this might be the case, we measured rRNA levels in blastocysts as compared with somatic tissues, such as brain or spleen. This was carried out by quantitative RT–PCR analysis using a set of three housekeeping genes to normalize the data. These studies show that there is approximately twofold more rRNA expression in the early embryo (Fig. 6C). This is consistent with studies showing that rRNA levels are elevated in undifferentiated ES cells, and are then down-regulated approximately twofold following differentiation to neural precursors (Efroni et al. 2008). In confirmation of this idea, we also found that, as opposed to somatic cells, >90% of the rDNA loci in ES cells are marked with both H3Ac and H3K4me. These experiments suggest that, during very early development, rRNA may be initially transcribed from all 10 loci and then undergo allelic repression at some later stage of differentiation.
Discussion
Although it has been known for a long time that a large percentage of the rDNA genes are inactivated in every somatic cell, the fundamental mechanism and molecular logic for this developmental process have remained elusive, mainly because it is difficult to study the epigenetics of repeated sequences. In order to overcome this problem, we developed a new cytological technique (ReTiSH) for measuring the time of replication at any sequence in the genome. This method is highly accurate, can be carried out on a very small number of cells in vivo, and, unlike the FISH method, is not affected by chromatid segregation (Azuara et al. 2003), so it yields easily interpretable results. In addition, this approach can be used to relate gene-specific replication timing to other chromosome markers.
Our studies indicate that rDNA repression takes place in an ordered, preprogrammed manner as part of a developmentally regulated pathway for distinguishing between chromosomal homologs in each cell. Thus, rDNA loci appear to be controlled in a manner similar to the X chromosome in female embryos. In the blastocyst, individual clusters (NORs) on both parental alleles replicate synchronously in early S phase and are all probably in an open conformation that allows transcription. At about the time of implantation, as seen in ES cells, one allele of each locus is stochastically chosen to undergo late replication. In some cells it is the paternal allele that is chosen, while in others it is the maternal copy, but each of the rDNA-containing chromosomes makes this decision independently. As demonstrated previously, asynchronous replication of multiple markers is coordinated on each chromosome (Singh et al. 2003; Ensminger and Chess 2004). Once these multichromosomal asynchronous replication patterns are established, they are then maintained in a clonal manner in differentiated cells. DNA methylation of rDNA appears to represent a secondary event that occurs post-implantation in a manner similar to what happens to the inactive X chromosome (Lock et al. 1987; Keohane et al. 1998) as well as genes responsible for pluripotency (Feldman et al. 2006; Epsztejn-Litman et al. 2008).
It should be noted that although the rDNA loci replicate synchronously in the blastula embryo, replication is seen to be asynchronous in ES cells that have been shown to have a developmental epigenetic pattern representative of the implantation stage. Despite this major change in the replication timing profile, almost all of these loci are packaged with H3Ac and H3K4me, and are probably fully expressed in these cells (Efroni et al. 2008). It thus appears that the switch to asynchronous replication occurs prior to the silencing of rDNA. Since there is a clear-cut allelic association between late replication timing and repressed chromatin structure in somatic cells (Fig. 4), these results suggest, but do not prove, that replication timing patterns at an early stage may serve as an epigenetic template for choosing which allele is marked for inactivation.
This concept may also be true for other examples of monoallelic choice. Several studies, for example, have demonstrated that the X chromosomes in female ES cells replicate asynchronously prior to inactivation (Gribnau et al. 2005; Dutta et al. 2009), and the same is true for Igκ in pre-B cells, where this locus replicates asynchronously in a manner that predicts which allele is ultimately selected for the first rearrangement reaction (Goldmit et al. 2005). Differential replication timing of the parental alleles at imprinted gene loci is set up in the gametes (Simon et al. 1999), and this appears to occur prior to allele-specific methylation, which many times is actually established following fertilization (Birger et al. 1999; El-Maarri et al. 2001). All of these examples clearly indicate that asynchronous replication may represent an early epigenetic mark that can be recognized and used for allelic choice.
While this regional mechanism may be involved in establishing the initial epigenetic state of rDNA loci during development, it is clear that ribosomal RNA synthesis may also be controlled in a dynamic manner (Hammond and Bowman 1988). It is known that NORs may be subject to physiological influences mediated by both positive and negative transcription factors that operate within the nucleolus (Grummt 2007). Interestingly, overexpression of TIP5, a key component of the nucleolar remodeling complex (NoRC), is actually capable of altering the epigenetic structure of individual rDNA copies by its ability to reorganize nucleosome positioning and to recruit Dnmts that methylate key sites in the promoter region, and through the action of histone deacetylases that may be responsible for shifting the DNA to a late replication profile, perhaps by modifying nucleosomes at nearby replication origins (Santoro et al. 2002; Santoro and Grummt 2005). Recent studies further suggest that inactive copies of rDNA can even be induced to undergo demethylation (Schmmitz et al. 2009). Other factors may be involved in maintenance of rDNA epigenetic structure (McStay and Grummt 2008).
It should be noted that this allelic mode of rDNA inactivation clearly provides a molecular mechanism for ensuring that the same number of rDNA loci are repressed in each cell. Furthermore, since each individual locus carries variable numbers of rDNA gene copies, this system also guarantees that ∼50% of the genes are repressed—the same number in each cell. Although rRNA levels can certainly be regulated by a number of different nucleolar transcription factors, maximal production of nontranslated RNA products of this nature is often controlled by gene copy number (Brown and Dawid 1968; Gall 1968). Our studies now demonstrate, for the first time, that mammalian ribosomal genes may indeed be subject to an epigenetic mechanism that allows for an increase in effective copy number during early embryogenesis. Considering the strict coordination between rRNA and ribosomal protein levels (LaMarca and Wassarman 1979; Hammond and Bowman 1988), this could play a role in providing additional ribosomes to keep up with the increase in protein synthesis that occurs during blastulation (LaMarca and Wassarman 1979).
Materials and methods
Cell culture
Primary lymphocytes were prepared from peripheral blood of YC (Kitsberg et al. 1993a) and transformed by EBV. Single-cell clones were then isolated by serial limiting dilutions. Mouse ES cells (ESD3) were grown in DMEM supplemented with 1000 U/mL LIF on 0.1% gelatin-coated plates. MEFs were cultured from 13.5-d embryos. All cells were grown in DMEM or RPMI medium supplemented with 20% inactivated fetal calf serum (HyClone), 2 mM glutamine, 1000 U/mL penicillin, 100 μg/mL streptomycin, 0.1 μg/mL β-mercaptoethanol, 5 μg/mL Na pyruvate, and 5 μg/mL nonessential amino acids.
ReTiSH
Unsynchronized, exponentially growing cells were labeled for various times with 3 × 10−5 M BrdU (Sigma), and Colcemid (Bet Haemek) was then added to a final concentration of 0.1 μg/mL for 1 h at 37°C. Cells were harvested and treated with prewarmed hypotonic KCl solution (0.075 M) for 30 min at 37°C, fixed, washed with methanol-glacial acetic acid (3:1), dropped gently on cold slides, and air-dried. Slides were then washed with PBS, fixed again with 2% formaldehyde for 2 min, washed in PBS (3×), and treated with pepsin (1 mg/mL in 2N HCl) for 10 min at 37°C. After additional washes with PBS and 2× SSC, the slides were then treated with Hoechst 33258 (0.5 μg/μL) for 15 min, exposed to 365-nm UV light for 20 min using a UV Stratalinker 2400 transilluminator (Stratagene), washed again in 2× SSC, and dried. ExoIII (0.1 U per slide) (Promega) digestion was carried out in buffer for 10 min at 37°C and the slides were then washed in 2× SSC, dehydrated through a series of cold ethanol washes, and air-dried. In BrdU pulse-chase experiments, cells were labeled with 1 × 10−6 M BrdU for 1 h, washed three times in PBS, and then grown for 10–12 h before treatment with Colcemid and fixation.
BAC probes were labeled directly with DIG or Biotin-conjugated nucleotides using a nick translation mix (Roche). The rDNA probe was labeled using a standard nick translation protocol substituting dTTP with dioxigenin-11-dUTP in a ratio of 2:1 (Selig et al. 1992). Labeled DNA probes (20–50 ng) were mixed with human placental DNA or mouse genomic sonicated DNA (10 μg) and human or mouse Cot-1 DNA (10 μg; GIBCO), ethanol-precipitated, and resuspended in Hybridization Buffer (50% deionized formamide, 10% dexstran sulfate, 2× SSC). After denaturation (5 min at 80°C), the probe mixture was preannealed for 10–30 min at 37°C in order to eliminate repeated sequences and then applied to undenatured specimens. Following hybridization overnight and subsequent washes, the slides were treated as described (Selig et al. 1992) with blocking solution (3% BSA, 4× SSC) for 10–30 min at 37°C; incubated with detection reagents for 30 min at 37°C in 1% BSA, 4× SSC, and 0.1% Tween 20; and then washed three times for 3 min each in 4× SSC and 0.1% Tween 20. Biotin-labeled probes were detected with rhodamine-conjugated avidin DCS (1:500 dilution; Vector Laboratories), and digoxigenin-labeled probes were detected with an anti-digoxigenin antibody conjugated to FITC (1:100 dilution; Roche). Counterstaining was done using diamidinophenylidole (DAPI; 200 ng/mL) in VectaShield. Visualization was accomplished by conventional fluorescence microscopy, and images were captured using a Nikon 90i C1 fluorescence microscope at 1000× magnification.
Standard FISH was performed as described previously (Lichter et al. 1990; Selig et al. 1992) by culturing cells for 1 h with BrdU (3 × 10−5 M), isolating nuclei using hypotonic KCl (0.5%) treatment, and fixing them with methanol:acetic acid (3:1). Briefly, denaturation was carried out by incubation in 70% formamide and 2× SSC for 2 min at 72°C, and then slides were dehydrated by a series of ice-cold ethanol washes (70%, 90%, and 100% for 5 min each). Probes were labeled as described above. BrdU was detected by an anti-BrdU antibody (1:100; NeoMarkers) followed by Amca or rhodamine-conjugated anti-mouse antibody (1:100; Jackson Immunoresearch Laboratories).
Preparation of blastocysts for ReTiSH
Superovulated C57Black BALB/C F1 female mice were mated with males, and zygotes were removed from the oviducts ∼14–20 h after hormone injection. Zygotes were incubated at 37°C in M16 medium (Sigma) to obtain preimplantation embryos at the morula–blastula stage. Embryos were incubated in culture with 1 × 10−6 M BrdU for different times, and Colcemid (0.1 μL/mL) was added prior to collection. Collected embryos were washed with PBS (without calcium or magnesium), counted, and transferred into a fresh 1% Na citrate solution for 7 min and fixed to a poly-L-lysine slide (Sigma) by putting them into a drop of methanol-glacial acetic acid (3:1). After drying, the slide was washed, dehydrated, permeabilized for 20 min in pepsin (0.1 mg/mL in 2 N HCl), and fixed for 10 min in 1% formaldehyde at 4°C.
Immunofluorescence
Colcemid-treated cell pellets were washed in PBS and then suspended in hypotonic KCl solution (0.075 M) for 10 min at a concentration of 0.2 × 10 to 1 × 106 cells per milliliter. Cytospin preparations were made with no more than 0.5 × 105 cells per slide by centrifuging for 7 min at 1000 rpm using a Shandon Cytospin Cytocentrifuge and SUPERFROST PLUS slides (Menzel-Glaser). Slides were air-dried and immersed in PBS for at least 10 min at room temperature, and nuclei were then fixed with 2% formaldehyde in PBS for 15 min, followed by blocking with 3% BSA/0.1% TWEEN for 15 min in 37°C. Anti-H3Ac, anti-H4Ac, or anti-H3K4me3 antibodies (Upstate Biotechnologies) were diluted 1:100 in PBT (PBS/0.1% TWEEN) and incubated for 0.5–1 h at 37°C in a humid chamber. After washing with PBT, slides were incubated with anti-rabbit Cy3, diluted 1:100 in PBS for 10–30 min at 37°C, and washed with PBS. Post-fixation was accomplished by treating with 2% formaldehyde for 10 min, washing with H2O, and then fixing again for 15 min in 3:1 methanol:acetic acid. Sequential FISH was done after complete drying of the fixed slides. In this procedure, denaturation (72°C) prior to hybridization was expanded to 5–7 min.
Bisulfite analysis
Bisulfite conversion of genomic DNA was carried out using the Qiagen EpiTect Bisulfite Kit according to the manufacturer's instructions. PCR primers were designed using Methyl Primer Express software version 1.0 (https://www2.appliedbiosystems.com). Primer sequences are available on request. After PCR amplification, products were extracted from gels using the Qiagen MinElute Gel Extraction Kit, cloned into the Promega pGEM-T vector system, transfected into Promega JM109 competent bacteria, and plated on LB-Agar + Amp + X-gal and IPTG. The DNA from 10–12 white clones of each ligation was amplified using the Illustra Templiphi 100 amplification kit and subjected to sequencing with T7 or Sp6 primers.
Expression
Total RNA (250 ng) was prepared with the TriPure isolation reagent (Roche) and converted to cDNA using the M-MLV reverse transcriptase (Promega) and random hexanucleotide pd(N)6 primers (GE Healthcare) in a reaction volume of 20 μL under conditions recommended by the manufacturer. In some cases, RNA was first treated with DNase I (Promega). Control reactions lacking reverse transcriptase were systematically verified for the absence of products. Quantitative real-time PCR (Q-PCR) was performed (Finnzymes) in a 20-μL reaction mix, and primer–dimer products were distinguished from specific products by dissociation analysis. For each primer pair designed for Q-PCR, a no-template control was also included. Measurements were performed with SYBR Green Universal Mix in triplicate, and three control genes were used to normalize between samples. For each cDNA sample, cycle thresholds for the target gene were calculated relative to that of the three control genes (HP1-γ, Cyclophilin-A, and Cyclophilin-B) (see Supplemental Table 1).
Acknowledgments
We are grateful to B. McStay for the gift of the human rDNA intergenic spacer probe, to A. Rahat for the chromosome 15 α satellite probe, and to Y. Kaufman for his assistance in analyzing blastocyst DNA methylation. This work was supported by grants from the Israel Academy of Sciences (to Y.B. and H.C.), the NIH, European Community 5th Framework Quality of Life Program (to Y.B.), the Israel Cancer Research Fund (to Y.B. and H.C.), and a research award from Lew Sanders (to H.C.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.544509.
Supplemental material is available at http://www.genesdev.org.
References
- Azuara V, Brown KE, Williams RR, Webb N, Dillon N, Festenstein R, Buckle V, Merkenschlager M, Fisher AG. Heritable gene silencing in lymphocytes delays chromatid resolution without affecting the timing of DNA replication. Nat Cell Biol. 2003;5:668–674. doi: 10.1038/ncb1006. [DOI] [PubMed] [Google Scholar]
- Birger Y, Shemer R, Perk J, Razin A. The imprinting box of the mouse Igf2r gene. Nature. 1999;397:84–88. doi: 10.1038/16291. [DOI] [PubMed] [Google Scholar]
- Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- Brown DD, Dawid IB. Specific gene amplification in oocytes. Oocyte nuclei contain extrachromosomal replicas of the genes for ribosomal RNA. Science. 1968;160:272–280. doi: 10.1126/science.160.3825.272. [DOI] [PubMed] [Google Scholar]
- Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- Cornforth MN, Eberle RL. Termini of human chromosomes display elevated rates of mitotic recombination. Mutagenesis. 2001;16:85–89. doi: 10.1093/mutage/16.1.85. [DOI] [PubMed] [Google Scholar]
- Dutta D, Ensminger AW, Zucker JP, Chess A. Asynchronous replication and autosome-pair non-equivalence in human embryonic stem cells. PLoS One. 2009;4:e4970. doi: 10.1371/journal.pone.0004970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RD, Buetow KH, et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Maarri O, Buiting K, Peery EG, Kroisel PM, Balaban B, Wagner K, Urman B, Heyd J, Lich C, Brannan CI, et al. Maternal methylation imprints on human chromosome 15 are established during or after fertilization. Nat Genet. 2001;27:341–344. doi: 10.1038/85927. [DOI] [PubMed] [Google Scholar]
- Ensminger AW, Chess A. Coordinated replication timing of monoallelically expressed genes along human autosomes. Hum Mol Genet. 2004;13:651–658. doi: 10.1093/hmg/ddh062. [DOI] [PubMed] [Google Scholar]
- Epsztejn-Litman S, Feldman N, Abu-Remaileh M, Shufaro Y, Gerson A, Ueda J, Deplus R, Fuks F, Shinkai Y, Cedar H, et al. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol. 2008;15:1176–1183. doi: 10.1038/nsmb.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, Cedar H, Bergman Y. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8:188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- Gall JG. Differential synthesis of the genes for ribosomal RNA during amphibian oogenesis. Proc Natl Acad Sci. 1968;60:553–560. doi: 10.1073/pnas.60.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshal K, Majumder S, Datta J, Motiwala T, Bai S, Sharma SM, Frankel W, Jacob ST. Role of human ribosomal RNA (rRNA) promoter methylation and of methyl-CpG-binding protein MBD2 in the suppression of rRNA gene expression. J Biol Chem. 2004;279:6783–6793. doi: 10.1074/jbc.M309393200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimelbrant AA, Ensminger AW, Qi P, Zucker J, Chess A. Monoallelic expression and asynchronous replication of p120 catenin in mouse and human cells. J Biol Chem. 2005;280:1354–1359. doi: 10.1074/jbc.M411283200. [DOI] [PubMed] [Google Scholar]
- Goldmit M, Bergman Y. Monoallelic gene expression: A repertoire of recurrent themes. Immunol Rev. 2004;200:197–214. doi: 10.1111/j.0105-2896.2004.00158.x. [DOI] [PubMed] [Google Scholar]
- Goldmit M, Yanhong J, Skok J, Roldan E, Jung S, Cedar H, Bergman Y. Epigenetic ontogeny of the κ locus during B cell development. Nat Immunol. 2005;6:198–203. doi: 10.1038/ni1154. [DOI] [PubMed] [Google Scholar]
- Gribnau J, Luikenhuis S, Hochedlinger K, Monkhorst K, Jaenisch R. X chromosome choice occurs independently of asynchronous replication timing. J Cell Biol. 2005;168:365–373. doi: 10.1083/jcb.200405117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I. Different epigenetic layers engage in complex crosstalk to define the epigenetic state of mammalian rRNA genes. Hum Mol Genet. 2007;16:R21–R27. doi: 10.1093/hmg/ddm020. [DOI] [PubMed] [Google Scholar]
- Grummt I, Rosenbauer H, Niedermeyer I, Maier U, Ohrlein A. A repeated 18 bp sequence motif in the mouse rDNA spacer mediates binding of a nuclear factor and transcription termination. Cell. 1986;45:837–846. doi: 10.1016/0092-8674(86)90558-1. [DOI] [PubMed] [Google Scholar]
- Haaf T. Analysis of replication timing of ribosomal RNA genes by fluorescence in situ hybridization. DNA Cell Biol. 1997;16:341–345. doi: 10.1089/dna.1997.16.341. [DOI] [PubMed] [Google Scholar]
- Hammond ML, Bowman LH. Insulin stimulates the translation of ribosomal proteins and the transcription of rDNA in mouse myoblasts. J Biol Chem. 1988;263:17785–17791. [PubMed] [Google Scholar]
- Hiratani I, Gilbert DM. Replication timing as an epigenetic mark. Epigenetics. 2009;4:93–97. doi: 10.4161/epi.4.2.7772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen P, Turner BM. The inactive X chromosome in female mammals is distinguished by a lack of histone H4 acetylation, a cytogenetic marker for gene expression. Cell. 1993;74:281–289. doi: 10.1016/0092-8674(93)90419-q. [DOI] [PubMed] [Google Scholar]
- Keohane AM, Lavender JS, O'Neill LP, Turner BM. Histone acetylation and X inactivation. Dev Genet. 1998;22:65–73. doi: 10.1002/(SICI)1520-6408(1998)22:1<65::AID-DVG7>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Kitsberg D, Selig S, Brandeis M, Simon I, Keshet I, Driscoll DJ, Nicholls RD, Cedar H. Allele-specific replication timing of imprinted gene regions. Nature. 1993a;364:459–463. doi: 10.1038/364459a0. [DOI] [PubMed] [Google Scholar]
- Kitsberg D, Selig S, Keshet I, Cedar H. Replication structure of the human β-globin gene domain. Nature. 1993b;366:588–590. doi: 10.1038/366588a0. [DOI] [PubMed] [Google Scholar]
- LaMarca MJ, Wassarman PM. Program of early development in the mammal: Changes in absolute rates of synthesis of ribosomal proteins during oogenesis and early embryogenesis in the mouse. Dev Biol. 1979;73:103–119. doi: 10.1016/0012-1606(79)90141-6. [DOI] [PubMed] [Google Scholar]
- Li J, Santoro R, Koberna K, Grummt I. The chromatin remodeling complex NoRC controls replication timing of rRNA genes. EMBO J. 2005;24:120–127. doi: 10.1038/sj.emboj.7600492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichter P, Tang CC, Call K, Hermanson G, Evans GA, Housman D, Ward DC. High resolution mapping of human chromosome 11 by in situ hybridization with cosmid clones. Science. 1990;247:64–69. doi: 10.1126/science.2294592. [DOI] [PubMed] [Google Scholar]
- Lock LF, Takagi N, Martin GR. Methylation of the HPRT gene on the inactive X occurs after chromosome inactivation. Cell. 1987;48:39–46. doi: 10.1016/0092-8674(87)90353-9. [DOI] [PubMed] [Google Scholar]
- Mayer C, Schmitz KM, Li J, Grummt I, Santoro R. Intergenic transcripts regulate the epigenetic state of rRNA genes. Mol Cell. 2006;22:351–361. doi: 10.1016/j.molcel.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Mayer C, Neubert M, Grummt I. The structure of NoRC-associated RNA is crucial for targeting the chromatin remodelling complex NoRC to the nucleolus. EMBO Rep. 2008;9:774–780. doi: 10.1038/embor.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay B, Grummt I. The epigenetics of rRNA genes: From molecular to chromosome biology. Annu Rev Cell Dev Biol. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- Mostoslavsky R, Singh N, Tenzen T, Goldmit M, Gabay C, Elizur S, Qi P, Reubinoff BE, Chess A, Cedar H, et al. Asynchronous replication and allelic exclusion in the immune system. Nature. 2001;414:221–225. doi: 10.1038/35102606. [DOI] [PubMed] [Google Scholar]
- Santoro R, Grummt I. Molecular mechanisms mediating methylation-dependent silencing of ribosomal gene transcription. Mol Cell. 2001;8:719–725. doi: 10.1016/s1097-2765(01)00317-3. [DOI] [PubMed] [Google Scholar]
- Santoro R, Grummt I. Epigenetic mechanism of rRNA gene silencing: Temporal order of NoRC-mediated histone modification, chromatin remodeling, and DNA methylation. Mol Cell Biol. 2005;25:352539–352546. doi: 10.1128/MCB.25.7.2539-2546.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R, Li J, Grummt I. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet. 2002;32:393–396. doi: 10.1038/ng1010. [DOI] [PubMed] [Google Scholar]
- Schmmitz KM, Schmitt N, Hoffmann-Rohrer U, Schafer A, Grummt I, Mayer C. TAF12 recruits Gadd45a and the nucleotide excision repair complex to the promoter of rRNA genes leading to active DNA demethylation. Mol Cell. 2009;33:344–353. doi: 10.1016/j.molcel.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Selig S, Okumura K, Ward DC, Cedar H. Delineation of DNA replication time zones by fluorescence in situ hybridization. EMBO J. 1992;11:1217–1225. doi: 10.1002/j.1460-2075.1992.tb05162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon I, Tenzen T, Reubinoff BE, Hillman D, McCarrey JR, Cedar H. Asynchronous replication of imprinted genes is established in the gametes and maintained during development. Nature. 1999;401:929–932. doi: 10.1038/44866. [DOI] [PubMed] [Google Scholar]
- Singh N, Ebrahimi FA, Gimelbrant AA, Ensminger AW, Tackett MR, Qi P, Gribnau J, Chess A. Coordination of the random asynchronous replication of autosomal loci. Nat Genet. 2003;33:339–341. doi: 10.1038/ng1102. [DOI] [PubMed] [Google Scholar]
- Tesar PJ, Chenoweth JG, Brook FA, Davies TJ, Evans EP, Mack DL, Gardner RL, McKay RD. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature. 2007;448:196–199. doi: 10.1038/nature05972. [DOI] [PubMed] [Google Scholar]