Abstract
Common lymphoid progenitors (CLPs) clonally produce both B- and T-cell lineages, but have little myeloid potential in vivo. However, some studies claim that the upstream lymphoid-primed multipotent progenitor (LMPP) is the thymic seeding population, and suggest that CLPs are primarily B-cell-restricted. To identify surface proteins that distinguish functional CLPs from B-cell progenitors, we used a new computational method of Mining Developmentally Regulated Genes (MiDReG). We identified Ly6d, which divides CLPs into two distinct populations: one that retains full in vivo lymphoid potential and produces more thymocytes at early timepoints than LMPP, and another that behaves essentially as a B-cell progenitor.
Keywords: B-cell development, T-cell development, fate decisions, hematopoiesis, lymphopoiesis
In embryonic and adult tissues, stem cells differentiate into their terminal progeny through a series of progenitor intermediates. Within these developmental intermediates, the expression signature of the mature lineage initiates (specification) and the potential to develop into alternative lineages is lost (commitment). In the hematopoietic hierarchy, such intermediates can be viewed as nodes or branchpoints between the different lineages. Cell fate decisions within these intermediates occur through a network of gene expression and epigenetic changes. Elucidating the molecular mechanisms that underlie developmental fate decisions has been the subject of intense research, and progress has depended on the ability to identify, isolate, and characterize the lineage potential and gene expression patterns of these intermediate populations.
However, even within the well-studied hematopoietic system, the identities of many developmental intermediates and the precise mechanisms that initiate a lineage choice remain unclear. For example, there is currently debate surrounding the identity of the cellular intermediate that serves as the branchpoint between the B- and T-lymphocyte lineages. The classical model of hematopoiesis contends that the common lymphoid progenitor (CLP) is the hub from which all lymphoid decisions are made (Kondo et al. 1997). Consistent with this model, we reported previously that the Flk2+ subset of CLPs has robust T-cell potential (Karsunky et al. 2008), and that it is the predominant source of T cells within the bone marrow in transplantation experiments (Serwold et al. 2008). However, other studies have suggested that CLPs predominantly differentiate into B cells, and that a population upstream of CLPs, the multipotent progenitor (MPP, sometimes called LMPP [lymphoid-primed MPP]), is the major source of T-cell progenitors from the bone marrow (Schwarz and Bhandoola 2004; Perry et al. 2006; Lai and Kondo 2007; Wada et al. 2008). These contradictory findings may have resulted from B-cell-biased progenitors that copurify and contaminate preparations of CLPs. Therefore, we sought to determine whether separate B-cell-biased and unbiased cells exist within the CLP population.
In a related study, we presented a new bioinformatics method called Mining Developmentally Regulated Genes (MiDReG), which mines the massive repertoire of publically available microarray data to identify genes that are up-regulated or down-regulated within a developmental pathway (D Sahoo, J Seita, D Bhattacharya, M Inlay, I Weissman, S Plevritis, and D Dill, in prep.). MiDReG does not require that arrays of the intermediate populations exist, only the knowledge of two (or more) genes within a developmental pathway: one that is expressed exclusively in the stem or progenitor population, and one that is expressed exclusively in the mature lineage. We validated MiDReG for B-cell development using two seed genes: Kit for stem and progenitor cells, and Cd19 for mature B cells. Here, we apply MiDReG to the B fate versus T fate decision. In doing so, we identified Ly6d, a surface marker that bisects CLPs into two distinct populations. The Ly6d− subset, called ALP (all-lymphoid progenitor), retains full lymphoid potential and early thymic seeding activity, whereas the Ly6d+ subset, called BLP (B-cell-biased lymphoid progenitor), up-regulates the B-cell-specifying factors Ebf1 and Pax5 and behaves essentially as a B-cell progenitor. Thus, Ly6d identifies the first stage of B-cell development (BLP), and the population residing at the branchpoint between B- and T-cell development (ALP).
Results and Discussion
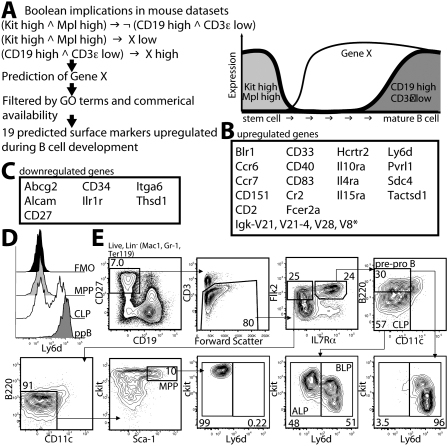
CLPs and the earliest B-cell progenitors reside at the transition from Kit-expressing progenitors to Cd19-expressing B-cell precursors, and thus represent the ideal intermediates to test MiDReG using the established seed genes. To identify markers that could potentially separate B-lineage-committed from uncommitted progenitors within the CLP population, we used the following seed conditions for MiDReG (Fig. 1). For the first seed, we used the logical combination “Kit high AND Mpl high” to represent the progenitors, as both are expressed on early hematopoietic cells but not mature B cells. For the second seed, we used “Cd19 high AND Cd3ɛ low,” as CD3ɛ is expressed only on T cells and therefore this combination would eliminate arrays of heterogeneous populations (e.g., whole-tissue arrays). We focused exclusively on genes encoding cell surface proteins, as designated by the Gene Ontology (GO) database, with commercially available antibodies suitable for flow cytometry. From this analysis, MiDReG identified 26 genes encoding cell surface proteins that were predicted to be differentially expressed during B-cell development: 19 up-regulated (Fig. 1B) and seven down-regulated (Fig. 1C).
Figure 1.
Prediction of surface markers up-regulated or down-regulated during B-cell development. (A) Prediction of genes encoding cell surface molecules up-regulated in B-cell development. The MiDReG algorithm uses Boolean implications from mouse data sets only. The first seed condition is Kit high AND Mpl high, and the second seed condition is CD19 high AND CD3ɛ low. The algorithm predicted gene X such that “(Kit high AND Mpl high) ⇒ X low” and “(Cd19 high AND Cd3ɛ low) ⇒ X high.” The gene list was filtered using membrane Gene Ontology classification and commercially available antibodies suitable for flow cytometry. (∧) AND, (¬) NOT, (→) “implies.” A diagram of the expression changes between Kit and Mpl, Gene X, and Cd19 and Cd3ɛ are depicted on the right. (B) Nineteen genes encoding cell surface molecules were predicted in this analysis. (*) Gene symbol truncated for brevity. Details of the MiDReG algorithm are described elsewhere (D Sahoo, J Seita, D Bhattacharya, M Inlay, I Weissman, S Plevritis, and D Dill, in prep.). (C) Prediction of down-regulated genes. Here, the algorithm predicted gene X such that “(Kit high AND Mpl high) ⇒ X high” and “(Cd19 high AND Cd3ɛ low) ⇒ X low.” Seven genes encoding cell surface molecules were identified after applying the membrane Gene Ontology and commercial antibodies filtrations. (D) Ly6d expression profile of MPPs (light gray), CLPs (white), and pre-pro-Bs (ppB, dark gray). Fluorescence minus one (FMO, black) control is shown for Lin− cells. (E) Kit/Ly6d contour plot of MPPs, CLPs, and pre-pro-Bs. The stains and gating strategy leading to each of these populations are shown. Percentages of displayed cells within each gate are shown.
We focused on four genes—Cd34, Cd27, Il1r1, and Ly6d, as antibodies to these proteins were readily available—and examined their surface expression during the progression through the MPP, CLP, pre-pro-B, and Fraction B (Fr. B) stages. Because of the known presence of non-B-lineage cells within the pre-pro-B-cell population (Li et al. 1996; Nikolic et al. 2002), we first recharacterized this population as described in Supplemental Figure S1 to isolate only the B-cell progenitors within this population. Despite MiDReG's prediction of down-regulation, CD34 protein levels were not markedly different between the MPP, CLP, pre-pro-B, and Fr. B stages (data not shown). The other surface proteins, however, revealed interesting expression patterns. CD27 is a known marker for early hematopoietic stem and progenitor cells, but is not expressed in mature naïve B cells. As we summarize in Supplemental Figure S2, CD27 is expressed on MPP, CLP, and pre-pro-B cells, but separates Fr. B cells into CD27+ and CD27− subsets. Further characterization reveals that these subsets, which we call Fr. B1 and Fr. B2, are functionally distinct and represent early and late stages of Fr. B, respectively. Interleukin-1 receptor 1 (Il1r1) is accurately predicted by MiDReG to be down-regulated during B-cell development (Supplemental Fig. S3). Furthermore, we find that CLP, pre-pro-B, and Fr. B cells all respond differently to IL-1 in vitro. While CLPs develop exclusively into dendritic cells (DCs) in IL-1 cultures, pre-pro-B cells die by apoptosis and Fr. B cells are unaffected (Supplemental Fig. S3).
The fourth marker, Ly6d, was predicted by MiDReG to be up-regulated during B-cell development (Fig. 1B). Ly6d, also known as ThB (Eckhardt and Herzenberg 1980), is expressed in all mature B cells and plasmacytoid DCs, as well as developing, but not mature, thymocytes (Supplemental Fig. S4; Reese et al. 2001). We examined Ly6d expression in MPP, CLP, and pre-pro-B cells and found that MPPs were uniformly Ly6d−, as were all hematopoietic stem cells, while Ly6d was highly expressed on pre-pro-B cells (Fig. 1D). Interestingly, Ly6d expression divided the CLP population into two approximately equal subpopulations (Fig. 1D). Ly6d− CLPs expressed higher levels of Kit than Ly6d+ CLPs, similar to the level of ckit expression observed for MPPs, consistent with a progenitor/progeny relationship between these three populations (Fig. 1E; Supplemental Fig. S5). Ly6d− CLPs also expressed lower levels of IL7Rα than Ly6d+ CLPs (Supplemental Fig. S5). We also observed that Ly6d− CLPs developmentally precede Ly6d+ CLPs in vitro (Supplemental Fig. S6). As we show below, only the Ly6d− subset of CLPs possesses full lymphoid potential. To distinguish this population from the original CLPs, we designate this population ALPs. As the Ly6d+ subset is almost totally B-cell-committed, we hereafter refer to this population as BLPs.
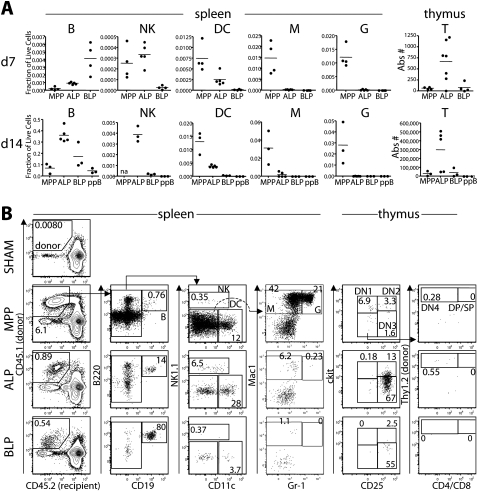
We next examined the in vivo lymphoid potential of ALPs and BLPs, as well as the stage that precedes them (MPP) and the stage that follows (pre-pro-B) (Fig. 2). We sorted and transplanted MPP, Ly6d− ALP, Ly6d+ BLP, and pre-pro-B cells intravenously into sublethally irradiated recipients at physiologic proportions (∼10,000–20,000 cells per transplant). At day 7, BLPs produced more B cells in the spleen than did ALPs. However, by day 14, ALPs surpassed BLPs and produced more B cells. Given the in vitro lineage relationship between ALPs and BLPs (Supplemental Fig. S6), these data suggest that BLPs are the more immediate precursor to B cells, and that ALPs produce B cells through a BLP intermediate. In contrast, at both time points, ALPs produced more DCs and natural killer (NK) cells in the spleen, and more T cells in the thymus, than did BLPs (Fig. 2A). While ALPs produced a low number of macrophages and granulocytes, MPPs produced ∼50-fold and 200-fold more, respectively. These data indicate retention of lymphoid potential during the LMPP-to-ALP transition with a substantial, but not total, loss of myeloid potential. Similar to pre-pro-B cells, BLPs generated almost exclusively B cells. Consistent with previous reports by our laboratory (Serwold et al. 2008), ALPs produced more T-lineage cells in the thymus than MPPs at these early time points.
Figure 2.
In vivo lineage potential of Ly6d− and Ly6d+ bone marrow progenitors by intravenous transplantation. (A) MPPs, Ly6d− CLPs (ALP), Ly6d+ CLPs (BLP), and pre-pro-Bs (ppB) were sorted and transplanted in physiologic proportions (∼10,000–20,000 cells per transplant) intravenously into sublethally irradiated recipients. At days 7 (top) and 14 (bottom), spleens and thymuses were harvested and analyzed for donor output in each of the four lymphoid lineages, as well as macrophages (M) and granulocytes (G). T-cell output was measured in the thymus (shown in the right column) and all other lineages in the spleen. Donor cells were congenic for two markers: (Donor) CD45.1, Thy1.2; (Host) CD45.2, Thy1.1. For spleen cells, the lineage output is shown as a fraction of total chimerism, with means indicated by horizontal bars. Thymuses were depleted of Thy1.1+ host cells prior to staining, and the absolute number of recovered T cells (DN1, DN2, DN3, DN4, DP, and SP) per thymus is shown in the right column. Data shown are a merge of four independent experiments. (na) Not analyzed. (B) Representative plots and gating strategy of donor lineages from the spleens and thymuses of intravenously transplanted MPPs, ALPs, and BLPs analyzed at day 7. The left column identifies the splenic chimerism for each sample, listed as the percentage of live cells. All other values are listed as a percentage of donor cells. Only the donor gate for the PBS-injected control (SHAM) is shown. Splenic and thymic lineages are defined as shown. In the thymus, cells shown are pregated as donor, and B220−CD19−CD11c−Mac1−Gr1−. Values of each thymus population are listed as a percentage of the total thymic donor cells (not shown).
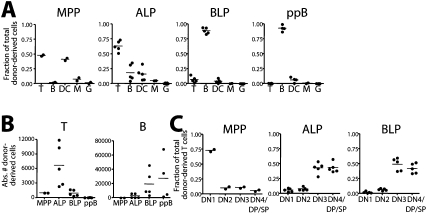
To directly assess the T-lineage potential of ALPs and BLPs in the absence of homing requirements, we transplanted these populations along with MPP and pre-pro-B cells intrathymically into nonirradiated recipients in the same proportions as above and analyzed T-cell output at day 9 (Fig. 3; Supplemental Fig. S7). Approximately 65% of donor cells derived from ALPs were of the T-cell lineage (Fig. 3A), and in 10-fold greater numbers than the T cells produced by either MPPs or BLPs (Fig. 3B). In contrast, BLPs, like pre-pro-B cells, produced almost exclusively B cells, comprising nearly 90% of donor-derived cells (Fig. 3A), and in numbers far greater than ALPs (Fig. 3B), suggesting that BLPs are almost fully committed to the B-cell fate even when placed directly into a strongly T-cell-inducing environment in vivo. However, those few BLPs that developed into T cells did so with similar kinetics as ALPs, as most were at the DN3 stage and beyond (Fig. 3C). In contrast, MPP-derived T cells did not differentiate significantly, and remained mostly at the DN1 stage (Fig. 3C). ALPs did produce a minor but detectable B-cell output (Fig. 3A,B), reminiscent of the lineage potential of the earliest Flk2+ thymic progenitor (Sambandam et al. 2005), and consistent with the low output of B cells from the thymus (Akashi et al. 2000). Thus, it appears Ly6d expression marks an important developmental transition, when CLPs lose T-, DC-, and NK-cell potential and commit to the B-cell lineage.
Figure 3.
In vivo lineage potential of Ly6d− and Ly6d+ bone marrow progenitors by intrathymic transplantation. (A) Intrathymic transplants (i.t.) of MPPs, ALPs, BLPs, and pre-pro-Bs (ppB) into nonirradiated recipients. Similar proportions of cells were transplanted as in Figure 2. Thymuses were harvested at day 9, and the distribution of donor cells within each lineage is shown. Displayed are the combined results from two independent experiments. (B) Absolute number of donor T-cell output (left) and B-cell output (right) recovered from the intrathymic transplants shown in A. (C) Distribution of donor T-lineage cells from MPPs, ALPs, and BLPs, listed as a fraction of total donor T cells. Representative stains for intrathymic analyses and definitions of all lineages can be found in Supplemental Figure S7.
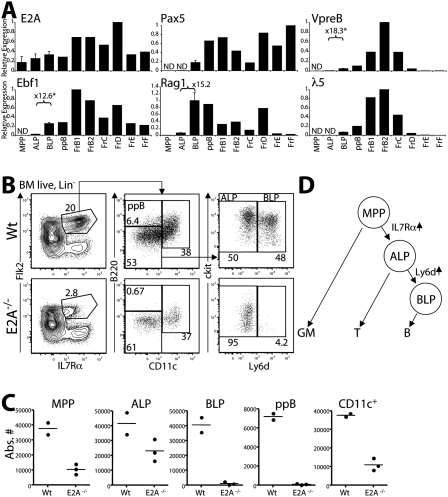
The transcription factor E2A is known to initiate the global gene expression profile of B-lineage specification through the up-regulation of genes such as Ebf1 and Pax5 (Kee and Murre 1998; Murre 2007). If Ly6d expression correlates with the earliest step in B-cell specification, we would predict that expression of these B-cell-specifying factors would begin during the transition from Ly6d− ALPs to Ly6d+ BLPs. Consistent with our hypothesis, expression of the B-lineage transcription factors Ebf1 and Pax5 increased dramatically during this transition, as did other E2A target genes involved in early B-cell development: Rag1, VpreB, and λ5 (Fig. 4A). To examine expression changes across a broader spectrum of genes, we compared microarrays of MPP, ALP, BLP, and pre-pro-B cells (Supplemental Fig. S8). In addition to the aforementioned B-cell factors, we identified a number of other factors including OCA-B (Pou2af1), Igα (Cd79a), Igβ (Cd79b), Blk, SpiB, and FoxO1 that also became highly up-regulated during the ALP-to-BLP transition.
Figure 4.
B-cell specification occurs during the ALP-to-BLP transition. (A) Quantitative RT–PCR of B-lineage genes in hematopoietic progenitors and B-committed populations in wild-type bone marrow. Samples were normalized to β-actin transcription and shown relative to the population with the highest expression of each gene. (ND) Not detected (Ct > 32 cycles). The fold change between ALPs and BLPs is shown for select genes, with an asterisk indicating statistical significance (unpaired t-test, n = 2, P < 0.05). Error bars are shown for the MPP, ALP, and BLP samples. (B) Wild-type and age-matched E2A−/− bone marrow were stained to examine changes in the proportions of MPP, ALP, BLP, and pre-pro-B cells. Only live lin− (Mac1−Ter119−Gr1−CD3−) and CD27+ cells are shown. Percentages within each gate are shown. (C) Absolute numbers of progenitor populations in wild-type and E2A−/− bone marrow. Absolute numbers are estimated for the two femurs, tibias, and hips of 6-wk-old mice. (D) Proposed model for the branching of the GM-cell, T-cell, and B-cell lineages from hematopoietic progenitors.
E2A functions throughout hematopoiesis, with established roles in lymphoid and myeloid development (Kee 2009), and even hematopoietic stem cell self-renewal (Yang et al. 2008; Semerad et al. 2009). However, only B-cell development is absolutely blocked in mice lacking E2A (Bain et al. 1994). While we would expect E2A-deficient mice to be blocked at the ALP–BLP transition, initial characterization of E2A knockout mice suggested that B-cell development was blocked at the pre-pro-B-cell stage, which is after this transition (Bain et al. 1997). We therefore re-examined E2A-deficient animals to determine the precise stage at which B-cell development is blocked (Fig. 4B,C). While the overall number of CLPs is reduced in these mice, consistent with previous reports (Borghesi et al. 2005; Kee 2009), we found no detectable pre-pro-B cells or BLPs (Fig. 4B,C). All B220+ cells we identified (traditionally a marker for the B-cell lineage) coexpressed the DC marker CD11c, and are likely DC-committed cells (Fig. 4B; Supplemental Fig. S2; Nikolic et al. 2002). There were still substantial, though reduced, numbers of ALPs detected, roughly equivalent to the known reduction in MPPs (Dias et al. 2008; Yang et al. 2008), indicating that the complete block in development occurs during the ALP-to-BLP transition. These data suggest that E2A activates a program of B-cell specification during the transition from ALP to BLP. Given that ALPs are the major thymic seeding population from the bone marrow (Fig. 2A; Serwold et al. 2008), and that, within the thymus, ALPs produce mainly T cells, we therefore conclude that ALPs reside at the branchpoint of B- and T-cell development (Fig. 4D).
In this report, we applied a novel algorithm called MiDReG to address an unresolved stage in hematopoietic development: the branchpoint between B- and T-cell development. MiDReG successfully predicted many developmentally regulated genes in B-cell development, and three surface markers functionally relevant to B-cell biology. One of these markers, Ly6d, separates CLPs into two distinct populations: one that contains full lymphoid potential (ALP) and another that is almost exclusively B-cell restricted in vivo (BLP). A similar finding using a λ5 reporter transgene has demonstrated that a small subset of CLPs is B-cell-restricted, although only 5% of CLPs expressed this reporter (Mansson et al. 2008). We did observe a large increase in λ5 expression between the ALP and BLP stages by both real-time RT–PCR and microarray analyses, suggesting that λ5 expression initiates within the BLP population. However, the BLP population encompasses a much larger fraction of CLPs than is indicated by λ5 reporter expression, demonstrating that Ly6d up-regulation is the earliest mark of B-cell specification identified to date, prior to B220, CD19, or λ5 expression. As shown by others (Zandi et al. 2008), a significant fraction of CLPs express Ebf1, which may play a role in initiating B-cell specification, perhaps by suppressing Id2/Id3 expression (Thal et al. 2009). As Ebf1 expression increases dramatically between the ALP and BLP populations, we suspect that the majority of Ebf1-expressing cells are contained within the BLP population.
Some groups have reported the existence of B220+ T-cell progenitors in the bone marrow (Martin et al. 2003). In contrast to these studies, we never observed in vivo T-cell potential from any B220+ population (Serwold et al. 2008). While Ly6d+ BLPs retain some residual T-cell potential, this may reflect a general property of development: that permanent commitment to a lineage does not occur until the chromatin of genes driving alternative lineages is completely shut (King et al. 2002). Likewise, we found a low level of myeloid output from ALP, particularly in the thymus. As a technical note, because of the lower level of IL7Rα expression in ALP, we find that the separation of ALPs from upstream MPPs depends critically on bright IL7Rα staining. Furthermore, Kit expression is higher in ALPs than bulk CLPs, and partially overlaps with the level of Kit expression in MPPs. These overlap Kit levels suggests that many previous studies implicating MPPs as proximate T-cell progenitors used preparations of MPPs that were contaminated with ALPs. The use of Ly6d greatly improves the ability to isolate full lymphoid progenitors from both downstream B-cell progenitors and upstream MPPs. These data also strongly suggest that the ALP is the central hub from which each lymphoid lineage fate restriction occurs. From a clinical standpoint, this is an important finding, as infusion of the most developmentally proximal transplantable T-cell progenitors can rapidly reconstitute immunity and prevent fatal infections following chemotherapy or irradiation (Arber et al. 2003).
Materials and methods
MiDReG
Details of the mathematics underlying MiDReG is described elsewhere (D Sahoo, J Seita, D Bhattacharya, M Inlay, I Weissman, S Plevritis, and D Dill, in prep.). For the prediction of genes encoding cell surface molecules developmentally regulated in B-cell development, two seed conditions were used: Seed A (Kit high and Mpl high) and Seed B (Cd19 high and Cd3ɛ low). For the up-regulated genes, MiDReG predicted gene X such that (Kit high AND Mpl high) ⇒ X low ⇒ ¬ (Cd19 high AND Cd3ɛ low). The down-regulated genes are predicted using (Kit high AND Mpl high) ⇒ X high ⇒ ¬ (Cd19 high AND Cd3ɛ low). Genes encoding surface proteins were identified using GO terms “membrane” and “membrane fraction.” The list of genes was further filtered using commercially available antibodies that are suitable for flow cytometry applications on Biocompare.
Animals
All animal procedures were approved by the International Animal Care and Use Committee, and the Stanford Administrative Panel on Laboratory Animal Care. C57Bl/Ka-Thy1.1 mice were derived and maintained at Stanford University. Bone marrow and spleen cells were obtained from mice aged 10–12 wk. C57Bl/Ka-Thy1.1 CD45.2+ (HZ) and C57Bl/Ka-Thy1.1 CD45.1 (BA) strains were derived and maintained in our laboratory. Bone marrow from E2A−/− mice and wild-type littermates were generously provided by the C. Murre laboratory. Peripheral blood was sampled from the tail vein and all intravenous cell transplants were performed by injection into the retroorbital sinus of isoflurane-anesthetized mice. Intrathymic injections were performed as described previously (Jerabek and Weissman 2001). B-1 cells were isolated via peritoneal lavage of euthanized animals. Donor mice were 4–8 wk old and recipient mice ranged from 6 to12 wk of age.
Antibodies
A complete list of all antibodies used in the study is shown in Supplemental Table S1.
Bone marrow preparation and staining
Bone marrow was harvested from donor mice by crushing bones and removing debris on density gradient using Histopaque 1119 or 1077, or a 50/50 mix (Sigma). Where indicated, bone marrow was lineage-depleted by adding lineage antibodies (Mac-1, Gr-1, Ter119, and, optionally, CD19), then adding dynabeads (Invitrogen) and removing bound cells via magnetic field according to the manufacturer's instructions. Depleted bone marrow was stained in PBS with 2% fetal calf serum (FCS) for 15 min on ice. Stained cells were resuspended in PBS with 10% FCS prior to sorting. For fluorescence minus one (FMO) controls, cells were stained with all antibodies except for the one in question.
Fluorescence-activated cell sorting (FACS)
All cells were sorted and data were collected on a BD FACS-Aria (Becton Dickinson). FlowJo software (TreeStar) was used for flow cytometric data analysis. Cells were sorted into ice-cold PBS with 10% FCS, or into tissue culture medium.
Cell cultures
Cells were cultured in Iscove's Modified Dulbecco's Eagle Medium (Invitrogen) with 10% FCS (Omega Scientific), 50 μM β-mercaptoethanol, sodium pyruvate, L-glutamine, and nonessential amino acids for the indicated time period in the presence of 10 ng/mL each Flt3L (R&D Systems), SCF (R&D Systems), and IL-7 (eBiosciences), and in the presence of OP9 stromal cells when indicated. IL-1α (Peprotech) was included in cultures as indicated at doses ranging from 1 pg/mL to 100 ng/mL as indicated.
Engraftment analysis
Mature thymocytes were depleted for host mature T cells using an anti-Thy1.1 (19XE5) antibody toxic to Thy1.1+ cells as described (Serwold et al. 2008). Briefly, single-cell suspensions of thymuses were incubated with 40 μg of anti-Thy1.1 for 1 h on ice. Dead cells and debris were separated by density gradient using Histopaque 1119. Spleens were harvested and made into single-cell suspensions, then treated with ACK lysis buffer (150 mM NH4Cl, 1 mM KHCO3, 0.1 mM EDTA) to remove red blood cells. For absolute cell number estimates of spleen and bone marrow, cells were counted using a Vi-CELL XR cell counter (Beckman Coulter) to determine total viable cells. The percentage of each population among live cells within each sample was calculated, then multiplied by the number of viable cells. For thymus analyses, after depletion, the entirety of remaining cells were analyzed.
Quantitative PCR for B-cell genes and microarray analyses
A complete list of primers sequences and quantitative real-time PCR protocol is shown in Supplemental Table S2. Microarray preparation and analysis are described in Supplemental Figure S8.
Acknowledgments
We thank L. Jerabek for laboratory management, C. Richter for antibody production, A. Mosley, J. Dollaga, and D. Escoto for animal care, E. Zuo and the Stanford PAN facility for microarray hybridization, and J. Tung and L. Herzenberg for the Ly6d antibody and technical advice. D.S. and D.L.D. designed MiDReG with assistance from S.K.P. D.S. and D.B. adapted MiDReG to look for surface markers with assistance from J.S. M.A.I. and D.B. characterized Ly6d, CD27, and IL1r1. T.S. and H.K. assisted with early characterization of CLP and pre-pro-B cells. M.A.I. and D.B. wrote the manuscript. We all edited the manuscript. All experiments were conducted in the laboratory of I.L.W. This investigation was supported by National Institutes of Health grants 5U56CA112973 (to S.K.P.) and 5R01AI047457 and 5R01AI047458 (to I.L.W.), and a grant from the Thomas and Stacey Siebel Foundation (to I.L.W). D.S., D.D., and S.K.P. were supported by National Institutes of Health grant 5U56CA112973 (to S.K.P.). M.A.I. is supported by PHS Grant number CA09151, awarded by the National Cancer Institute, and a fellowship from the California Institute for Regenerative Medicine (T1-00001); D.B. is supported by a fellowship from the Cancer Research Institute (T32AI0729022) and from the National Institutes of Health (K01DK078318); J.S. is supported by a fellowship from the California Institute for Regenerative Medicine (T1-00001); and T.S. is supported by a fellowship from the National Institutes of Health (F32AI058521).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1836009.
Supplemental material is available at http://www.genesdev.org.
References
- Akashi K, Richie LI, Miyamoto T, Carr WH, Weissman IL. B lymphopoiesis in the thymus. J Immunol. 2000;164:5221–5226. doi: 10.4049/jimmunol.164.10.5221. [DOI] [PubMed] [Google Scholar]
- Arber C, BitMansour A, Sparer TE, Higgins JP, Mocarski ES, Weissman IL, Shizuru JA, Brown JM. Common lymphoid progenitors rapidly engraft and protect against lethal murine cytomegalovirus infection after hematopoietic stem cell transplantation. Blood. 2003;102:421–428. doi: 10.1182/blood-2002-12-3834. [DOI] [PubMed] [Google Scholar]
- Bain G, Maandag EC, Izon DJ, Amsen D, Kruisbeek AM, Weintraub BC, Krop I, Schlissel MS, Feeney AJ, van Roon M. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–892. doi: 10.1016/0092-8674(94)90077-9. [DOI] [PubMed] [Google Scholar]
- Bain G, Robanus Maandag EC, te Riele HP, Feeney AJ, Sheehy A, Schlissel M, Shinton SA, Hardy RR, Murre C. Both E12 and E47 allow commitment to the B cell lineage. Immunity. 1997;6:145–154. doi: 10.1016/s1074-7613(00)80421-5. [DOI] [PubMed] [Google Scholar]
- Borghesi L, Aites J, Nelson S, Lefterov P, James P, Gerstein R. E47 is required for V(D)J recombinase activity in common lymphoid progenitors. J Exp Med. 2005;202:1669–1677. doi: 10.1084/jem.20051190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias S, Mansson R, Gurbuxani S, Sigvardsson M, Kee BL. E2A proteins promote development of lymphoid-primed multipotent progenitors. Immunity. 2008;29:217–227. doi: 10.1016/j.immuni.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckhardt LA, Herzenberg LA. Monoclonal antibodies to ThB detect close linkage of ly-6 and a gene regulating ThB expression. Immunogenetics. 1980;11:275–291. doi: 10.1007/BF01567794. [DOI] [PubMed] [Google Scholar]
- Jerabek L, Weissman IL. Intrathymic injection for analysis of T-cell progenitor activity. In: Klug CA, Jordan CT, editors. Hematopoietic stem cell protocols. Humana Press; Totowa, NJ: 2001. pp. 161–165. [DOI] [PubMed] [Google Scholar]
- Karsunky H, Inlay MA, Serwold T, Bhattacharya D, Weissman IL. Flk2+ common lymphoid progenitors possess equivalent differentiation potential for the B and T lineages. Blood. 2008;111:5562–5570. doi: 10.1182/blood-2007-11-126219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee BL. E and ID proteins branch out. Nat Rev Immunol. 2009;9:175–184. doi: 10.1038/nri2507. [DOI] [PubMed] [Google Scholar]
- Kee BL, Murre C. Induction of early B cell factor (EBF) and multiple B lineage genes by the basic helix–loop–helix transcription factor E12. J Exp Med. 1998;188:699–713. doi: 10.1084/jem.188.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King AG, Kondo M, Scherer DC, Weissman IL. Lineage infidelity in myeloid cells with TCR gene rearrangement: A latent developmental potential of proT cells revealed by ectopic cytokine receptor signaling. Proc Natl Acad Sci. 2002;99:4508–4513. doi: 10.1073/pnas.072087899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- Lai AY, Kondo M. Identification of a bone marrow precursor of the earliest thymocytes in adult mouse. Proc Natl Acad Sci. 2007;104:6311–6316. doi: 10.1073/pnas.0609608104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YS, Wasserman R, Hayakawa K, Hardy RR. Identification of the earliest B lineage stage in mouse bone marrow. Immunity. 1996;5:527–535. doi: 10.1016/s1074-7613(00)80268-x. [DOI] [PubMed] [Google Scholar]
- Mansson R, Zandi S, Anderson K, Martensson IL, Jacobsen SE, Bryder D, Sigvardsson M. B-lineage commitment prior to surface expression of B220 and CD19 on hematopoietic progenitor cells. Blood. 2008;112:1048–1055. doi: 10.1182/blood-2007-11-125385. [DOI] [PubMed] [Google Scholar]
- Martin CH, Aifantis I, Scimone ML, von Andrian UH, Reizis B, von Boehmer H, Gounari F. Efficient thymic immigration of B220+ lymphoid-restricted bone marrow cells with T precursor potential. Nat Immunol. 2003;4:866–873. doi: 10.1038/ni965. [DOI] [PubMed] [Google Scholar]
- Murre C. Regulation and function of the E2A proteins in B cell development. Adv Exp Med Biol. 2007;596:1–7. doi: 10.1007/0-387-46530-8_1. [DOI] [PubMed] [Google Scholar]
- Nikolic T, Dingjan GM, Leenen PJ, Hendriks RW. A subfraction of B220+ cells in murine bone marrow and spleen does not belong to the B cell lineage but has dendritic cell characteristics. Eur J Immunol. 2002;32:686–692. doi: 10.1002/1521-4141(200203)32:3<686::AID-IMMU686>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Perry SS, Welner RS, Kouro T, Kincade PW, Sun XH. Primitive lymphoid progenitors in bone marrow with T lineage reconstituting potential. J Immunol. 2006;177:2880–2887. doi: 10.4049/jimmunol.177.5.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese JT, Mehta H, Chappell CH, Bamezai A. Downregulated expression of ly-6-ThB on developing T cells marks CD4+CD8+ subset undergoing selection in the thymus. Dev Immunol. 2001;8:107–121. doi: 10.1155/2001/63451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambandam A, Maillard I, Zediak VP, Xu L, Gerstein RM, Aster JC, Pear WS, Bhandoola A. Notch signaling controls the generation and differentiation of early T lineage progenitors. Nat Immunol. 2005;6:663–670. doi: 10.1038/ni1216. [DOI] [PubMed] [Google Scholar]
- Schwarz BA, Bhandoola A. Circulating hematopoietic progenitors with T lineage potential. Nat Immunol. 2004;5:953–960. doi: 10.1038/ni1101. [DOI] [PubMed] [Google Scholar]
- Semerad CL, Mercer EM, Inlay MA, Weissman IL, Murre C. E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors. Proc Natl Acad Sci. 2009;106:1930–1935. doi: 10.1073/pnas.0808866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serwold T, Ehrlich LI, Weissman IL. Reductive isolation from bone marrow and blood implicates common lymphoid progenitors as the major source of thymopoiesis. Blood. 2008;113:807–815. doi: 10.1182/blood-2008-08-173682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thal MA, Carvalho TL, He T, Kim HG, Gao H, Hagman J, Klug CA. Ebf1-mediated down-regulation of Id2 and Id3 is essential for specification of the B cell lineage. Proc Natl Acad Sci. 2009;106:552–557. doi: 10.1073/pnas.0802550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H, Masuda K, Satoh R, Kakugawa K, Ikawa T, Katsura Y, Kawamoto H. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–772. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- Yang Q, Kardava L, St Leger A, Martincic K, Varnum-Finney B, Bernstein ID, Milcarek C, Borghesi L. E47 controls the developmental integrity and cell cycle quiescence of multipotential hematopoietic progenitors. J Immunol. 2008;181:5885–5894. doi: 10.4049/jimmunol.181.9.5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandi S, Mansson R, Tsapogas P, Zetterblad J, Bryder D, Sigvardsson M. EBF1 is essential for B-lineage priming and establishment of a transcription factor network in common lymphoid progenitors. J Immunol. 2008;181:3364–3372. doi: 10.4049/jimmunol.181.5.3364. [DOI] [PubMed] [Google Scholar]