Abstract
HepA-related protein (HARP) (also known as SMARCAL1) is an ATP-driven annealing helicase that catalyzes the formation of dsDNA from complementary Replication protein A (RPA)-bound ssDNA. Here we find that HARP contains a conserved N-terminal motif that is necessary and sufficient for binding to RPA. This RPA-binding motif is not required for annealing helicase activity, but is essential for the recruitment of HARP to sites of laser-induced DNA damage. These findings suggest that the interaction of HARP with RPA increases the concentration of annealing helicase activity in the vicinity of ssDNA regions to facilitate processes such as DNA repair.
Keywords: HARP, annealing helicase, RPA, SMARCAL1, SIOD
Proteins in the SNF2 family of ATPases participate in a variety of nuclear processes, such as chromatin assembly and remodeling, transcription, DNA repair, and recombination (Gorbalenya and Koonin 1993; Eisen et al. 1995; Flaus et al. 2006). The HepA-related protein (HARP, also known as SMARCAL1) is a distant member of the SNF2 family (Coleman et al. 2000; Flaus et al. 2006). HARP is present in many eukaryotes but appears to be absent in fungi. In humans, mutations in HARP contribute to the pleiotropic disorder known as Schimke immunoosseous dysplasia (SIOD) (Boerkoel et al. 2002).
HARP is an ATP-dependent annealing helicase (Yusufzai and Kadonaga 2008). Specifically, HARP is able to rewind complementary ssDNA that is bound by the ssDNA-binding protein Replication protein A (RPA). HARP binds preferentially to fork DNA relative to ssDNA or to dsDNA (Yusufzai and Kadonaga 2008). The ATPase activity of HARP is also stimulated preferentially by fork DNA relative to ssDNA or dsDNA (Yusufzai and Kadonaga 2008). This property is consistent with the observation that the ATPase activity of HARP is enhanced by DNA species with ssDNA–dsDNA junctions (Hockensmith et al. 1986; Muthuswami et al. 2000). These findings suggest a model wherein the binding of HARP to a DNA fork activates its ATP-driven motor with which it catalyzes the rewinding of DNA.
The DNA rewinding activity of HARP could potentially participate in many different processes such as transcription, DNA replication, and DNA repair, in which ssDNA regions are generated by the action of helicases or polymerases (for example, see Liu and Wang 1987; Kowalski et al. 1988; Havas et al. 2000; Pyle 2008). These ssDNA regions can be stabilized by ssDNA-binding proteins (SSBs) such as RPA, the major SSB in eukaryotes (for example, see Wold 1997; Iftode et al. 1999; Zou et al. 2006). RPA is a heterotrimer (RPA1 [70 kDa], RPA2 [32 kDa], and RPA3 [14 kDa]) that binds stably to ssDNA and prevents complementary DNA from reannealing. RPA is required for many cellular processes, including replication, recombination, and repair, during which it stabilizes ssDNA intermediates. Because HARP catalyzes the regeneration of dsDNA from complementary RPA-bound ssDNA, it is possible that there is a specific link between HARP and RPA. In this study, we identify and characterize a direct interaction between these factors.
Results and Discussion
HARP is present in a stable complex with RPA
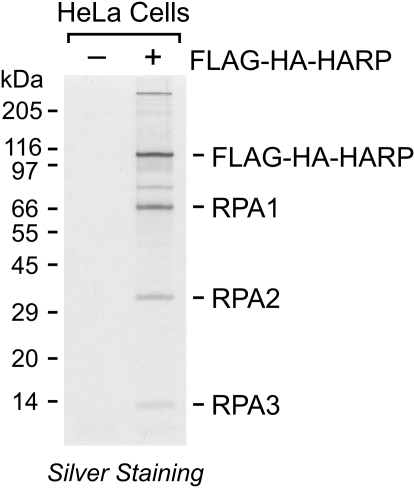
To gain a better understanding of the function of HARP, we created a stable HeLa cell line that expresses a transgene encoding Flag-HA-tagged HARP protein, and then purified Flag-HA-HARP and associated proteins from these cells by sequential Flag and HA immunoaffinity chromatography. As a control, the same procedures were performed in parallel with normal HeLa cells lacking the HARP transgene. The resulting samples were analyzed by SDS–polyacrylamide gel electrophoresis, which revealed several polypeptides that copurify with Flag-HA-HARP. Several independent repetitions of this procedure showed that three polypeptides consistently copurified with HARP. Analysis of these three polypeptides by mass spectrometry indicated that they are the three subunits of RPA (Fig. 1).
Figure 1.
Purification of a HARP-containing complex. Nuclear extracts were prepared from normal HeLa S3 cells (−) or HeLa cells expressing Flag-HA-tagged HARP (+). The proteins were subjected to sequential anti-Flag and anti-HA immunoaffinity purification, and then analyzed by SDS–polyacrylamide gel electrophoresis and silver staining. The polypeptides that consistently copurified with Flag-HA-HARP were identified by mass spectroscopy and are indicated.
To investigate whether the interaction between HARP and RPA occurs in other cell types, we tested the association of the native proteins in M059K human glioblastoma cells. In these experiments, we observed that RPA coimmunoprecipitates with HARP in whole-cell extracts from M059K cells (Supplemental Fig. S1). We also observed coimmunoprecipitation of RPA and HARP in cell extracts from human Jurkat and MCF7 cells (data not shown). Thus, HARP forms a stable complex with RPA in different cell types.
HARP interacts with RPA via a conserved N-terminal motif
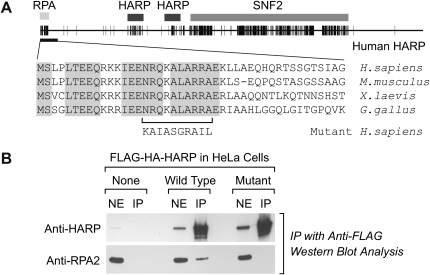
We next sought to identify the region of HARP that is responsible for its interaction with RPA. First, we examined the conserved regions of vertebrate HARP proteins. This analysis revealed the previously described SNF2 ATPase domain and HARP motifs of unknown function (Fig. 2A; Coleman et al. 2000). In addition, we observed that the N terminus of HARP contains a highly charged conserved region (Fig. 2A). We investigated whether other proteins contain sequence motifs that resemble the conserved N-terminal region of HARP, and found that a related sequence, which binds to the RPA2 subunit of RPA, is present in several DNA repair factors that are known to interact with RPA (Mer et al. 2000). These findings suggested that the N-terminal region of HARP is an RPA interaction motif.
Figure 2.
The conserved N-terminal region of HARP is required for the interaction of HARP with RPA. (A) Schematic representation of the conserved regions of HARP. The human, mouse, frog, and chicken HARP proteins were aligned by using the Clustal W algorithm, and conserved amino acid residues with 100% identity are represented by black hatches. The amino acid sequences of the N-terminal region are shown below, and the substitutions in the mutant human HARP protein are indicated. (B) Coimmunoprecipitation of RPA and HARP. HARP was immunoprecipitated with anti-Flag resin from nuclear extracts of cells containing wild-type or mutant Flag-HA-HARP. The amino acid substitutions in the mutant HARP protein are as depicted in A. As a control, normal HeLa cells lacking Flag-HA-HARP were also treated in parallel. Western blot analysis of nuclear extract (NE) and immunoprecipitated samples (IP) was performed by using anti-HARP and anti-RPA2 antibodies.
To test this idea, we generated a mutant version of HARP containing amino acid substitutions in the region of the N terminus that corresponds to the putative RPA2-binding motif (Fig. 2A; Mer et al. 2000). We then created a stable HeLa cell line that expresses a transgene encoding the mutant HARP protein with Flag and HA tags. The effect of the N-terminal mutations on the binding of RPA to HARP was tested in coimmunoprecipitation experiments with HeLa cell lines containing wild-type versus mutant transgenic HARP proteins. The transgenic Flag-HA-HARP proteins were immunoprecipitated with anti-Flag antibodies, and the presence of RPA was detected by Western blot with antibodies against RPA2. These experiments revealed that RPA coimmunoprecipitates with wild-type HARP but not with mutant HARP (Fig. 2B). Western blot analyses with anti-HARP also confirmed the presence of the transgenic Flag-HA-HARP proteins in both cell lines. As a control, RPA was not detected in the anti-Flag immunoprecipitates of normal HeLa cells lacking transgenic Flag-HA-HARP. These results indicate that sequences in the conserved N terminus of HARP are necessary for the interaction of RPA with HARP.
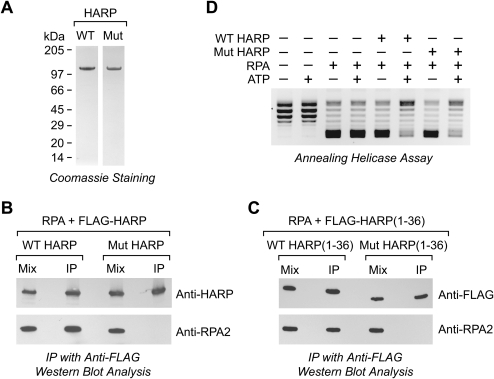
To determine whether RPA binds directly to HARP, we synthesized Flag-tagged wild-type and mutant HARP proteins in Sf9 cells with a baculovirus expression system, and purified the proteins to near homogeneity by Flag affinity chromatography (Fig. 3A). We also synthesized human RPA in Escherichia coli and purified the protein to near homogeneity (Henricksen et al. 1994; Yusufzai and Kadonaga 2008). Purified Flag-tagged wild-type or mutant HARP was incubated with purified RPA, and the resulting samples were subjected to immunoprecipitation with anti-Flag antibodies. The presence of HARP and RPA in the immunoprecipitates was then detected by Western blot analysis with anti-HARP or anti-RPA2 antibodies. These experiments revealed that RPA binds directly to wild-type HARP but does not interact with the mutant HARP (Fig. 3B). Hence, there is a direct interaction between HARP and RPA that is dependent on the conserved N-terminal region of HARP.
Figure 3.
The RPA interaction region of HARP is not required for annealing helicase activity. (A) Purification of wild-type and mutant HARP proteins. Recombinant Flag-HARP proteins were synthesized by baculovirus expression in Sf9 cells, purified by Flag affinity chromatography, and analyzed by SDS–polyacrylamide gel electrophoresis and staining with Coomassie Blue R-250. The amino acid substitutions in the mutant HARP protein are depicted in Figure 2A. (B) The conserved N-terminal region of HARP is necessary for the binding of purified HARP to purified RPA. Wild-type or mutant Flag-HARP was incubated with purified recombinant RPA, and then immunoprecipitated with anti-Flag resin. After the beads were washed, the bound proteins were eluted and detected by Western blot analysis. (C) The N-terminal 36-amino-acid residues of HARP are sufficient for binding to RPA. The wild-type and mutant versions of the N-terminal 36-amino-acid residues of HARP (fused to a C-terminal Flag tag) were synthesized in bacteria and purified to near homogeneity. The binding of these peptides to purified RPA was analyzed, as in B. (D) Both wild-type and mutant HARP proteins possess ATP-dependent annealing helicase activity. Annealing helicase assays were carried out as described by Yusufzai and Kadonaga (2008). HARP-mediated DNA rewinding is monitored by the ATP-dependent relaxation of RPA-unwound DNA in the presence of topoisomerase I. An equimolar amount of UTP was used as a control for the absence of ATP.
We then further investigated whether the N terminus of HARP is sufficient for binding to RPA. To this end, we synthesized and purified the wild-type and mutant versions of peptides that comprise the N-terminal 36-amino-acid residues of HARP along with a C-terminal Flag tag. Coimmunoprecipitation analyses with these peptides and purified RPA revealed that the wild-type peptide, but not the mutant peptide, binds to RPA (Fig. 3C). Thus, the collective results indicate that the N-terminal 36-amino-acid residues of HARP are necessary and sufficient for binding to RPA.
The N-terminal RPA-binding motif of HARP is not needed for annealing helicase activity
We then examined whether the conserved N-terminal region of HARP is needed for its annealing helicase activity. In the annealing helicase assay, HARP catalyzes the ATP-dependent formation of dsDNA from partially unwound plasmid DNA containing RPA-bound ssDNA bubbles (Supplemental Fig. S2; Yusufzai and Kadonaga 2008). In the presence of topoisomerase I, annealing helicase activity is detected by the conversion of the partially unwound DNA (containing the RPA-bound ssDNA bubbles) from a negatively supercoiled state to relaxed dsDNA circles. We observed that the wild-type and mutant HARP proteins exhibit approximately the same specific activity in the annealing helicase assay over a range of protein concentrations and reaction times (Fig. 3D; data not shown). These findings indicate that the binding of RPA to HARP is not required for the annealing helicase activity of HARP.
The RPA-binding motif is required for recruitment of HARP to sites of DNA damage
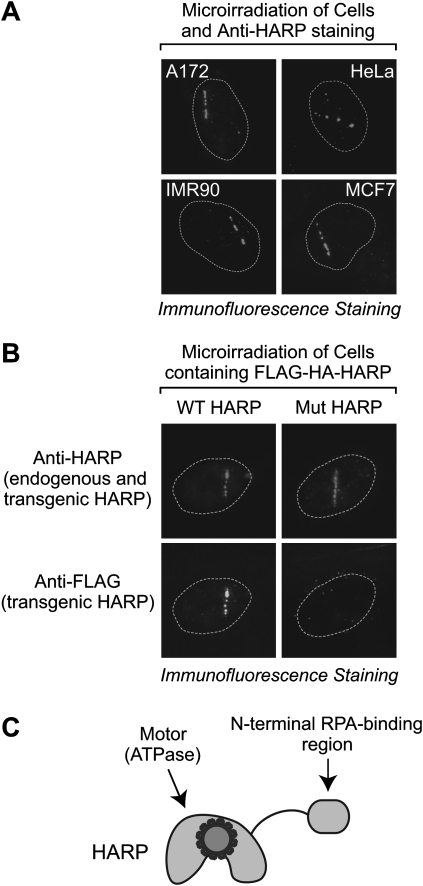
Because the intrinsic annealing helicase activity of HARP acts independently of its binding to RPA, we further investigated a potential function for the RPA–HARP interaction. RPA-bound ssDNA is a key intermediate in processes such as DNA replication and repair (for example, see Wold 1997; Iftode et al. 1999; Zou et al. 2006). We examined a potential function of HARP in DNA repair. To this end, we microirradiated several different human cell lines (A172, HeLa, IMR90, and MCF7) and determined whether HARP localizes to sites of DNA damage. In these studies, we used a highly focused nanosecond green laser that mostly generates DNA double-strand breaks (DSBs) in defined regions of nuclei (for example, see Kim et al. 2002, 2007). The endogenous HARP protein was visualized by immunofluorescence with anti-HARP antibodies. We observed that HARP localizes to sites of laser-induced DNA damage in each of the cell lines tested (Fig. 4A). These findings suggest a role of HARP in DSB repair.
Figure 4.
Wild-type but not RPA-binding-defective mutant HARP localizes to laser-induced damage sites. (A) Endogenous HARP protein is localized to sites of DNA damage in several different cell lines. The indicated cells were microirradiated with a nanosecond green laser. After 1 h, the cells were fixed and stained with anti-HARP antibodies. The nuclei are outlined with a dotted line. (B) Cells containing wild-type or mutant Flag-HA-HARP were microirradiated, as in A. After 30 min, cells were fixed and stained with anti-HARP and anti-Flag antibodies. The anti-HARP antibodies recognize both the endogenous and transgenic HARP, whereas the anti-Flag antibodies recognize only the transgenic HARP. All of the wild-type HARP-containing cells (N = 8) exhibited specific localization of HARP to the DNA damage sites, whereas none of the mutant HARP-containing cells (N = 8) showed localization of the mutant HARP to the DNA damage sites. (C) A simple view of HARP protein.
To assess the relative timing of the recruitment of HARP and RPA to laser-induced DNA damage sites, we microirradiated HeLa cells and then fixed the cells at specific time points subsequent to irradiation. These experiments revealed that the timing of the localization of HARP to damaged DNA is similar to that of RPA (Supplemental Fig. S3A). The early response factor Mre11 is recruited to DNA damage sites prior to RPA (Kim et al. 2005). Similarly, we found that Mre11 is localized to microirradiated DNA before HARP (Supplemental Fig. S3B).
To test the role of the HARP–RPA interaction in the recruitment of HARP to damaged DNA, we compared the properties of wild-type versus RPA-binding mutant HARP. We created stable cell lines that express wild-type or mutant Flag-HA-HARP proteins. Then, the localization of total HARP proteins (both endogenous HARP and transgenic Flag-HA-HARP) was visualized by immunofluorescence with anti-HARP antibodies, whereas the localization of transgenic HARP proteins was determined by immunofluorescence with anti-Flag antibodies.
These experiments revealed that wild-type Flag-HA-HARP localizes to the sites of laser-induced DNA damage (Fig. 4B, bottom left) in a manner similar to endogenous HARP (Fig. 4A,B [top left]). We also confirmed the localization of wild-type Flag-HA-HARP to sites of DNA damage that are generated with a lower-intensity 405-nm blue laser (Supplemental Fig. S4, top left), which creates DSBs in DNA that is presensitized by the incorporation of BrdU (Kong et al. 2009).
Next, we analyzed the localization of mutant Flag-HA-HARP to damaged DNA. As shown in Figure 4B (bottom right), mutant Flag-HA-HARP (as detected with anti-Flag antibodies) is not localized to sites of green laser-induced DNA damage, whereas, in contrast, endogenous HARP (as detected with anti-HARP antibodies) is recruited to the DNA damage regions (Fig. 4B, top right). The inability of the mutant Flag-HA-HARP to localize to damaged DNA was also observed upon microirradiation of BrdU-treated cells with the blue laser (Supplemental Fig. S4, top right). These results indicate that the conserved N-terminal RPA-binding motif is essential for the recruitment of HARP to sites of DNA damage.
Summary and conclusions
In this study, we found that HARP binds stably to RPA via a conserved N-terminal RPA interaction region (Fig. 4C). The binding of HARP to the N-terminal region is not required for its annealing helicase activity, but is essential for the recruitment of HARP to sites of laser-induced DNA damage. In addition, the ATPase activity of HARP is not stimulated by ssDNA or RPA (Yusufzai and Kadonaga 2008). These findings suggest a model in which the interaction between RPA and HARP serves to recruit HARP to regions of ssDNA that are formed during DNA damage and repair. This work has focused on the role of HARP in the response to laser-induced DNA damage, which mainly involves DSBs. It is also possible, however, that HARP participates in other processes, such as DNA replication. For instance, during the replication of damaged DNA, the presence of a lesion can uncouple the replicating helicase from the polymerase and lead to the formation of an extended ssDNA bubble (Byun et al. 2005). Thus, in a more general sense, the recruitment of HARP to ssDNA via RPA might provide a concentrated source of annealing helicase activity that acts to rewind complementary ssDNA regions into dsDNA.
Materials and methods
Purification of the HARP complex
The HARP complex was purified by sequential anti-Flag and anti-HA immunoaffinity steps from HeLa S3 cells that stably express a cDNA encoding wild-type human HARP with C-terminal Flag and HA tags. The generation of the stable HeLa S3 cell line and the affinity purification were carried out by using previously described methods (Nakatani and Ogryzko 2003). The specific procedures are described in detail in the Supplemental Material.
Mutant HARP protein
The human HARP cDNA was modified by overlapping PCR with primers that convert the coding sequence from AATCGACAAAAGGCTCTGGCCCGCAGAGCT to AAGGCAATTGCCTCAGGAAGAGCAATCCTG from +46 to +75 relative to A + 1 in the ATG-initiating codon. These mutations result in the substitution of 10 consecutive amino acid residues in the HARP protein, as depicted in Figure 2A. The integrity of the sequence of the mutant HARP cDNA was verified by DNA sequencing. A stable HeLa S3 cell line expressing the cDNA encoding the mutant HARP with C-terminal Flag and HA tags was constructed in an analogous manner as for wild-type HARP.
Coimmunoprecipitation of HARP and RPA
Nuclear extracts were prepared from HeLa cells containing wild-type or mutant HARP protein with C-terminal Flag and HA tags. Flag-HA-HARP and associated proteins were immunoprecipitated with anti-Flag resin, as described for the purification of the HARP complex. Proteins were detected by Western blot analysis by using anti-HARP (rabbit polyclonal antibodies raised against a recombinant C-terminal fragment comprising amino acid residues 820–954 along with a His6 tag) or anti-RPA2 (gift from Bruce Stillman, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY) antibodies. Methods for the coimmunoprecipitation of purified HARP and RPA proteins are described in the Supplemental Material.
Annealing helicase assay
The annealing helicase assay was performed as described previously (Yusufzai and Kadonaga 2008).
Green laser microirradiation
Laser microirradiation and immunofluorescence staining were performed as described previously (Kong et al. 2009). In these experiments, a small region inside the cell nucleus was irradiated by focusing a second harmonic (532 nm) of a pulsed neodymium:ytrium aluminum garnet laser within the selected Z plane through an epifluorescence port of a Zeiss LSM 410 confocal system. An ∼10-μm line was scanned continuously for 2 min. After microirradiation, cells were fixed and stained with anti-HARP or anti-Flag (M2; Sigma) antibodies. Typically, four to six cells per plate, using up to seven plates, were microirradiated and then fixed within 30–60 min post-irradiation. Experiments were repeated at least three times and consistent results were obtained. Cell viability was confirmed by morphology.
Acknowledgments
We thank Tammy Juven-Gershon, Jer-Yuan Hsu, Barbara Rattner, Josh Theisen, Sharon Torigoe, and Moriah Eustice for critical reading of the manuscript. We are grateful to Dr. Bruce Stillman (Cold Spring Harbor Laboratory) for the generous gift of antibodies against human RPA2. This work was supported by grants from the National Institutes of Health to J.T.K. (GM058272) and to K.Y. (CA100710), and the Department of Defense Breast Cancer Research Program to K.Y. (DAMD17-03-1-0436). In addition, T.Y. received support from an NIH National Research Service Award Fellowship (F32 GM76936).
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1831509.
Supplemental material is available at http://www.genesdev.org.
References
- Boerkoel CF, Takashima H, John J, Yan J, Stankiewicz P, Rosenbarker L, André JL, Bogdanovic R, Burguet A, Cockfield S, et al. Mutant chromatin remodeling protein SMARCAL1 causes Schimke immuno-osseous dysplasia. Nat Genet. 2002;30:215–220. doi: 10.1038/ng821. [DOI] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes & Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman MA, Eisen JA, Mohrenweiser HW. Cloning and characterization of HARP/SMARCAL1: A prokaryotic HepA-related SNF2 helicase protein from human and mouse. Genomics. 2000;65:274–282. doi: 10.1006/geno.2000.6174. [DOI] [PubMed] [Google Scholar]
- Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: Subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbalenya AE, Koonin EV. Helicases: Amino acid sequence comparisons and structure–function relationships. Curr Opin Struct Biol. 1993;3:419–429. [Google Scholar]
- Havas K, Flaus A, Phelan M, Kingston R, Wade PA, Lilley DM, Owen-Hughes T. Generation of superhelical torsion by ATP-dependent chromatin remodeling activities. Cell. 2000;103:1133–1142. doi: 10.1016/s0092-8674(00)00215-4. [DOI] [PubMed] [Google Scholar]
- Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A: Expression, complex formation, and functional characterization. J Biol Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
- Hockensmith JW, Wahl AF, Kowalski S, Bambara RA. Purification of a calf thymus DNA-dependent adenosinetriphosphatase that prefers a primer–template junction effector. Biochemistry. 1986;25:7812–7821. doi: 10.1021/bi00372a005. [DOI] [PubMed] [Google Scholar]
- Iftode C, Daniely Y, Borowiec JA. Replication protein A (RPA): The eukaryotic SSB. Crit Rev Biochem Mol Biol. 1999;34:141–180. doi: 10.1080/10409239991209255. [DOI] [PubMed] [Google Scholar]
- Kim JS, Krasieva TB, LaMorte V, Taylor AM, Yokomori K. Specific recruitment of human cohesin to laser-induced DNA damage. J Biol Chem. 2002;277:45149–45153. doi: 10.1074/jbc.M209123200. [DOI] [PubMed] [Google Scholar]
- Kim JS, Krasieva TB, Kurumizaka H, Chen DJ, Taylor AM, Yokomori K. Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells. J Cell Biol. 2005;170:341–347. doi: 10.1083/jcb.200411083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Heale JT, Zeng W, Kong X, Krasieva TB, Ball AR, Jr, Yokomori K. In situ analysis of DNA damage response and repair using laser microirradiation. Methods Cell Biol. 2007;82:377–407. doi: 10.1016/S0091-679X(06)82013-3. [DOI] [PubMed] [Google Scholar]
- Kong X, Mohanty SK, Stephens J, Heale JT, Gomez-Godinez V, Shi LZ, Kim JS, Yokomori K, Berns MW. Comparative analysis of different laser systems to study cellular responses to DNA damage in mammalian cells. Nucleic Acids Res. 2009;37:e68. doi: 10.1093/nar/gkp221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski D, Natale DA, Eddy MJ. Stable DNA unwinding, not ‘breathing,’ accounts for single-strand-specific nuclease hypersensitivity of specific A+T-rich sequences. Proc Natl Acad Sci. 1988;85:9464–9468. doi: 10.1073/pnas.85.24.9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mer G, Bochkarev A, Gupta R, Bochkareva E, Frappier L, Ingles CJ, Edwards AM, Chazin WJ. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell. 2000;103:449–456. doi: 10.1016/s0092-8674(00)00136-7. [DOI] [PubMed] [Google Scholar]
- Muthuswami R, Truman PA, Mesner LD, Hockensmith JW. A eukaryotic SWI2/SNF2 domain, an exquisite detector of double-stranded to single-stranded DNA transition elements. J Biol Chem. 2000;275:7648–7655. doi: 10.1074/jbc.275.11.7648. [DOI] [PubMed] [Google Scholar]
- Nakatani Y, Ogryzko V. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 2003;370:430–444. doi: 10.1016/S0076-6879(03)70037-8. [DOI] [PubMed] [Google Scholar]
- Pyle AM. Translocation and unwinding mechanisms of RNA and DNA helicases. Annu Rev Biophys. 2008;37:317–336. doi: 10.1146/annurev.biophys.37.032807.125908. [DOI] [PubMed] [Google Scholar]
- Wold MS. Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- Yusufzai TM, Kadonaga JT. HARP is an ATP-dependent annealing helicase. Science. 2008;322:748–750. doi: 10.1126/science.1161233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y, Liu Y, Wu X, Shell SM. Functions of human replication protein A (RPA): From DNA replication to DNA damage and stress responses. J Cell Physiol. 2006;208:267–273. doi: 10.1002/jcp.20622. [DOI] [PMC free article] [PubMed] [Google Scholar]