Abstract
Mutations in SMARCAL1 (HARP) cause Schimke immunoosseous dysplasia (SIOD). The mechanistic basis for this disease is unknown. Using functional genomic screens, we identified SMARCAL1 as a genome maintenance protein. Silencing and overexpression of SMARCAL1 leads to activation of the DNA damage response during S phase in the absence of any genotoxic agent. SMARCAL1 contains a Replication protein A (RPA)-binding motif similar to that found in the replication stress response protein TIPIN (Timeless-Interacting Protein), which is both necessary and sufficient to target SMARCAL1 to stalled replication forks. RPA binding is critical for the cellular function of SMARCAL1; however, it is not necessary for the annealing helicase activity of SMARCAL1 in vitro. An SIOD-associated SMARCAL1 mutant fails to prevent replication-associated DNA damage from accumulating in cells in which endogenous SMARCAL1 is silenced. Ataxia-telangiectasia mutated (ATM), ATM and Rad3-related (ATR), and DNA-dependent protein kinase (DNA-PK) phosphorylate SMARCAL1 in response to replication stress. Loss of SMARCAL1 activity causes increased RPA loading onto chromatin and persistent RPA phosphorylation after a transient exposure to replication stress. Furthermore, SMARCAL1-deficient cells are hypersensitive to replication stress agents. Thus, SMARCAL1 is a replication stress response protein, and the pleiotropic phenotypes of SIOD are at least partly due to defects in genome maintenance during DNA replication.
Keywords: SMARCAL1, HARP, replication, DNA damage response, RPA, checkpoint
Genome maintenance requires the coordinated activities of cell cycle, DNA repair, and DNA replication proteins. Defects in any of these activities can cause genome instability and disease, including developmental disorders, premature aging, and cancer predisposition (Hartwell and Kastan 1994; Kastan and Bartek 2004; McKinnon and Caldecott 2007; Barzilai et al. 2008). The ataxia-telangiectasia mutated (ATM), ATM and Rad3-related (ATR), and DNA-dependent protein kinase (DNA-PK) are apical protein kinases activated by DNA damage or replication stress that function to coordinate cell cycle transitions with DNA metabolism, including DNA repair and replication (Abraham 2004; Cimprich and Cortez 2008). These kinases are activated in premalignant lesions presumably due to replication stress caused by activated oncogenes or inactivation of tumor suppressors (Bartkova et al. 2005, 2006; Gorgoulis et al. 2005).
Accurate replication of the genome and continuous surveillance of its integrity are essential for cell survival and the avoidance of diseases such as cancer. The genome is constantly exposed to environmental and endogenous genotoxic insults that challenge DNA replication. The replication stress response is a subset of the DNA damage response that acts during every cell division cycle to deal with these challenges, and promotes the faithful duplication of the genome. The accumulation of ssDNA at stalled replication forks as a consequence of polymerase and helicase uncoupling is an important signal to activate replication stress response pathways, including the ATR checkpoint (Byun et al. 2005). The heterotrimeric ssDNA-binding protein Replication protein A (RPA) is a mediator of multiple protein–protein interactions at stalled replication forks that promote signaling and repair (Binz et al. 2004; Fanning et al. 2006; Cimprich and Cortez 2008).
All nucleic acid metabolism, including replication, happens in the context of chromatin. Chromatin regulation is an integral part of the DNA damage and replication stress responses. Perhaps the best understood example is phosphorylation of histone H2AX by the ATM/ATR kinases (Fillingham et al. 2006). This histone modification regulates the recruitment of both checkpoint and repair proteins to DSBs. In addition, everything from the location of replication origins to the elongation rate of replication forks is influenced by chromatin structure (Tabancay and Forsburg 2006).
The SNF2 family of ATPases acts in the context of chromatin to regulate transcription, replication, repair, and recombination. Sequence analysis has defined 24 SNF2 subfamilies (Flaus et al. 2006). Many of these proteins act as chromatin remodelers to alter DNA–protein interactions. In yeast, the activities of SNF2 family members are important for replication initiation and replication through heterochromatin (Flanagan and Peterson 1999; Papamichos-Chronakis and Peterson 2008; Vincent et al. 2008). They are also directly involved in responding to replication stress. For example, mutations in the yeast Ino80 complex cause hypersensitivity to replication stress agents such as hydroxyurea (HU). Furthermore, this complex accumulates at stalled replication forks and is essential to restart replication (Shimada et al. 2008).
Defects in SNF2 genes cause many human diseases, including Schimke immunoosseous dysplasia (SIOD) due to loss-of-function mutations in the SNF2 protein SMARCAL1 (also called HARP and DNA-dependent ATPase A) (Boerkoel et al. 2002). SIOD is a multisystem disorder characterized by renal failure, growth defects, immune deficiencies, and other complex phenotypes.
The cellular function of the SMARCAL1 protein has not been described. In vitro, SMARCAL1 is a DNA-dependent ATPase that binds and is stimulated by forked DNA structures (Muthuswami et al. 2000). SMARCAL1 has not been shown to remodel chromatin; however, it was demonstrated recently to be an ATP-dependent annealing helicase in vitro (Yusufzai and Kadonaga 2008). SMARCAL1 reanneals ssDNA bubbles in plasmid DNA that were stabilized by RPA. The cellular context for where and when this activity might be important has not been described.
We identified SMARCAL1 in two ongoing functional genetic screens to identify novel genome maintenance activities in human cells. These screens used markers of activated DNA damage response pathways to find genes whose deregulation by either RNAi or overexpression cause spontaneous DNA damage in the absence of any genotoxic agents. SMARCAL1 was identified in both screens. Subsequent experiments have defined SMARCAL1 as a replication stress response protein that acts to maintain genome integrity at stalled replication forks.
Results
Changes in SMARCAL1 expression cause activation of the DNA damage response
We identified SMARCAL1 in two ongoing functional genomic screens using RNAi and cDNA overexpression to identify novel genome maintenance activities in human cells. The complete results of these screens will be presented elsewhere. In the RNAi screen, cells were transfected with one RNAi molecule per well in 96-well plates. Initially, DNA damage response activation resulting from RNAi-mediated gene silencing in HeLa cells was assayed by immunofluorescence staining using a phospho-peptide-specific antibody to the ATM substrate KAP1 (KRAB domain-associated protein 1). The damage-induced phosphorylation of this transcriptional corepressor is necessary for ATM-mediated chromatin relaxation following DNA double-strand break (DSB) formation (Ziv et al. 2006). To confirm the DNA damage response activation phenotype and ensure it was neither cell type-specific nor marker-specific, four individual siRNA oligonucleotides for each candidate gene were transfected into a second cell type (U2OS), and DNA damage response activation was monitored by examining H2AX S139 phosphorylation (γH2AX). The ATM/ATR family of kinases phosphorylate γH2AX at sites of DSBs resulting in the formation of discrete, nuclear foci (Fernandez-Capetillo et al. 2004; Stucki and Jackson 2006).
A cDNA overexpression screen was completed using a similar methodology using cDNAs expressed from a CMV promoter. In this case, small pools of cDNA expression vectors were cotransfected with a GFP vector to mark successfully transfected cells and DDR activation was monitored with antibodies to phosphorylated KAP1. Positive pools were then deconvoluted and tested in both HeLa and U2OS cells using both the KAP1 and γH2AX markers.
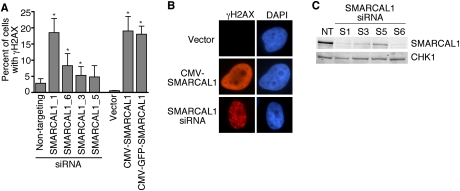
SMARCAL1 was identified in both the RNAi and cDNA overexpression screens. In the RNAi screen, three of four siRNAs targeting SMARCAL1 caused substantial activation of the DNA damage response in the absence of any added genotoxic agents (Fig. 1A,B). We confirmed the siRNAs successfully silenced SMARCAL1 expression, and the amount of H2AX phosphorylation correlated with the level of knockdown (Fig. 1C).
Figure 1.
Deregulation of SMARCAL1 expression causes activation of the DNA damage response. (A) U2OS cells were transfected with siRNA targeting SMARCAL1 or expression vectors encoding either SMARCAL1 or GFP-SMARCAL1. Three days after transfection, cells were stained with antibodies to γH2AX and appropriate secondary antibodies. The percentage of cells with γH2AX staining was scored. Error bars are standard deviation (n ≥ 3). (*) P < 0.05. (B) Representative images of γH2AX staining. (C) Immunoblots of U2OS cell lysates with antibodies to SMARCAL1 or CHK1 after transfection with the indicated SMARCAL1 siRNAs. (NT) Nontargeting.
SMARCAL1 silencing and overexpression cause distinct γH2AX phenotypes. γH2AX localizes to foci in SMARCAL1-silenced cells but exhibits pan-nuclear staining in cells overexpressing SMARCAL1 (Fig. 1B). γH2AX foci are thought to represent sites of DSBs, and the foci induced in SMARCAL1-silenced cells did colocalize with other markers of DSBs including Rad51 (data not shown). The cause of the pan-nuclear γH2AX staining is unclear, but we also observed this staining pattern in a large percentage of cells transfected with RNAi to silence the ssDNA-binding protein RPA. However, we were unable to detect a significant increase in Rad51 foci formation or evidence of DSBs using Comet assays in the overexpression context, raising the possibility that the DNA damage response activation is not associated with DSBs in this case. Neither of the staining patterns was associated with apoptotic nuclei, and both patterns were due to chromatin-associated γH2AX (data not shown). Neither overexpression nor siRNA silencing of SMARCAL1 caused a discernable change in cell proliferation rates, but we did observe a slight increase in the percentage of cells in the G2/M phase of the cell cycle (Supplemental Fig. 1).
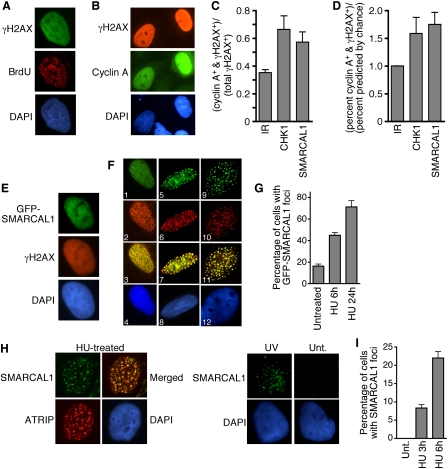
Approximately 20% of the SMARCAL1-overexpressing cells exhibited the pan-nuclear γH2AX staining pattern. These cells were exclusively in S phase since they incorporated BrdU and were cyclin A-positive (Figs. 2A,B). RNAi silencing of SMARCAL1 also caused DNA damage predominantly in replicating cells. Costaining SMARCAL1-silenced cells with antibodies to both γH2AX and cyclin A demonstrated that 60% of the γH2AX+ cells stained strongly for cyclin A (Fig. 2C). This is 1.75-fold higher than predicted by chance and similar to what is observed when the replication checkpoint protein CHK1 is silenced (Fig. 2D). In contrast, there is no preference for γH2AX staining in any phase of the cell cycle after ionizing irradiation, which causes DNA breaks irrespective of cell cycle position. We also did not observe DNA damage response activation in G0-arrested, SMARCAL1-depleted cells (data not shown). Thus, the DNA damage response activation caused by both overexpression or silencing of SMARCAL1 is associated with cells undergoing DNA replication.
Figure 2.
SMARCAL1 functions at stalled replication forks. (A,B) U2OS cells were transfected with a SMARCAL1 expression vector then fixed and stained with the indicated antibodies and appropriate fluorophore-conjugated secondary antibodies. In A, the cells were pulsed with BrdU for 20 min prior to staining. (C,D) U2OS cells transfected with siRNA targeting CHK1 or SMARCAL1 were stained with antibodies to γH2AX and cyclin A. Cells were scored as positive for one or both proteins by immunofluorescence imaging. Nontransfected cells were also irradiated with 5 Gy of IR for comparison. (E–G) U2OS cells were transfected with a GFP-SMARCAL1 expression vector, then fixed and stained. Staining in F is as follows: (Panels 1–4) SMARCAL1 (green) and BrdU (red); (panels 5–8) SMARCAL1 (green) and RPA (red); (panels 9–12) SMARCAL1 (green) and PCNA (red). Panels 3, 7, and 11 are merged images, and panels 4, 8, and 12 were stained with DAPI. In G, the cells containing SMARCAL1 localized to foci were scored after addition of 2 mM HU. (H) Localization of endogenous SMARCAL1 was examined by indirect immunofluorescence with affinity-purified anti-SMARCAL1 antibody in HU or UV radiation-treated U2OS cells. These cells also stably express HA-ATRIP, which permitted analysis of colocalization using anti-HA monoclonal antibody. Specificity of the SMARCAL1 antibody was confirmed in SMARCAL1-silenced cells. (Unt) Untreated. Cells were extracted with Triton X-100 prior to staining, so only chromatin-bound SMARCAL1 is observed. (I) The percentage of cells containing endogenous SMARCAL1 in foci was scored in cells treated with HU for the indicated times. Error bars in all graphs are standard deviation (n = 3).
SMARCAL1 localizes to stalled replication forks via an interaction with RPA34
Adding an N-terminal green fluorescent protein (GFP) tag to SMARCAL1 did not alter the γH2AX phenotype caused by overexpression (Fig. 1A), but did allow us to correlate the γH2AX with both SMARCAL1 expression levels and localization. GFP-SMARCAL1 accumulated in nuclear foci in 20% of the GFP-SMARCAL1-overexpressing cells (Fig. 2E). The GFP-SMARCAL1 overexpression foci correlated perfectly with the pan-nuclear γH2AX phenotype. Cells in which GFP-SMARCAL1 did not accumulate into foci did not have detectable γH2AX staining.
The focal localization pattern and its restriction to S-phase cells suggested SMARCAL1 might be localized to replication factories. Indeed, labeling replication factories with BrdU, RPA, or PCNA demonstrated colocalization of SMARCAL1 foci with sites of DNA replication (Fig. 2F). Treating cells with HU induced a marked increase in the percentage of cells containing SMARCAL1 foci (Fig. 2G). Importantly, we also found that endogenous SMARCAL1 localizes to stalled replication forks (marked by the ATR-interacting protein ATRIP) following treatment with HU or ultraviolet (UV) radiation (Fig. 2H). The percentage of cells containing SMARCAL1 foci increased with the time of HU exposure (Fig. 2I). Endogenous SMARCAL1 foci were rarely observed in undamaged cells. These localization data place SMARCAL1 at sites of replication stress. We also observed SMARCAL1 foci in response to ionizing radiation (IR) treatment. These foci appeared several hours after irradiation in only the S/G2-phase cells that contain cyclin A (Supplemental Fig. 2). This pattern is consistent with a requirement for end-resection at a DSB.
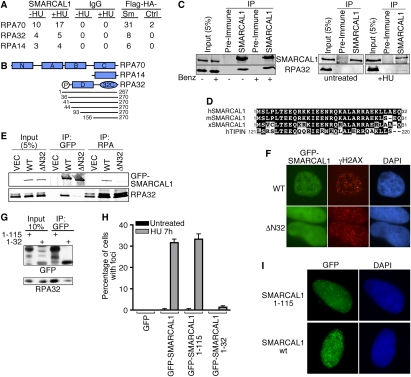
To identify SMARCAL1-interacting proteins, we performed both immunopurifications combined with mass spectrometry and a two-hybrid screen. Immunopurifying endogenous SMARCAL1 complexes with antibodies to the C terminus of SMARCAL1 or using a tandem epitope approach combined with mass spectrometry identified peptides from all three subunits of the ssDNA-binding protein RPA (Fig. 3A). The two-hybrid screen using a full-length SMARCAL1 protein bait identified 10 interacting fragments of the 32-kDa subunit of RPA. The minimal interacting region contained RPA32 amino acids 156–267, corresponding to the winged-helix protein interaction domain (32C) (Fig. 3B). This region of RPA32 binds to several DNA repair proteins, including UNG2, XPA, and RAD52 (Mer et al. 2000). Coimmunoprecipitation of endogenous proteins confirmed the SMARCAL1–RPA interaction, and indicated it is stimulated by replication stress (Fig. 3C).
Figure 3.
An interaction with RPA is necessary and sufficient to localize SMARCAL1 to stalled replication forks. (A) Endogenous SMARCAL1 or Flag-HA-SMARCAL1 was immunopurified from nuclear cell extracts and the resulting protein complexes were analyzed by mass spectrometry. The table indicates the number of peptides identified for each RPA protein subunit. (IgG) Control immunoprecipitation; (Sm) =Flag-HA-SMARCAL1; (Ctrl) Flag-HA empty vector cells. Where indicated, the cells were treated with 1 mM HU for 16 h prior to the purification. (B) A schematic diagram of RPA subunits. The lines below RPA32 indicate the sizes (with amino acid numbers) of the different RPA32 protein fragments identified in the unbiased two-hybrid screen using full-length SMARCAL1 as a bait. (C) HeLa cell nuclear extracts from untreated cells or cells treated with HU for 8 h were used for immunoprecipitation with anti-SMARCAL1 or preimmune antibodies. In the left panel, the lysates were treated with benzonase nuclease (Benz.) as indicated to ensure the interaction is not dependent on DNA. Immunoprecipitated proteins were separated by SDS-PAGE and immunoblotted with RPA or SMARCAL1 antibodies. (D) Sequence alignment of human, mouse, and Xenopus SMARCAL1 with human TIPIN. (E) HEK293 cells were transfected with wild-type GFP-SMARCAL1 (WT) or GFP-SMARCAL1 lacking the first 32 amino acids (Δ32). Cells transfected with an empty vector (VEC) were also prepared as a control. Anti-GFP or anti-RPA immunoprecipitated proteins were separated by SDS-PAGE and blotted with anti-GFP or anti-RPA2 antibodies. (F) U2OS cells were transfected with GFP-SMARCAL1-WT or GFP-SMARCAL1-Δ32 vectors and treated with HU for 6 h. Approximately 30% of the wild-type SMARCAL1-expressing cells had SMARCAL1 foci, while we never observed the SMARCAL1-Δ32 protein in foci. Representative images are shown. (G) GFP-SMARCAL1 containing only the first 32 or 115 amino acids of SMARCAL1 was transfected into cells, immunoprecipitated, and immunoblotted as in E. (H) The indicated expression vectors (pLL5.0-GFP-GW backbone for attenuated expression levels) were transfected into HeLa cells. The cells were treated for 7 h with 1 mM HU or left untreated and the percentage of cells containing foci of the indicated proteins was scored. Error bars are standard deviation (n = 3). (I) Representative images of the localization of SMARCAL1-1-115 and wild-type SMARCAL1.
While a C-terminal antibody to SMARCAL1 efficiently coimmunoprecipitates all three RPA subunits (Figs. 3A,C), an antibody raised to the N terminus of SMARCAL1 fails to coimmunoprecipitate RPA (data not shown), suggesting it may compete for the same binding surface as RPA. Sequence analysis of this SMARCAL1 region identified a highly evolutionarily conserved sequence (amino acids 1–32) that has similarity to the Timeless-Interacting Protein (TIPIN) (Fig. 3D). Notably, this region of TIPIN was demonstrated previously to bind RPA32 (Unsal-Kacmaz et al. 2007), suggesting it might be the RPA32-interacting surface on SMARCAL1. Indeed, deletion of the first 32 amino acids of SMARCAL1 severely impairs its ability to interact with RPA (Fig. 3E). Furthermore, this SMARCAL1-ΔN32 protein fails to localize to intranuclear foci in either the absence or presence of HU (Fig. 3F).
To determine whether this N-terminal region of SMARCAL1 is also sufficient to bind RPA32 and localize SMARCAL1 to stalled forks, we fused it to GFP. A GFP protein containing only the first 32 amino acids of SMARCAL1 (GFP-SMARCAL1 1–32) bound poorly to RPA and localized to foci in only a small percentage of HU-treated cells (Fig. 3G,H). However, fusion of a slightly longer portion of the SMARCAL1 N terminus (amino acids 1–115) to GFP created a protein that binds to RPA and localizes to stalled replication forks as efficiently as full-length SMARCAL1 (Fig. 3G–I). In these experiments, we used a GFP expression vector (pLL5.0-GFP-GW) with attenuated expression levels to avoid spontaneously activating the DNA damage response in the absence of HU that is observed with wild-type GFP-SMARCAL1 expressed at higher levels. Thus, the N terminus of SMARCAL1 encodes a binding surface for RPA32 that is both necessary and sufficient to localize SMARCAL1 to stalled replication forks.
An SIOD-associated patient mutation and an RPA-binding mutant are both defective in the cellular functions of SMARCAL1
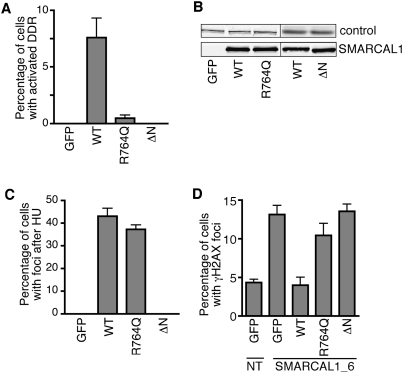
To determine whether the spontaneous DNA damage response activation observed after high levels of SMARCAL1 overexpression is due to too much enzymatic activity, we examined whether an SIOD-associated patient mutation (R764Q) in SMARCAL1 that perturbs its ATPase and annealing helicase activities alters the ability of overexpressed SMARCAL1 to cause DNA damage. We also tested the ability of SMARCAL1 lacking its RPA-binding domain to cause DNA damage response activation to determine whether localization to replication factories and/or RPA binding is essential. GFP-SMARCAL1-R764Q is deficient in activating the DNA damage response when overexpressed in cells compared with wild-type SMARCAL1 (Fig. 4A). Similarly, the GFP-SMARCAL1-Δ32 protein completely fails to activate the DNA damage response, although it is expressed at equal levels to wild-type SMARCAL1 (Fig. 4A,B). The SIOD mutant SMARCAL1 protein can be recruited to stalled replication forks as efficiently as wild-type SMARCAL1 in HU-treated cells (Fig. 4C). In contrast, as noted previously, the SMARCAL1-Δ32 protein does not localize to foci. These data indicate that both the RPA-binding and enzymatic activities of SMARCAL1 are necessary to cause replication-associated DNA damage response activation. Overexpression of SMARCAL1 likely deregulates its activity, perhaps allowing aberrant access to replication forks and causing problems during DNA replication.
Figure 4.
RPA binding and annealing helicase activities of SMARCAL1 are required for its cellular functions. (A) HeLa cells were transfected with expression vectors (pLEGFP backbone for high levels of expression) encoding GFP-tagged wild-type SMARCAL1 (WT), the annealing helicase-deficient SIOD patient mutant protein R764Q, or SMARCAL1-Δ32. The amount of DNA damage response activation was scored by monitoring KAP1 phosphorylation. (B) Expression levels of the GFP-SMARCAL1 proteins were determined by immunoblotting. The loading control was either ATM or RPA32. (C) The percentage of HU-treated cells in which GFP-SMARCAL1 wild-type or mutant proteins localized to intranuclear foci was scored. Cells were treated with HU for 8 h. (D) U2OS cells were cotransfected with the indicated pLL5.0-GFP-GW plasmids that encode siRNA-resistant SMARCAL1 cDNAs and siRNA targeting SMARCAL1. GFP-positive cells were scored for γH2AX staining. (GFP) GFP empty vector; (NT) nontargeting siRNA control. Error bars in all panels are standard deviation (n = 6).
We next asked whether the SIOD patient mutation or the RPA-binding mutation would impair the function of endogenous SMARCAL1 in cells. Specifically, we tested the ability of wild-type and mutant SMARCAL1 proteins to functionally complement the spontaneous appearance of γH2AX foci in SMARCAL1-silenced cells. U2OS cells cotransfected with siRNA-resistant pLL5.0-GFP-GW-SMARCAL1 cDNAs and either nontargeting siRNA as a control or SMARCAL1 siRNA were stained with antibodies to γH2AX. GFP-positive cells were scored for γH2AX foci. As expected, transfection of the GFP empty vector by itself did not prevent the appearance of γH2AX foci in SMARCAL1-silenced cells (Fig. 4D). Expression of wild-type GFP-SMARCAL1 completely suppressed this defect. However, neither expression of the R764Q SIOD mutant nor the ΔN RPA-binding mutant SMARCAL1 protein was capable of preventing the appearance of γH2AX foci when endogenous SMARCAL1 was silenced (Fig. 4D). Thus, RPA binding is essential for the function of SMARCAL1 in vivo. These data also link the SIOD phenotype caused by the R764Q mutation directly to a genome maintenance defect.
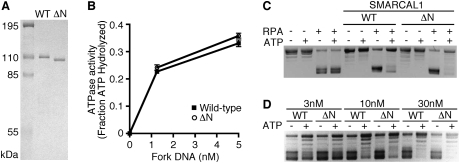
The ability of SMARCAL1 to bind to RPA raises the question of whether this binding serves only to regulate the localization of SMARCAL1 or if it also is important for the annealing helicase activity described previously for SMARCAL1 (Yusufzai and Kadonaga 2008). The annealing helicase assay uses the ability of RPA to bind to supercoiled plasmid DNA and form bubbles by melting the DNA strands. When SMARCAL1 is added, the DNA strands are reannealed in an ATP-dependent reaction, thereby displacing RPA. We tested SMARCAL1-Δ32 to determine if this RPA-binding mutant maintains its ATPase and annealing helicase activities. We found no difference between wild-type and mutant SMARCAL1 in either assay (Fig. 5). Fork DNA stimulates the ATPase activity of SMARCAL1-Δ32 equivalently to wild-type SMARCAL1 (Fig. 5B). Furthermore, it acts as an annealing helicase as efficiently as wild-type SMARCAL1 (Fig. 5C,D). The small differences observed at some concentrations were not reproducible. Thus, RPA binding serves primarily to regulate the localization of SMARCAL1, which is critical for SMARCAL1 function in cells but is not essential for its enzymatic activity.
Figure 5.
The SMARCAL1 RPA32-binding domain is not necessary for ATPase or annealing helicase activities in vitro. (A) Coomassie-stained gel of the Flag-SMARCAL1 and Flag-SMARCAL1-Δ32 purified from baculovirus-infected insect cells. (B) ATPase activity of the wild-type and Δ32 SMARCAL1 proteins was measured in the presence of the indicated concentrations of fork DNA. Error bars are standard deviation (n = 3). (C,D) Annealing helicase activities of wild-type and Δ32 SMARCAL1 proteins. The concentration of SMARCAL1 protein in C is 300 nM.
SMARCAL1 is regulated by the DNA damage response
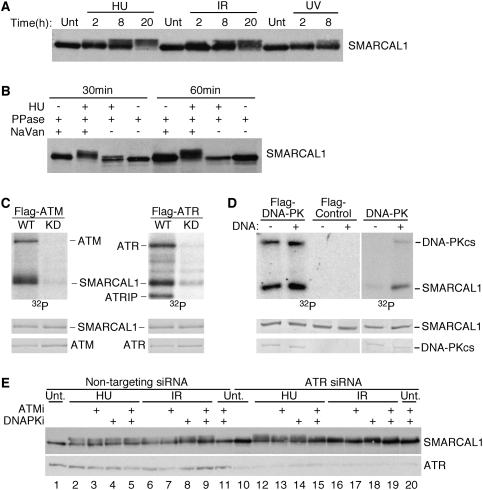
In the course of studying SMARCAL1, we noticed that it often migrates as multiple bands on SDS-PAGE gels. Closer examination indicated that SMARCAL1 exhibits a phosphorylation-dependent gel mobility shift when cells are treated with replication stress and DNA-damaging agents including HU, IR, and UV radiation (Fig. 6A,B).
Figure 6.
ATM, ATR, and DNA-PK phosphorylate SMARCAL1. (A) Cell lysates from cells treated with 1 mM HU, 10 Gy of IR, or 50 J/m2 of UV radiation were separated by SDS-PAGE and immunoblotted. The time points represent the duration of the HU treatment or the length of time the cells were allowed to recover following the radiation exposure. (Unt) Untreated. (B) Cell lysates from untreated or HU-treated cells were incubated in the presence of λ phosphatase and the phosphatase inhibitor sodium vanadate as indicated for 30 or 60 min prior to separation by SDS-PAGE and immunoblotting. (C,D) Flag-ATM or Flag-ATR wild-type (WT) or kinase-dead (KD) proteins or wild-type Flag-DNA-PKcs were immunopurified from transfected cells. (Right panel) Wild-type DNA-PK was also purchased from Promega. The kinases were incubated with purified SMARCAL1 in the presence of γ-32P-ATP. Sonicated salmon sperm DNA was added to the DNA-PK reactions as indicated. Reactions were separated by SDS-PAGE and subjected to autoradiography (32P). The levels of SMARCAL1, ATM, ATR, and DNA-PKcs proteins in each reaction were visualized by Coomassie staining. (E) 293T cells were transfected with either nontargeting (NT) or ATR siRNA. Cells were also treated with specific ATM (KU55933, 10 μM) (Hickson et al. 2004) or DNA-PK (KU57788, 1 μM) (Leahy et al. 2004) inhibitors as indicated and exposed to 1 mM HU or 10 Gy of IR. Cell lysates were separated by SDS-PAGE and immunoblotted for SMARCAL1 or ATR.
SMARCAL1 contains several consensus motifs (SQ/TQ) for phosphorylation by the DNA damage and replication stress-activated ATM/ATR family of kinases. To determine whether these kinases can phosphorylate SMARCAL1, we used in vitro kinase assays. Purified wild-type ATM and ATR but not kinase-dead proteins efficiently phosphorylated SMARCAL1 in vitro (Fig. 6C). Purified DNA-PK also phosphorylated SMARCAL1 in a DNA-stimulated reaction (Fig. 6D).
To determine which of these kinases may be responsible for the phosphorylation of SMARCAL1 in cells, each was depleted using siRNA. However, we failed to detect any difference in the HU-dependent phosphorylation of SMARCAL1 when individual kinases were silenced (Supplemental Fig. 3A; data not shown). We then examined combinations of specific kinase inhibitors and RNAi. Specific ATM and DNA-PK inhibitors had no appreciable effect on SMARCAL1 phosphorylation after HU even when used together (Fig. 6E, cf. lanes 2 and 5). The inhibitors also had only a minimal effect after IR (Fig. 6E, cf. lanes 6 and 9). Silencing ATR by itself also had minimal effects after HU treatment, but it did reduce phosphorylation after IR (Fig. 6E, cf. lanes 6 and 16). Silencing ATR and treating cells with the specific ATM and DNA-PK inhibitors significantly reduced the phosphorylation of SMARCAL1 after both HU and IR treatment (Fig. 6E, cf. lanes 5 and 15, lanes 9 and 19). The residual phosphorylation of SMARCAL1 under these circumstances, in which all three kinases are inhibited, may be due to incomplete silencing of ATR, inhibition of ATM or DNA-PK, or, potentially, the involvement of additional kinases.
This type of phosphorylation pattern is common to many substrates of the ATM/ATR family of kinases—such as RPA32—that are phosphorylated by multiple family members. SMARCAL1 is likely phosphorylated on several sites since the banding pattern on SDS-PAGE gels is complex. To identify sites of phosphorylation, we immunopurified SMARCAL1 from HU-treated cells and subjected it to mass spectrometric analysis. Several phosphorylation sites were observed including S173, which lies in a good consensus sequence for the ATM/ATR family (SQE) (Supplemental Fig. 3B). Further analysis will be required to identify all of the damage-dependent phosphorylation events and their functional significance. However, our data indicate that SMARCAL1 functions downstream from the ATM/ATR/DNA-PK kinases in response to replication stress.
Loss of Smarcal1 function causes persistent RPA phosphorylation, RPA loading onto chromatin, and hypersensitivity to replication stress agents
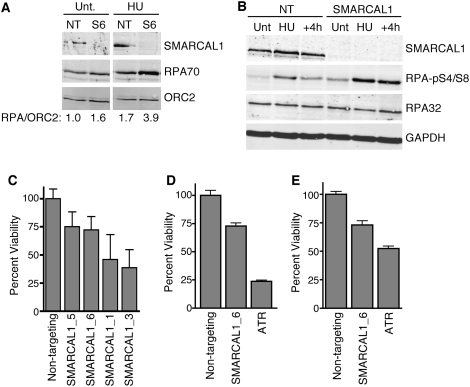
The localization of SMARCAL1 to stalled replication forks, appearance of γH2AX foci in S phase of SMARCAL1-silenced cells, phosphorylation by the ATM/ATR/DNA-PK kinases, and its ability to act as an annealing helicase in vitro suggest that SMARCAL1 may have a critical function to anneal DNA strands at stalled replication forks. If this were true, we would expect that SMARCAL1-deficient cells should accumulate more RPA bound to ssDNA than control cells. Consistent with this hypothesis, more RPA is loaded onto chromatin in SMARCAL1-deficient cells than control cells, and this difference is even more pronounced after exposure to HU (Fig. 7A). SMARCAL1 is also loaded onto chromatin under these conditions. RPA loaded at stalled replication forks is rapidly phosphorylated by the ATM/ATR family of kinases (Binz et al. 2004). SMARCAL1-silenced cells treated with HU exhibited hyperphosphorylation of RPA that also persisted after the HU was removed (Fig. 7B). These data are consistent with a model in which the annealing helicase activity of SMARCAL1 functions to reduce the amount of ssDNA available for RPA binding at stalled forks.
Figure 7.
SMARCAL1-deficient cells exhibit increased RPA-loading onto chromatin and persistent RPA phosphorylation after replication stress, and are hypersensitive to replication stress agents. (A) The chromatin fraction of cells transfected with nontargeting (NT) or SMARCAL1 (S6) siRNA was isolated and immunoblotted with the indicated antibodies. Quantitation of the amount of RPA versus ORC2 in the chromatin fraction was performed with an Odyssey instrument (arbitrary units). (B) Cells transfected with nontargeting or SMARCAL1 siRNA were treated with HU for 5 h (HU) then the HU was removed and the cells were allowed to recover for 4 h (+4h). Total cell lysates were prepared and immunoblotted with the indicated antibodies. (Unt) Untreated. (C) Cells transfected with the indicated siRNA were split into four plates, two of which were treated with 2 mM HU for 24 h. The HU was removed and replaced with standard growth media for an additional 24 h before cell viability was quantitated using the WST-1 cell proliferation reagent (Roche). Reported values are the mean and standard deviation of three experiments. All four SMARCAL1 siRNAs yielded significant hypersensitivity to HU with P values ranging from 0.02 to 10−4. (D,E) Cells transfected with the indicated siRNAs were treated with 5 μM aphidicolin or 150 nM camptothecin and cell viability compared with untreated controls. The P value of nontargeting compared with SMARCAL1-silenced for both aphidicolin (n = 12) and camptothecin (n = 24) treatments are <10−5.
If SMARCAL1 activity is important for proper nucleic acid metabolism at sites of replication stress, then cells lacking SMARCAL1 should be hypersensitive to replication stress agents. Indeed, silencing SMARCAL1 causes a marked hypersensitivity to HU, the DNA polymerase inhibitor aphidicolin, and the topoisomerase I inhibitor camptothecin (Fig. 7C,D). Thus, SMARCAL1 activity is important to maintain viability when cells are challenged with drugs that induce uncoupling of helicase and polymerases at replication forks.
Discussion
The SIOD disease is caused by loss-of-function mutations in SMARCAL1 (Boerkoel et al. 2002). This disease has a complex phenotype affecting multiple organs and tissues. There is also variability in phenotypic severity among patients. These characteristics have made it difficult to propose a unifying model for the molecular functions of SMARCAL1 that would explain the disease. Our mechanistic analysis of SMARCAL1 suggests at least part of the underlying molecular cause of SIOD is a defect in cellular responses to replication stress.
SMARCAL1 is a member of the SNF2 family of ATPases that function in a variety of chromatin-associated processes, including transcription, replication, and DNA repair (Flaus et al. 2006). These proteins often alter DNA–protein interactions as a consequence of hydrolyzing ATP. A single SMARCAL1 protein is found in humans, and it is evolutionarily conserved in invertebrates, including Caenorhabditis elegans and Drosophila melanogaster. However, yeast orthologs have not been identified.
A major advance in understanding SMARCAL1 activity was made with the identification of its annealing helicase activity in vitro (Yusufzai and Kadonaga 2008). While many helicases can act to reverse a fork under some in vitro conditions, SMARCAL1 is the only protein identified thus far that acts primarily to anneal two DNA strands without exhibiting any helicase activity. The context of where and when this activity is important in the cell has not been defined.
We defined SMARCAL1 as a replication stress response protein that acts at stalled replication forks to limit replication-associated DNA damage. SMARCAL1 contains an RPA32-interacting domain at its N terminus similar to a domain in the replication stress response protein TIPIN (Unsal-Kacmaz et al. 2007). This domain is both necessary and sufficient to localize SMARCAL1 to stalled forks. RPA binding is required for the genome maintenance activity of SMARCAL1 during DNA replication, but it is not required for its annealing helicase activity in vitro.
Like many replication stress response proteins, SMARCAL1 is phosphorylated by the ATM/ATR/DNA-PK family of protein kinases. SMARCAL1 contains multiple consensus sequences for phosphorylation, and its complex gel mobility pattern indicates that several may be phosphorylated. Our mass spectrometry analysis indicates one of these is S173. The ATM/ATR/DNA-PK kinases have somewhat overlapping and redundant activities on SMARCAL1 in response to replication stress, since all three kinases must be inhibited to observe a significant reduction in phosphorylation after HU. This is not unusual for substrates of these kinases, since HU can activate all three kinases and there is significant cross-talk between them.
Both silencing and overexpression of SMARCAL1 causes activation of the DNA damage response kinases in replicating cells. Activation of the DNA damage response upon overexpression of SMARCAL1 is strictly limited to S-phase cells. The pattern of γH2AX staining in these SMARCAL1-overexpressing cells is unusual in that it is pan-nuclear and never localized in discrete foci. This pattern has been noted previously in cells in which ATR is activated in the absence of DNA damage (Kumagai et al. 2006; Ball et al. 2007). However, we found no evidence that SMARCAL1 functions as a direct regulator of ATR activation. We also observed this pan-nuclear pattern when RPA is depleted from cells with RNAi. However, in this circumstance, a significant portion of cells in the population also exhibit focal γH2AX staining. As yet, the molecular basis of the SMARCAL1 overexpression-associated pan-nuclear γH2AX staining is unclear, but our data do indicate that it is dependent on both the ATPase- and RPA-binding activities of SMARCAL1. Multiple proteins bind to the same region of RPA32 as SMARCAL1 (Mer et al. 2000). A competition between interacting proteins is likely to prevent SMARCAL1 from gaining access to the limited amount of RPA at normal elongating forks. However, when SMARCAL1 is overexpressed, it may outcompete other proteins, gain access to normal replication forks, and then use its enzymatic activity to reanneal DNA strands inappropriately. RPA silencing may phenocopy SMARCAL1 overexpression in some cells when RPA levels are insufficient to stabilize the ssDNA at elongating forks.
Silencing SMARCAL1 increases both the amount of RPA bound to chromatin and its phosphorylation following a challenge with HU. RPA phosphorylation also persists after removing HU from the growth media in SMARCAL1-silenced cells. These results are consistent with the idea that SMARCAL1 acts as an annealing helicase at stalled forks to limit the amount of ssDNA that accumulates. Since the DNA at a replication fork is not bound into nucleosomes, it may have an increased propensity to form bubbles or flaps that would be substrates for the annealing helicase activity of SMARCAL1 (Supplemental Fig. 4). We envision at least three possible substrates for the annealing helicase activity of SMARCAL1 at a stalled fork. (1) The uncoupled replication fork itself could be a substrate, in which case SMARCAL1 may limit the amount of helicase–polymerase uncoupling. This activity would presumably require dissociation of the MCM helicase complex for SMARCAL1 to obtain access. However, dissociation of the MCMs would be problematic, since they cannot be reloaded during S phase. (2) Single-stranded bubbles may form in the dsDNA at the stalled fork, since it is not stabilized into nucleosome structures. The positive supercoiling ahead of the fork would likely inhibit bubble formation in that location. However, the precatanenes behind the fork may not prevent ssDNA bubbles from forming, especially in genomic sequences that are AT-rich or prone to secondary structure formation. (3) SMARCAL1 may anneal DNA flaps that could form when replicative polymerases dissociate from the DNA. Flaps could form on either the leading strand or lagging strand. They could also form at the 5′ end of an Okazaki fragment. If SMARCAL1 prevents flap formation, then it may act to inhibit a process of fork reversal (Michel et al. 2004; Lambert et al. 2007) that is energetically favorable but thought to be inhibited by the tethering of the nascent strand ends to their parental templates by the replisome during normal replication (Postow et al. 2001). Finally, it is possible that SMARCAL1 acts at replication origins to limit replication origin firing. Of course, these potential activities are not mutually exclusive.
SMARCAL1 deficiency causes the appearance of DSB markers at replication factories, increased RPA loading and phosphorylation in response to replication stress, and decreased viability after treatment of cells with agents that stall replication forks. Cells expressing only an SIOD-associated mutant SMARCAL1 protein accumulate γH2AX foci even in the absence of a replication stress agent. While it is difficult to link these effects directly to the symptoms in SIOD patients, these data suggest that loss of SMARCAL1 function in patients may cause DNA replication-associated genome instability that contributes to the pleiotropic phenotypes of this disease.
Materials and methods
Cell culture
Cells were cultured in DMEM + FBS. Plasmid transfections were performed by Lipofectamine 2000 (Invitrogen) or Fugene (Roche). siRNA transfections were performed with HiPerfect (Qiagen) using siRNAs purchased from Qiagen or Dharmacon.
Vectors
The details of all vector constructs are available on request. pLEGFP was used for high levels of GFP-SMARCAL1 expression sufficient to cause DNA damage. pLL5.0-GFP-GW was used for attenuated expression levels that did not induce DNA damage.
Antibodies and immunofluorescence
The antibodies used were as follows: KAP1 pS824 and SMARCAL1 (Bethyl Laboratories), γH2AX (Upstate Biotechnologies), HA (Covance), PCNA (Santa Cruz Biotechnologies), cyclin A and RPA (Neomarkers), and BrdU (Becton Dickenson). Additionally, custom SMARCAL1 antibodies targeting the N or C terminus of SMARCAL1 were produced by Covance and Open Biosystems. Immunofluorescent images were obtained with a Zeiss Axioplan microscope equipped with a Zeiss camera. The percent of cells staining with both cyclin A and γH2AX predicted by chance was calculated as follows: (percent of cyclin A+ and γH2AX+ cells) ÷ (percent cyclin A+ cells × percent γH2AX+ cells).
Protein interactions
The two-hybrid screen was performed by Hybrigenics using their ULTImate Y2H methodology. Immunopurifications of endogenous SMARCAL1 complexes from nuclear extracts was performed with a peptide antibody to the C terminus of SMARCAL1. Associated proteins were identified by tandem mass spectrometry by the Vanderbilt Mass Spectrometry Center. Flag-HA dual epitope purifications were performed according to published protocols (Nakatani and Ogryzko 2003) using 293T cells stably expressing Flag-HA-SMARCAL1.
ATPase and annealing helicase assays
Flag-SMARCAL1 and Flag-SMARCAL1-Δ32 were purified from baculovirus-infected insect cells essentially as described previously (Yusufzai and Kadonaga 2008), with the following modifications. Cells were lysed in 20 mM Tris (pH 7.5), 150 mM NaCl, 0.1 mM EDTA, 1 mM DTT 0.2 mM PMSF, 1 μg/mL leupeptin, 1 μg/mL aprotinin, and 0.1% Triton. A final CM-sepharose step was added after elution of protein from the Flag beads. The ATPase assay was performed as described (Yusufzai and Kadonaga 2008). The annealing helicase assay was also performed as described (Yusufzai and Kadonaga 2008), with the following modifications: Topoisomerase I was purchased (Invitrogen) and pBluescript was used as the plasmid substrate.
Chromatin fractionation
Chromatin fractionation was performed as described previously (Mendez and Stillman 2000).
Replication stress agent sensitivity assays
Cells transfected with siRNAs using HiPerfect (Qiagen) were treated with drug for 24 h, washed, and then allowed to recover for 24 h. Cell viability was measured using the WST-1 reagent (Roche). All statistical tests were performed on the log of the ratio of treated/untreated measurements using a two-tailed unpaired t-test.
Acknowledgments
We thank Shawn Levy, David Cappel, Daniel Kaplan, and Chelsea Cupp for technical assistance, and Cornelius Boerkoel, Scott Hiebert, Karlene Cimprich, Christine Eischen, and Brandt Eichman for helpful discussions. We also thank David Friedman of the Vanderbilt Proteomics Laboratory for the mass spectrometry analyses. This work was supported by National Cancer Institute grant R21CA132010, a Susan G. Komen for the Cure grant, the Vanderbilt-Ingram Cancer Center SPORE in Breast Cancer (P50 CA098131), the Robert J. Kleberg Jr. and Helen C. Kleberg Foundation, the Ingram Charitable Fund, and the Vanderbilt Center for Molecular Toxicology (P30 ES000267). C.A.L. was supported by a Department of Defense Breast Cancer Research Program predoctoral fellowship (W81XWH-06-1-0528).
Footnotes
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1839909.
Supplemental material is available at http://www.genesdev.org.
References
- Abraham RT. PI 3-kinase related kinases: ‘Big’ players in stress-induced signaling pathways. DNA Repair (Amst) 2004;3:883–887. doi: 10.1016/j.dnarep.2004.04.002. [DOI] [PubMed] [Google Scholar]
- Ball HL, Ehrhardt MR, Mordes DA, Glick GG, Chazin WJ, Cortez D. Function of a conserved checkpoint recruitment domain in ATRIP proteins. Mol Cell Biol. 2007;27:3367–3377. doi: 10.1128/MCB.02238-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- Barzilai A, Biton S, Shiloh Y. The role of the DNA damage response in neuronal development, organization and maintenance. DNA Repair (Amst) 2008;7:1010–1027. doi: 10.1016/j.dnarep.2008.03.005. [DOI] [PubMed] [Google Scholar]
- Binz SK, Sheehan AM, Wold MS. Replication Protein A phosphorylation and the cellular response to DNA damage. DNA Repair (Amst) 2004;3:1015–1024. doi: 10.1016/j.dnarep.2004.03.028. [DOI] [PubMed] [Google Scholar]
- Boerkoel CF, Takashima H, John J, Yan J, Stankiewicz P, Rosenbarker L, Andre JL, Bogdanovic R, Burguet A, Cockfield S, et al. Mutant chromatin remodeling protein SMARCAL1 causes Schimke immuno-osseous dysplasia. Nat Genet. 2002;30:215–220. doi: 10.1038/ng821. [DOI] [PubMed] [Google Scholar]
- Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes & Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich KA, Cortez D. ATR: An essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Capetillo O, Lee A, Nussenzweig M, Nussenzweig A. H2AX: The histone guardian of the genome. DNA Repair (Amst) 2004;3:959–967. doi: 10.1016/j.dnarep.2004.03.024. [DOI] [PubMed] [Google Scholar]
- Fillingham J, Keogh MC, Krogan NJ. γH2AX and its role in DNA double-strand break repair. Biochem Cell Biol. 2006;84:568–577. doi: 10.1139/o06-072. [DOI] [PubMed] [Google Scholar]
- Flanagan JF, Peterson CL. A role for the yeast SWI/SNF complex in DNA replication. Nucleic Acids Res. 1999;27:2022–2028. doi: 10.1093/nar/27.9.2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Jr, Kastrinakis NG, Levy B, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Kastan MB. Cell cycle control and cancer. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- Hickson I, Zhao Y, Richardson CJ, Green SJ, Martin NM, Orr AI, Reaper PM, Jackson SP, Curtin NJ, Smith GC. Identification and characterization of a novel and specific inhibitor of the ataxia-telangiectasia mutated kinase ATM. Cancer Res. 2004;64:9152–9159. doi: 10.1158/0008-5472.CAN-04-2727. [DOI] [PubMed] [Google Scholar]
- Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR–ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- Lambert S, Froget B, Carr AM. Arrested replication fork processing: Interplay between checkpoints and recombination. DNA Repair (Amst) 2007;6:1042–1061. doi: 10.1016/j.dnarep.2007.02.024. [DOI] [PubMed] [Google Scholar]
- Leahy JJ, Golding BT, Griffin RJ, Hardcastle IR, Richardson C, Rigoreau L, Smith GC. Identification of a highly potent and selective DNA-dependent protein kinase (DNA-PK) inhibitor (NU7441) by screening of chromenone libraries. Bioorg Med Chem Lett. 2004;14:6083–6087. doi: 10.1016/j.bmcl.2004.09.060. [DOI] [PubMed] [Google Scholar]
- McKinnon PJ, Caldecott KW. DNA strand break repair and human genetic disease. Annu Rev Genomics Hum Genet. 2007;8:37–55. doi: 10.1146/annurev.genom.7.080505.115648. [DOI] [PubMed] [Google Scholar]
- Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: Assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mer G, Bochkarev A, Gupta R, Bochkareva E, Frappier L, Ingles CJ, Edwards AM, Chazin WJ. Structural basis for the recognition of DNA repair proteins UNG2, XPA, and RAD52 by replication factor RPA. Cell. 2000;103:449–456. doi: 10.1016/s0092-8674(00)00136-7. [DOI] [PubMed] [Google Scholar]
- Michel B, Grompone G, Flores MJ, Bidnenko V. Multiple pathways process stalled replication forks. Proc Natl Acad Sci. 2004;101:12783–12788. doi: 10.1073/pnas.0401586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthuswami R, Truman PA, Mesner LD, Hockensmith JW. A eukaryotic SWI2/SNF2 domain, an exquisite detector of double-stranded to single-stranded DNA transition elements. J Biol Chem. 2000;275:7648–7655. doi: 10.1074/jbc.275.11.7648. [DOI] [PubMed] [Google Scholar]
- Nakatani Y, Ogryzko V. Immunoaffinity purification of mammalian protein complexes. Methods Enzymol. 2003;370:430–444. doi: 10.1016/S0076-6879(03)70037-8. [DOI] [PubMed] [Google Scholar]
- Papamichos-Chronakis M, Peterson CL. The Ino80 chromatin-remodeling enzyme regulates replisome function and stability. Nat Struct Mol Biol. 2008;15:338–345. doi: 10.1038/nsmb.1413. [DOI] [PubMed] [Google Scholar]
- Postow L, Crisona NJ, Peter BJ, Hardy CD, Cozzarelli NR. Topological challenges to DNA replication: Conformations at the fork. Proc Natl Acad Sci. 2001;98:8219–8226. doi: 10.1073/pnas.111006998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K, Oma Y, Schleker T, Kugou K, Ohta K, Harata M, Gasser SM. Ino80 chromatin remodeling complex promotes recovery of stalled replication forks. Curr Biol. 2008;18:566–575. doi: 10.1016/j.cub.2008.03.049. [DOI] [PubMed] [Google Scholar]
- Stucki M, Jackson SP. γH2AX and MDC1: Anchoring the DNA-damage-response machinery to broken chromosomes. DNA Repair (Amst) 2006;5:534–543. doi: 10.1016/j.dnarep.2006.01.012. [DOI] [PubMed] [Google Scholar]
- Tabancay AP, Jr, Forsburg SL. Eukaryotic DNA replication in a chromatin context. Curr Top Dev Biol. 2006;76:129–184. doi: 10.1016/S0070-2153(06)76005-7. [DOI] [PubMed] [Google Scholar]
- Unsal-Kacmaz K, Chastain PD, Qu PP, Minoo P, Cordeiro-Stone M, Sancar A, Kaufmann WK. The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement. Mol Cell Biol. 2007;27:3131–3142. doi: 10.1128/MCB.02190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent JA, Kwong TJ, Tsukiyama T. ATP-dependent chromatin remodeling shapes the DNA replication landscape. Nat Struct Mol Biol. 2008;15:477–484. doi: 10.1038/nsmb.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusufzai T, Kadonaga JT. HARP is an ATP-driven annealing helicase. Science. 2008;322:748–750. doi: 10.1126/science.1161233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv Y, Bielopolski D, Galanty Y, Lukas C, Taya Y, Schultz DC, Lukas J, Bekker-Jensen S, Bartek J, Shiloh Y. Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat Cell Biol. 2006;8:870–876. doi: 10.1038/ncb1446. [DOI] [PubMed] [Google Scholar]