Abstract
The paths that hematopoietic stem cells take to develop from multipotent, self-renewing cells into committed lymphocytes has been a topic of debate for some time. During early hematopoiesis, multiple branchpoints have been described in which progeny cells segregate into cell lineages with distinct developmental potentials. In this issue of Genes & Development, Inlay and colleagues (pp. 2376–2381) identify novel intermediate stages through which hematopoietic progenitor cells travel.
Keywords: B-cell development, T-cell development, fate decisions, hematopoiesis, lymphopoiesis
Hematopoietic development is initiated from long-term hematopoietic stem cells (LT-HSCs) that are multipotent and have the potential to self-renew (Spangrude et al. 1988). In the adult bone marrow, LT-HSCs differentiate into multipotent progenitor cells (MPPs). The initial view was that MPPs are precursors for the two major lineage-restricted progenitors: a common lymphoid progenitor (CLP) and a common myeloid progenitor (CMP) population (Kondo et al. 1997; Akashi et al. 2000). Progeny derived from CLPs segregate into either the B-cell or the T-cell lineage (Kondo et al. 1997). The CMPs give rise to either the granulocyte/macrophage progenitors (GMP) or megakaryocytic/erythrocyte progenitors (MEPs) (Akashi et al. 2000).
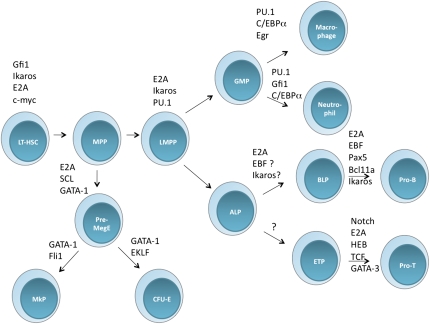
The view of the road map for early hematopoiesis has been modified recently. MPPs can give rise to erythroid and megakaryocytic progeny through various intermediate stages that include the premegakaryocytic/erythroid (Pre-MegE) compartment (Fig. 1; Adolfsson et al. 2005; Pronk et al. 2007). The adaptive and innate immune system arise from a common hub, named the lymphoid-primed MPP compartment (LMPP). Specifically, LMPPs give rise primarily to lymphoid as well as macrophage and granulocytic cell lineages. Multiple intermediate stages have been identified in progeny derived from the LMPPs (Fig. 1). These include the CLPs, GMPs, and early thymic progenitors (ETPs). Overall, both the classical and revised pathways of early hematopoiesis indicate that as cells become specified toward the lymphoid and myeloid cell lineage, they progressively lose their ability to develop into alternative cell lineages. In this issue of Genes & Development, Inlay et al. (2009) identify novel intermediates within the CLP compartment.
Figure 1.
Development of hematopoietic progenitors from the LT-HSC. The diagram shows the road map of early hematopoiesis. The revised map shows the newly defined and characterized ALP and BLP compartments. Transcription factors that act at distinct stages or transition during early hematopoiesis are indicated. (MkP) megakaryocytic progenitor; (CFU-E) colony-forming unit erythroid progenitor. Note that the CLP population is segregated into two distinct stages, named the ALP and the BLP compartments.
Determinants that control hematopoiesis
Numerous studies have revealed the activities of transcription factors during early hematopoiesis, commitment, and maintenance. In vascular endothelial cells, the Runt domain-containing factor Runx1 plays a critical role during the developmental progression from hemogenic endothelial cells to LT-HSCs (Chen et al. 2009). Within the LT-HSC compartment, Ikaros primes a lymphoid-specific program of gene expression at the LT-HSC cell stage (Ng et al. 2009). Gfi-1 and the E2A proteins act at the LT-HSC stage to maintain the LT-HSC pool by restricting cellular proliferation (Hock et al. 2004; Yang et al. 2008; Semerad et al. 2009).
Beyond the LT-HSC compartment, GATA-1 and PU.1 act at the branchpoint that segregates the erythroid/megakaryocytic from the myeloid/lymphoid cell lineages (Fig. 1; Scott et al. 1994; Shivdasani et al. 1997; Laslo et al. 2006). Both factors antagonize each other, and it has been shown that the induction of GATA-1 and PU.1 expression permits the development into either the erythroid or the myeloid cell lineage (Rekhtman et al. 1999). In the LMPP compartment, PU.1 acts with E2A and Ikaros to promote development toward the lymphoid/myeloid pathway (Dias et al. 2008; Ng et al. 2009; Semerad et al. 2009). Since they share a large subset of common target genes, E2A and Ikaros likely act in parallel to induce differentiation along the LMPP trajectory. Among the subset of common E2A and Ikaros targets in the LMPP compartment are IL7Rα, Notch1, Socs2, Socs3, TdT, and CCR9 (Ikawa et al. 2006; Schwartz et al. 2006; Dias et al. 2008; Ng et al. 2009). As mentioned previously, LMPPs have the ability to developmentally progress into GMPs, CLPs, and ETPs. In the GMP compartment, relatively high levels of PU.1 act in concert with C/EBPα to promote a myeloid cell fate (Laslo et al. 2008). In CLPs, PU.1 and E47, and possibly Ikaros, in conjunction with IL7Rα-mediated signaling, activate the expression of EBF (Kee and Murre 1998; Ikawa et al. 2004; Medina et al. 2004; Roessler et al. 2007; Reynaud et al. 2008). Finally, recent data have established that the E2A proteins act, directly or indirectly, to activate the expression of Bcl11a, another key player in early B-lineage maturation (Ikawa et al. 2006). In sum, these observations indicate that PU.1, Ikaros, and E2A act in concert to induce a lymphoid-specific program of gene expression in early hematopoietic progenitors (Fig. 1).
B-lineage specification and commitment
EBF is a critical determinant involved in B-cell commitment. Once activated, EBF induces the expression of Pax-5, which, in turn, in a feedback loop, elevates EBF transcription (Sigvardsson et al. 1997; Kee and Murre 1998; Roessler et al. 2007). EBF and Pax5 then act in concert to suppress a program of T-lineage- and myeloid-specific gene expression, contributing to the commitment process that underpins the development of B-lineage cells (Nutt et al. 1999; Pongubala et al. 2008). EBF and Pax5 expression is maintained at the pro-B-cell stage by the activities of E2A (Lazorchak et al. 2006; Kwon et al. 2008). Additionally, E2A, EBF, and Pax5 act together to induce the expression of a subset of B-lineage-specific genes (Sigvardsson et al. 1997; Kee and Murre 1998; Nutt et al. 1999; Maier et al. 2004). EBF, Pax5, Ikaros, and YY1 also function in pro-B cells, directly or indirectly, to activate Igh V(D)J gene rearrangement (Fuxa et al. 2004; Seet et al. 2004; Liu et al. 2007; Kwon et al. 2008; Reynaud et al. 2008). From these observations, a network of transcriptional regulators has been assembled that provides a framework for B-cell specification and commitment (Laslo et al. 2008).
T-lineage specification and commitment
Distinct progenitors have been described that have the ability to develop toward the T-cell lineage. They potentially include the CLP, CLP2, ETP, and early progenitors with lymphoid and myeloid potential (EPLM) compartments (Kondo et al. 1997; Martin et al. 2003; Wada et al. 2008). It seems likely that these intermediate populations originate in LMPPs. LMPPs are primed to undergo T-lineage specification by the E2A proteins that induce Notch-1 expression (Ikawa et al. 2006; Dias et al. 2008). In the thymus, progeny derived from LMPPs develop into ETPs upon Notch-mediated signaling. Developing thymocytes continue to require the expression of E2A and its heterodimeric partner, HEB (Bain et al. 1997a; Barndt et al. 2000; Ikawa et al. 2006). Specifically, they regulate a subset of genes involved in Notch-mediated signaling, including Notch-1, Hes-1, Hes-5, Grg3, and Grg6 (Ikawa et al. 2006). Forced expression of Notch-1 in E2A-ablated fetal thymocytes overrides the arrest in thymocyte development, providing physiological support for an important role for E2A in modulating the expression of genes encoding components of the Notch-1 ensemble (Ikawa et al. 2006). In addition, the E2A proteins also regulate pTα transcription as well as TCRβ V(D)J locus rearrangement (Agata et al. 2007). Once a pre-TCR has been generated, E2A DNA-binding activity declines to promote developmental progression and cellular expansion, and to ensure allelic exclusion (Engel and Murre 2004; Agata et al. 2007). The decrease in E2A DNA-binding activity during β-selection results in a decline of Notch-1 abundance, consistent with an important role for the E2A proteins in modulating Notch-1 gene expression (Yashiro-Ohtani et al. 2009).
In developing thymocytes, the E2A proteins form heterodimers with HEB (Murre 2005). Two isoforms of HEB, HEBcan and HEBalt, have been described (Wang et al. 2006). The two HEB isoforms are generated by alternative promoter usage. HEBcan contains two transactivation domains, whereas HEBalt lacks the most N-terminal-located transactivation domain (Wang et al. 2006). HEBalt is expressed exclusively in the DN compartment, and its expression is regulated, at least in part, by Notch signaling (Wang et al. 2006). Thus, it is conceivable that E2A and HEBcan in LMPPs act to induce the expression of Notch-1 as well as other components of the Notch signaling cascade. Upon arriving in the thymus, Notch-1-mediated signaling, in conjunction with HEBcan, would then induce HEBalt expression. How HEBalt acts with E2A and HEBcan in DN thymocytes to promote developmental progression is an important question that remains to be addressed. In addition to Notch signaling and E-protein activity, GATA-3, Myb, Runx1-CBFβ, Gfi-1, and TCF1, as well as Ikaros, are essential to promote the developmental progression of T-lineage progenitors toward maturity. From these observations, a regulatory network has been assembled that underpins the commitment and developmental progression of early T-cell progenitors (Georgescu et al. 2008).
A new strategy to identify developmentally regulated genes
As discussed above, there are conflicting data regarding the pathways that hematopoietic progenitors take in order to reach their final destination. For example, in the original model, the CLP compartment was viewed as the branchpoint from which B- and T-lineage cells diverge. However, this view has been challenged (Lai and Kondo 2008; Wada et al. 2008). Other studies have indicated that the CLP compartment is a heterogeneous population containing B-cell-committed cells (Mansson et al. 2008). In an attempt to further characterize the CLP compartment, Inlay et al. (2009) used a novel approach that permits the identification of markers that identify intermediate developmental stages. The computational approach is named “Mining Developmentally Regulated Genes” (MiDReG) (D Sahoo, J Seita, D Bhattacharya, MA Inlay, I Weissman, S Plevritis, and D Dill, in prep.). MiDReG explores the entire set of publicly accessible microarray data to permit the identification of genes whose expression patterns are modulated during developmental progression. It is not essential upon applying MiDReG that microarray data are available for intermediate compartments. Rather, the expression patterns of only two genes are required in order to establish the boundaries between developmental stages for which novel markers are to be identified. Such “boundary” genes are termed seed genes. As a strategy to identify markers that are expressed at intermediate developmental stages, Boolean implications were used. Boolean implications indicate that, for example, if gene X expression is high, then gene B expression levels are low (for further details, see Inlay et al. 2009). In principle, the approach can be used for any developmental system. Upon mining the databases that are available, markers can be identified whose expression patterns are changed between the developmental stages that are marked by the seed genes. Using MiDReG, 26 genes were identified that encode for transmembrane proteins and were predicted to be differentially expressed during B-cell maturation. One of these, Ly6D, turned out to be particularly interesting. MPPs lacked detectable Ly6D expression, whereas pre-pro-B cells showed relatively high Ly6D levels. Most telling, the CLPs showed a bimodal pattern of Ly6d expression. The Ly6D− subset contained full lymphoid developmental capacity, termed “ALPs” (all lymphoid progenitor). However, the Ly6D+ compartment was primarily committed to the B-cell lineage, and appropriately called BLPs (B-cell-biased lymphoid progenitors). Thus, these data identified a population of cells within the CLP compartment that lacks B220 and CD19 expression but are fully committed to the B-cell lineage. Furthermore, the study showed that Ly6D is a cell surface marker that permits the identification and isolation of progenitors that are committed to the B-cell lineage.
In sum, these observations indicate that the CLP compartment is a heterogeneous population, consisting of progenitors with full lymphoid potential as well as B-cell-committed progenitors. As discussed previously, this notion is not entirely novel. Using transgenic reporter mice that use the λ5 promoter as a B-lineage-specific marker, a fraction of CLPs was shown to be specified toward the B-cell lineage with little T-cell potential, whereas the population that lacks λ5 promoter activity was shown to contain both B- and T-lineage potential (Mansson et al. 2008).
As described above, other intermediate stages have been identified with common lymphoid potential, termed the CLP2 compartment (Martin et al. 2003). CLP2 cells express B220 but retain both B- and T-lineage potential. How CLP2s and ALPs relate to each other remains an interesting but open question. The road leading from LMPPs to thymic progenitors still contains several holes, but it seems likely that these will be fixed in the near future.
Defining the developmental arrest in E2A-deficient mice
Previous studies have demonstrated that B-cell development in E2A-deficient mice is blocked at a stage called Hardy Fraction A (Bain et al. 1997b). While B220-positive cells were detectable, Igh DJ rearrangements were absent. Previous observations also have demonstrated a partial block at the CLP cell stage in E2A-deficient mice (Borghesi et al. 2005). These data have raised the possibility that B-cell development in E2A-deficient mice is blocked at an intermediate stage, positioned between the CLP and the pre-pro-B-cell compartment. Using Ly6d as a marker, Inlay et al. (2009) convincingly demonstrate that B-cell development in E2A-deficient mice is arrested at the ALP cell stage.
The E2A proteins have been demonstrated previously to act upstream of EBF and Pax5 to regulate B-cell commitment (Kee and Murre 1998; Seet et al. 2004; Roessler et al. 2007). Inlay et al. (2009) also demonstrate that during the transition from the ALP to the BLP cell stage, both EBF and Pax5 levels are elevated, consistent with a critical role for E2A in activating the expression of EBF and Pax5. We are now faced with the question as to how E2A becomes activated during the transition from the ALP to the BLP cell stage. It seems plausible that the Id gene products are involved in this process, since overall E2A abundance does not appear to differ significantly between the early hematopoietic compartments (Semerad et al. 2009). Another mechanism may involve an exchange of E2A partners, involving HEB and/or E2-2. Alternatively, differential post-translational marking of E2A proteins, perhaps induced by cytokine-mediated signaling, may promote the transition from the ALP to the BLP cell stage. This is a critical issue that needs to be resolved in order to achieve a better understanding of how progenitor cells become committed to the B-cell lineage.
The findings reported by Inlay et al. (2009) also raise the question as to whether the arrest in B-cell development in EBF-null mutant mice also occurs during the transition from the ALP to the BLP. It seems likely, since B-lineage priming is substantially decreased in EBF-ablated CLPs (Zandi et al. 2008). Using Ly6D as a marker, this remaining question can now be resolved easily.
The data also bring into question where thymic progenitors come from. Are they derived from both the LMPP and/or ALP compartments through independent pathways? What is the mechanism that promotes T-cell versus B-cell bifurcation at the ALP cell stage? As described above, E2A proteins in the LMPP and DN compartment regulate the expression of Notch-1. However, since the E2A proteins act in both early B- and T-cell progenitors, it seems unlikely that they by themselves are the critical determinants that determine B-cell versus T-cell fate. The E2A partner HEB might play a critical role, modulating the T-cell versus B-cell choice at the ALP cell stage.
Conclusion
During the past two decades, enormous progress has been made in determining the pathways that underlie early hematopoiesis. Many steps in the pathways remain to be resolved both at the cellular and molecular levels. Nevertheless, through concerted activities by so many in the field, the basic framework that underpins the development of the adaptive immune system is being revealed at a rapid pace.
Acknowledgments
I thank members of my laboratory for comments on the manuscript. I apologize to my colleagues for not referencing literature covering the entire field because of either space limitations or ignorance.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1861709.
References
- Adolfsson J, Mansson R, Buza-Vidas N, Hultquist A, Liuba K, Jensen CT, Bryder D, Yang L, Borge OJ, Thoren LA, et al. Identification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential a revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Agata Y, Tamaki N, Sakamoto S, Ikawa T, Masuda K, Kawamoto H, Murre C. Regulation of T cell receptor b gene rearrangements and allelic exclusion. Immunity. 2007;27:871–884. doi: 10.1016/j.immuni.2007.11.015. [DOI] [PubMed] [Google Scholar]
- Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- Bain G, Engel I, Robanus Maandag EC, te Riele HPJ, Voland JR, Sharp LL, Chun J, Murre C. E2A deficient mice show abnormalities in T cell development and rapidly develop malignant T cell lymphomas. Mol Cell Biol. 1997a;17:4782–4791. doi: 10.1128/mcb.17.8.4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain G, Robanus Maandag EC, te Riele HP, Feeney AJ, Sheehy A, Schlissel M, Shinton SA, Hardy RR, Murre C. Both E12 and E47 allow commitment to the B cell lineage. Immunity. 1997b;6:145–154. doi: 10.1016/s1074-7613(00)80421-5. [DOI] [PubMed] [Google Scholar]
- Barndt RJ, Dai M, Zhuang Y. Functions of E2A-HEB heterodimers in T-cell development revealed by a dominant negative mutation of HEB. Mol Cell Biol. 2000;20:6677–6685. doi: 10.1128/mcb.20.18.6677-6685.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borghesi L, Aites J, Nelson S, Lefterov P, James P, Gerstein R. E47 is required for V(D)J recombinase activity in common lymphoid progenitors. J Exp Med. 2005;202:1669–1677. doi: 10.1084/jem.20051190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MJ, Yokomizo T, Brandon MZ, Dzierzak E, Speck NA. Runx1 is required for the endothelial to haematopoietic cell transition but not thereafter. Nature. 2009;457:887–891. doi: 10.1038/nature07619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias S, Mansson R, Gurbuxani S, Sigvardsson M, Kee BL. E2A proteins promote development of lymphoid-primed multipotent progenitors. Immunity. 2008;29:217–227. doi: 10.1016/j.immuni.2008.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel I, Murre C. E2A proteins enforce a proliferation checkpoint in developing thymocytes. EMBO J. 2004;23:202–211. doi: 10.1038/sj.emboj.7600017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldan E, Busslinger M. Pax5 induces V-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes & Dev. 2004;18:411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgescu C, Longabaugh WJ, Scripture-Adams DD, David-Fung ES, Yui MA, Zarnegar MA, Bolouri H, Rothenberg EV. A gene regulatory network armature for T lymphocyte specification. Proc Natl Acad Sci. 2008;105:20100–20105. doi: 10.1073/pnas.0806501105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hock H, Hamblen MJ, Rooke HM, Schindler JW, Saleque S, Fujiwara Y, Orkin SH. Gfi-1 restricts proliferation and preserves functional integrity of haematopietic stem cells. Nature. 2004;431:1002–1007. doi: 10.1038/nature02994. [DOI] [PubMed] [Google Scholar]
- Ikawa T, Kawamoto H, Wright LY, Murre C. Long-term cultured E2A-deficient hematopoietic progenitor cells are pluripotent. Immunity. 2004;20:349–360. doi: 10.1016/s1074-7613(04)00049-4. [DOI] [PubMed] [Google Scholar]
- Ikawa T, Kawamoto H, Goldrath AW, Murre C. E proteins and Notch signaling cooperate to promote T cell lineage specification and commitment. J Exp Med. 2006;203:1329–1342. doi: 10.1084/jem.20060268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inlay MA, Bhattacharya D, Sahoo D, Serwold T, Seita J, Karsunky H, Plevritis SK, Dill DL, Weissman IL. Ly6d marks the earliest stage of B-cell specification and identifies the branchpoint between B-cell and T-cell development. Genes & Dev. 2009 doi: 10.1101/gad.1836009. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee BL, Murre C. Induction of early B cell factor (EBF) and multiple B lineage genes by the basic helix–loop–helix transcription factor E12. J Exp Med. 1998;188:699–713. doi: 10.1084/jem.188.4.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- Kwon K, Hutter C, Sun Q, Bilic I, Cobaleda C, Malin S, Busslinger MA. Instructive role of the transcription factor E2A in early B lymphopoiesis and germinal center B cell development. Immunity. 2008;28:751–762. doi: 10.1016/j.immuni.2008.04.014. [DOI] [PubMed] [Google Scholar]
- Lai AY, Kondo M. T and B lymphocyte differentiation from hematopoietic stem cells. Semin Immunol. 2008;20:207–212. doi: 10.1016/j.smim.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laslo P, Spooner CJ, Warmflash A, Lancki DW, Lee HJ, Sciammas R, Gantner BN, Dinner AR, Singh H. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126:755–766. doi: 10.1016/j.cell.2006.06.052. [DOI] [PubMed] [Google Scholar]
- Laslo P, Pongubala JMR, Lancki DW, Singh H. Gene regulatory networks directing myeloid and lymphoid cell fates within the immune system. Semin Immunol. 2008;20:228–235. doi: 10.1016/j.smim.2008.08.003. [DOI] [PubMed] [Google Scholar]
- Lazorchak AS, Wojciechowski J, Dai M, Zhuang Y. E2A promotes the survival of precursor and mature B lymphocytes. J Immunol. 2006;177:2495–2504. doi: 10.4049/jimmunol.177.4.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Schmidt-Supprian M, Shi M, Hobeika E, Barteneva N, Jumaa H, Pelanda R, Reth M, Skok J, Rajewsky K, et al. Yin Yang 1 is a critical regulator of B cell development. Genes & Dev. 2007;21:1179–1189. doi: 10.1101/gad.1529307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier H, Ostraat R, Gao H, Fields S, Shinto SA, Medina KL, Ikawa T, Murre C, Singh H, Hardy RR, et al. Early B cell factor cooperates with Runx1 and mediates epigenetic changes associated with mb-1 transcription. Nat Immunol. 2004;5:1069–1077. doi: 10.1038/ni1119. [DOI] [PubMed] [Google Scholar]
- Mansson R, Zandi S, Andersson IL, Jacobsen SE, Bryder D, Sigvardsson M. B-lineage commitment prior to surface expression of B220 and CD19 on hematopoietic progenitor cells. Blood. 2008;15:1048–1055. doi: 10.1182/blood-2007-11-125385. [DOI] [PubMed] [Google Scholar]
- Martin CH, Aifantis I, Scimone ML, von Adrian UH, Reizis B, von Boehmer H, Gounari F. Efficient thymic migration of B220+ lymphoid-restricted bone marrow cells with T precursor potential. Nat Immunol. 2003;4:866–873. doi: 10.1038/ni965. [DOI] [PubMed] [Google Scholar]
- Medina KL, Pongubala JM, Reddy KL, Lancki DW, Dekoter R, Kieslinger M, Grosschedl R, Singh H. Assembling a gene regulatory network for specification of the B cell fate. Dev Cell. 2004;7:607–617. doi: 10.1016/j.devcel.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Murre C. Helix–loop–helix proteins and lymphocyte development. Nat Immunol. 2005;6:1079–1086. doi: 10.1038/ni1260. [DOI] [PubMed] [Google Scholar]
- Ng SY, Yoshida T, Zhang J, Georgopoulos K. Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity. 2009;30:493–507. doi: 10.1016/j.immuni.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562. doi: 10.1038/44076. [DOI] [PubMed] [Google Scholar]
- Pongubala JM, Northrup DL, Lancki DW, Medina KL, Treiber T, Bertolino E, Thomas M, Grosschedl R, Allman D, Singh H. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat Immunol. 2008;9:203–215. doi: 10.1038/ni1555. [DOI] [PubMed] [Google Scholar]
- Pronk CJ, Rossi DJ, Mansson R, Attema JL, Norddahl GL, Chan CK, Sigvardsson M, Weissman IL, Bryder D. Elucidation of the phenotypic, function and molecular topography of a myeloperythorid progenitor cell hierarchy. Cell Stem Cell. 2007;11:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- Rekhtman N, Radparvar F, Evans T, Skoultchi AI. Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: Functional antagonism in erythroid cells. Genes & Dev. 1999;13:1398–1411. doi: 10.1101/gad.13.11.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud D, Demarco IA, Reddy KL, Schjerven H, Bertolino E, Chen Z, Smale ST, Winandy S, Singh H. Regulation of B cell commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat Immunol. 2008;9:927–936. doi: 10.1038/ni.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler S, Gyory I, Imhof S, Spivakov M, Williams RR, Busslinger MA, Fisher AG, Grosschedl R. Distinct promoters mediate the regulation of Ebf1 gene expression by interleukin-7 and Pax5. Mol Cell Biol. 2007;2:579–594. doi: 10.1128/MCB.01192-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz R, Engel I, Fallahi-Sichani M, Petrie HT, Murre C. Gene expression patterns define novel roles for E47 in cell cycle progression, cytokine-mediated signaling, and T lineage development. Proc Natl Acad Sci. 2006;103:9976–9981. doi: 10.1073/pnas.0603728103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- Seet CS, Brumbaugh RL, Kee BL. Early B cell factor promotes B lymphopoiesis with reduced interleukin 7 responsiveness in the absence of E2A. J Exp Med. 2004;199:1689–1700. doi: 10.1084/jem.20032202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semerad CL, Mercer EM, Inlay MA, Weissman IL, Murre C. E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphid and myeloerythorid progenitors. Proc Natl Acad USA. 2009;106:1930–1935. doi: 10.1073/pnas.0808866106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivdasani RA, Fujiwara Y, McDevitt MA, Orkin SH. A lineage-selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet development. EMBO J. 1997;16:3965–3973. doi: 10.1093/emboj/16.13.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigvardsson M, O'Riordan M, Grosschedl R. EBF and E47 collaborate to induce expression of the endogenous surrogate light chain genes. Immunity. 1997;7:25–36. doi: 10.1016/s1074-7613(00)80507-5. [DOI] [PubMed] [Google Scholar]
- Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- Wada H, Masuda K, Satoh R, Kakugawa K, Ikawa T, Katsura Y, Kawamoto H. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–772. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- Wang D, Claus CL, Vaccarelli G, Braunstein M, Schmitt TM, Zuniga-Pflucker JC, Rothenberg EV, Andersson MK. The basic helix–loop–helix transcription factor HEBAlt is expressed in pro-T cells and enhances the generation of T cell precursors. J Immunol. 2006;117:109–119. doi: 10.4049/jimmunol.177.1.109. [DOI] [PubMed] [Google Scholar]
- Yang Q, Kardava L, St Leger A, Martincic K, Varnum-Finney B, Bernstein ID, Milcarek C, Borghesi L. E47 controls the developmental integrity and cell cycle quiescence of multipotential hematopoietic progenitors. J Immunol. 2008;181:5885–5894. doi: 10.4049/jimmunol.181.9.5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashiro-Ohtani Y, He Y, Ohtani T, Jones ME, Shestova O, Xu L, Fang TC, Chiang MY, Blacklow SC, Zhuang Y, et al. Pre-TCR signaling inactivates Notch1 transcription by antagonizing E2A. Genes & Dev. 2009;23:1665–1676. doi: 10.1101/gad.1793709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zandi S, Mansson R, Tsapogas P, Zetterblad J, Bryder D, Sigvardsson M. EBF1 is essential for B-lineage priming and establishment of a transcription factor network in common lymphoid progenitors. J Immunol. 2008;181:3364–3372. doi: 10.4049/jimmunol.181.5.3364. [DOI] [PubMed] [Google Scholar]