Abstract
Rare individuals report repeated unprotected HIV-1 sexual exposures, yet remain seronegative for years. We investigated the possibility that reduced in vitro CD4+ T cell susceptibility to HIV-1 infection protects such highly exposed seronegative (ES) individuals. Susceptibility to three R5-tropic HIV-1 isolates, regardless of inoculating dose, was remarkably similar between 81 ES and 33 low-risk controls. In 94% (99/105) of donors, we observed a 1.36 log-unit range in HIV-1JR-CSF production, with similar results for HIV-11192. The median frequency of intracellular Gag+ T cells after single-round infection was similar in ES (5.2%) and controls (7.2%), p = 0.456. However, in repeated testing, CD4+ T cells from two controls (6.1%) and four ES (4.9%) exhibited a 10- to 2500-fold reduction in HIV-1 production and required 5- to 12-fold greater HIV-11192 and HIV-1JR-CSF inocula to establish infection (TCID50). Reduced viral entry cannot explain the low producer phenotype; no differences in CCR5 receptor density or β-chemokine production were observed. In conclusion, we have identified a remarkably narrow range of HIV-1 susceptibility in seronegative donors regardless of risk activity, which can be applied as a benchmark to assess vaccine-induced antiviral effector activities. However, CD4+ T cells from a subset of individuals demonstrated reduced HIV-1 susceptibility unexplained by impaired entry, lending support to the possibility that cellular restriction of HIV-1 may account for continued seronegativity in some of those having repeated sexual exposure. Identifying the host-virus interactions responsible for diminished in vitro susceptibility may contribute to the development of novel therapeutic strategies.
Introduction
Rare individuals remain human immunodeficiency virus type 1 (HIV-1) seronegative despite repeated unprotected sexual exposures.1–5 Apart from the relatively low transmission frequency during sexual contact, mechanisms of protection for exposed seronegative (ES) persons are largely undefined and are likely to be multifactorial. The role of HIV-1-specific adaptive immunity and immune activation has received significant consideration in evasion of infection by ES.1,3,4,6–23 Further, homozygosity for a 32-base pair deletion in the HIV-1 coreceptor CCR5 (CCR5Δ32) confers strong protection against R5-dependent strains, and CCR5Δ32 homozygous persons are frequently overrepresented in ES cohorts of European descent.24 Nonetheless, persistent host immunity and coreceptor polymorphisms cannot explain why the majority of ES worldwide resist infection.
That CD4+ T cells and monocyte-derived macrophages from individual donors differ in their permissiveness to HIV-1 infection in vitro25–28 is well recognized. We hypothesized that diminished target cell susceptibility to incoming HIV-1 can confer relative protection in ES, much as in vitro CD4+ T cell susceptibility to simian immunodeficiency virus (SIV) in rhesus macaques correlates closely with their in vivo set-point viremia after viral challenge.29 Previous efforts to address CD4+ T cell susceptibility in ES led to the identification of the CCR5 coreceptor and the CCR5Δ32 mutation that results in its loss at the cell surface.30,31 Furthermore, reduced CD4+ T cell susceptibility and the ES phenotype have been associated with lower CCR5 coreceptor surface expression or elevated production of its natural ligands, the β-chemokines.18,32–39 Recently, variable CCL3L1 copy number resulting in increased MIP-1αP production has been associated with HIV-1 seronegativity,40 although the importance of this finding remains in dispute.41
Whether ES demonstrate diminished target cell permissiveness to HIV-1 exclusive of coreceptor defects is not clear, in part because systematic analyses in sufficient numbers of ES and relevant control populations are lacking.24,31,33,35,36,42 In one cohort of serodiscordant men who have sex with men (MSM), lack of transmission was correlated with reduced HIV-1 susceptibility in peripheral blood mononuclear cells (PBMC) from the uninfected partner.18,24 By contrast, investigations of highly exposed female commercial sex workers and intravenous drug users were unable to associate continued seronegativity with altered PBMC susceptibility35,36,42 or enhanced β-chemokine production.35 Notably, in many studies,18,24,33,35,36 the antiviral activities of natural killer cells and CD8+ T cells contained within PBMC may have obscured differences in CD4+ T cell susceptibility to HIV-1 infection. Therefore, the role of CD4+ T cell permissiveness in defining an individual's propensity to become infected during a given sexual exposure remains uncertain.
Since 1995, we have followed an ES cohort reporting repeated high-risk sexual behavior with at least one known HIV-1-infected partner. Only 3.7% (3/81) of volunteers have the CCR5Δ32/Δ32 genotype. Examination of viral isolates from the infected sexual partners of the ES revealed no infectivity defects and viral loads in infected sexual partners are similar to a matched control population.3,22 Additionally, cytolytic or IFN-γ-secreting HIV-1-specific CD8+ cells in this ES cohort have been either undetectable or sustained longitudinally in only a minority of volunteers.3,14,22,43 Therefore, the basis for HIV-1 resistance in the majority of participants remains unidentified. To explore factors that may confer relative resistance to infection, we analyzed CD4+ T cell susceptibility to HIV-1 in ES and low-risk controls.
Materials and Methods
Study population
Enrollment criteria and study procedures have been described for 81 ES subjects in Seattle.3 The institutional review board approved the study protocol, and each subject provided written informed consent prior to enrollment. All ES reported here remained HIV-1 seronegative throughout follow-up. Sexual contacts for low-risk control volunteers (n = 33) were known to be HIV-1 uninfected.
HIV-1 viral isolates
HIV-1JR-CSF was generated by 293T cell transfection44 with proviral plasmid pYK-JRCSF [contributed by I.S.Y. Chen and Y. Koyanagi45 and provided by the NIH AIDS Research and Reference Reagent Program (NARRRP)]. Primary isolates HIV-11192 and HIV-11196 were obtained from PBMC of Seattle area HIV-1-infected, treatment-naive participants. HIV-11192 was isolated 3 months postinfection and HIV-11196 4.5 months postinfection. Both primary isolates were expanded only twice on pooled CD8-depleted PBMC from 10 HIV-1-uninfected donors. Titers for all HIV-1 stocks were determined using the MAGI-CCR5 assay46,47 (UT3-MAGI-CCR5 cells kindly provided by M. Emerman).
Infection kinetics assay
We investigated 10 different multiplicities of infection (MOI) ranging from 3.84 × 10−8 to 3.0 × 10−2 in each viral strain to select test inocula. The following MOI were investigated: HIV-11192, MOI 0.003 and 0.0006; HIV-11196, MOI 0.015 and 0.003; and HIV-1JR-CSF, MOI 0.003 and 0.006.
For all experiments described, primary CD4+ T cells were isolated from cryopreserved PBMC by negative selection using magnetic antibody bead separation (Miltenyi, Auburn, CA) and stimulated by 1.5 μg/ml phytohemagluttinin (Remel, Lenexa, KS) for 3 days at 37°C and 5% CO2. CD4+ lymphoblasts were incubated with virus supernatant at 37°C for 4 h, washed, and cultured in four replicates of 2 × 105 each in HEPES-buffered RPMI 1640 (Gibco, Carlsbad, CA) supplemented with penicillin (50 U/ml, Gibco), streptomycin (50 μg/ml, Gibco), l-glutamine (2 mM, Gibco), 10% heat-inactivated fetal bovine serum (Gemini Bio-Products, Woodland, CA), and 100 U/ml interleukin-2 (Chiron, Emeryville, CA). At 3, 5, 7, 10, and 12 days postinfection (dpi), 100 μl of supernatant was harvested and stored at −70°C for batch HIV-1 quantitation.
HIV-1 in harvested supernatants was quantified by either p24 ELISA (Perkin Elmer, Boston) or MAGI-CCR5 assay,46,47 with results reported as total pg p24 Ag or blue-forming units (BFU), respectively, in culture. Between 3 and 12 replicates were measured for viral production. Statistically outlying results (less than the 25th percentile minus 1.5 times the interquartile range) were defined as corresponding to reduced HIV-1 productive capacity.48,49
Characterization of uninfected primary CD4+ lymphoblasts
Viability of purified uninfected CD4+ lymphoblasts maintained under the same conditions as infected cultures was characterized by trypan blue exclusion using phase-contrast light microscopy. Infected culture supernatants whose corresponding uninfected cultures had <60% viability at 7 dpi were excluded from further analysis.
On the day of infection, uninfected cells were characterized by fluorescent monoclonal antibody (mAb) staining for CD3, CD4, CD8, and CCR5 expression (all BD Biosciences, San Diego, CA). Cultures with CD8+ cells in excess of 3% of CD3+ T lymphocytes were excluded from further analysis. Anti-CD14 mAb PE staining in a subset of 20 samples verified the absence of CD14+ cells at the time of infection.
Quantitation of first-round HIV-1 infection
Frequency of first-round infection was quantified as previously described.50 Purified CD4+ lymphoblasts were infected with HIV-1JR-CSF (MOI 0.75) in the presence of 1 μM indinavir (NARRRP). Forty-eight hours postinfection harvested cells were stained by fluorescent mAb for CD3, CD4, and HLA Class I expression (all BD Biosciences), followed by fixation, permeabilization, and staining with anti-Gag mAb FITC (KC57-FITC; Beckman Coulter, Miami, FL). Infections were performed in duplicate or triplicate, and reported with the percentage of mock-infected CD3+CD4lo-Gag+ cells (less than 0.2%, median 0.01%) subtracted.
Pseudotyped HIV-1ΔenvGFP infection assay
For single-round infection using a GFP-expressing pseudotype, an HIV-1SF162 or HIV-1HXB2 envelope was used to package vector HIV-1LAIΔenvGFP51 (provided by M. Emerman, FHCRC) by 293T cotransfection. Purified CD4+ lymphoblasts (1 × 105) were infected with serial viral dilutions by spinoculation52 for 1 h at 1900 × g and 30°C. After 40–44 h infection in medium containing recombinant interleukin (IL)-2 (final concentration, 50 U/ml), cells were fixed in 2% paraformaldehyde and assessed for Tat-mediated green fluorescent protein (GFP) expression by flow cytometry using FlowJo software (Tree Star, Inc., Ashland, OR) for analysis.
TCID50 determination
For TCID50 determination, six replicates of 5 × 104 purified CD4+ lymphoblasts were incubated with one of ten 2.5-fold HIV-1 serial dilutions (37°C, 5% CO2, 4 h), washed, and maintained in culture media containing 50 U/ml IL-2. The presence of p24 antigen in the supernatant was tested at 7 dpi, and wells containing >2.5 pg/ml (the limit of detection) were considered positive for virus replication. TCID50 was calculated according to the Reed and Muench formula,53 with results expressed as the number of virions required to establish infection in 50% of cultures, based on titer of the virus stock on PBMC from four HIV-1 low-risk individuals.
CCR5 genotyping, expression, and ligand levels
CCR5Δ32 genotype30 was determined using DNA restriction fragment length polymorphism (RFLP) analysis.54 CCR5 coreceptor surface expression among resting, cryopreserved PBMC was characterized using a previously described two-step staining protocol.55 MIP-1β and RANTES in culture supernatants at 7 dpi were quantified by ELISA (R&D Systems), used according to the manufacturer's instructions. For quantitation of MIP-1α, an in-house ELISA was employed. MIP-1αP copy number was determined by quantitative PCR, as described previously.40
Statistical analyses
Demographic and genotypic comparison of ES volunteers and low-risk controls was performed using Fisher's exact test. First-round HIV-1 infection data and log-transformed MAGI assay and p24 ELISA results were assessed by Student's t test. When no HIV-1 production was detectable, the value of 10 pg p24 Ag or 10 BFU, was used to represent the limit of detection. Mann–Whitney ranked comparison was employed to contrast low producers to as many as nine normally susceptible individuals. Data from CCR5Δ32 homozygous individuals were excluded.
To estimate the basic reproductive number in vitro, R0, we employed a four-compartment rate-equation (ODE) model. The model contains uninfected target cells (T), infected cells in the eclipse phase (I), infected cells in the productive phase (P), and free infectious virions (V). Target cells were infected at rate ιTV and progressed to production at rate ηI. Productively infected cells produced virus at rate ρP and died at rate δPP. Free virions lost infectivity at rate δVV. To convert between p24 and infectious virions, we used an empirical regression relation: log(V) = 0.97 log(p24). We fixed η = 1.0, δP = 1.0, and δV = 2.8 (all inverse days), leaving two free parameters in the model: ι (infectivity) and ρ (productivity). R0 is related to these parameters by R0 = (ιρT0)/(δPδv), where T0 is the initial target cell density. From the branching process version of the same model, we derived the expression for the extinction probability, p, given ρ and R0. The relation TCID50 × log(p) = log(0.5) eliminated one parameter. We fit the remaining one-parameter ODE model to the growth rate data (mean of observations of each subject) by least-squares, equivalent to maximum likelihood for a measurement-error model. We report log likelihood-based confidence intervals.
Results
Study population
ES and low-risk control volunteers were well matched by gender, sexual orientation, and CCR5 genotype (Table 1). ES subjects were slightly older than low-risk volunteers. More non-European Americans were in the low-risk group, but this was unrelated to CCR5Δ32 genotype (Table 1). Excluding CCR5Δ32 homozygous negative controls, the frequency of the CCR5Δ32 allele was 0.115 (18/156) in ES and 0.125 (8/64) in low-risk participants (p-value = 1.0).
Table 1.
Demographic and Genotypic Comparison of Exposed Seronegative and Low-Risk Study Groups
| Low producers (n = 6) | ES (n = 81) | Low-risk controls (n = 33) | p-valuea | |
|---|---|---|---|---|
| Gender | 0.359 | |||
| Male | 4 (67%) | 71 (88%) | 26 (79%) | |
| Female | 2 (33%) | 10 (12%) | 7 (21%) | |
| Sexual activity | 0.668 | |||
| Homosexual | 4 (67%) | 67 (83%) | 26 (79%) | |
| Bisexual | 0 (0%) | 1 (1.2%) | 0 (0%) | |
| Heterosexual | 2 (33%) | 13 (16%) | 7 (21%) | |
| Ethnicity | 0.054b | |||
| European-American | 5 (83%) | 76 (94%) | 26 (79%) | |
| African-American | 1 (17%) | 1 (1.2%) | 4 (12%) | |
| Latino | 0 (0%) | 3 (3.7%) | 2 (6.1%) | |
| Other | 0 (0%) | 1 (1.2%) | 1 (3.0%) | |
| Age (median, range) | 38 (25–51) | 41 (25–68) | 37 (26–55) | 0.028c |
| CCR5 genotype | 0.961d | |||
| Δ32/Δ32 | 0 (0%) | 3 (3.7%) | 1 (3.0%) | |
| WT/Δ32 | 3 (50%) | 18 (22%) | 8 (24%) | |
| WT/WT | 3 (50%) | 60 (74%) | 24 (76%) |
Fisher exact probability test unless otherwise noted.
European Americans vs. all others.
Mann–Whitney ranked comparison.
WT/Δ32 vs. WT/WT. All comparisons between ES and low-risk controls.
CD4+ T cell susceptibility screen results
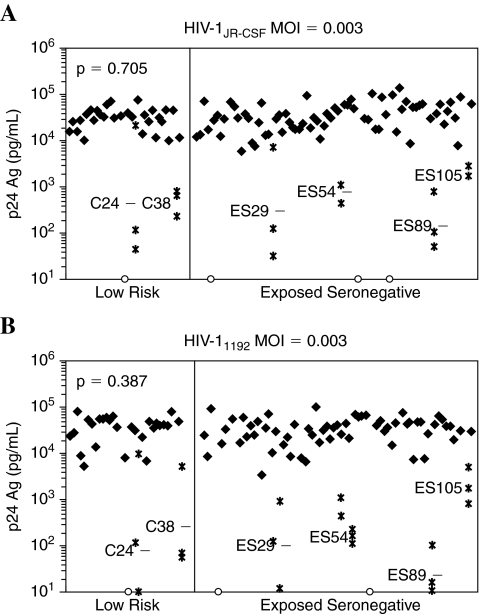
To determine whether decreased CD4+ T cell susceptibility contributes to relative resistance to HIV-1 infection among ES, we quantified HIV-1 production during extended culture of infected CD4+ T cells from 81 ES and 33 low-risk controls. Cells were challenged with two different MOI for each of three R5-tropic viral strains, including the well-described molecular clone HIV-1JR-CSF and two local primary isolates, HIV-11192 and HIV-11196. We excluded data for nine study participants from the analysis because CD8+ T cell contamination exceeded 3% (n = 3), cell viability in day 7 mock-infected cultures was less than 60% (n = 2), or CCR5Δ32 homozygosity was demonstrated (n = 4). We observed no overall differences in CD4+ T cell susceptibility between the ES and control groups regardless of the viral isolate or inoculating dose (Fig. 1 and Table 2). This was true whether HIV-1 was quantified by production of p24 Ag (ELISA) or infectious virions (MAGI assay), or whether the comparison was made in supernatants harvested at 5 dpi (p > 0.288) or 7 dpi (p > 0.316, Table 2).
FIG. 1.
Rare individuals display markedly reduced capacity to sustain HIV-1 infection in vitro. Supernatants harvested at 5 and 7 days postinfection (dpi) were analyzed for p24 content. Results were similar for 5 and 7 dpi and for all HIV-1 strains and MOI examined. Data for 7 dpi are shown. (A) Screen results when CD4+ T cells were infected with HIV-1JR-CSF; (B) screen results when CD4+ T cells were infected with HIV-11192. Closed symbols represent the log-transformed mean of repeated experiments conducted in cells from venipuncture dates spanning a median of 9.1 months, where applicable (see text). Open symbols denote data from CCR5Δ32 homozygotes. Volunteers having reduced CD4+ T cell infection as measured by p24 Ag ELISA (defined as statistical outliers, less than the 25th percentile minus 1.5 times the interquartile range) are shown as asterisks and with the log-transformed mean of three separate experiments noted as a dashed line.
Table 2.
Comparison of CD4+ T Cell Capacity to Sustain HIV-1 Infection between ES and Low-Risk Individuals
|
p24 antigen production | ||||
|---|---|---|---|---|
| |
|
Median log10 pg p24/ml |
||
| |
|
Study population |
|
|
| HIV-1 strain | MOI | ES | Low risk | p-value |
| 5 days postinfection | ||||
| JR-CSF | 0.003 | 4.65 | 4.80 | 0.712 |
| 0.0006 | 4.28 | 4.62 | 0.577 | |
| 1192 | 0.003 | 4.68 | 4.79 | 0.461 |
| 0.0006 | 4.18 | 4.15 | 0.531 | |
| 1196 | 0.015 | 4.74 | 4.71 | 0.451 |
| 0.003 | 4.63 | 4.12 | 0.288 | |
| 7 days postinfection | ||||
| JR-CSF | 0.003 | 4.49 | 4.50 | 0.705 |
| 0.0006 | 4.41 | 4.37 | 0.928 | |
| 1192 | 0.003 | 4.57 | 4.62 | 0.387 |
| 0.0006 | 4.40 | 4.36 | 0.316 | |
| 1196 | 0.015 | 4.29 | 4.31 | 0.655 |
| 0.003 | 4.25 | 4.37 | 0.945 | |
|
Infectious virion production | ||||
|---|---|---|---|---|
| |
|
Median log10 BFU/ml |
||
| Study population | ||||
| HIV-1 strain | MOI | ES | Low risk | p-value |
| 5 days postinfection | ||||
| JR-CSF | 0.003 | 4.68 | 4.65 | 0.859 |
| 0.0006 | 3.22 | 3.69 | 0.864 | |
| 1192 | 0.003 | 4.69 | 4.74 | 0.782 |
| 0.0006 | 4.00 | 4.15 | 0.475 | |
| 1196 | 0.015 | 4.52 | 4.47 | 0.759 |
| 0.003 | 3.84 | 3.57 | 0.969 | |
| 7 days postinfection | ||||
| JR-CSF | 0.003 | 4.37 | 4.28 | 0.225 |
| 0.0006 | 4.72 | 4.76 | 0.565 | |
| 1192 | 0.003 | 4.56 | 4.47 | 0.475 |
| 0.0006 | 4.70 | 4.65 | 0.637 | |
| 1196 | 0.015 | 4.48 | 4.46 | 0.332 |
| 0.003 | 4.26 | 4.12 | 0.578 | |
Furthermore, with the exception of rare individuals having substantially reduced in vitro HIV-1 productive capacity (see below), we observed remarkably uniform HIV-1 production across each group (Fig. 1). In 99/105 (94%) of donors, we observed only a 1.36 log-unit range in HIV-1JR-CSF production by CD4+ lymphoblasts at MOI 0.003. Similarly, CD4+ lymphoblast infection with local primary isolate HIV-11192 (MOI 0.003) yielded a 1.47 log-unit range in p24 production in 88/94 (94%) of donors tested. Thus, CD4+ T cells from volunteers reporting either high- or low-risk behaviors were equally able to sustain HIV-1 production during in vitro infection, and the conspicuously narrow range of HIV-1 production observed at all inocula and viral strains tested permitted identification of outlying individuals having profound defects in CD4+ T cell susceptibility.
Identification of subjects whose activated CD4+ T cells produce low-level HIV-1
Strikingly, we identified 6 of 105 (5.7%) volunteers in whom CD4+ T cell cultures produced a statistically outlying reduction in HIV-1 during extended in vitro culture (Table 3). These low producers were not restricted to the ES cohort; we identified two low-risk and four ES individuals in this class. HIV-1 production in infected cultures from these individuals commonly was 100-fold lower (range, 10- to 2500-fold decrease) than that observed in the remaining 99 individuals examined, suggesting that reduced CD4+ T cell susceptibility is not a broadly acting mechanism of resistance in our ES cohort. However, select individuals in both risk groups exhibited remarkably reduced susceptibility to HIV-1 infection among circulating CD4+ T cells. Markedly diminished HIV-1 productive capacity may provide a means of in vivo protection from infection in these volunteers, and thus we explored further the characteristics of HIV-1 susceptibility in these subjects, designated “low producers.”
Table 3.
p24 Ag Production in CD4+ T Cell Cultures from Six Low Producers vs. All Normally Susceptible Study Participants, 7 Days Postinfection
| |
log10 pg p24/ml in supernatant |
||||
|---|---|---|---|---|---|
| Expt 1a | Expt 2 | Expt 3 | Mean | Median | |
| HIV-1JR-CSF, MOI 0.003 | |||||
| C24 | 4.27 | 1.64 | 2.13 | 2.68 | 2.13 |
| C38 | 2.50 | 2.56 | 2.83 | 2.63 | 2.56 |
| ES29 | 4.08 | 1.58 | 2.05 | 2.57 | 2.05 |
| ES54 | 2.81 | 2.61 | 3.05 | 2.82 | 2.81 |
| ES89 | 3.09 | 1.74 | 2.01 | 2.28 | 2.01 |
| ES105 | 3.62 | 3.41 | 3.63 | 3.55 | 3.62 |
| Other ES | 4.51 | 4.50 | |||
| Other low risk | 4.49 | 4.52 | |||
| HIV-11192, MOI 0.003 | |||||
| C24 | 1.00 | ndb | 1.00 | 1.00 | 1.00 |
| C38 | 1.97 | 2.67 | 1.55 | 2.06 | 1.97 |
| ES29 | nd | 2.67 | 1.08 | 1.88 | 1.88 |
| ES54 | 1.93 | 2.67 | 1.51 | 2.04 | 1.93 |
| ES89 | 2.67 | 1.05 | 1.40 | 1.71 | 1.40 |
| ES105 | 2.90 | 3.08 | 3.91 | 3.30 | 3.08 |
| Other ES | 4.40 | 4.56 | |||
| Other low risk | 4.40 | 4.62 | |||
PBMC from independent venipuncture dates used in each experiment.
nd, not done.
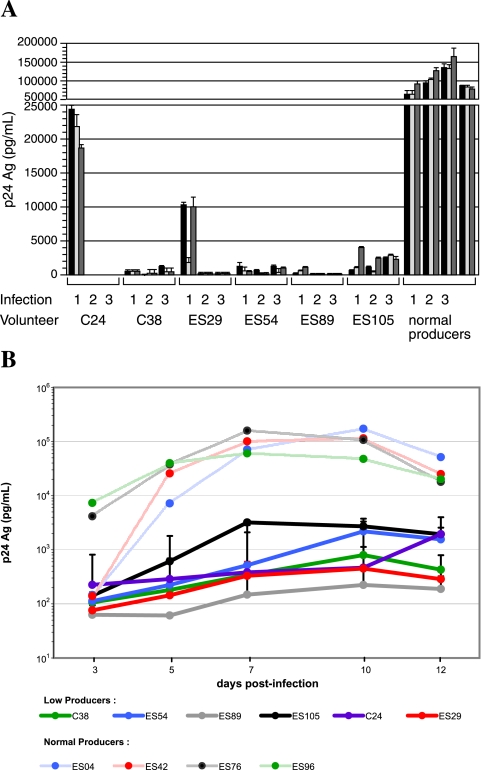
Low-producer phenotype is reproducible and stable over time
To confirm our observation of diminished HIV-1 production in CD4+ T cells from six low producers, we twice repeated the in vitro infection experiment in cells from separate venipuncture dates. We quantified HIV-1 production in supernatants from three separate infected replicates from each experiment. We included as controls samples from volunteers who had previously demonstrated normal HIV-1 production (between the 25th and 90th percentiles) in each infection experiment. In total, extended in vitro infection with HIV-1JR-CSF MOI 0.003 was repeated in CD4+ T cells from 32 individuals (four independent experiments conducted in six subjects, three experiments in six, and two experiments in 20 volunteer samples), in addition to the one experiment conducted in the remaining 77 subjects (including four CCR5Δ32 homozygotes exempted from analysis). Similarly, 123 observations of HIV-11192 infection at MOI 0.003 were made in 97 volunteers (including three CCR5Δ32 homozygotes exempted from analysis).
Replicate wells from a single infection produced strongly concordant results (Fig. 2A). In addition, independent experiments using CD4+ T cells isolated from different venipuncture dates were highly reproducible (Fig. 2A and Table 3). Infection assay in low-producer CD4+ T cells was repeated on either three (C24, C38, ES29, ES54) or four (ES89, ES105) occasions, employing cells from venipuncture dates with a median span of 9.1 months. Likewise, normally susceptible cells queried in independent experiments (n = 32) corresponded to venipuncture dates with a median span of 11.7 months. Results from independent experiments in low producers and normally susceptible cells were similarly reproducible. The normally susceptible phenotype was stable over time, with the median experimental range of 0.22 log-units. In no instance did data from a single experiment conducted in cells having normal susceptibility fall into the range of the low producers. Two exceptions to this trend were seen in the low producers C24 and ES29. Experiments using these subjects' cells from two different visit dates produced minimally detectable HIV-1 p24 antigen, while HIV-1 production fell within the lowest quartile of the median response in an experiment from a third venipuncture date. Taken together, the six low producers maintained lower levels of HIV-1 production during repeated testing of cells isolated at different time points. These findings demonstrate that in vitro susceptibility to infection is highly reproducible between replicates and is stable over time.
FIG. 2.
Low-producer phenotype is consistent over time. (A) HIV-1 core antigen production in three different replicate cultures from three separate CD4+ T cell infection experiments with HIV-1JR-CSF (MOI 0.003) is shown, with data from different replicates within each experiment clustered together. (B) Infection kinetics curves for six low-producer individuals and four individuals having normal capacity for productive in vitro infection with HIV-1JR-CSF (MOI 0.003). A solid line indicates low producers; a dashed line indicates normally susceptible. For HIV-1 low producers, lines represent the log-transformed mean of results from three separate experiments involving cells isolated from different venipuncture dates. SEM are shown.
Peak HIV-1 levels are diminished in low producers
To distinguish between delayed versus lower peak HIV-1 production, we contrasted HIV-1 infectivity from serial culture time points in four individuals having normal p24 production in the initial screen (mean 80th percentile) and six low-producer subjects. In supernatants harvested at 3, 5, 7, 10, and 12 dpi, we consistently observed markedly reduced HIV-1 in the low producers, with p24 Ag reaching a plateau between 7 and 10 dpi in all cases (Fig. 2B). Of note, the peak and plateau phase of infection was similar between the low and normal producers. With the exception of the two low producers noted previously, C24 and ES29, the kinetics and level of HIV-1 production in volunteers having diminished susceptibility to HIV-1 infection were similar between experiments conducted independently (Fig. 2B). The reduced peak HIV-1 production achieved by infected CD4+ T cells in these volunteers was profound and reproducible.
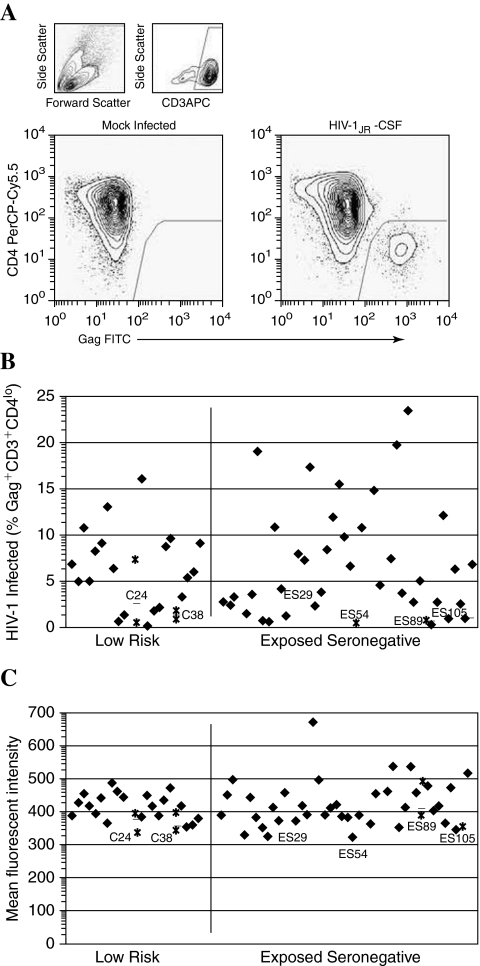
Screening by single-round infection
We also investigated the frequency and quantity of Gag production following a single round of HIV-1 infection in CD4+ T cells from 46 ES and 20 low-risk controls, from which four CCR5Δ32 homozygotes were removed from further analysis (Fig. 3A). The protease inhibitor indinivir was added to achieve single cycle replication without secondary spread. As in the extended infection assay, we observed no differences between ES and low-risk groups in the susceptibility of CD4+ lymphoblasts to HIV-1 infection (Fig. 3B), with the median frequency of CD3+ Gag+ T cells in ES and controls being 5.61% and 6.30%, respectively (p-value = 0.456). Significantly lower percentages of Gag+ cells were observed in CD4+ T cells from low producers (median, 1.20%) than in the remaining samples (median, 6.0%), p-value = 0.007. We observed a modest, but statistically significant, decrease in Gag expression per infected cell (Fig. 3C) among low producers relative to normally susceptible cells (median mean fluorescent intensity 178 vs. 208, respectively; p-value = 0.010). In addition, in a subset of volunteers, CD3+Gag+ percentages were quantified following in vitro infection in the presence of the reverse transcriptase inhibitor AZT to confirm that any differences in Gag positivity were not due to a reduction in intracellular Gag from the infecting virions. For this assay, CD4+ T lymphoblasts from four low producers in whom cell numbers were not limiting were compared to eight volunteers having normally susceptible cells (data not shown). In this case, percentages of CD3+Gag+ cells did not differ between low producers (median, 0.53%; range, 0–1.05%) and susceptible cells (median, 0.81%; range, 0.18–3.72%), p-value = 0.222.
FIG. 3.
Single-round infection of primary CD4+ T cell blasts with HIV-1JR-CSF. (A) Infected cells are identified by p55 Gag expression and CD4 downregulation in the CD3+ population. (B) Low-producer cells demonstrate lower infected cell percentages during the first round of infection, p-value = 0.007. (C) Gag+ cells from low producers show a modest, but significant, reduction in mean Gag expression, p-value = 0.010. Closed symbols represent the mean of repeated experiments, where applicable. Open symbols denote data from CCR5Δ32 homozygotes. Data from infection of low-producer cells are shown as asterisks, with the mean of separate experiments noted as a dashed line, where applicable. Gag densities for CCR5Δ32 homozygotes were set to zero.
Despite the statistical significance of our findings following CD4+ lymphoblast infection in the presence of indinavir, the wide range of Gag+ T cell frequencies impeded de novo identification of volunteers having a low-producer phenotype by this single-round infection assay using the criterion that they be statistical outliers, as in the extended infection assay. Indeed, 16/56 volunteers having normal HIV-1 p24 production in the extended infection assay demonstrated single-round CD3+Gag+ percentages similar to the low producers. Likewise, the difference in intracellular Gag expression was not complemented by a marked difference in the values observed (mean fluorescent intensity range, 160–211 in low producers vs. 162–334 in normally susceptible cells). The broad range of percentage of infected cells and minimal change in Gag expression per cell obscures interpretation of the biological relevance of these statistically significant results.
Increased HIV-1 inoculum required to establish infection in low-producer cells
To ascertain whether diminished capacity to produce HIV-1 in vitro was correlated with an increase in the viral inoculum required to establish infection, we determined relative HIV-1JR-CSF TCID50 in CD4+ T cells from six low producers and four volunteers with normal HIV-1 production. The median HIV-1JR-CSF TCID50 for low producers was 16.4 (range, 9.6–81.43) compared to 1.3 (range, 1.0–1.8) for normally susceptible subjects (Table 4). TCID50 values for low and normal producers were nonoverlapping, with the six low producers requiring a one-log increase in HIV-1JR-CSF inoculum required to establish infection. The large magnitude of this increase in TCID50 was strongly significant (p-value = 0.010). Furthermore, this finding was not limited to the molecular clone HIV-1JR-CSF. Although samples from ES54, ES89, and ES105 were not available, we tested relative TCID50 of the local primary isolate HIV-11192 in three low producers compared to five volunteers with normal HIV-1 production. Median TCID50 for the three low producers assessed was 3.2 (range, 1.9–9.4) compared to 0.6 (range, 0.3–0.8) for five normal producers. Again, TCID50 values for the two groups were nonoverlapping, and despite the very small numbers of samples available for this experiment, this difference was statistically significant (p-value = 0.025), reflecting a strong biological difference in HIV-11192 susceptibility between low and normal producers.
Table 4.
Minimum Infecting Inoculum Required to Establish Infection Is Increased in Low Producers Relative to Normally Susceptible Volunteers
| |
TCID50 |
|
|---|---|---|
| |
HIV-1 strain |
|
| JR-CSF | 1192 | |
| Low producers | ||
| C24 | 7.07 | 1.88 |
| C38 | 81.43 | 9.38 |
| ES29 | 17.68 | 3.21 |
| ES54 | 24.00 | nda |
| ES89 | 9.60 | nd |
| ES105 | 15.18 | nd |
| Median | 16.43 | 3.21 |
| Normally susceptible | ||
| ES31 | nd | 0.84 |
| ES36 | 0.97 | 0.84 |
| ES42 | nd | 0.29 |
| ES76 | 1.13 | 0.64 |
| ES96 | 1.79 | 0.64 |
| ES102 | 1.54 | nd |
| Median | 1.33 | 0.64 |
| p-valueb | 0.0095 | 0.025 |
nd, not done.
Mann-Whitney statistical comparison.
Low-producer phenotype not dependent on CCR5-mediated entry
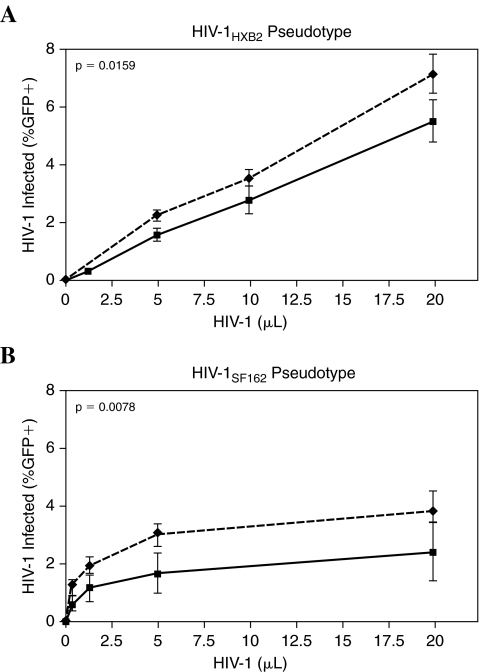
We evaluated whether diminished infection in low producers is dependent on the coreceptor usage of the infecting HIV-1 strain. We assessed the frequency of infection by a GFP-encoding HIV-1LAI vector pseudotyped with the HIV-1SF162 or HIV-1HXB2 envelopes to measure HIV-1 susceptibility based on viral entry alone. Infection frequency was quantified over a range of viral inputs, and the area under the curve was determined for each of 15 volunteers (six low producers and nine normally susceptible volunteers) over the dynamic range of the assay. We observed a modest reduction in infection among low producers regardless of whether the HIV-1SF162 or HIV-1HXB2 pseudotype was used (Fig. 4). Infection with the HIV-1HXB2 pseudotype resulted in 1.4-fold greater infection (mean area under the curve) in normal producers than in low producers (p-value = 0.016), whereas the HIV-1SF162 pseudotype produced 1.8-fold greater infection in normal producers than in low producers (p-value = 0.008). In addition, among infected cells there was a modest but significant reduction in GFP production observed in low producers (data not shown). Infection with the HIV-1HXB2 pseudotype resulted in median 1.3-fold greater intracellular GFP in normal producers than in low producers (p-value = 0.003), while infection with the HIV-1SF162 pseudotype yielded a median 1.2-fold greater GFP expression in normal producers than in low producers (p-value = 0.002). While the impairment in Tat-driven GFP expression in first-round HIV-1 infection was small in magnitude, the strong statistical significance of this reduction mirrors that detected in our assessment of Gag expression following first-round infection with native virus, above. The reduction in low-producer infection observed during single-round infection with HIV-1-enveloped pseudotypes may be amplified over extended infection to yield the marked reduction in HIV-1 susceptibility apparent in TCID50 or p24 production following challenge with either HIV-1JR-CSF or HIV-11192. Because this effect is observed when either the R5 or X4 HIV-1 envelope is used for entry, the low-producer phenotype cannot be attributed to a reduction in viral entry due to coreceptor tropism.
FIG. 4.
Coreceptor tropism only partially contributes to the low-producer phenotype. The frequency of infection is shown in six low producers (solid lines) and nine normally susceptible subjects (dashed lines) when infected with HIV-1LAIΔenvGFP vector pseudotyped with (A) HIV-1HXB2 or (B) HIV-1SF162, envelope. Infection with an HIV-1HXB2 pseudotype was 1.4-fold greater in normal producers than in low producers. Infection with HIV-1SF162 pseudotype was 1.8-fold greater in normal producers than in low producers. Mean background-subtracted results are shown. The experiment was performed in duplicate and was repeated in three low producers and four normally susceptible volunteers.
Similarly, surface expression of CCR5 on resting CD4+ T cells also did not differ between five of six low-producer volunteers (C38 not assessed due to lack of cells) and 73/99 volunteers having normal susceptibility. Normally susceptible individuals who were tested for CCR5 expression exhibited the same range of p24 production for infection with HIV-1JR-CSF at an MOI of 0.003 (median log10 4.47) as all 99 normal producers screened (median log10 4.49). The frequency of CCR5 expression among CD3+CD4+ T cells did not differ between samples from low producers (median 13.4%, range, 8.6–31.8%) and normal producers (median 20.0%, range 4.5–62.9%), with low producer CCR5+ cell frequency falling between the 15th and 86th percentiles (p-value = 0.557, data not shown). The median CCR5 density in low producers was 2547 (range, 1198–2751) compared with the median of 3761 (range, 1205–8859) in normally susceptible individuals, a trend that did not reach statistical significance (p-value = 0.08). Twenty-nine volunteers having similar or less CCR5 surface expression than the low producers were capable of producing as much as 3 logs greater p24 Ag during in vitro infection. Thus, low CCR5 expression may contribute to the low-producer phenotype, but cannot fully explain it.
Finally, we investigated the level of β-chemokines, the natural ligands of CCR5, in the culture supernatants following infection with either HIV-1JR-CSF or HIV-11192 in the six low producers and in 13 participants having normal in vitro susceptibility. No differences in the concentration of MIP-1α, MIP-1β, or RANTES were observed in supernatants harvested at 7 dpi between the two groups (data not shown). Furthermore, the MIP-1αP copy number was unexceptional in the four low-producer volunteers in whom it was determined (ES29, one copy/cell; ES54, ES89, ES105, two copies/cell). Taken together, we conclude that the low-producer phenotype observed in these six individuals cannot be attributed to diminished viral entry into CD4+ T cells, in contrast to previous findings in other ES cohorts.18,32–36
Basic reproductive number is strongly diminished in HIV-1 low producers
To quantify the potential effect of diminished CD4+ T cell susceptibility on the probability of in vivo infection, we fit our data to a rate-equation (ODE) model of HIV-1 infection of CD4+ T cells in order to estimate the basic reproductive number in vitro (R0) for six low producers and four normally susceptible volunteers. Because data were most complete for experiments employing HIV-1JR-CSF, we used this virus strain in our rate-equation model. The R0 values show a clear distinction between these two groups (Table 5). The low-producer phenotype was associated with a substantial reduction in R0, with a median R0 of 1.575 (range, 1.13–1.8) compared to a median R0 of 5.45 (range, 5.2–6.6) in volunteers having normal susceptibility. The difference in R0 values between these two groups was strongly significant, with p-value = 0.010.
Table 5.
Basic Reproductive Number in Vitro is Significantly Reduced in Six Low Producers Relative to Four Normally Susceptible Volunteers
| R0 | 95% CIa | p-valueb | |
|---|---|---|---|
| Low producers | |||
| C24 | 1.7 | (1.5, 1.7) | |
| C38 | 1.45 | (1.33, 1.53) | |
| ES29 | 1.25 | (1.15, 1.33) | |
| ES54 | 1.7 | (1.6, 1.8) | |
| ES89 | 1.13 | (1.11, 1.18) | |
| ES105 | 1.8 | (1.4, 2.0) | |
| Median | 1.58 | ||
| Normally susceptible | 0.095 | ||
| ES04 | 5.2 | (3.2, 7.3) | |
| ES42 | 5.7 | (4.7, 6.7) | |
| ES76 | 6.6 | (5.7, 7.5) | |
| ES96 | 5.2 | (4.6, 5.7) | |
| Median | 5.45 | ||
Log-likelihood ratio test.
Mann-Whitney statistical comparison.
Discussion
While the role of host proteins affecting viral entry has been vigorously investigated in ES cohorts, the importance of downstream events in maintaining prolonged seronegativity has been largely overlooked. Recent identification of cellular antiviral proteins such as TRIM5α, which disrupts reverse transcription,56,57 and APOBEC3G and APOBEC3F, which induce G to A hypermutation in viral genomes,58–64 suggests that postentry restriction of HIV-1 replication may explain the ES phenotype. Resequencing of TRIM5α in this ES cohort did not identify any polymorphisms resulting in resistance to HIV-1 infection in vitro.65 While APOBEC3 genetic polymorphisms have not been thoroughly examined in ES cohorts, such groups may be enriched for rare variants having increased deamination activity or the ability to escape HIV-1 Vif-induced degradation. Likewise, the ability of Vpu to counteract endocytosis of nascent virions66 might be diminished by as-yet unidentified mutations within the CD317 tetherin gene that allowed CD317 escape from binding by Vpu.67 Certain ES individuals may be protected from HIV-1 infection via postentry restriction due to alteration of these host-virus interactions.
Alternatively, continued seronegativity in ES may result from a mutation abrogating interaction between a required host protein and its viral partner. Myriad host proteins are complicit in viral disassembly,68,69 nuclear import,70 genomic integration,71 assembly,72 and budding.73,74 The level of genetic variation in host proteins required for late events, such as ABCE1, remains unknown. Whether polymorphisms in required host proteins result in abortive infection in select ES requires investigation. Elucidation of the heterogeneous mechanisms by which ES volunteers evade infection may have profound impact on the treatment of HIV-1-infected individuals. While CCR5Δ32 homozygosity provides protection for only a minority of ES persons, their identification has contributed to our understanding of HIV-1 entry requirements and the eventual development of entry inhibitors such as maraviroc and vicriviroc.
In this study, we investigated the possibility that diminished CD4+ T cell susceptibility to productive HIV-1 infection may contribute to continued seronegativity among highly exposed persons. Despite the commonly held view that primary CD4+ T cell susceptibility varies substantially between subjects, we found that cells from 94% (99/105) of volunteers supported HIV-1JR-CSF infection similarly, with only a 1.36 log-unit difference in p24 produced at 7 dpi. Similar results were observed following infection with HIV-11192, with 94% (88/94) of volunteers demonstrating a 1.47 log-unit range in p24 production. Importantly, our findings indicate that with rare exceptions, exposed seronegative CD4+ T cells, upon activation in vitro, can efficiently support HIV-1 replication, and the levels of viral production and susceptibility are similar to those from normal low-risk control donors. This confirms some previous findings28,31,35,36 but stands in contrast to others.18,42,75 However, here we have undertaken more extensive analysis using a well-defined target cell population in greater numbers of ES and low-risk control volunteers than has been performed previously.
The pattern of viral replication was consistently observed with a panel of R5 HIV-1 strains, including the molecular clone HIV-1JR-CSF and two local primary isolates, tested at two or more MOIs. The remarkably narrow range of permissivity to viral infection permitted recognition of volunteers having profound defects in CD4+ T cell susceptibility. Indeed, we identified six volunteers, unrelated to HIV-1 risk, with significantly diminished capacity to sustain HIV-1 infection in vitro. In these six individuals, HIV-1 production by CD4+ T cells during extended infection with HIV-1JR-CSF or HIV-11192 was markedly reduced (up to 2500-fold). This finding was reproducible and persistent, with similar observations made in cells isolated on different venipuncture dates spanning a median of 9.1 months. While low producers repeatedly demonstrated a statistically significant decrease both in percent HIV-1-infected cells and in Tat-mediated protein expression following single-round infection assays involving both native HIV-1 and pseudotype infection, the modest size of these differences impedes interpretation of their biological importance. Finally, a substantially larger viral inoculum was required to establish infection in activated CD4+ T cells, with a median 12-fold increase in HIV-1JR-CSF TCID50 in low-producer volunteers compared to those with normal susceptibility (p-value = 0.010), and a median 5-fold increase in TCID50 when HIV-11192 was used to challenge (p-value = 0.025). Although variation in donor cell permissivity has been reported in several investigations,25–27 this very low-level HIV-1 production unrelated to CCR5 or β-chemokine genotypic polymorphisms or expression is striking and suggests that alterations in the activities of other host factors supporting the HIV-1 life cycle may contribute to HIV-1 resistance.
Differences in viral entry cannot explain the low producer phenotype. No differences in β-chemokine expression, frequency of CCR5+CD4+ T cells, or density of CCR5 surface expression were identified in the low producers. In addition, low producers did not differ from normal producers in Gag positivity following exposure to HIV-1JR-CSF in the presence of AZT. These findings are consistent with the report of a postentry block in a single volunteer in a Vietnamese ES cohort, whereas barriers to CCR5-mediated viral entry were found in four additional low producers.76 Furthermore, we observed a strongly significant reduction in infection when either R5 (p-value = 0.008) or X4 (p-value 0.015) HIV-1-enveloped pseudotypes were used to challenge, replicating our earlier single-round infection assay results while holding the viral backbone constant and varying only the viral coreceptor utilized. The consistent observation of a modest but reproducible and statistically significant reduction in Tat-mediated gene expression may indicate the mechanism responsible for the low-producer phenotype, but the biological significance of this observation is unclear and requires further investigation. Additional studies, including quantitation of late reverse transcripts and integrated viral DNA, are needed to identify the step in viral replication restricted in these cells. Furthermore, restricted HIV-1 replication allowing viral gene expression but leading to abortive infection might be associated with HIV-1-specific immune responses. While ES29 was one of the few volunteers in whom we observed broad, strong virus-specific immune responses, similar findings were not obtained in ES54.14 Investigation of HIV-1-specific immune responses in the remaining low-producer ES volunteers is needed.
Taken together, these findings suggest that a small percentage of persons (6% in this study) may have the ability to acquire HIV-1 infection but not support sufficiently high levels of replication and cell-to-cell spread. Of note, this may be the situation occurring in those ES previously identified as having very low level HIV-1 in CD4+ T cells without seroconversion.77,78 This hypothesis is further supported by the calculated 3.5-fold decrease in the in vitro basic reproductive number, R0 (p-value = 0.010). In ODE models, R0 > 1.0 implies that infection will spread within a community, whereas R0 < 1.0 predicts that an infection will become extinct. We acknowledge that the in vitro experimental conditions, such as strong CD4+ T cell activation and sustained proliferation mediated by relatively high IL-2 concentration, do not permit direct extrapolation of the reported values of R0 to those in vivo. However, the relative values of R0 may reflect significant biological differences between the two groups (normal and low producers). HIV-1 is poorly transmissible, and it has been suggested that the remarkably low rate of HIV-1 transmission by sexual exposure may be explained by extinction.79 Lower R0 values increase the probability that infection will fail to propagate in a community, or fail to transmit to an exposed individual. Therefore, the low in vitro R0 calculated here may provide an explanation as to why a small number of ES individuals in our cohort remain uninfected.
In conclusion, while altered CD4+ T cell susceptibility to HIV-1 infection does not mediate continued seronegativity for the majority of participants in this ES cohort, a minority of ES persons appear to restrict HIV-1 infection following HIV-1 entry. Among these select volunteers, diminished CD4+ T cell susceptibility to HIV-1 might contribute to continued seronegativity despite frequent sexual exposures. Further dissection of the mechanisms responsible for HIV-1 restriction in these select volunteers will elucidate postentry host-viral interactions, which may identify novel targets for future antiretroviral therapeutics and genetic polymorphisms contributing to relative HIV-1 resistance.
Acknowledgments
We are indebted to the volunteers whose participation in this study made these investigations possible. Michael Emerman provided invaluable scientific discussion and reagents. We are grateful to John Mascola for his help in implementing the protocol for first-round infection in our laboratory. We thank Jean Lee and Julie Cooper for volunteer recruitment, Deborah Lee for database management, and Phyllis Stegall for editorial assistance. This work was supported by National Institutes of Health Grants AI47086, AI35605, and AI057005. E.C.S. is supported by National Institutes of Health T32 Grants AI007140 and GM0726, with funding from the Seattle Chapter of ARCS (Achievement Rewards for College Scientists) and the Poncin Scholarship Fund. M.J.M. is a recipient of the Burroughs Wellcome Clinical Scientist Award for Translational Research.
Disclosure Statement
No competing financial interests exist.
References
- 1.Beyrer c. Artenstein Aw. Rugpao S, et al. Epidemiologic and biologic characterization of a cohort of human immunodeficiency virus type 1 highly exposed, persistently seronegative female sex workers in northern Thailand. Chiang Mai HEPS Working Group. J Infect Dis. 1999;179:59–67. doi: 10.1086/314556. [DOI] [PubMed] [Google Scholar]
- 2.Fowke KR. Nagelkerke NJ. Kimani J, et al. Resistance to HIV-1 infection among persistently seronegative prostitutes in Nairobi, Kenya. Lancet. 1996;348:1347–1351. doi: 10.1016/S0140-6736(95)12269-2. [DOI] [PubMed] [Google Scholar]
- 3.Goh WC. Markee J. Akridge RE, et al. Protection against human immunodeficiency virus type 1 infection in persons with repeated exposure: Evidence for T cell immunity in the absence of inherited CCR5 coreceptor defects. J Infect Dis. 1999;179:548–557. doi: 10.1086/314632. [DOI] [PubMed] [Google Scholar]
- 4.Rowland-Jones S. Sutton J. Ariyoshi K, et al. HIV-specific cytotoxic T-cells in HIV-exposed but uninfected Gambian women. Nat Med. 1995;1:59–64. doi: 10.1038/nm0195-59. [DOI] [PubMed] [Google Scholar]
- 5.Follezou JY. Lan NY. Lien TX, et al. Clinical and biological characteristics of human immunodeficiency virus-infected and uninfected intravascular drug users in Ho Chi Minh City, Vietnam. Am J Trop Med Hyg. 1999;61:420–424. doi: 10.4269/ajtmh.1999.61.420. [DOI] [PubMed] [Google Scholar]
- 6.Alimonti JB. Kimani J. Matu L, et al. Characterization of CD8 T-cell responses in HIV-1-exposed seronegative commercial sex workers from Nairobi, Kenya. Immunol Cell Biol. 2006;84:482–485. doi: 10.1111/j.1440-1711.2006.01455.x. [DOI] [PubMed] [Google Scholar]
- 7.Ball TB. Ji H. Kimani J. McLaren P. Marlin C. Hill AVS. Plummer FA. Polymorphisms in IRF-1 associated with resistance to HIV-1 infection in highly exposed uninfected Kenyan sex workers. AIDS. 2007;21:1091–1101. doi: 10.1097/QAD.0b013e3280ef6ae1. [DOI] [PubMed] [Google Scholar]
- 8.Bégaud E. Chartier L. Marechal V, et al. Reduced CD4 T cell activation and in vitro susceptibility to HIV-1 infection in exposed uninfected Central Africans. Retrovirology. 2006;3:35. doi: 10.1186/1742-4690-3-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belec L. Ghys PD. Hocini H, et al. Cervicovaginal secretory antibodies to human immunodeficiency virus type 1 (HIV-1) that block viral transcytosis through tight epithelial barriers in highly exposed HIV-1-seronegative African women. J Infect Dis. 2001;184:1412–1422. doi: 10.1086/324375. [DOI] [PubMed] [Google Scholar]
- 10.Bernard NF. Yannakis CM. Lee JS. Tsoukas CM. Human immunodeficiency virus (HIV)-specific cytotoxic T lymphocyte activity in HIV-exposed seronegative persons. J Infect Dis. 1999;179:538–547. doi: 10.1086/314621. [DOI] [PubMed] [Google Scholar]
- 11.Clerici M. Giorgi JV. Chou CC, et al. Cell-mediated immune response to human immunodeficiency virus (HIV) type 1 in seronegative homosexual men with recent sexual exposure to HIV-1. J Infect Dis. 1992;165:1012–1019. doi: 10.1093/infdis/165.6.1012. [DOI] [PubMed] [Google Scholar]
- 12.Clerici M. Levin JM. Kessler HA, et al. HIV-specific T-helper activity in seronegative health care workers exposed to contaminated blood. JAMA. 1994;271:42–46. [PubMed] [Google Scholar]
- 13.Eyeson J. King D. Boaz MJ, et al. Evidence for Gag p24-specific CD4 T cells with reduced susceptibility to R5 HIV-1 infection in a UK cohort of HIV-exposed-seronegative subjects. AIDS. 2003;17:2299–2311. doi: 10.1097/00002030-200311070-00004. [DOI] [PubMed] [Google Scholar]
- 14.Hladik F. Desbien A. Lang J, et al. Most highly exposed seronegative men lack HIV-1-specific, IFN-gamma-secreting T cells. J Immunol. 2003;171:2671–2683. doi: 10.4049/jimmunol.171.5.2671. [DOI] [PubMed] [Google Scholar]
- 15.Kaul R. Dong T. Plummer FA, et al. CD8(+) lymphocytes respond to different HIV epitopes in seronegative and infected subjects. J Clin Invest. 2001;107:1303–1310. doi: 10.1172/JCI12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaul R. Plummer FA. Kimani J, et al. HIV-1-specific mucosal CD8+ lymphocyte responses in the cervix of HIV-1-resistant prostitutes in Nairobi. J Immunol. 2000;164:1602–1611. doi: 10.4049/jimmunol.164.3.1602. [DOI] [PubMed] [Google Scholar]
- 17.Kebba A. Kaleebu P. Rowland S, et al. Distinct patterns of peripheral HIV-1-specific interferon-gamma responses in exposed HIV-1-seronegative individuals. J Infect Dis. 2004;189:1705–1713. doi: 10.1086/383227. [DOI] [PubMed] [Google Scholar]
- 18.Koning FA. Jansen CA. Dekker J, et al. Correlates of resistance to HIV-1 infection in homosexual men with high-risk sexual behaviour. AIDS. 2004;18:1117–1126. doi: 10.1097/00002030-200405210-00005. [DOI] [PubMed] [Google Scholar]
- 19.Mazzoli S. Trabattoni D. Lo Caputo S, et al. HIV-specific mucosal and cellular immunity in HIV-seronegative partners of HIV-seropositive individuals. Nat Med. 1997;3:1250–1257. doi: 10.1038/nm1197-1250. [DOI] [PubMed] [Google Scholar]
- 20.Nicastri E. Sarmati L. Ercoli L, et al. Reduction of IFN-gamma and IL-2 production by peripheral lymphocytes of HIV-exposed seronegative subjects. AIDS. 1999;13:1333–1336. doi: 10.1097/00002030-199907300-00009. [DOI] [PubMed] [Google Scholar]
- 21.Rowland-Jones SL. Dong T. Fowke KR, et al. Cytotoxic T cell responses to multiple conserved HIV epitopes in HIV-resistant prostitutes in Nairobi. J Clin Invest. 1998;102:1758–1765. doi: 10.1172/JCI4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmechel SC. Russell N. Hladik F, et al. Immune defence against HIV-1 infection in HIV-1-exposed seronegative persons. Immunol Lett. 2001;79:21–27. doi: 10.1016/s0165-2478(01)00262-0. [DOI] [PubMed] [Google Scholar]
- 23.Tran HK. Chartier L, et al. Systemic immune activation in HIV-1-exposed uninfected Vietnamese intravascular drug users. AIDS Res Hum Retroviruses. 2006;22:255–261. doi: 10.1089/aid.2006.22.255. [DOI] [PubMed] [Google Scholar]
- 24.Blaak H. van't Wout AB. Brouwer M, et al. Infectious cellular load in human immunodeficiency virus type 1 (HIV-1)-infected individuals and susceptibility of peripheral blood mononuclear cells from their exposed partners to non-syncytium-inducing HIV-1 as major determinants for HIV-1 transmission in homosexual couples. J Virol. 1998;72:218–224. doi: 10.1128/jvi.72.1.218-224.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spira AI. Ho DD. Effect of different donor cells on human immunodeficiency virus type 1 replication and selection in vitro. J Virol. 1995;69:422–429. doi: 10.1128/jvi.69.1.422-429.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ciuffi A. Bleiber G. Munoz M, et al. Entry and transcription as key determinants of differences in CD4 T-cell permissiveness to human immunodeficiency virus type 1 infection. J Virol. 2004;78:10747–10754. doi: 10.1128/JVI.78.19.10747-10754.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eisert V. Kreutz M. Becker K, et al. Analysis of cellular factors influencing the replication of human immunodeficiency virus type I in human macrophages derived from blood of different healthy donors. Virology. 2001;286:31–44. doi: 10.1006/viro.2001.0940. [DOI] [PubMed] [Google Scholar]
- 28.Koning FA. Otto SA. Hazenberg MD, et al. Low-level CD4+ T cell activation is associated with low susceptibility to HIV-1 infection. J Immunol. 2005;175:6117–6122. doi: 10.4049/jimmunol.175.9.6117. [DOI] [PubMed] [Google Scholar]
- 29.Goldstein S. Brown CR. Dehghani H. Lifson JD. Hirsch VM. Intrinsic susceptibility of rhesus macaque peripheral CD4(+) T cells to simian immunodeficiency virus in vitro is predictive of in vivo viral replication. J Virol. 2000;74:9388–9395. doi: 10.1128/jvi.74.20.9388-9395.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu R. Paxton WA. Choe S, et al. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 31.Paxton WA. Martin SR. Tse D, et al. Relative resistance to HIV-1 infection of CD4 lymphocytes from persons who remain uninfected despite multiple high-risk sexual exposure. Nat Med. 1996;2:412–417. doi: 10.1038/nm0496-412. [DOI] [PubMed] [Google Scholar]
- 32.Paxton WA. Liu R. Kang S, et al. Reduced HIV-1 infectability of CD4+ lymphocytes from exposed-uninfected individuals: Association with low expression of CCR5 and high production of beta-chemokines. Virology. 1998;244:66–73. doi: 10.1006/viro.1998.9082. [DOI] [PubMed] [Google Scholar]
- 33.John R. Arango-Jaramillo S. Finny GJ. Schwartz DH. Risk associated HIV-1 cross-clade resistance of whole peripheral blood mononuclear cells from exposed uninfected individuals with wild-type CCR5. J Acquir Immune Defic Syndr. 2004;35:1–8. doi: 10.1097/00126334-200401010-00001. [DOI] [PubMed] [Google Scholar]
- 34.Paxton WA. Kang S. Liu R, et al. HIV-1 infectability of CD4+ lymphocytes with relation to beta-chemokines and the CCR5 coreceptor. Immunol Lett. 1999;66:71–75. doi: 10.1016/s0165-2478(98)00154-0. [DOI] [PubMed] [Google Scholar]
- 35.Fowke KR. Dong T. Rowland-Jones SL, et al. HIV type 1 resistance in Kenyan sex workers is not associated with altered cellular susceptibility to HIV type 1 infection or enhanced beta-chemokine production. AIDS Res Hum Retroviruses. 1998;14:1521–1530. doi: 10.1089/aid.1998.14.1521. [DOI] [PubMed] [Google Scholar]
- 36.Messele T. Rinke de Wit TF. Brouwer M, et al. No difference in in vitro susceptibility to HIV type 1 between high-risk HIV-negative Ethiopian commercial sex workers and low-risk control subjects. AIDS Res Hum Retroviruses. 2001;17:433–441. doi: 10.1089/088922201750102526. [DOI] [PubMed] [Google Scholar]
- 37.Iqbal SM. Ball TB. Kimani J. Kiama P. Thottingal P. Embree JE. Fowke KR. Plummer FA. Elevated T cell counts and RANTES expression in the genital mucosa of HIV-1-resistant Kenyan commercial sex workers. J Infect Dis. 2005;192:728–738. doi: 10.1086/432482. [DOI] [PubMed] [Google Scholar]
- 38.Suresh M. Wanchu A, et al. Spontaneous and antigen-induced chemokine production in exposed but uninfected partners of HIV type 1-infected individuals in North India. AIDS Res Hum Retroviruses. 2007;23:261–268. doi: 10.1089/aid.2006.0146. [DOI] [PubMed] [Google Scholar]
- 39.Thomas SM. Tse DB, et al. CCR5 expression and duration of high risk sexual activity among HIV-seronegative men who have sex with men. AIDS. 2007;20:1879–1883. doi: 10.1097/01.aids.0000244207.49123.ff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gonzalez E. Kulkarni H. Bolivar H, et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science. 2005;307:1434–1440. doi: 10.1126/science.1101160. [DOI] [PubMed] [Google Scholar]
- 41.Shao W TJ. Song W. Wang C. Li Y. Wilson CM. Kaslow RA. CCL3L1 and CCL4L1: Variable gene copy number in adolescents with and without human immunodeficiency virus type 1 (HIV-1) infection. Genes Immun. 2007;8:224–231. doi: 10.1038/sj.gene.6364378. [DOI] [PubMed] [Google Scholar]
- 42.Truong LX. Luong TT. Scott-Algara D, et al. CD4 cell and CD8 cell-mediated resistance to HIV-1 infection in exposed uninfected intravascular drug users in Vietnam. AIDS. 2003;17:1425–1434. doi: 10.1097/00002030-200307040-00002. [DOI] [PubMed] [Google Scholar]
- 43.Akridge R. Hladik F. Markee J, et al. Cellular immunity and target cell susceptibility in persons with repeated HIV-1 exposure. Immunol Lett. 1999;66:15–19. doi: 10.1016/s0165-2478(98)00180-1. [DOI] [PubMed] [Google Scholar]
- 44.Bartz SR. Vodicka MA. Production of high-titer human immunodeficiency virus type 1 pseudotyped with vesicular stomatitis virus glycoprotein. Methods. 1997;12:337–342. doi: 10.1006/meth.1997.0487. [DOI] [PubMed] [Google Scholar]
- 45.Cann AJ. Zack JA. Go AS, et al. Human immunodeficiency virus type 1 T-cell tropism is determined by events prior to provirus formation. J Virol. 1990;64:4735–4742. doi: 10.1128/jvi.64.10.4735-4742.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kimpton J. Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vodicka MA. Goh WC. Wu LI, et al. Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology. 1997;233:193–198. doi: 10.1006/viro.1997.8606. [DOI] [PubMed] [Google Scholar]
- 48.Deep R. Probability and Statistics: Sampling, Data Displays, Measure of Central Tendencies. Academic Press; San Diego: 2006. [Google Scholar]
- 49.Mendenhall W. Beaver R. Beaver B. Describing data with numerical measures. In: Mendenhall W, editor; Beaver R, editor; Beaver B, editor. Introduction to Probability and Statistics. Duxbury Press; Pacific Grove, CA: 2005. [Google Scholar]
- 50.Mascola JR. Louder MK. Winter C, et al. Human immunodeficiency virus type 1 neutralization measured by flow cytometric quantitation of single-round infection of primary human T cells. J Virol. 2002;76:4810–4821. doi: 10.1128/JVI.76.10.4810-4821.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamashita M. Emerman M. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J Virol. 2004;78:5670–5678. doi: 10.1128/JVI.78.11.5670-5678.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O'Doherty U. Swiggard WJ. Malim MH. Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J Virol. 2000;74:10074–10080. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reed LJ. Muench H. A simple method of estimating fifty per cent endpoint. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 54.Quillent C. Oberlin E. Braun J, et al. HIV-1-resistance phenotype conferred by combination of two separate inherited mutations of CCR5 gene. Lancet. 1998;351:14–18. doi: 10.1016/S0140-6736(97)09185-X. [DOI] [PubMed] [Google Scholar]
- 55.Hladik F. Liu H. Speelmon E, et al. Combined effect of CCR5-delta32 heterozygosity and the CCR5 promoter polymorphism: 2459 A/G on CCR5 expression and resistance to human immunodeficiency virus type 1 transmission. J Virol. 2005;79:11677–11684. doi: 10.1128/JVI.79.18.11677-11684.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stremlau M. Owens CM. Perron MJ. Kiessling M. Autissier P. Sodroski J. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427:848–853. doi: 10.1038/nature02343. [DOI] [PubMed] [Google Scholar]
- 57.Munk C. Brandt SM. Lucero G. Landau NR. A dominant block to HIV-1 replication at reverse transcription in simian cells. Proc Natl Acad Sci USA. 2002;99:13843–13848. doi: 10.1073/pnas.212400099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lecossier D. Bouchonnet F. Clavel F. Hance AJ. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science. 2003;300:1112. doi: 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- 59.Sheehy AM. Gaddis NC. Choi JD. Malim MH. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- 60.Zheng YH. Irwin D. Kurosu T. Tokunaga K. Sata T. Peterlin BM. Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J Virol. 2004;78:6073–6076. doi: 10.1128/JVI.78.11.6073-6076.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang H. Yang B. Pomerantz RJ. Zhang C. Arunachalam SC. Gao L. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mangeat B. Turelli P. Caron G. Friedli M. Perrin L. Trono D. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 63.Harris RS. Bishop KN. Sheehy AM, et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 64.Wiegand HL. Doehle BP. Bogerd HP. Cullen BR. A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J. 2004;23:2451–2458. doi: 10.1038/sj.emboj.7600246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Speelmon EC. Livingston-Rosanoff D. Li SS, et al. Genetic association of the antiviral restriction factor TRIM5alpha with human immunodeficiency virus type 1 infection. J Virol. 2006;80:2463–2471. doi: 10.1128/JVI.80.5.2463-2471.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Neil SJ ES. Jouvenet N. Bieniasz PD. HIV-1 Vpu promotes release and prevents endocytosis of nascent retrovirus particles from the plasma membrane. PLoS Pathog. 2006;2:e39. doi: 10.1371/journal.ppat.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Neil SJD. Zang T. Bieniasz PD. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 68.Franke EK. Yuan HE. Luban J. Specific incorporation of cyclophilin A into HIV-1 virions. Nature. 1994;372:359–362. doi: 10.1038/372359a0. [DOI] [PubMed] [Google Scholar]
- 69.Cartier C. Sivard P. Tranchat C. Decimo D. Desgranges C. Boyer V. Identification of three major phosphorylation sites within HIV-1 capsid. Role of phosphorylation during the early steps of infection. J Biol Chem. 1999;274:19434–19440. doi: 10.1074/jbc.274.27.19434. [DOI] [PubMed] [Google Scholar]
- 70.Llano M. Vanegas M. Fregoso O, et al. LEDGF/p75 determines cellular trafficking of diverse lentiviral but not murine oncoretroviral integrase proteins and is a component of functional lentiviral preintegration complexes. J Virol. 2004;78:9524–9537. doi: 10.1128/JVI.78.17.9524-9537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen H. Engelman A. The barrier-to-autointegration protein is a host factor for HIV type 1 integration. Proc Natl Acad Sci USA. 1998;95:15270–15274. doi: 10.1073/pnas.95.26.15270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zimmerman C. Klein KC. Kiser PK, et al. Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature. 2002;415:88–92. doi: 10.1038/415088a. [DOI] [PubMed] [Google Scholar]
- 73.Martin-Serrano J. Zang T. Bieniasz PD. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001;7:1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- 74.Garrus JE. von Schwedler UK. Pornillos OW, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- 75.Williams L. Cloyd M. Polymorphic human gene(s) determines differential susceptibility of CD4 lymphocytes to infection by certain HIV-1 isolates. Virology. 1991;184:723–728. doi: 10.1016/0042-6822(91)90442-e. [DOI] [PubMed] [Google Scholar]
- 76.Sáez-Cirión A. Versmisse P. Truong LX. Chakrabarti LA. Carpentier W. Barré-Sinoussi F. Scott-Algara D. Pancino G. Persistent resistance to HIV-1 infection in CD4 T cells from exposed uninfected Vietnamese individuals is mediated by entry and post-entry blocks. Retrovirology. 2006;8:81. doi: 10.1186/1742-4690-3-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhu T. Corey L. Hwangbo Y, et al. Persistence of extraordinarily low levels of genetically homogeneous human immunodeficiency virus type 1 in exposed seronegative individuals. J Virol. 2003;77:6108–6116. doi: 10.1128/JVI.77.11.6108-6116.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koning FA. van der Vorst TJ. Schuitemaker H. Low levels of human immunodeficiency virus type 1 DNA in high-risk seronegative men. J Virol. 2005;79:6551–6553. doi: 10.1128/JVI.79.10.6551-6553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wick D. Self SG. Early HIV infection in vivo: Branching-process model for studying timing of immune responses and drug therapy. Math Biosci. 2000;165:115–134. doi: 10.1016/s0025-5564(00)00013-4. [DOI] [PubMed] [Google Scholar]