Abstract
Two different ligand occupancy structures of cytochrome P450 2B4 (CYP2B4) in complex with 1-biphenyl-4-methyl-1H–imidazole (1-PBI) have been solved by x-ray crystallography. 1-PBI belongs to a series of tight binding, imidazole-based CYP2B4 inhibitors. 1-PBI binding to CYP2B4 yields a type II spectrum with a Ks value of 0.23 µM and inhibits enzyme activity with an IC50 value of 0.035 µM. Previous CYP2B4 structures have shown a large degree of structural movement in response to ligand size. With two phenyl rings, 1-PBI is larger than 1-(4-chlorophenyl)imidazole (1-CPI) and 4-(4-chlorophenyl)imidazole (4-CPI) but smaller than bifonazole, which is branched and contains three phenyl rings. The CYP2B4:1-PBI complex is a structural intermediate to the closed CPI and the open bifonazole structures. The B/C loop reorganizes itself to include two short partial helices while closing one side of the active site. The F-G helix cassette pivots over the I-helix in direct response to the size of the ligand in the active site. A cluster of Phe residues at the fulcrum of this pivot point allows for dramatic repositioning of the cassette with only a relatively small amount of secondary structure rearrangement. Comparisons of ligand bound CYP2B4 structures reveal trends in plastic region mobility that could allow for predictions of their position in future structures based on ligand shape and size.
Keywords: cytochrome P450 2B4, 1-biphenyl-4-methyl-1H–imidazole, 1-PBI, x-ray crystallography, plastic regions, F-G helix cassette
Cytochromes P450 (P450s) are a superfamily of heme-containing monooxygenase enzymes that are involved in the biogenesis of sterols and hormones and the oxidation of numerous endogenous and xenobiotic compounds (1). However, it is the crucial role of P450s in phase I metabolism of drugs and drug candidates that has led to intense interest from the healthcare and pharmaceutical industries (2). By hydroxylating largely hydrophobic drugs, P450s reduce the lipid solubility of these compounds and generate water soluble products that are suitable for excretion (3). Unlike the classical lock and key enzyme models, many P450s are able to incorporate a single oxygen atom into a large number of biological substrates of varying size and shape with a high degree of stereo- and regiospecificity. Determining the structural and mechanistic basis of this versatility has been the focus of numerous structural investigations (4–6).
Despite the large range of substrates, the large, single-domain P450 fold is remarkably conserved across families. Accordingly, the ability to accommodate molecules with such high variability reflects the remarkable flexibility of many secondary structure elements (7, 8). P450s are able to compact their structures in order to close around empty active sites or smaller ligands as seen in CYP2B4 with 4-CPI and 1-CPI (9, 10), CYP3A4 (11, 12), and numerous examples from the 2C subfamily (13–17). In contrast, P450s are also able to dramatically alter their structures to accommodate larger, bulkier ligands as seen in CYP2B4 with bifonazole (18).
Research in our laboratory has sought to gain insight into this flexibility through crystallographic studies of an N-terminal modified form of cytochrome P450 2B4 (CYP2B4dH) in complex with imidazole-based inhibitors of varying size and shape (9). The P450 2B subfamily exhibits a relatively low level of catalytic conservation across species, making these enzymes an ideal candidate for structure-function studies (19). Comparisons of the ligand free (20), 4-CPI and 1-CPI (9, 10), and bifonazole (18) bound structures reveal plastic regions that are able to reposition themselves in order to accommodate the different inhibitors while still maintaining the overall P450 fold. The two largest plastic regions are comprised of the B’- and C-helices and the region stretching from the N-terminus of the F-helix through the N-terminus of the I-helix (18). Other studies have shown that this ability is not exclusive to 2B4 (15, 21, 22).
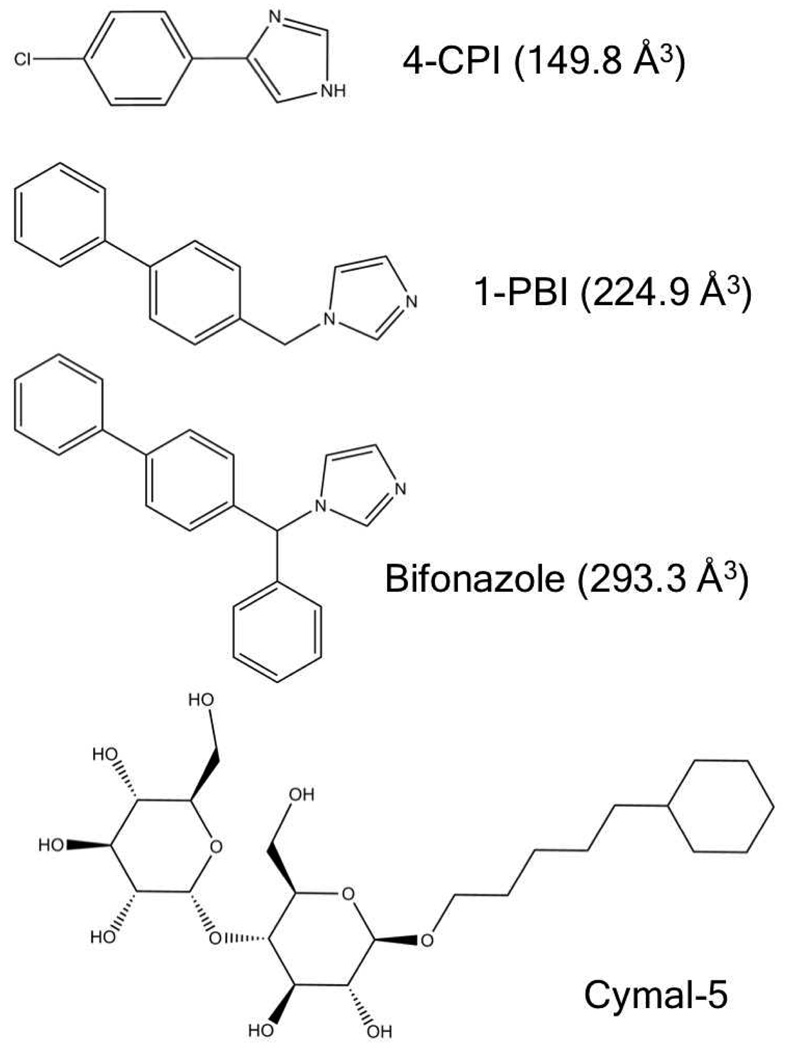
Previous CYP2B4 structures show snapshots at the apparent limits of P450 flexibility, but it has been unclear how the enzyme is able to make the transition from one state to the other, i.e. from wide-open to closed. Crystallization studies with a ligand of intermediate size could help to better understand this transition. For that purpose the inhibitor 1-biphenyl-4-methyl-1H–imidazole (1-PBI) was designed and synthesized, as it is larger than 1-CPI and 4-CPI, but smaller than the branched bifonazole by the loss of one phenyl ring (Fig. 1). Spectral binding and inhibition studies showed that the compound has sub-micromolar affinity for CYP2B4, indicating that it would be a good candidate for crystallization studies. The structure of the 1-PBI complex reveals an intermediate state compared to the open bifonazole complex and the closed CPI complexes. Previously defined plastic regions (18) adjust to accommodate the new inhibitor molecule, while still forming a similar dimer to the bifonazole bound 2B4 structure. The F-G helix cassette as well as the B/C loop reposition themselves in direct response to the size of the ligand bound in the active site. Analysis of these three CYP2B4 global conformations has given insight into the multiple ways this enzyme adapts to different ligands, which could be used to predict the relative position of the F-G cassette in future structures.
Figure 1.
Stick model structures of 1-PBI, 4-CPI, and bifonazaole inhibitor molecules and Cymal-5 detergent. Volumes for inhibitors calculated using http://www.molinspiration.com/cgi-bin/properties are listed in parantheses.
Materials and Methods
Materials
4-(Bromomethyl)biphenyl was used as received from Fisher Scientific (Pittsburgh, PA). Column chromatography was performed using Chromatographic Silica Gel 60–200 Mesh from Fisher Scientific (Pittsburgh, PA). Cymal-5 was from Anatrace (Maumee, OH). Protein concentration was performed using 50 kDa molecule weight cutoff Amicon Ultra filtration devices from Millipore (Billerica, MA).
Protein Expression and Purification
Cytochrome P450 2B4dH(H226Y) was expressed and purified as described previously (9, 20). This mutant is used to prevent dimerization of the ligand-free form of the enzyme (9). An overnight, Luria-Bertani broth culture of Escherichia coli containing the pKK2B4dH(H226Y) plasmid was used to inoculate Terrific broth. Terrific broth cultures were grown at 37° C until A600nm reached approximately 1. Protein expression was induced by the addition of isopropyl β-D-1-thiogalactopyranoside and δ-aminolevulinic acid. Protein expression continued for 48–68 h at 30 °C after which the cells were harvested by centrifugation and lysed. Cytochrome P450 was separated from the membrane by the addition of Cymal-5 in high salt buffer. After ultracentrifugation, the supernatant was purified using a Ni2+-NTA metal affinity column, followed by a carboxymethyl Sepharose ion-exchange column. The final protein buffer contained 50 mM potassium phosphate buffer pH 7.4, 20% (v/v) glycerol, 500 mM NaCl, 1 mM EDTA and 0.2 mM DTT. This preparation will be designated CYP2B4 for simplicity.
Synthesis of 1-biphenyl-4-methyl-1H–imidazole
This compound was synthesized in the Organic Chemistry Core, University of Texas Medical Branch, Galveston, TX. The reaction was carried out under argon in oven-dried glassware. 4-(Bromomethyl)biphenyl (4.21 g, 17.04 mmol) was dissolved in 80 mL of acetonitrile in a sealed 100 mL vessel charged with argon. Imidazole (3.45 g, 50.7 mmol) was weighed into a 250 mL three-necked round bottom flask, and 20 mL of acetonitrile was added under argon. Imidazole was predried by lyophilization for 24 hours before use. Acetonitrile was found to be dry by Karl Fischer Titration (37 ppm water). The flask was fitted with a condenser and the condenser was sealed with a rubber stopper and an argon balloon. The second neck was fitted with an addition funnel, sealed off with a rubber stopper. The third neck was capped off. The reaction was magnetically stirred at room temperature to dissolve the imidazole and then the 4-(bromomethyl)biphenyl solution was added dropwise through the addition funnel at room temperature. The reaction was refluxed for two hours with a heating mantle under magnetic stirring.
The mixture was cooled to room temperature and then diluted with 100 mL CHCl3 and washed twice with two consecutive 100 mL portions of 5% sodium bicarbonate solution. The organic phase was washed with 200 mL saturated sodium chloride solution. The organic layer was dried with magnetic stirring over a layer anhydrous sodium sulfate for five minutes and filtered, and solvents were removed by rotoevaporation.
The crude product (3.09 g) was purified by flash column chromatography with 300 g of silica gel, with a column load of CHCl3/hexane (9: 1), and eluted using a step gradient starting from hexane/CHCl3 (1: 9) in 100 mL increments, increasing CHCl3 at 10% per increment until the top impurity was removed. The eluent ratio was then increased to CHCl3/MeOH (9: 1) in 200 mL increments to elute the product. Straight MeOH was used to elute the final impurities. The product was a white solid (2.73 g, 68% yield); mp 141°C, 1H NMR (300 MHz d-DMSO) δ ppm: 5.223 (s, 1H, methyl); 6.897 (d, 1H, 4’ imidazole); 7.201 (d, 1H, 5’ imidazole); 7.330 (m, 4H, 2, 3, 5 & 6 phenyl); 7.430 (t, 1H, 10 phenyl); 7.620 (m, 8, 9, 11 & 12, phenyl); 7.762 (s, 1H, 2’ imidazole); MS-ES+ 235.41. 1H NMR spectra were recorded in d-DMSO at 300MHz on a Varian spectrometer. Thin layer chromatography was performed on Whatman Al Sil G/UV aluminum backed UV fluorescent plates in CHCl3/MeOH (19:1) and visualized in an iodine chamber.
Spectral Studies of 1-PBI Binding
Difference spectra were recorded using 1 µM CYP2B4 on a Shimadzu 2401 PC spectrophotometer at 25°C. A baseline was recorded between 350 and 500 nm with protein in 0.1 M HEPES pH 7.4 (1 mL). Difference spectra were recorded after the addition of inhibitor in methanol to the sample cuvette and the same amount of methanol to the reference cuvette. The spectral dissociation constant (Ks ) was obtained by fitting the data to the equation for “tight binding” 2ΔA = (ΔAmax/[E0]) ((KD + [I0] + [E0]) + ((K D + [I0] + [E0]))2 − 4[E0][I0])½). All data treatment and fitting of the inhibition and titration curves were performed with our SpectraLab software package (23).
Enzyme Inhibition Assay
Enzyme inhibition was measured using 7-EFC O -deethylation assay in a final reaction volume of 100 µL at 0.05 to 5 µM 1-PBI concentrations as described previously (24). In brief, the reaction mixture contained 150 µM 7-EFC in the standard reconstitution system (cytochrome P450: cytochrome P450 reductase: cytochrome b 5, 1:4:2) at 5 pmol P450 in 50 mM HEPES (pH 7.4), 15 mM MgCl2, and 2% MeOH. Samples were incubated at 37 °C for 10 min before the addition of NADPH (1 mM) to initiate the reaction. After 5 min the reaction was terminated by the addition of 50 µl of 20% trichloroacetic acid. Then, 2 ml of Tris-HCl buffer (pH 9.0) was added to the reaction mixture (50 µl), and the product was quantified by measuring 7-HFC at λex and λem of 410 and 530 nm, respectively. The turnover number (nmol/min/nmol P450) was determined using a standard curve of 7-HFC. Nonlinear regression analysis was performed to fit the data using a four-parameter logistic function to derive the IC 50 value for 1-PBI.
Crystallization and Data Collection
Prior to crystallization, CYP2B4:1-PBI complexes were prepared by one of two methods. In method A, 18 µM protein was mixed with a 10-fold excess of 1-PBI. This solution was incubated overnight at 4 °C and then concentrated until protein was 550–600 µM. At this point Cymal-5 was added to 4.8 mM and an additional 1mM 1-PBI was added. Crystals were grown by sitting drop vapor diffusion in drops containing 278 µM CYP2B4:1-PBI complex, 25 mM potassium phosphate buffer pH 7.4, 10% (v/v) glycerol, 250 mM NaCl, 0.5 mM EDTA, 0.1 mM DTT, 2.4 mM Cymal-5, 50 mM HEPES buffer pH 7.5, 5% (v/v) 2-methyl-2,4-pentanediol, and 5% (w/v) polyethylene glycol 6,000. Drops were equilibrated against well solutions containing 100 mM HEPES buffer pH 7.5, 10% (v/v) 2-methyl-2,4-pentanediol, and 10% (w/v) polyethylene glycol 6,000 at 18° C.
In method B, 18 µM protein was mixed with a 1.2-fold excess of 1-PBI. This solution was incubated and concentrated the same as in method A. At this point Cymal-5 was added to 4.8 mM and an additional 0.5 mM 1-PBI was added. Crystals were grown by hanging drop vapor diffusion in drops containing 278 µM cytochrome P450 2B4:1-PBI complex, 25 mM potassium phosphate buffer pH 7.4, 10% (v/v) glycerol, 250 mM NaCl, 0.5 mM EDTA, 0.1 mM DTT, 2.4 mM Cymal-5, 50 mM HEPES buffer pH 7.5, 4% (v/v) 2-methyl-2,4-pentanediol, and 6.5% (w/v) polyethylene glycol 6,000. Drops were equilibrated against well solutions containing 100 mM HEPES buffer pH 7.5, 8% (v/v) 2-methyl-2,4-pentanediol, and 13% (w/v) polyethylene glycol 6,000 at 18° C.
Crystals were transferred to mother liquor containing 15% (v/v) glycerol and 15% (v/v) ethylene glycol before being flash frozen in liquid nitrogen. 2.5 Å (method A) and 3.2 Å (method B) data sets were collected on beamlines 9-1 and 7-1 respectively of the Stanford Synchrotron Radiation Laboratory (Stanford, CA). Both data sets were collected over 90° at 100 K on a Quantum-315 CCD detector (Area Detector Equipment). Data were processed using MOSFLM (25) and SCALA (26). Data collection and diffraction statistics are summarized in Table 1.
Table 1.
X-ray Data Collection & Refinement Statistics
| Crystal | ||
| Ligand Occupancy | Triple | Single |
| Space Group | P212121 | P212121 |
| Unit Cell | ||
| a(Å) | 86.7 | 86.9 |
| b(Å) | 152.5 | 151.8 |
| c(Å) | 181.9 | 181.6 |
| α=β=γ (°) | 90 | 90 |
| Data Collection | ||
| X-ray source | SSRL BL 9–1 | SSRL BL 7–1 |
| Wavelength (Å) | 0.98 | 0.98 |
| Resolution Range (Å) | 48.46 (2.50) | 49.69 (3.20) |
| Total Observations | 283,391 | 162,407 |
| Unique Observations | 81,702 | 73,790 |
| Completeness (%) | 97.2 (93.3) | 99.0 (99.5) |
| Redundancy | 3.5 (3.4) | 4.1 (4.1) |
| I/σ | 5.6 (1.1) | 4.4 (1.4) |
| Rmerge (%)a | 8.3 (65.6) | 14.4 (50.6) |
| Refinement Statistics | ||
| R-factor (%)b | 21.84 | 23.74 |
| Rfree (%)b | 24.66 | 26.96 |
| RMS deviations | ||
| Bond lengths (Å) | 0.02 | 0.015 |
| Bond angles (°) | 1.842 | 1.467 |
| Ramachandran Plot | ||
| Residuesc | 1,782 | 1,778 |
| Preferred | 1,693 | 1,381 |
| Allowed | 85 | 135 |
| Disallowed | 4 | 4 |
| No. of Atoms | ||
| Protein | 14,219 | 13,435 |
| Heme | 172 | 172 |
| 1-PBI | 192 | 72 |
| Cymal-5 | 88 | 0 |
| Water | 180 | 36 |
Values for the highest resolution shell are in parentheses.
Rmerge = [ΣhΣi | Ih-Ihi|/ΣhΣiIhi] where Ih is the mean of Ihi observations of reflection h
R-factor & Rfree = Σ||Fobs|-|Fcalc||/Σ|Fobs| × 100 for 95% of the recorded data (R-factor) and 5% of data (Rfree)
Number non-glycine, non-proline residues used for calculation
Structure Determination and Refinement
The 2.5 Å CYP2B4:1-PBI structure was solved by molecular replacement using an ensemble of all four previously solved CYP2B4 structures (1SUO, 1PO5, 2BDM, 2Q6N) with residues 101–120 and 205–232 omitted as a search model in the program Phaser (27). The electron density map was 4-fold averaged according to the non-crystallographic symmetry using DM (26). The omitted chain segments were clear in the unbiased DM map, and their sequence registry was confirmed by the side chains of Phe 108, Trp 111, and Phe 115, and Phe 206, Phe 207, Phe 212, Phe 217, Phe 220, Phe 223, Tyr 226, and Phe 227. Initial rounds of refinement and model building were performed using Refmac (28) and MIFit (29) until an R-factor of ~25% was reached. After this point model building was performed in COOT (30) using 2Fo -Fc electron density maps contoured at 1-σ. 1-PBI and Cymal-5 molecules were modeled into unbiased difference electron density prior to the final stage of refinement. The final rounds of refinement were performed using Phenix (31) until a final R-factor of 21.8% was reached. The Rfree of the final model is 24.66%.
The 3.2 Å CYP2B4:1-PBI structure was solved by molecular replacement with Phaser using the 2.5 Å structure A chain as a search model. Following an initial rigid body refinement in Refmac, Phenix refinement was used to reach a final R-factor of 23.74% and an Rfree of 26.96%. Model building was performed in COOT using 2Fo -Fc electron density maps contoured at 1-σ.
All protein model figures were based on the higher resolution structure unless otherwise noted. All protein model figures were made using PyMOL (32).
Results
1-PBI Binding and Inhibition Studies
Spectral studies showed that 1-PBI binds to CYP2B4 with a Ksvalue of 0.23 µM. Inhibition assays showed that 1-PBI inhibits the 7-EFC O -deethylation activity of CYP2B4 with an IC 50 value of 0.035 µM (data not shown). For reference, 4-CPI binds to CYP2B4 with a Ks of 0.04 µM and inhibits activity with an IC 50 of 0.11 µM (33). The K s and IC 50 of bifonazole are 0.13 µM and 0.9 µM respectively (18).
Finished Model
The 1-PBI inhibitor complex at both high and low molar excess crystallized in space group P212121 with a Matthews coefficient of 2.8 Å3/Da, which corresponds to four molecules per asymmetric unit and a solvent content of ~56%. Except for two short surface loops, the model is complete for each of the four copies. The finished 2.5 Å protein model contains 14,219 atoms. Chain A is modeled from residues 27–134, 139–275, and 283–493. Chain B is modeled from residues 28–134, 139–273, and 284–492. Chain C is modeled from 26–134, 139–273, and 285–492. Chain D is modeled from residues 28–135, 140–273, and 284–492. Numbers correspond to the full-length enzyme, which has 491 residues; in some cases portions of the C-terminal His tag were visible in the electron density maps. Two disordered regions in each chain occur in the C/D and H/I loops. The coordinates were analyzed using PROCHECK (34) to confirm proper stereochemistry (Table 1).
The crystal obtained by method A contained 12 1-PBI molecules, 4 heme molecules, 4 Cymal-5 molecules, and 180 waters. The Cymal-5 molecules showed strong electron density for the cyclohexane ring, the aliphatic chain, and one of the two maltose sugars. The second sugar extends into bulk solvent and is not clearly defined in the electron density maps. This detergent molecule binds at a similar site in the bifonazole structure. In both structures, Cymal-5 interacts with hydrophobic residues in the C-helix. However, in the structure of the bifonazole complex, the detergent is in a different orientation and is close enough to the active site to pack against the inhibitor molecule. There are three 1-PBI molecules per protein chain in the current structure. One occupies the active site, with a coordinate bond between the imidazole group and the heme-iron. The other two inhibitor molecules sit inside a previously identified peripheral binding site (18) near the largely hydrophobic dimer interface. One inhibitor molecule does not have the imidazole chain modeled as it accesses a solvent channel and is not clearly defined. Similar to the bifonazole structure (18), all of the organic molecules found in the structure, except those bound to the heme, were found near the dimer interface and interact with residues from the opposite chain. Refinement and model statistics can be found in Table 1.
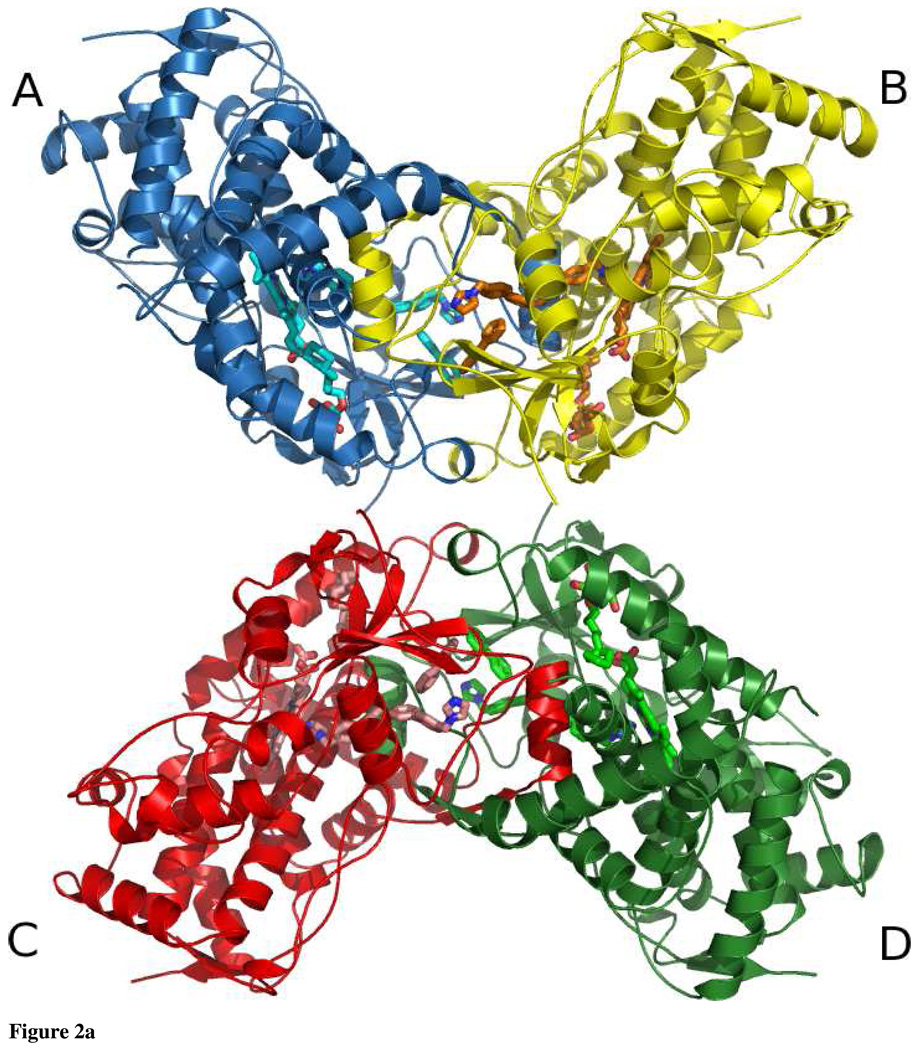
Tetramer
The asymmetric unit of the present structure is made up of a tetramer that is a dimer of dimers (Fig. 2a). The A and B chains form one dimer, and the C and D chains form the other. Overall, the tetramer buries ~6700 Å2 of solvent accessible surface area with ~2760 Å2 buried at the AB dimer interface, ~2790 Å2 buried at the CD dimer interface, and ~1150 Å2 at the interface of AB and CD dimers (35, 36). The tetramer is formed by a handful of intersubunit contacts. The carboxylate group of Asp 374 of the A chain makes a water mediated hydrogen bond to the hydroxyl group of Tyr 111 of the C chain. The same interaction is found between the B and D chains. In addition, Tyr 111 and Phe 115 of the A, B, C, and D chains pack against Val 381, Pro 383, and the aliphatic chains of Asp 374 and Gln 376 of the C, D, A, and B chains respectively.
Figure 2.
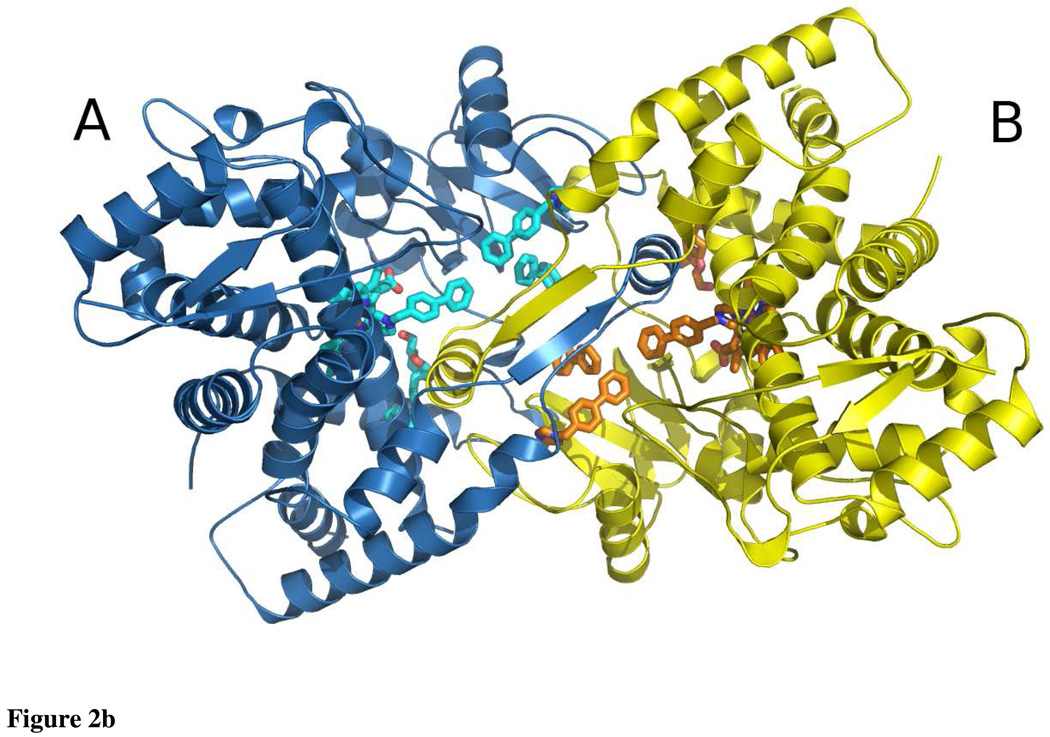
Ribbon diagram of CYP2B4:1-PBI complex asymmetric unit looking down a non-crystallographic two-fold axis. Chain A is colored blue, chain B is colored yellow, chain C is colored red and chain D is colored green. Heme, 1-PBI, and Cymal-5 are shown as cyan (A), orange (B), salmon (C) and light green (D) sticks. a: The four chains form a tetramer that is a dimer of dimers. The A and B chains form one dimer and the C and D chains form the other dimer. b: Ribbon diagram of CYP2B4:1-PBI complex dimer looking down the other non-crystallographic two-fold axis. Coloring is the same as Fig. 2a and is rotated 90° with respect to Fig. 2a. Organic molecules are shown as cyan and orange sticks for the A and B chains respectively. The F’-helix of each monomer reaches across the dimer interface to interact with the active site of the other chain. The peripheral 1-PBI molecules bind at the dimer interface.
Dimer
The CYP2B4:1-PBI complex dimer is similar to the 2-fold symmetric dimer found in crystals of the CYP2B4:bifonazole complex (18), but differs from that seen in the CYP2B4 structure in the absence of exogenous ligand (20), in which His 226 coordinates to the heme iron of the opposite chain. Except for this open CYP2B4 structure, all ligand bound 2B4 structures have been determined using H226Y mutant enzyme to eliminate this open dimer. In the present structure, the AB and CD dimers are interdigitated structures. In order to bury exposed hydrophobic residues, dimer formation occurs on the same surface that is proposed to interact with the ER membrane (37, 38).
Each intercalated dimer is created by the F’-helix of one monomer reaching across the dimer interface to interact with the C- and I-helices of the other monomer (Fig. 2b). Specifically, Val 216, Phe 217, and Phe 220 of the F’-helix pack against Leu 295 and the β-carbon of Ser 294 of the I-helix and to a lesser extent Trp 121 of the C-helix. At the N-terminus of the F’-helix, the Ser 211 hydroxyl hydrogen bonds to the carboxylate of Glu 301 of the I-helix. In this same region, Ser 213 makes contact with the main chain oxygen of Phe 202 of the F-helix. A portion (206FSLIS210) of the F/F’ loop forms a short anti-parallel β-sheet with the same residues of the other monomer. One final set of interactions occurs between the loop connecting β-strands 1 and 2 and the B/C loop of the opposite chain. Here, the main chain oxygen of Ser 72 hydrogen bonds to the amide nitrogen of Gln 109. The main chain nitrogen of Ser 72 is also in position to contact the main chain oxygen of Ile 101. In addition, one of the nitrogens of the guanidinium group of Arg 73 hydrogen bonds to the main chain oxygen of Val 103.
Single Ligand Occupancy Structure
In order to investigate any role that non-active site 1-PBI molecules might play in stabilizing the interdigitated dimer, an additional structure was also solved at a much lower inhibitor to protein ratio (method B). The low inhibitor concentration structure is virtually identical to that of the high inhibitor concentration structure, except that it lacks the Cymal-5 and the two peripheral 1-PBI molecules. The complex crystallized in the same space group with essentially the same unit cell dimensions (Table 1). An overlay of the two structures (Fig. S1) yields an RMSD of ~0.6 Å for the dimer and an RMSD less than 0.5 Å for each of the monomers. The C/D loop is ordered in this structure, but a portion of the loop between β-strands 8 and 9 becomes disordered. The latter loop interacts with one of the extra 1-PBI molecules in the high inhibitor concentration structure. The lack of this interaction likely leads to increased flexibility in the loop and weak electron density. Hence, the monomer conformation appears to be induced by 1-PBI binding in the active site and stabilized by dimerization, while the peripheral sites are adventitious.
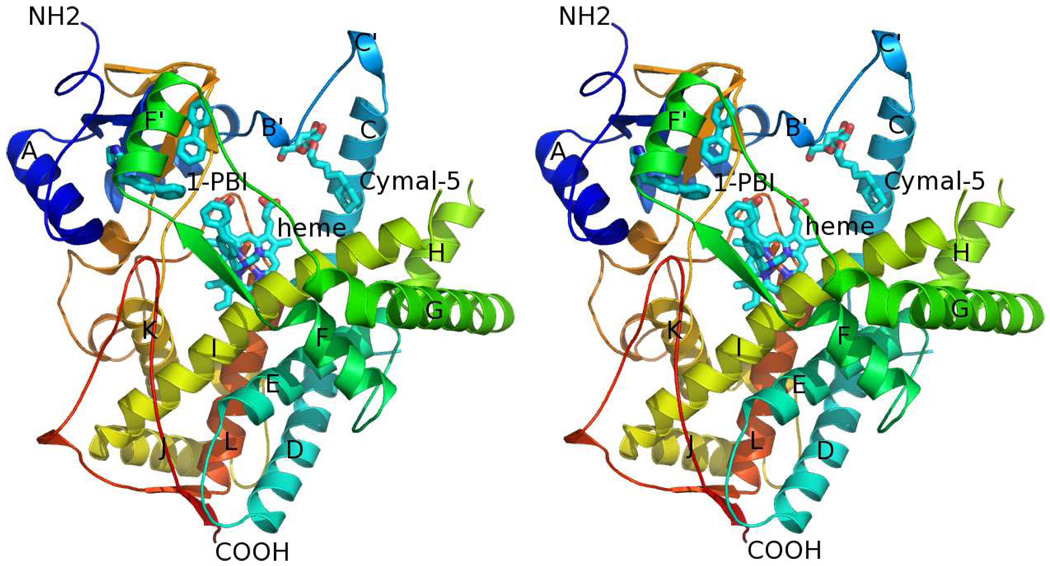
2B4:1-PBI Complex Monomer
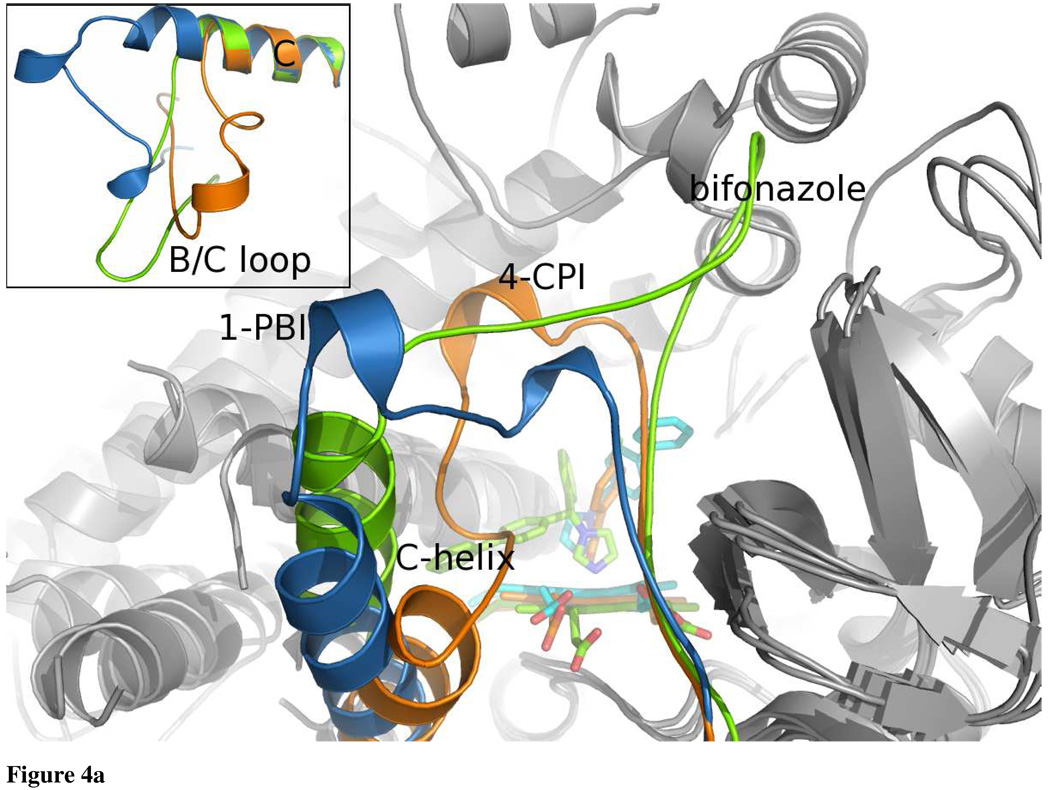
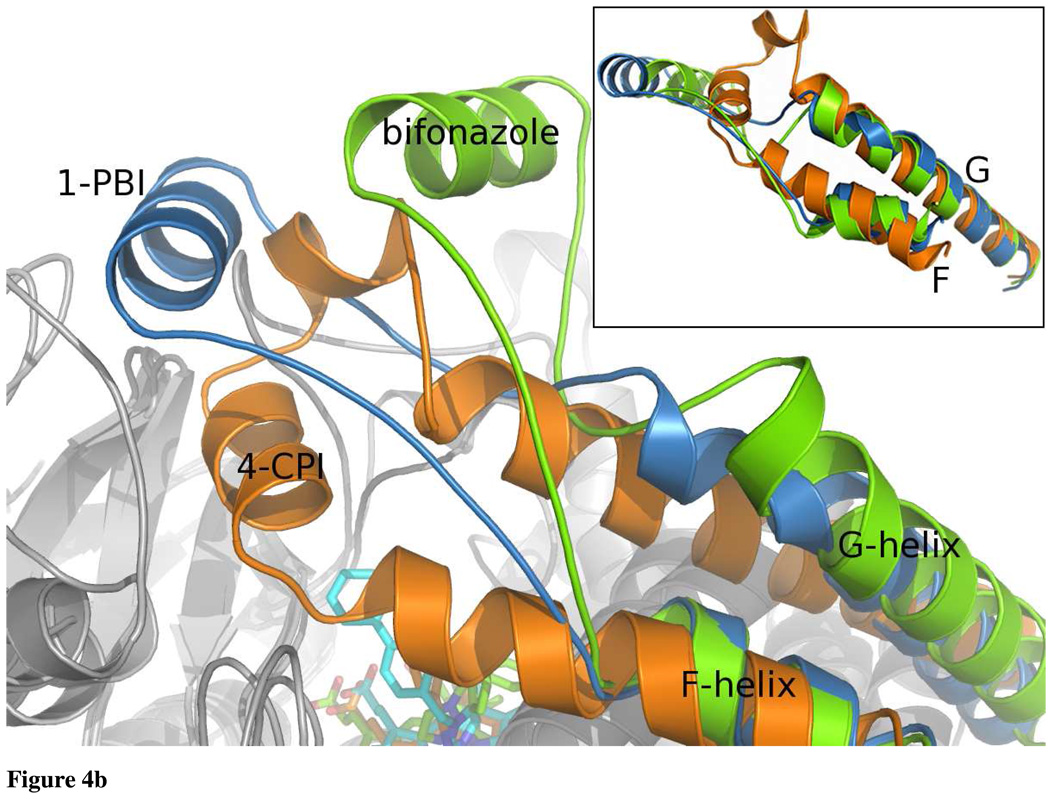
The CYP2B4:1-PBI complex retains the overall P450 fold, while molding to the 1-PBI inhibitor (Fig. 3). Each of the four monomers is in essentially the same conformation with an RMSD of less than 0.2 Å for each chain. The core of the P450 fold, which includes the C-terminus of helix A through helix B, helices D and E, the C-terminus of helix I through helix K, and all the β-sheets, remains relatively unchanged compared with other CYP2B4 structures. Differences found between the current and previous CYP2B4 structures echo prior definitions of mobile, plastic regions (18). Major changes occur in two main locations in the protein. The B/C loop makes a dramatic shift toward the F’/G loop and also contains two small helices (B’ and C’) to compact the structure in this region (Fig. 4a). This is in contrast to the CPI structures, where a longer B’ helix actually enters the active site to close around the smaller CPI inhibitors, and to the bifonazole structure, where the B/C loop is completely extended toward the region where the 1-PBI F’-helix sits. The second region with significant differences is the ‘roof’ of the active site. Similar to the bifonazole structure, in the 1-PBI complex the F-helix begins to unwind at Gln 204 to form a more extended structure when compared with the 4-CPI structure. The active site lid formed by residues that form the F/F’ loop through the G-helix, adopts a conformation that is intermediate to that seen in the bifonazole and CPI structures (Fig. 4b). In addition to making a large portion of the dimer contacts, the F’-helix also interacts significantly with the non-structured, N-terminal region that precedes the A-helix.
Figure 3.
Divergent stereoview of CYP2B4:1-PBI complex monomer A chain ribbon diagram. The chain is colored from blue to red from the N-terminus to the C-terminus respectively. Heme, 1-PBI, and Cymal-5 molecules are shown as cyan sticks. The F-, F’-, and G-helices rest across the I-helix forming a lid over the active site.
Figure 4.
Ribbon diagram of CYP2B4:1-PBI, 4-CPI, and bifonazole complex overlays. a: The 1-PBI, 4-CPI, and bifonazole B/C loop and C-helix are colored blue, orange, and green respectively. The B/C loop rearranges itself in response to the various ligands bound to CYP2B4. Inset shows the B/C loop and C-helix overlaid apart from the overall protein structure. The C-helix is essentially identical in each, but large differences are seen in the position of the B/C loop. b: The 1-PBI, 4-CPI, and bifonazole F–G helix cassettes are colored coded as in Fig. 4a. The F-, F’-, and G-helices cassette pivots over the I-helix to accommodate the various inhibitor molecules bound to the active site heme. The size of the inhibitor (4-CPI<1-PBI<bifonazole) is somewhat proportional to the angle the cassette makes with repect to the I-helix. Inset shows the F-, F’-, and G-helices cassette overlaid apart from the overall protein structure. The N-terminus of the F-helix and the G-helix retain very similar structures; however the point at which the C-terminus of the F-helix unwinds as well as the secondary structure elements between the F- and G-helices changes in each structure.
Inhibitor Binding Site
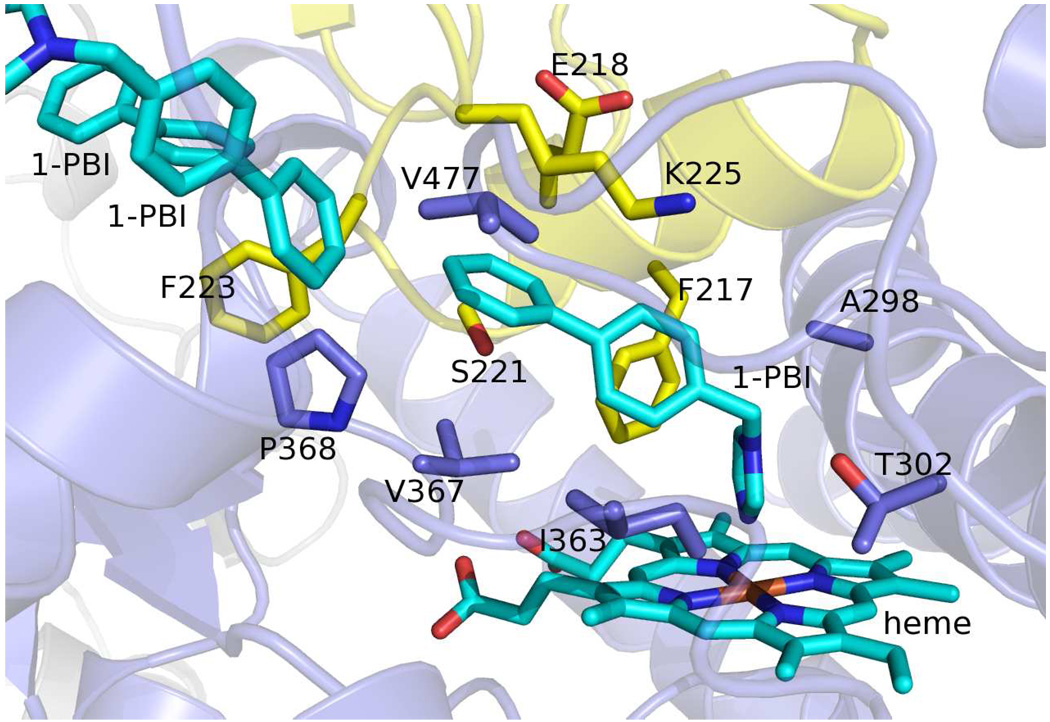
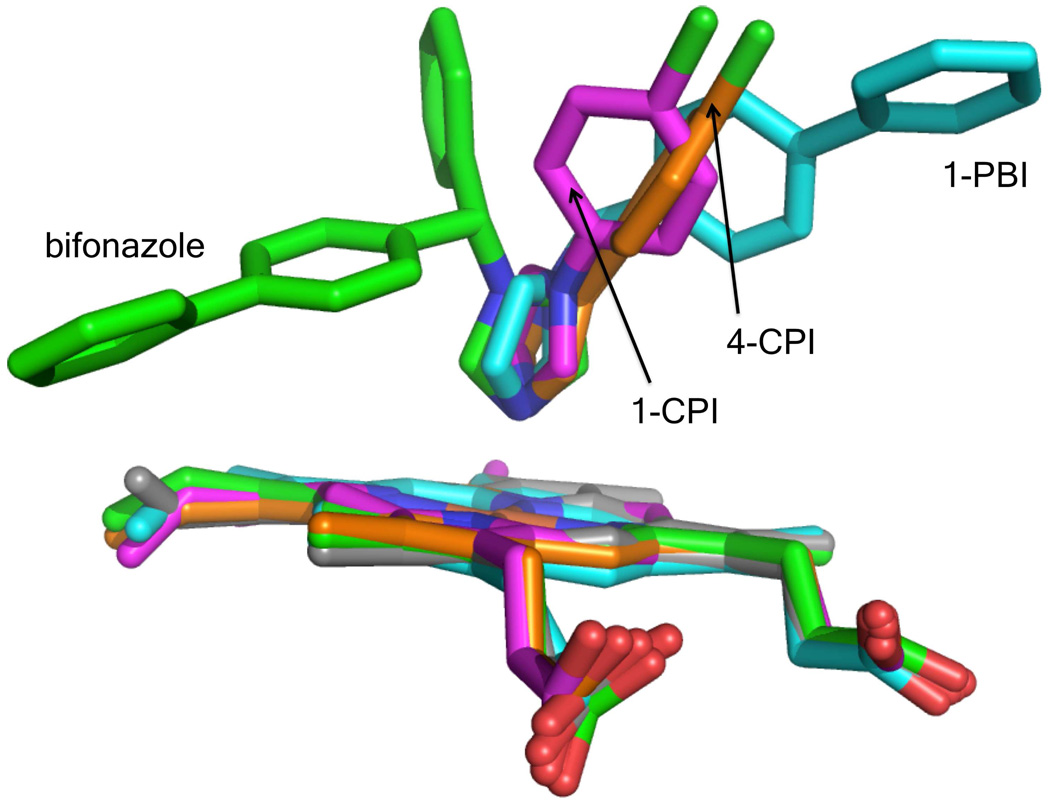
There are three 1-PBI molecules bound per monomer in the 2.5 Å crystal structure. One is bound in the active site with an imidazole nitrogen-heme iron bond length of 2.19 Å (Fig. 5). As with other CYP2B4 structures, the imidazole ring is vertical with respect to the plane of the heme. This ring is almost parallel with respect to the I-helix and resembles the bifonazole structure more than the two CPI structures, where the imidazole ring makes a ~60° with the I-helix. In contrast to the bifonazole structure where the biphenyl moiety of the inhibitor is pointed towards the C-helix and is almost parallel to the I-helix, the 1-PBI biphenyl moiety points towards the dimer interface and is perpendicular to the I-helix (Fig. 6).
Figure 5.
Semi-transparent ribbon and stick diagram of the CYP2B4:1-PBI complex active site. Chain A is colored blue and chain B is colored yellow. Heme and 1-PBI molecules are shown as cyan sticks. Side chains of residues identified within a 5 Å radius of the 1-PBI coordinated to the heme iron are shown as blue and yellow sticks.
Figure 6.
Stick diagram of CYP2B4 heme and ligand overlay. The ligand free (gray) , 1-CPI (magenta), 4-CPI (orange), bifonazole (green), and 1-PBI (cyan) structures are shown. The active site heme remains in same orientation in each structure with only small changes. Each inhibitor molecule changes orientation in the different structures. The imidazole ring of 1-CPI and 4-CPI are in similar orientations, but the angle the ligand makes with respect to the heme differs as well as the conformation of the phenyl ring. The 1-PBI biphenyl moiety points in the same direction as the heme A ring propionate, but in bifonazole it is oriented directly between the C and D rings of the heme.
Like the overall structure of the 1-PBI complex, the inhibitor interactions (Fig. 5) are also a hybrid of the bifonazole and CPI structures. The imidazole ring of 1-PBI makes van der Waals contacts with Ala 298 and Thr 302 from substrate recognition site (SRS) 4 and Ile 363 from SRS 5 (39), interactions common to all other CYP2B4 inhibitor structures. Val 367, Pro 368 (SRS 5), and Val 477 (SRS 6) make contact with the 1-PBI biphenyl group (Fig. 5). Val 367 and Val 477 were previously identified in CPI binding, but Pro 368 is a newly established ligand-binding residue. This reflects the fact that the biphenyl group in 1-PBI group reaches towards the dimer interface. Importantly, the 3.2 Å 1-PBI structure in the absence of excess 1-PBI shows the active site inhibitor binding in the same orientation, being held into place through interactions with the same residues.
Numerous residues were identified to interact with CPI (9, 10) and bifonazole (18), but do not play a role in 1-PBI binding. Residues that do interact with 1-PBI are mostly in similar orientations in other 2B4 structures. Only Ala 298 and Val 477 shift position significantly due to their placement in plastic regions 4 and 5 respectively (18). Ile 101, Val 104, Ile 114, Phe 115, Phe 297, and Glu 301 interact with CPI, but do not make any contacts with 1-PBI. Ile 101, Val 104, Ile 114, and Phe 115 are part of the B/C loop and C-helix region that has moved away from the active site in the CYP2B4:1-PBI complex. Phe 297 is facing away from the active site in the 1-PBI structure due to changes in the I-helix. The side chain of Glu 301 has rotated to hydrogen bond with Thr 305. Variations in the orientation of the biphenyl group of 1-PBI and bifonazole also result in different interactions. Ser 128, Met 132, Val 292, Leu 295, and Phe 296 all make contact with bifonazole because its biphenyl group points towards the C-helix. None of these residues are involved in 1-PBI binding.
Significant interactions with the opposite chain in the dimer make the remaining 1-PBI contacts (Fig. 5). These are from Ser 214, Phe 217, Phe 223, and the aliphatic chains of Glu 218 and Lys 225. The bifonazole structure also includes significant ligand interactions from a symmetry related protein molecule, but the 1-PBI structure only retains Phe 217 among these contacts. These differences result from changes in the orientation of the biphenyl group of the bifonazole and 1-PBI inhibitors (Fig. 6).
In contrast to 1-PBI in the active site, it appears that the two peripheral 1-PBI molecules at the subunit interface are merely filling residual surface pockets in the structure, as opposed to forcing the enzyme into a new conformation, because they are only observed at a large molar excess (Fig. S2). Their contacts include Leu 43, Leu 51, Phe 55, Leu 70, Arg 73, Ile 101, Phe 368, Phe 389, Val 477, and from the F’/G loop of the opposite monomer, Phe 223, Phe 227, and Thr 230. The CYP2B4:bifonazole structure also exhibits an extra inhibitor molecule binding at nearly the same site, although mostly different residues make up the majority of the contacts.
Heme Binding Site
Comparison of multiple CYP2B4 structures shows that the C-terminus of the I-helix and the entirety of the L-helix maintain a rigid conformation, while there is larger flexibility in the opposite side of the heme binding pocket along the C-helix and the N-terminus of the I-helix. The reactive center of cytochrome P450 hinges on proper orientation of a heme molecule, which is anchored by Cys 436 coordinating the heme iron as well as multiple hydrogen bonds to the A and D ring propionate groups (Fig. S3). In the present structure, the hydrogen bonding partners of the A and D ring propionates are most closely related to those of the 4-CPI (9) and 1-CPI (10) CYP2B4 structures (Table 2). The A ring propionates maintain contacts with Ser 430 and His 369 as seen in all other CYP2B4 structures. In addition, the A ring also receives a hydrogen bond from the guanidinium group of Arg 434. A higher degree of variability exists in the D ring interactions. Here, Arg 98, Trp 121, Arg 125, and Arg 434 secure the propionate carboxylate. The large degree of variability in these contacts when compared with previously published CYP2B4 structures probably occurs due to the high number of rotamer conformations that Arg 434 may adopt, as well as mobility of the C-helix and B/C loop.
Table 2.
Number of hydrogen bonds to heme propionate groups in cytochrome P450 2B4
| Crystal Structure | |||||
|---|---|---|---|---|---|
| Residue | Bifonazole | 4-CPI | 1-PBIa | 1-CPIa | Ligand Free |
| Arg 98 | 2 | 2 | 2 | 2 | |
| Trp 121 | 1 | 1 | 1 | ||
| Arg 125 | 1 | 1 | 1 | ||
| His 369 | 1 | 1 | 1 | 1 | 1 |
| Ser 430 | 1 | 1 | 1 | 1 | 1 |
| Arg 434 | 1 | 1 | 2 | 1 | |
| Waters | 3 | 4 | |||
| Cymal-5 | 1 | ||||
A chain
Discussion
A new conformation of CYP2B4, in complex with the tight binding, designed inhibitor 1-PBI has been captured in a crystal structure. Two structures, determined at very different inhibitor to enzyme molar ratios, establish that the change in conformation is driven by heme bound 1-PBI. Previous structures (9, 10, 18, 20) explored the extremes of CYP2B4 flexibility, but the mechanism of the open to closed transition was unclear. Based on these structures, an inhibitor of intermediate molecular size, 1-PBI, was synthesized. The resulting 1-PBI-bound CYP2B4 structure provides insight into the secondary structural repositioning of the P450 fold necessary to transition from the open (bifonazole), to intermediate (1-PBI), and to closed (4-CPI, 1-CPI) protein states. Defined plastic regions (B’-, C-, F-, F’-, and G-helices and the F/F’ and F’/G loops) are repositioned without significant changes to secondary structure elements to bind 1-PBI, while the B/C loop undergoes dramatic refolding (Fig. 4a and 4b). Comparison of all three CYP2B4 conformations reveals that a cluster of Phe residues, packed at the junction of the F-, G-, and I-helices, allows the F-G helix cassette to pivot on the I-helix in direct response to the size of the ligand bound in the active site.
The CYP2B4:1-PBI complex crystallizes with two dimers per asymmetric unit (Fig. 2a) where the dimers exploit a similar subunit interface as in the CYP2B4:bifonazole dimer. This dimer formation buries the mostly hydrophobic F’-helix in the active site cleft of the opposite monomer (Fig. 2b). Small differences in monomer orientation at the dimer interface may reflect variability in how individual monomers orient themselves with respect to the membrane of the endoplasmic reticulum (18, 37). This orientation would implant the A- and F’-helices and the B/C loop into the membrane (18), regions that have been previously identified to associate with the membrane (38, 40). In this case, the opening and closing of the active site cleft, which exposes hydrophobic residues, would not carry a large thermodynamic penalty due to the environment of the phospholipid core of the bilayer.
Movement of the B/C loop and C-helix aids in accommodating the active site in response to ligand binding (Fig 4a). In the current structure, the C-helix pivots on its C-terminus, pushing it further away from the active site than in other CYP2B4 structures. The B’-helix changes position to partially fill the void left behind, but not to the extent seen in the 4-CPI structure (Fig. 4a). Residues on the C-helix have been identified in cytochrome b5 and NADPH-cytochrome P450 reductase binding (41), and flexibility of these structural elements may be central to the mechanism by which P450s bind to redox partners (15, 20, 41, 42). Heme binding also seems to be affected by structural variability in this region. Arg 98, Trp 121, and Arg 125 all make varying contributions to heme propionate hydrogen bonding in each CYP2B4 structure (Table 2).
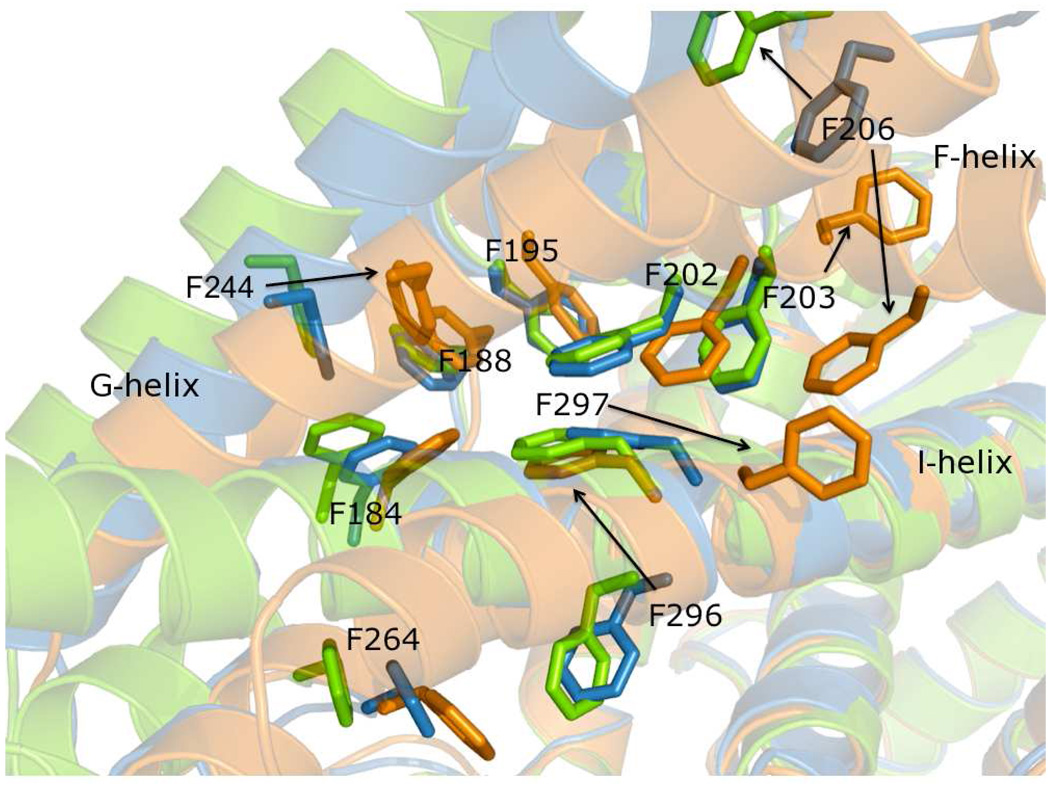
The F-G helix cassette appears to be key to adopting new conformations to bind various ligands. It is able to pivot across the I-helix at a cluster of phenylalanines (Fig. 4b and Fig. 7). At the point where the F-, G-, and I- helices intersect, there are 10 phenylalanine residues. These are Phe 184, Phe 188, Phe 195, Phe 202, Phe 203, Phe 206, Phe 244, Phe 264, Phe 296, and Phe 297 (Fig. 7). Based on a sequence alignment of seven other 2 family P450s (not shown), eight of these residues are either completely or partially conserved. Two of these, Phe 206 and Phe 297, were identified previously as highly mobile residues (10). Superposition of the CYP2B4 4-CPI, bifonazole, and 1-PBI structures reveals a complex rearrangement of these residues in concert with different rotamer conformations of the Phe side chains in the cluster. Viewed from the perspective of the F-G helix cassette with respect to the I-helix, there are three conformations: open (bifonazole), intermediate (1-PBI), and closed (4-CPI) (Fig. 7). Prior to the current 1-PBI structure, it was not entirely clear how CYP2B4 accomplished the dramatic transition from the tightly packed 4-CPI structure to the wide-open bifonazole conformation. This constellation of phenylalanines appears to be central to CYP2B4’s ability to flex this region through a large range of conformations. The large phenyl side chains are able to repack themselves in numerous conformations, excluding solvent as the F-G helix cassette repositions itself with respect to the I-helix in response to ligand binding. In the current intermediate structure, there are no voids remaining as the protein has shifted into position to bind 1-PBI. However in the bifonazole structure, the resultant packing of Phe side chains does leave open space between the G- and I-helices that was filled by a peripheral inhibitor molecule (18). Hence, the intermediate conformation appears to provide a low-energy pathway between the open and closed forms of CYP2B4.
Figure 7.
Transparent ribbon and stick diagram of CYP2B4 overlays highlighting the Phe cluster at the F-, G-, and I-helix intersection. The 4-CPI, 1-PBI, and bifonazole strucutres are shown in orange, blue, and green respectively. The cluster of Phe residues at this helical intersection reposition their side chains in each structure to fill void volumes as the F–G helix cassette pivots across the I-helix. Black arrows indicate large shifts in Phe location. The largest changes come at residue numbers 203, 206, 244, 296, and 297. The position of Phe 203 is conserved in the 1-PBI and bifonazole strucutres, but this residue swings around to the other side of the F-helix in the 4-CPI structure. Phe 206 diverges significantly in all structures based on the unraveling of the F-helix. The 4-CPI Phe 296 occupies the same place as Phe 297 in the 1-PBI and bifonazole structures. Phe 297 of the 4-CPI structure rotates around to interior of the active site.
As in the bifonazole bound structure, crystallization of the CYP2B4:1-PBI complex by method A (large molar excess of 1-PBI) resulted in inhibitor molecules binding to peripheral sites. There is precedent from structures of other P450 families for ligand binding at distal sites away from the active site. In some cases, this can influence enzyme function through cooperatively like in CYP3A4 (11, 21) and P450eryF (43). Non-active site ligand binding is also observed without functional consequences as in CYP2C8 (13) and CYP158A and A1 (44).
Compared with the bifonazole complex, the orientation of 1-PBI in the active site was somewhat surprising (Fig. 6), suggesting difficulty in predicting the placement of bound ligand molecules binding in CYP2B4. Bifonazole binds to CYP2B4 in a manner that pushes the biphenyl moiety towards the C-helix, but the biphenyl group of 1-PBI is turned more than 90° towards the dimer interface. Even the isomers 1-CPI and 4-CPI bind with some variability. However, there are trends in the plastic regions of the enzyme in response to the size of the inhibitor found in the active site. Given the four ligand bound structures of CYP2B4, it is evident that the F-G helix cassette repositions itself manner consistent with ligand size (Fig. 1, Fig. 4b). Smaller ligands (4-CPI, 1-CPI) result in an overall closed structure, with the F-G helix cassette creating a small angle (18.7°) with the I-helix and active site. 1-PBI, which is larger than CPI by one phenyl group, creates a slightly larger angle of 29.4°. In the complex with bifonazole, the largest of the inhibitors crystallized with CYP2B4, the F-G cassette moves upward at an even larger angle (47.4°) resulting in a markedly open structure. The plastic region that includes the B/C loop and C-helix is not as predictable, but there does appear to be a trend in the proximity of the B/C loop to the active site and the size of the ligand (Fig. 4a). Increasing ligand size pushes the B/C loop away from the active site, but there are secondary structural elements (the B’- and C’-helix) that fold and unfold with little correlation to the size of the ligand.
Variations in the F-G helix cassette in response to ligand size were first recognized in CYP102A1 (BM3) (42, 45) and have also been noted in mammalian CYP2C5 (14, 22) and bacterial CYP119 (46, 47) and CYP154C1 (48). CYP3A4 is curiously able to bind a wide variety of ligand sizes with no apparent shifts in the F-G helix cassette (11, 12, 21). It should be noted that while CYP3A4 lacks a Phe cluster where the F- and G-helices intersect the I-helix, it does have 7 Phe residues that sit above the active site. The CYP3A4 Phe cluster is able to reposition side chains in order the alter the active site volume (21) and also change the length of the F-helix (11). The repacking of these residues could partially explain how CYP3A4 accommodates so many different ligands without large structural rearrangements. It is possible that P450s in general are able to utilize Phe clusters near α-helix bundles as in the CYP2B4 complexes to move large protein secondary structural elements in order to accommodate different ligands.
Supplementary Material
Acknowledgements
We thank Dr. P. Ross Wilderman for assistance using ChemDraw. We also thank the staff at the Stanford Synchrotron Radiation Laboratory for assistance with data collection. The Stanford Synchrotron Radiation Laboratory is operated by Stanford University on behalf of the United States Department of Energy, Office of Basic Energy Sciences. The Stanford Synchrotron Radiation Laboratory is supported by the National Institute of Health, the National Center for Research Resources, the Biomedical Technology Program and the U.S. Department of Energy of Biological and Environmental Research.
Abbreviations and textual footnotes
- CYP2B4dH(H226Y)
an N-terminal truncated and modified and C- terminal His-tagged form of cytochrome P450 2B4 with an internal mutation
- HEPES
N-[2-hydroxyethylpiperazine-N'-[2-ethanesulfonic acid]
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- 7-EFC
7-ethoxy-4-(trifluoromethyl)coumarin
- 7-HFC
7-hydroxy-4-trifluoromethylcoumarin
- 4-CPI
4-(4-chlorophenyl)imidazole
- 1-CPI
1-(4-chlorophenyl)imidazole
- 1-PBI
1-biphenyl-4-methyl-1H–midazole
- DMSO
dimethyl sulfoxide
- Ni2+-NTA
nickel- nitrilotriacetic acid
- SSRL
Stanford Synchrotron Radiation Laboratory
- BL
Beamline
- RMSD
root mean square deviation.
Footnotes
This research was supported by NIH grants ES003619 and ES006676.
Atomic coordinates have been deposited in the Protein Data Bank (accession codes 3G5N, 3G93)
Supporting Information
Structural overlays of the dimer and monomer of the single and triple occupancy structures. Electron density maps showing the peripheral binding site for both the single and triple occupancy structures. Heme propionate interactions with protein side chains. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Johnson EF, Stout CD. Structural diversity of human xenobiotic-metabolizing cytochrome P450 monooxygenases. Biochem Biophys Res Commun. 2005;338:331–336. doi: 10.1016/j.bbrc.2005.08.190. [DOI] [PubMed] [Google Scholar]
- 2.Al Omari A, Murry DJ. Pharmacogenetics of the Cytochrome P450 Enzyme System: Review of Current Knowledge and Clinical Significance. J Pharm Pract. 2007;20:206–218. [Google Scholar]
- 3.Jabri E. P450 on drugs. Nat Struct Biol. 2003;10:587. doi: 10.1038/nsb0803-587. [DOI] [PubMed] [Google Scholar]
- 4.Li H, Poulos TL. Crystallization of cytochromes P450 and substrate-enzyme interactions. Curr Top Med Chem. 2004;4:1789–1802. doi: 10.2174/1568026043387205. [DOI] [PubMed] [Google Scholar]
- 5.McLean KJ, Dunford AJ, Neeli R, Driscoll MD, Munro AW. Structure, function and drug targeting in Mycobacterium tuberculosis cytochrome P450 systems. Arch Biochem Biophys. 2007;464:228–240. doi: 10.1016/j.abb.2007.03.026. [DOI] [PubMed] [Google Scholar]
- 6.Otyepka M, Skopalik J, Anzenbacherova E, Anzenbacher P. What common structural features and variations of mammalian P450s are known to date? Biochim Biophys Acta. 2007;1770:376–389. doi: 10.1016/j.bbagen.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 7.Poulos TL. Structural and functional diversity in heme monooxygenases. Drug Metab Dispos. 2005;33:10–18. doi: 10.1124/dmd.104.002071. [DOI] [PubMed] [Google Scholar]
- 8.Poulos TL. Structural biology of heme monooxygenases. Biochem Biophys Res Commun. 2005;338:337–345. doi: 10.1016/j.bbrc.2005.07.204. [DOI] [PubMed] [Google Scholar]
- 9.Scott EE, White MA, He YA, Johnson EF, Stout CD, Halpert JR. Structure of mammalian cytochrome P450 2B4 complexed with 4-(4-chlorophenyl)imidazole at 1.9Å resolution: Insight into the range of P450 conformations and coordination of redox partner binding. J Biol Chem. 2004;279:27294–27301. doi: 10.1074/jbc.M403349200. [DOI] [PubMed] [Google Scholar]
- 10.Zhao Y, Sun L, Muralidhara BK, Kumar S, White MA, Stout CD, Halpert JR. Structural and Thermodynamic Consequences of 1-(4-Chlorophenyl)imidazole Binding to Cytochrome P450 2B4. Biochemistry. 2007;46:11559–11567. doi: 10.1021/bi7011614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams PA, Cosme J, Vinkovic DM, Ward A, Angove HC, Day PJ, Vonrhein C, Tickle IJ, Jhoti H. Crystal structures of human cytochrome P450 3A4 bound to metyrapone and progesterone. Science. 2004;305:683–686. doi: 10.1126/science.1099736. [DOI] [PubMed] [Google Scholar]
- 12.Yano JK, Wester MR, Schoch GA, Griffin KJ, Stout CD, Johnson EF. The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05-A resolution. J Biol Chem. 2004;279:38091–38094. doi: 10.1074/jbc.C400293200. [DOI] [PubMed] [Google Scholar]
- 13.Schoch GA, Yano JK, Wester MR, Griffin KJ, Stout CD, Johnson EF. Structure of human microsomal cytochrome P450 2C8: Evidence for a peripheral fatty acid binding site. J Biol Chem. 2004;279:9497–9503. doi: 10.1074/jbc.M312516200. [DOI] [PubMed] [Google Scholar]
- 14.Wester MR, Johnson EF, Marques-Soares C, Dansette PM, Mansuy D, Stout CD. Structure of a substrate complex of mammalian cytochrome P450 2C5 at 2.3 A resolution: Evidence for multiple substrate binding modes. Biochemistry. 2003;42:6370–6379. doi: 10.1021/bi0273922. [DOI] [PubMed] [Google Scholar]
- 15.Wester MR, Yano JK, Schoch GA, Yang C, Griffin KJ, Stout CD, Johnson EF. The structure of human cytochrome P450 2C9 complexed with flurbiprofen at 2.0-A resolution. J Biol Chem. 2004;279:35630–35637. doi: 10.1074/jbc.M405427200. [DOI] [PubMed] [Google Scholar]
- 16.Williams PA, Cosme J, Sridhar V, Johnson EF, McRee DE. Mammalian Microsomal Cytochrome P450 Monooxygenase: Structural Adaptations for Membrane Binding and Functional Diversity. Mol Cell. 2005;5:121–131. doi: 10.1016/s1097-2765(00)80408-6. [DOI] [PubMed] [Google Scholar]
- 17.Williams PA, Cosme J, Ward A, Angove HC, Matak Vinkovic D, Jhoti H. Crystal structure of human cytochrome P450 2C9 with bound warfarin. Nature. 2003;424:464–468. doi: 10.1038/nature01862. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Y, White MA, Muralidhara BK, Sun L, Halpert JR, Stout CD. Structure of microsomal cytochrome P450 2B4 complexed with the antifungal drug bifonazole: insight into P450 conformational plasticity and membrane interaction. J Biol Chem. 2006;281:5973–5981. doi: 10.1074/jbc.M511464200. [DOI] [PubMed] [Google Scholar]
- 19.Zhao Y, Halpert JR. Structure-function analysis of cytochromes P450 2B. Biochim Biophys Acta. 2006;1770:402–412. doi: 10.1016/j.bbagen.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 20.Scott EE, He YA, Wester MR, White MA, Chin CC, Halpert JR, Johnson EF, Stout CD. An open conformation of mammalian cytochrome P450 2B4 at 1.6-A resolution. Proc Natl Acad Sci USA. 2003;100:13196–13201. doi: 10.1073/pnas.2133986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ekroos M, Sjogren T. Structural basis for ligand promiscuity in cytochrome P450 3A4. Proc Natl Acad Sci USA. 2006;103:13682–13687. doi: 10.1073/pnas.0603236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wester MR, Johnson EF, Marques-Soares C, Dijols S, Dansette PM, Mansuy D, Stout CD. Structure of mammalian cytochrome P450 2C5 complexed with diclofenac at 2.1 A resolution: evidence for an induced fit model of substrate binding. Biochemistry. 2003;42:9335–9345. doi: 10.1021/bi034556l. [DOI] [PubMed] [Google Scholar]
- 23.Davydov DR, Deprez E, Hoa GH, Knyushko TV, Kuznetsova GP, Koen YM, Archakov AI. High-pressure-induced transitions in microsomal cytochrome P450 2B4 in solution: evidence for conformational inhomogeneity in the oligomers. Arch Biochem Biophys. 1995;320:330–344. doi: 10.1016/0003-9861(95)90017-9. [DOI] [PubMed] [Google Scholar]
- 24.Scott EE, Spatzenegger M, Halpert JR. A truncation of 2B subfamily cytochromes P450 yeilds increased expression levels, increased solubility, and decreased aggregation while retaining function. Arch Biochem Biophys. 2001;395:57–68. doi: 10.1006/abbi.2001.2574. [DOI] [PubMed] [Google Scholar]
- 25.Leslie AG. Integration of macromolecular diffraction data. Acta Crystallogr Sect D Biol Crystallogr. 1999;55:1696–1702. doi: 10.1107/s090744499900846x. [DOI] [PubMed] [Google Scholar]
- 26.Bailey S. The Ccp4 suite: Programs for protein crystallography. Acta Crystallogr Sect D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 27.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murshudov GN, Vagin, Alexei A, Dodson, Eleanor J. Refinement of Macromolecular Structures by the Maximum-Likelihood Method. Acta Crystallogr Sect D Biol Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 29.McRee DE. Differential evolution for protein crystallographic optimizations. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2276–2279. doi: 10.1107/S0907444904025491. [DOI] [PubMed] [Google Scholar]
- 30.Emsley PaC, Kevin Coot: Model-Building Tools for Molecular Graphics. Acta Crystallogr Sect D Biol Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 31.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr Sect D Biol Crystallogr. 2002;D58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 32.DeLano WL. The PyMOL Molecular Graphics System, MacPyMOL ed. Palo Alto, CA, USA.: DeLano Scientific; 2002. [Google Scholar]
- 33.Muralidhara BK, Negi S, Chin CC, Braun W, Halpert JR. Conformational flexibility of mammalian cytochrome P450 2B4 in binding imidazole inhibitors with different ring chemistry and side chains. Solution thermodynamics and molecular modeling. J Biol Chem. 2006;281:8051–8061. doi: 10.1074/jbc.M509696200. [DOI] [PubMed] [Google Scholar]
- 34.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 35.Briggs PJ. CCP4 Newsletter. Daresbury, UK: 2000. AREAIMOL. [Google Scholar]
- 36.Saff EBKABJ. Distributing Many Points on a Sphere. The Mathematical Intelligencer. 1997;19:5–11. [Google Scholar]
- 37.Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: Orientations of Proteins in Membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 38.Williams PA, Cosme J, Sridhar V, Johnson EF, McRee DE. Mammalian microsomal cytochrome P450 monooxygenase: structural adaptations for membrane binding and functional diversity. Mol Cell. 2000;5:121–131. doi: 10.1016/s1097-2765(00)80408-6. [DOI] [PubMed] [Google Scholar]
- 39.Gotoh O. Substrate recognition sites in cytochrome P450 family 2 (CYP 2) proteins inferred from comparative analyses of amino acid and coding nucleotide sequences. J Biol Chem. 1992;267:83–90. [PubMed] [Google Scholar]
- 40.de Lemos-Chiarandini C, Frey AB, Sabatini DD, Kreibich G. Determination of the membrane topology of the phenobarbital-inducible rat liver cytochrome P-450 isoenzyme PB-4 using site-specific antibodies. J Cell Biol. 1987;104:209–219. doi: 10.1083/jcb.104.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bridges A, Gruenke L, Chang YT, Vakser IA, Loew G, Waskell L. Identfication of the binding site on cytochrome P450 2B4 for cytochrome b5 and cytochrome P450 reductase. J Biol Chem. 1998;273:17036–17049. doi: 10.1074/jbc.273.27.17036. [DOI] [PubMed] [Google Scholar]
- 42.Haines DC, Tomchick DR, Machius M, Peterson JA. Pivotal role of water in the mechanism of P450BM-3. Biochemistry. 2001;40:13456–13465. doi: 10.1021/bi011197q. [DOI] [PubMed] [Google Scholar]
- 43.Cupp-Vickery J, Anderson R, Hatziris Z. Crystal structures of ligand complexes of P450eryF exhibiting homotropic cooperativity. Proc Natl Acad Sci USA. 2000;97:3050–3055. doi: 10.1073/pnas.050406897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao B, Lamb DC, Lei L, Kelly SL, Hachy DL, Waterman MR. Different Binding Modes of Two Flaviolin Substrate Molecules in Cytochrome P450 158A1 (CYP158A1) Compared to CYP158A. Biochemistry. 2007;46:8725–8733. doi: 10.1021/bi7006959. [DOI] [PubMed] [Google Scholar]
- 45.Li H, Poulos TL. Fatty acid metabolism, conformational change, and electron transfer in P-450BM-3. Biochim Biophys Acta. 1999;1441:141–149. doi: 10.1016/s1388-1981(99)00161-4. [DOI] [PubMed] [Google Scholar]
- 46.Park SY, Yamane K, Adachi S, Shiro Y, Weiss KE, Maves SA, Sligar SG. Thermophilic cytochrome P450 (CYP119) from Sulfolobus solfataricus: high resolution structure and functional properties. J Inorg Biochem. 2002;91:491–501. doi: 10.1016/s0162-0134(02)00446-4. [DOI] [PubMed] [Google Scholar]
- 47.Yano JK, Koo LS, Schuller DJ, Li H, Ortiz de Montellano PR, Poulos TL. Crystal structure of a thermophilic cytochrome P450 from the archaeon Sulfolobus solfataricus. J Biol Chem. 2000;275:31086–31092. doi: 10.1074/jbc.M004281200. [DOI] [PubMed] [Google Scholar]
- 48.Podust LM, Kim Y, Arase M, Neely BA, Beck BJ, Bach H, Sherman DH, Lamb DC, Kelly SL, Waterman MR. The 1.92-A structure of Streptomyces coelicolor A3(2) CYP154C1. A new monooxygenase that functionalizes macrolide ring systems. J Biol Chem. 2003;278:12214–12221. doi: 10.1074/jbc.M212210200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.