Abstract
Protein X-ray crystallography recently celebrated its 50th anniversary. The structures of myoglobin and hemoglobin determined by Kendrew and Perutz provided the first glimpses into the complex protein architecture and chemistry. Since then, the field of structural molecular biology has experienced extraordinary progress and now over 53,000 proteins structures have been deposited into the Protein Data Bank. In the past decade many advances in macromolecular crystallography have been driven by world-wide structural genomics efforts. This was made possible because of third-generation synchrotron sources, structure phasing approaches using anomalous signal and cryo-crystallography. Complementary progress in molecular biology, proteomics, hardware and software for crystallographic data collection, structure determination and refinement, computer science, databases, robotics and automation improved and accelerated many processes. These advancements provide the robust foundation for structural molecular biology and assure strong contribution to science in the future. In this report we focus mainly on reviewing structural genomics high-throughput X-ray crystallography technologies and their impact.
Introduction
In the 21st century, with the advent of new astonishingly efficient genome sequencing techniques[1], microarray experiments[2], and new proteomics technologies, biology has seen huge increases in DNA sequence, gene expression and proteomics data. The Human Genome and other genome sequencing projects continue to deliver new protein sequences at an astounding pace. New protein families are being discovered in newly sequenced genomes[3–6*] [www.jgi.doe.gov/programs/GEBA/pilot.html] or are overrepresented in specific environments such as human microbiomes [7]. For a large fraction of these proteins families we have virtually no functional or structural data[8*].
Since its inception in the 1950’s of the 20th century, structural molecular biology provided critical observations in biology, and it continues to contribute greatly to the understanding of many biological processes. Structures helped to define biological concepts, explain molecular and biochemical function, and facilitate understanding underlying biochemical mechanisms (see human aldose reductase data[9]). Structural molecular biology helped to decipher basic principles of protein structure and assembly, mechanisms of biochemical reactions, and details of macromolecular interactions, and has contributed to developing new pharmaceuticals. At present, the majority of proteins associated with many key cellular processes have structural representatives known[3]. The atomic resolution structures explained basic principles of proteins three-dimensional structures at a very high level of detail. High-quality structural models are available for the majority of important protein families. Moreover, the structures of numerous complexes and multi-component assemblies have been also determined, and in the past few years, excellent progress has been made for several classes of membrane proteins, including GPCRs[10**]. Structural coverage of several important drug targets is remarkable (kinases, phosphatases, proteases)[11]. From its inception, structural molecular biology had accomplished this quest by focusing on specific systems and individual proteins. At the same time it is worth noticing that some very large and important protein families are still poorly structurally studied[4]. Moreover, many biologists would like to have a structure of their favorite protein available and several lists of the most desired structures have been compiled[12]. Structural genomics researchers’ intention was to fill this major gap by developing high throughput (HTP) methods, improving quality of structures and reducing the cost of structure determination.
The structural genomics (SG) programs were initiated in 1990’s. For the first time, these programs attempted to use available comprehensive genomic information to select protein targets for structure determination. SG’s quest is to determine rapidly a large number of novel structures in order to expand structural and functional knowledge for proteins found in genomes. This approach is based on the notion that a structure available for one member of a protein sequence family will provide structural information for the whole family and will help explain the function of the majority of members of the family. In the past 10 years, SG has significantly expanded its contribution of structures (Table 1). Because SG uses genomic data to select proteins for structure determination and avoids proteins with known structural homologues, SG has become the most important source of structural novelty. This is especially true for the NIH-funded Protein Structure Initiative[13] (PSI), which contributed 1520 novel structures in the second phase of this program[14*], 39% of all novel structures deposited during this period of time (http://targetdb.pdb.org/Metrics/MilestonesTables.html).
Table 1.
World-wide structural genomics centers.
| Center | Structures in PDB |
|---|---|
| RIKEN Structural Genomics/Proteomics Initiative | 2650 |
| Midwest Center for Structural Genomics (PSI) | 997 |
| Joint Center for Structural Genomics (PSI) | 839 |
| New York SGX Research Center for Structural Genomics (PSI) | 733 |
| Structural Genomics Consortium | 681 |
| Northeast Structural Genomics Consortium (PSI) | 667 |
| TB Structural Genomics Consortium | 184 |
| Center for Eukaryotic Structural Genomics (PSI) | 180 |
| Southeast Collaboratory for Structural Genomics (PSI) | 117 |
| Structural Proteomics in Europe | 114 |
| Berkeley Structural Genomics Center (PSI) | 95 |
| Seattle Structural Genomics Center for Infectious Disease | 83 |
| Montreal-Kingston Bacterial Structural Genomics Initiative | 76 |
| Structural Genomics of Pathogenic Protozoa Consortium | 66 |
| Center for Structural Genomics of Infectious Diseases | 66 |
| Structure 2 Function Project | 52 |
| Mycobacterium Tuberculosis Structural Proteomics Project | 21 |
| Ontario Centre for Structural Proteomics | 18 |
| Oxford Protein Production Facility | 17 |
| Israel Structural Proteomics Center | 16 |
| Center for Structures of Membrane Proteins | 11 |
| Integrated Center for Structure and Function Innovation (PSI) | 11 |
| Marseilles Structural Genomics Program @ AFMB | 10 |
| Medical Structural Genomics of Pathogenic Protozoa (PSI) | 5 |
| Scottish Structural Proteomics Facility | 5 |
| Center for High-Throughput Structural Biology (PSI) | 5 |
| Paris-Sud Yeast Structural Genomics | 4 |
| Bacterial targets at IGS-CNRS, France | 4 |
| Accelerated Technologies Center for Gene to 3D Structure (PSI) | 3 |
| Structural Proteomics in Europe 2 | 3 |
| Protein Structure Factory | 1 |
| New York Structural GenomiX Research Consortium (PSI) | 1 |
| New York Consortium on Membrane Protein Structure (PSI) | 1 |
| Total (source PDB) | 7733 |
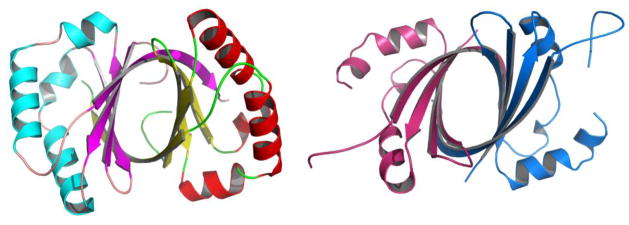
From structural genomic and other data it has become apparent that remarkably, proteins with the same fold and nearly identical structures show no sequence identity. Interestingly, these proteins may perform the same, similar (Fig. 1) or different functions[15*]. This clearly underlies the importance of, and the need for, structural information. At the same time, structures alone are not always sufficient to decipher specific biochemical protein function, although the progress in identifying function has been significant[15*,16*]. It has also become evident that proteins are built from smaller domains that are being reused for performing different functions and this modularity of proteins may be the most important characteristic of protein functional and structural design[5,8]. Moreover, knowing the structures of domains can be highly valuable in interpreting lower resolution data for large macromolecular assemblies[17].
Figure 1.
Structures of two novel monooxygenases. ActVA-Orf6, PDB id 1LQ9 (left), involved in actinorhodin biosynthesis and IsdI PDB id 1SQE (right), involved in heme degradation. These proteins show virtually no sequence similarity, but they share a ferredoxin-like fold - an α+β sandwich with an anti-parallel β-sheet. The structures can be superimposed with rmsd 1.92 Å over 107 residues. These proteins function as monooxygenases on quite different substrates.
In this review we will attempt to summarize how SG has impacted the progress in structural molecular biology, where we are now, and what directions structural molecular biology may take in the future. Although SG has contributed strongly to both X-ray crystallography and NMR, in this review we will focus exclusively on progress in X-ray crystallography. Because of the huge contribution SG has brought to the development of methods, this review can’t be comprehensive and will be limited to selected technologies, and also because we are associated with the PSI SG effort, this review will focus mainly on PSI contributions, although significant progress has been made by SG efforts world-wide.
SG contributed many technological advances that are directly applicable to structural molecular biology
The scale (7733 structures, including 4054 from the PSI alone in past 9 years, Table 1 (http://targetdb.pdb.org/Metrics/MilestonesTables.html)) and comprehensives in the SG approach from gene to structure [18**,19**,20*,21*,22*] permitted very rigorous testing and advancing methods and technologies for structural molecular biology. Methods that have been tested on one or few proteins now could be verified with hundreds and thousands of highly diverse samples passed through the same experimental protocol and using the same equipment. To analyze multiple targets efficiently it is clear that parallel process must be employed[21*,23,24**,25]. Many steps in molecular biology are simple but labor intense and are repeated hundreds of times; therefore such steps can strongly benefit by applying automation and robotics. Therefore, for the first time, SG provided a large-scale parallel platform to thoroughly evaluate structural molecular biology methods. Large data sets are generated and can be mined to extract significant trends to identify what works, what does not and therefore what to avoid[21**,26**]. Moreover, because of the open policy, SG has shared these improvements with the biological community. As a result, today we have many robust, efficient and cost effective approaches in molecular biology and protein production, crystallization, data collection and structure determination using X-ray crystallography, structural model generation, structure refinement and validation. Moreover, the cost has been significantly reduced (5–6 fold) and quality of structure determination has been improved. The methods developed in SG have wide applicability in structural molecular biology and many components of the HTP pipelines have been already adopted world-wide, many are freely accessible to the scientific community at the structural genomics and synchrotron beamlines facilities[27]. Furthermore, these methods are also being broadly disseminated; for example, the PSI SG efforts contributed thus far 720 manuscripts describing a wide range of technologies. The biology community can also take advantage of HTP pipelines and nominate proteins for structure determination through the PSI KB website[28*] to use these pipelines. In addition, the PSI technology portal lists information about hundreds different technologies developed by PSI SG centers (http://kb.psi-structuralgenomics.org/KB/).
Structural genomics contributed significantly to four major challenges in protein structural molecular biology: 1) it has developed several HTP methods for improving proteins and producing high quality samples for structure determination, 2) it has established and verified general and robust structure phasing approaches using synchrotron radiation, 3) it has reduced errors and increased the speed and efficiency of structure determination, and 4) it has large scale tested numerous approaches that may serve as salvage pathways to improve the success rate in structure determination of recalcitrant proteins.
Is a protein suitable for structure determination?
Not all proteins are designed for crystallization or are compatible with the structure determination process using X-ray crystallography. SG developed methods to assess suitability of protein for structural biology pipelines, especially suitable for protein domains.
Data mining to optimize protein sequence for structure determination
The scale of SG efforts now allows for identification of certain protein features and biophysical parameters that indicate higher propensity to crystallize. Therefore, one can analyze protein family sequences or sequence constructs and identify those that are more likely to result in structure. For example, Godzik and colleagues from Joint Center for Structural Genomics (JCSG) designed a “crystallization feasibility” score indicating which protein sequence shows higher propensity to advance to three-dimensional structure[26**]. Analyses of PDB depositions suggest that these same features and properties are present for proteins with known structures, suggesting that this approach should help to evaluate crystallization feasibility and success in structure determination and it should be of high interest to all structural biologists, as well as to molecular and biochemistry laboratories[26**]. This resource is freely available to the scientific community (ffas.burnham.org/XtalPred). A similar approach for designing domain constructs has been developed by Babnigg and colleagues at the MCSG and is available to the scientific community (MCSG, bioinformatics.anl.gov/cgi-bin/tools/PDpredictor). Multiple proteins (orthologues) and/or multiple constructs (mutants or length variants) of the same protein can be screened in order to identify suitable protein constructs that can be produced at a structural molecular biology scale and quality[29,30].
Employing enhanced hydrogen/deuterium exchange MS for refinement of crystallographic protein construct
The JCSG has developed a number of enhancements for high-resolution deuterium exchange MS (DXMS) technology that allows rapid identification of unstructured regions in proteins. Structural comparisons showed that the DXMS method can correctly localize even small internal regions of disorder that correlated extraordinarily well with lack of density in the crystallographic maps. The DXMS analysis identified truncations that greatly improved crystallization and have been used for structure determination. This approach represents a rapid and generalized method that can be applied to any structural molecular biology protein of interest[31**]. The method is now used broadly to study protein structure and dynamics and ligand binding[32].
In structural genomics, the process of solving structures from clone to deposit provides a unique knowledge base. As the experimental data accumulates rapidly in the databases, new experiments can be planned and executed more efficiently based on past successes and failures. This enables the evaluation of each step’s effectiveness and allows the elimination of approaches that are less effective or produce sub-optimal results (at the same time promoting more effective approaches). This guides the decision making process whether to stop or continue or to apply a particular salvage approach to “challenging” targets. For specific classes of proteins and recalcitrant targets, an alternative set of “salvage” protocols and methods must be identified as early as possible and applied in order to increase success rate. This process enables the development of methods that allow the SG community to continuously redefine valid targets and create new, improved protocols[25,33].
Gene cloning and protein expression
Obtaining high-quality protein samples for structure determination is a not a trivial task, especially if one has to do it for many different proteins, mutants or different constructs. Wide application of recombinant technology for gene cloning and protein expression in vivo and in vitro has been critical to production of high quality samples for structural molecular biology. Two leading approaches have emerged in gene cloning:
Ligation Independent Cloning (LIC)
The use of LIC vectors allows rapid progression from gene to expression clone of a complete gene or its fragment using polymerase chain reaction (PCR). LIC eliminates restriction enzymes and DNA ligase components of traditional cloning protocols. LIC provides unique cloning sites, is directional, high efficiency, simple, rapid, inexpensive, and low-background. The LIC method can rapidly generate multiple constructs from a single template, and is compatible with multiple vectors. The LIC approach can be implemented readily in a highly parallel format with minimal optimization and is well suited to both robotic and manual cloning and expression. For example, the Midwest Center for Structural Genomics (MCSG) developed a series of LIC vectors, “pMCSG”, and a set of semi-automated protocols which can be applied to different proteins and used in several applications. Some vectors allow the addition of affinity tags to a protein of interest, some provide enhancers of protein solubility (like maltose binding protein), and some can be used for protein co-expression experiments or track protein behavior during expression using fluorescence tags. These new protein expression vectors were tested on nearly 40,000 gene constructs and also can be applied to large proteins and eukaryotic domains[34**,35]. The MCSG vectors are available through the PSI Material Repository (http://www.hip.harvard.edu/PSIMR/) and a wide variety of LIC compatible vectors are available commercially. LIC vectors are also being applied for expression of membrane associated and transmembrane proteins.
Polymerase Incomplete Primer Extension (PIPE)
In the alternative approach, the PIPE method was developed by the Joint Center for Structural Genomics (JCSG) for HTP cloning of many protein constructs. PIPE combines cloning and mutagenesis into a simple two-step protocol with high efficiency and flexibility. With the PIPE protocol, all major cloning operations are achieved by transforming competent cells with PCR products immediately following amplification. Using straightforward primer design conventions and PCR, short, overlapping sequences are introduced at the ends of these incomplete extension mixtures. These extensions allow complementary strands to anneal and produce a hybrid vector/insert permutation. The hybrids are then directly transformed into recipient cells without any post-PCR enzymatic manipulations. Similarly to the LIC approach, all major cloning operations are achieved by transforming competent cells with PCR products. The method is robust and amenable to automation as only a few, simple processing steps are needed. Using this approach, researchers in the JCSG have cloned thousands of genes in parallel using minimum effort[24**].
High-throughput sample preparation for structural biology
Expression of proteins from cloned genes in E. coli has been optimized in the SG pipelines to produce more soluble proteins in large quantities using high-density cultures. These new approaches are fully compatible with efficient in vivo incorporation of selenium atoms to aid phasing.
New high-density media for protein production of native and SeMet-labeled proteins
A drive to improve protein expression resulted in the development of new media that are compatible with the introduction of selenomethionine (SeMet) to proteins. Studier lab has been at the forefront of developing vectors using a T7lac promoter for efficient production of a wide variety of proteins in E. coli. Systematic analysis of bacterial growth allowed for the development of reliable non-inducing and auto-inducing media in which batch cultures can grow to high densities. Expression strains grown to saturation in non-inducing media retain plasmids and remain fully viable for an extended period of time. Auto-induction allows for the efficient screening of many clones in parallel for expression and solubility, as cultures only have to be inoculated and grown to saturation, and yields of target protein are typically several-fold higher than obtained by conventional IPTG induction. Auto-inducing media have been developed for labeling proteins with SeMet and for production of target proteins by the arabinose induction of T7 RNA polymerase from the pBAD promoter in BL21-AI[36**]. Similarly, high efficiency minimal media was developed at the MCSG by optimizing the media composition. This new “pink” version of the M9 medium permits E. coli to grow at low temperature to high densities and is fully compatible with incorporation of SeMet to proteins[34**].
In SG effort, the majority of protein expression is done in E. coli, but expressing proteins in bacteria in a soluble form is often a major challenge. Low temperature expression of proteins in many cases improves their solubility and stability. The Inouye Laboratory at the Northeast Structural Genomics Consortium developed a set of expression vectors, termed pCold vectors, which drive the high expression of cloned genes by cold-shock promoter. Proteins from both microbial and eukaryotic sources can be produced with very high yields[37*].
Yeast strain for expression of SeMet-labeled proteins
Malkowski and colleagues at the Center for High-Throughput Structural Biology engineered Saccharomyces cerevisiae to accept SeMet, therefore making it possible to label proteins expressed in yeast with SeMet. By deleting SAM1 and SAM2 genes encoding AdoMet synthetase which converts methionine to S-adenosylmethionine, they created a strain with reduced SeMet toxicity[38*]. The strain requires AdoMet for growth but it can grow on high concentrations of SeMet. Proteins expressed in this strain of yeast are labeled with SeMet and are suitable for structure determination.
Cell-free protein expression
Expressing proteins in bacterial systems has certain drawbacks and the use of cell-free protein expression has been shown to provide good alternative. The E. coli and wheat germ based systems have been used for a very long time, but only after SG programs started using these expression systems have they been tested vigorously and compared[39–41]. The progress in optimizing particularly the wheat germ expression system is encouraging. The quality of extracts and new instrumentation caused successful applications of these systems to produce proteins for functional and structural molecular biology studies ref]. In general, two level expression and purification protocols are recommended: a small microgram-scale is used for rapid evaluation, and large milligram-scale protein production of tagged proteins is used for to obtain both unlabeled and labeled proteins required for structure-based determinations by X-ray crystallography.
Optimizing protein for structure determination
Obtaining suitable protein for structure determination often requires the modification of the protein itself. For example removing unstructured regions of proteins can provide a considerable advantage in obtaining high-quality crystals.
In situ proteolysis to aid crystallization
Partial proteolysis combined with mass spectrometry has been used for a long time to define the optimal protein construct for structure determination. Now SG expanded this method forward with in situ proteolysis during crystallization. The MCSG and Structural Genomics Consortium (SGC) in a collaborative effort systematically study in situ proteolysis of proteins to promote crystallization. Typically in situ proteolysis removed residues at either the N- or C-termini, or both. The use of in situ proteolysis provides a path to significantly increase the success rate of protein structure determination particularly for recalcitrant proteins[42,43*] and it appears, with over 12% success rate, to be the most efficacious crystallization salvage strategy.
Modification of the protein surface has been shown to aid crystallization. The SG programs have extensively tested several of these approaches with two briefly described below.
Surface entropy reduction to promote protein crystallization
Derewenda’s laboratory at the Integrated Center for Structure and Function Innovation developed a method to engineer a protein surface designed to form intermolecular contacts that could support crystal packing. This approach is based on the concept of surface entropy reduction (SER), i.e., the replacement of small clusters of two to three solvent-exposed high conformational entropy residues with small residues such as alanine. The method has been successfully used to crystallize a number of novel proteins and many recalcitrant proteins. It has shown to be an effective salvage pathway for proteins that are difficult to crystallize[44*]. The surface entropy reduction prediction server (SERp), designed to identify mutations that may facilitate crystallization, has been developed and is available at www.doe-mbi.ucla.edu/Services/SER[45*].
Reductive methylation of protein surface
Similarly, at the MCSG, surface modification using reductive methylation of lysine residues in proteins has been tested on several hundred unique protein targets that failed to crystallize or produce X-ray quality crystals. The chemical modification is fast, specific, inexpensive, and requires few steps under relatively mild buffer and chemical conditions. Following the method described by I. Rayment[46] the proteins can be methylated and screened using the HTP crystallization pipeline. Reductive methylation of lysine residues alters protein surface properties and crystallization behavior. For 7% of screened target proteins crystal structures have been obtained, the methylated proteins tend to diffract to higher resolution as compared with native. The method is well suited to HTP projects as well as regular laboratories[47*].
Protein purification and crystallization
Recent advances in protein purification
For structural molecular biology applications the resulting protein samples must be compatible with the structure determination regime, specifically protein must comply with the crystallization process. The approaches for HTP protein purification and production of crystals for many proteins that are suitable for synchrotron-based X-ray crystallography have been developed and implemented in a number of SG centers[48*]. In general the quantities of proteins must achieve protein concentrations in the range of 5–25 mg/ml, allowing screening 500–1000 crystallization conditions, and produce X-ray-quality single crystals. A high protein purity of >95% is required. The only practical and HTP approach that is compatible with the above criteria is purification using affinity tags and semiautomated chromatographic workstations capable of performing minimum two consecutive chromatographic steps[49]. SG has contributed to the development of instrumentation and protocols for semi-automated multi-dimensional chromatography that are compatible with structural molecular biology requirements. New commercially available chromatographic workstations allow the development of advanced multi-step purification protocols and the implementation of these protocols for many proteins, protein constructs, or allow purification at milligram to gram quantities of proteins for HTP structure determination and other applications like screening for drug discovery. For example, the MCSG has developed and extensively tested several protocols for HTP automated protein purification using affinity and size exclusion chromatography, protein refolding, proteolytic cleavage on the column and others. These protocols implemented on AKTA EXPLORER 3D and AKTAexpress workstations are fast and produce many proteins in parallel in milligram quantities. Similar approaches are being developed for purification of small protein-protein complexes[50]. The automated chromatography has been successfully applied to several thousands of proteins of microbial and eukaryotic origin in many SG laboratories world-wide and now being also used in smaller laboratories.
Reducing the amount of protein for crystallization
In protein crystallography, the amount of material for crystallization is often a limiting factor. The SG efforts in crystallization have been focused, among other goals, on lowering the amount of sample needed and discovering the best crystallization formulations. New optimized crystallization screens are now available commercially (JCSG, JCSG1-4, ANL-1 and 2) that improve the success rate for many proteins. The use of very small volumes has been explored and extensively tested with over 50,000 proteins in the SG centers. Small crystallization volumes tend to reduce equilibration times and increase the success rate[48*,51*,52]. Optimization approaches to turn very small or low-quality crystals into useful diffracting ones must be taken into consideration and must be adapted to HTP. The use of automation in HTP screening with optimized custom screens increases the chance of improving crystals.
The use of a nanovolume microfluidic environment for plug-based and counterdiffusion methods in confined geometries (plastic labcards) is being developed by a number of SG centers[53*,54*]. These approaches are being developed for both in situ X-ray screening and data collection. Crystallization in a microfluidic environment has minimal sample requirements and may allow crystallization of proteins that can be obtained only in very small amounts. At the same time, rapid progress is being made on using mini-beams for data collection at the synchrotrons (see below).
The use of synchrotron facilities for protein structure determination
The SG effort in methods and technology-development has also made a major impact on the automation of crystal handling, data collection using cryo-crystallography and synchrotron X-ray sources, and structure determination using multi- or single-wavelength anomalous diffraction (MAD/SAD) phasing and automated approaches, automation of model building and structure refinement and verification.
High-throughput crystallography using synchrotron radiation
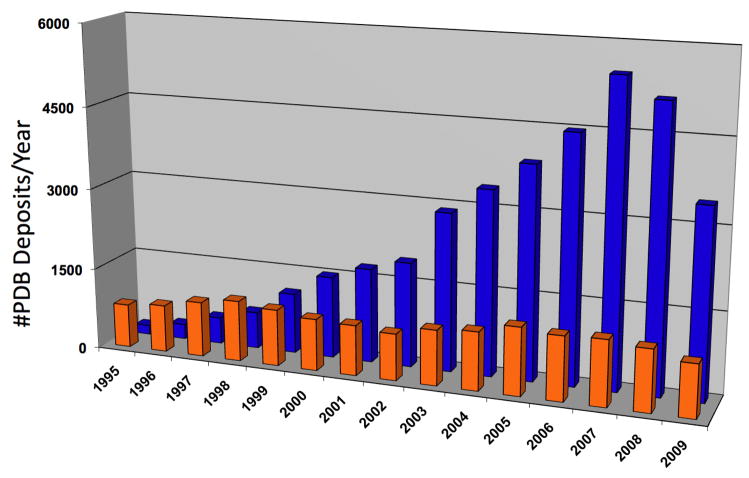
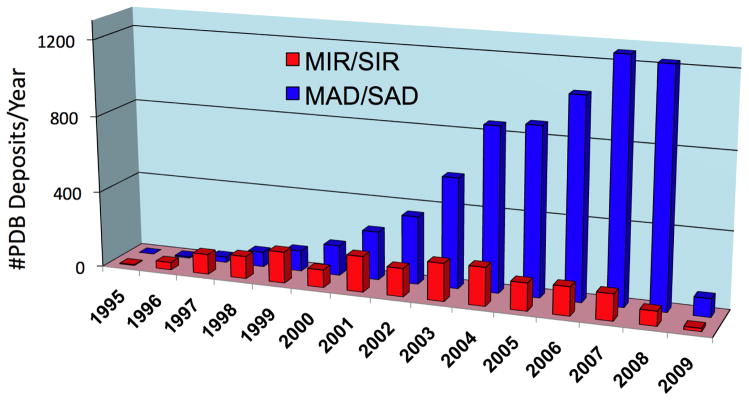
At the onset of SG programs it become evident that in order to accomplish the throughput needed to determine hundreds of protein structures, synchrotron facilities must be used. Fortunately, third generation synchrotron facilities were coming on line in Europe, the US and Japan. Now over one hundred twenty synchrotron beamlines are available world-wide for macromolecular X-ray crystallography (http://biosync.rcsb.org/). Moreover, the efficiency of many beamlines has increased with a number of them turning out over 100 and the best approaching 300 structures per year. The high flux, brilliance, and flexibility inherent in the design of the optics, coupled with a kappa-geometry goniometer and beamline control software, allows optimal strategies to be adopted in protein crystallographic experiments, thus maximizing the chances of their success. The synchrotron beamlines, when combined with crystal cryo-protection and robotic crystal handling, allowed for the optimal use of the “anomalous signal” for phasing structures. Data can be collected from a single crystal and the phases can be extracted semi-automatically[19**,55*]. The SG programs have significantly contributed to establishing MAD/SAD as a routine method of protein structure determination (Fig. 2). Moreover, ultrafast MAD/SAD data collection is now possible on a routine basis and with the widespread use of SeMet for phase determination the method has become the most prominent experimental approach in de novo determining structures of protein. Developments in crystallographic software are complementing these advances, paving the way for improving quality and accelerated protein structure determination[56]. For many proteins, data are being processed in near real time and structures are now determined at the synchrotron facility. This changed the approach and planning of the diffraction experiments and increased success rates. The use of synchrotron facilities has truly transformed SG and structural molecular biology research and contributed to the growth of structures in PDB (Fig. 3).
Figure 2.
Protein structural deposits with data obtained from home (orange) and synchrotron sources (blue).
Figure 3.
Structure determination using the traditional heavy atom (MIR/SIR) approach and using the anomalous signal (MAD/SAD) approach.
The use of SeMet for phasing structure
Synchrotrons allow the optimal use of anomalous scattering for protein structure determination. The applicability of this method has been widely extended by the ease of incorporation of Se atoms in the form of SeMet into the protein in vivo, providing a source of measurable anomalous signal[57]. SG programs have widely adopted this approach, combining it with cryo-crystallography at the synchrotrons often permits all data to be measured from a single crystal[58]. This approach significantly reduces systematic errors in the experiment and is truly HTP. It eliminates the arduous search for suitable heavy-atom derivatives and the problems of non-isomorphism between native and derivative crystals, as all data are collected from the same crystal. As the Se atoms are covalently bound, problems with poorly occupied sites are eliminated and often a single, well-ordered selenium site is sufficient to phase 25–35 kDa protein (Fig. 4) and produce an excellent experimental electron density maps[59].
Figure 4.
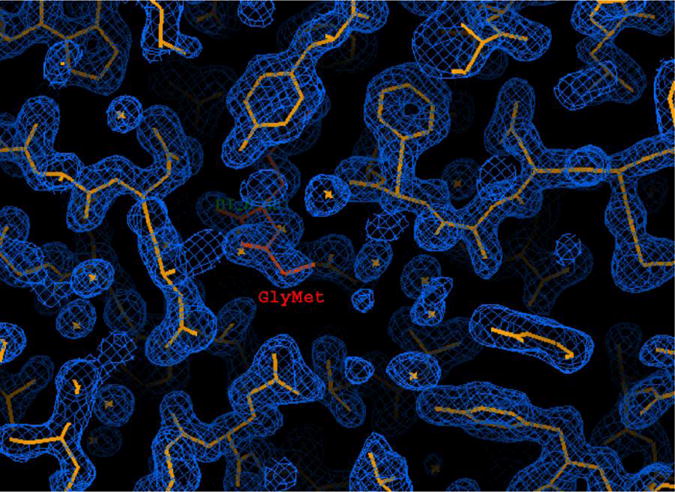
MAD/SAD phasing provides higher quality electron density maps, allows automated map interpretation and improves structure quality. The electron density of GmpC lipoprotein-9 from S. aureus was obtained with 1 selenium atom per 297 residues [59]. The phases were obtained from a SAD experiment near the selenium edge. The experimental maps were calculated at 1.7 Å and contoured at 1σ.
Use of X-ray minibeams
The use of mini-beams is a new exciting development in synchrotron research. Mini-beams (5–20 microns in diameter) provide several important benefits. Illuminating very small crystals with a small beam reduces the scatter from the matter surrounding the crystal. Low background scattering from a minibeam can lead to significant improvements in the signal-to-noise ratio, Rmerge and effective diffraction limit[60*]. Mini-beams can help with crystal inhomogeneity to identify best-ordered crystal regions. In addition, based on theoretical considerations and some recent experimental results[61,62], it appears that crystals only a few μm in size (microbeams) give usable data sets with lower radiation damage than projected from radiation damage studies with larger crystals. The use of nano-liter crystallization technology in combination with synchrotron micro-beams has the potential to revolutionize structural molecular biology.
Improving data for structure determination
A novel approach to scaling diffraction intensities has been developed by SG researchers. This method minimizes the disagreement among multiple measurements of symmetry-related reflections using a stable refinement procedure. The scale factors are described by a flexible exponential function that allows different scaling corrections to be chosen and combined according to the needs of the experiment. The scaling model includes: scale and temperature factor per batch of data; temperature factor as a continuous function of the radiation dose; absorption in the crystal; uneven exposure within a single diffraction image; and corrections for phenomena that depend on the diffraction peak position on the detector. This scaling model can be extended to include additional corrections for various instrumental and data-collection problems[63*].
Semi-automated structure determination using X-ray crystallography and synchrotron radiation
The development of several large-scale SG projects world-wide presented new challenges in the field of crystallographic macromolecular structure determination. The use of anomalous signal allowed standardization of data collection strategies and phasing approaches without compromising the structure quality; in fact, it improved the quality of experimental electron density maps. For example, the MCSG has integrated strategy with data collection, data reduction, phasing and model building. This significantly accelerated the process of structure determination and minimizes the number of data sets and synchrotron time required for structure solution. The new software suite HKL3000 for semi-automated structure determination was developed by Minor, Otwinowski and colleagues[19**]. The software attempts to solve the structure using different algorithms and approaches, rapidly converting diffraction data into an interpretable electron density map, and for smaller structures, into an initial model in near real time. The heuristics for choosing the best computational strategy at different data resolution limits of phasing signal and crystal diffraction are being optimized. The typical end result is an interpretable electron-density map with a partially built structure and, in some cases, an almost complete model. The system is combined with relational databases and linked to external web resources (MCSG, SGPDB, PDB, Swissprot, NCBI and others) and is easily accessible via HKL3000 GUI. The software has been successfully tested on several hundred novel proteins and has resulted in over 800 PDB deposits.
A novel software suite called PHENIX has been developed by Adams, Terwilliger and colleagues[18**,55*,64]. The software combines all the necessary algorithms to proceed from reduced intensity data to a refined molecular model and facilitate semi-automated structure solution. The PHENIX software suite is a highly automated (see Adams et al. this issue) and can rapidly arrive at an initial partial model of a structure without significant human intervention, given moderate resolution, and good quality data. The software is composed of several integrated modules for structure determination: maximum-likelihood molecular replacement (PHASER), heavy-atom search (HySS), template- and pattern-based automated model-building (RESOLVE, TEXTAL), automated macromolecular refinement (PHENIX. refine), and iterative model-building, density modification and refinement that can operate at moderate resolution (RESOLVE, AutoBuild). These algorithms are based on a highly integrated and comprehensive set of crystallographic libraries that have been built and made available to the community. The PHENIX modules are tightly linked and made easily accessible through the Wizards and the GUI.
Improving structure deposition
In the PSI all protein structures are made available to the public as soon as they are completed and deposited into the PDB. In order to expedite the deposition process, the Burley laboratory developed Deposit3D[65*]. This command-line script gathers all the required structure-deposition information and outputs this data into an mmCIF file for subsequent upload through the RCSB PDB ADIT interface. Deposit3D is very useful for SG pipeline projects because it allows workers involved with various stages of a structure-determination project to pool their different categories of annotation information before starting a deposition session. It also helps individual researchers to standardize files and help in the deposition process.
Structural genomics knowledge database
SG also pioneered the comprehensive approach to combining structural data with many other resources available on line. The recently established PSI Structural Genomics Knowledgebase (PSI SGKB) is a free, comprehensive resource produced in collaboration between the Protein Structure Initiative (PSI) and Nature Publishing Group. The PSI SGKB, (http://kb.psi-structuralgenomics.org) attempts to combine SG data into knowledge that can be used by the biological research community to understand living systems and disease[28*]. The PSI SGKB serves as a continually updated portal to research data and other resources from the PSI and links to structures in the Protein Data Bank (PDB), functional annotations, associated homology models, worldwide protein target tracking information, PSI technologies, available protocols and PSI Material Repository. By making these resources freely available to scientific community, the PSI SGKB serves as a bridge to connect the structural molecular biology and the greater biomedical communities.
The perspective
Structural genomics systematically addressed major challenges in structural molecular biology and substantially improved the methods of structure determination applicable to many proteins. As a result, several robust HTP structure determination pipelines have been established with the capacity to determine hundreds of protein structures per year. The cost of structure determination has been reduced substantially and the quality of structures has been significantly improved. SG has also changed the approach to structure determination by identifying bottlenecks, testing salvage approaches and evaluating their efficacy using a large set of novel proteins. Custom and commercial instrumentation integrated into the PSI pipelines are largely available to the scientific community. However, there are a number of challenges that need to be addressed. For example, the PSI technology centers focus their efforts on methods development in structure determination of membrane proteins. The new HTP technologies are being applied with encouraging success to membrane proteins and small complexes[66]. The Center for Structures of Membrane Proteins and the New York Consortium on Membrane Protein Structure have had good success expressing, purifying and determining the structures of representative members of membrane protein classes. Thus far, 12 structures of membrane proteins have been deposited to PDB. Similarly, new HTP technologies can accelerate expression, purification and structure determination of small protein/protein complexes. The use of mini- and micro-beams and small crystals offers new opportunities in determining structures of proteins and various complexes that were not possible just few years ago. Reducing X-ray radiation damage, improving data processing and better modeling damage may further enhance these opportunities. At the foundation of these new developments is significant investment in methods, technology, and access to the most challenging data. To assure healthy and robust structural molecular biology these developments must continue to address new challenges and advance biomedical research to the benefit the society. However, a number of questions remain. How can structural molecular biology cope with this enormity of genomic and proteomic data and contribute valuable knowledge to rapidly expanding biology? This and other questions will need to be addressed in the near future.
Acknowledgments
We thank all members of the Structural Biology Center and the Midwest Center for Structural Genomics at Argonne National Laboratory and Center for Structural Genomics of Infectious Diseases for their help in conducting experiments. We wish to thank the members of the PSI, both past and present, for all of their efforts on behalf of the PSI. Finally, we recognize the valuable contributions made by SG researchers throughout the world. We would like to thank Drs. T.A. Binkowski and A. Kouranov for providing data for Fig. 3, Dr. R-g. Zhang for help with Fig. 4, and Lindsey Butler for help in preparation of the manuscript for publication. This work was supported by National Institutes of Health Grant GM074942 and by the U.S. Department of Energy, Office of Biological and Environmental Research, under contract DE-AC02-06CH11357.
Footnotes
The submitted manuscript has been created by UChicago Argonne, LLC, Operator of Argonne National Laboratory (“Argonne”). Argonne, a U.S. Department of Energy Office of Science laboratory, is operated under Contract No. DE-AC02-06CH11357. The U.S. Government retains for itself, and others acting on its behalf, a paid-up nonexclusive, irrevocable worldwide license in said article to reproduce, prepare derivative works, distribute copies to the public, and perform publicly and display publicly, by or on behalf of the Government.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Voelkerding KV, Dames SA, Durtschi JD. Next-generation sequencing: from basic research to diagnostics. Clin Chem. 2009;55:641–658. doi: 10.1373/clinchem.2008.112789. [DOI] [PubMed] [Google Scholar]
- 2.Dufva M. Introduction to microarray technology. Methods Mol Biol. 2009;529:1–22. doi: 10.1007/978-1-59745-538-1_1. [DOI] [PubMed] [Google Scholar]
- 3.Marsden RL, Lewis TA, Orengo CA. Towards a comprehensive structural coverage of completed genomes: a structural genomics viewpoint. BMC Bioinformatics. 2007;8:86. doi: 10.1186/1471-2105-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mardsen RLOC. Target selection for structural genomics: an overview. Methods Mol Biol. 2008;426:3–25. doi: 10.1007/978-1-60327-058-8_1. [DOI] [PubMed] [Google Scholar]
- 5*.Orengo CA, Thornton JM. Protein families and their evolution-a structural perspective. Annu Rev Biochem. 2005;74:867–900. doi: 10.1146/annurev.biochem.74.082803.133029. [DOI] [PubMed] [Google Scholar]
- 6.Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K, Eisen JA, Heidelberg KB, Manning G, Li W, et al. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh AHDT, Letunic I, Raes J, Bork P. Discovering functional novelty in metagenomes: examples from light-mediated processes. J Bacteriol. 2009;191:32–41. doi: 10.1128/JB.01084-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8*.Levitt M. Nature of the protein universe. Proc Natl Acad Sci U S A. 2009;106:11079–11084. doi: 10.1073/pnas.0905029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blakeley MP, Ruiz F, Cachau R, Hazemann I, Meilleur F, Mitschler A, Ginell S, Afonine P, Ventura ON, Cousido-Siah A, et al. Quantum model of catalysis based on a mobile proton revealed by subatomic x-ray and neutron diffraction studies of h-aldose reductase. Proc Natl Acad Sci U S A. 2008;105:1844–1848. doi: 10.1073/pnas.0711659105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10**.Hanson MASR. Discovery of new GPCR biology: one receptor structure at a time. Structure. 2009;17:8–14. doi: 10.1016/j.str.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bakan A, Lazo JS, Wipf P, Brummond KM, Bahar I. Toward a molecular understanding of the interaction of dual specificity phosphatases with substrates: insights from structure-based modeling and high throughput screening. Curr Med Chem. 2008;15:2536–2544. doi: 10.2174/092986708785909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhattacharya A. Protein structures: Structures of desire. Nature. 2009;459:24–27. doi: 10.1038/459024a. [DOI] [PubMed] [Google Scholar]
- 13.Norvell JC, Berg JM. Update on the protein structure initiative. Structure. 2007;15:1519–1522. doi: 10.1016/j.str.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 14*.Nair R, Liu J, Soong TT, Acton TB, Everett JK, Kouranov A, Fiser A, Godzik A, Jaroszewski L, Orengo C, et al. Structural genomics is the largest contributor of novel structural leverage. J Struct Funct Genomics. 2009;10:181–191. doi: 10.1007/s10969-008-9055-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15*.Binkowski TA, Joachimiak A. Protein functional surfaces: global shape matching and local spatial alignments of ligand binding sites. BMC Struct Biol. 2008:8. doi: 10.1186/1472-6807-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16*.Addou SRR, Lee D, Orengo CA. Domain-based and family-specific sequence identity thresholds increase the levels of reliable protein function transfer. J Mol Biol. 2009;387:416–430. doi: 10.1016/j.jmb.2008.12.045. [DOI] [PubMed] [Google Scholar]
- 17.Ritchie DW. Recent progress and future directions in protein-protein docking. Curr Protein Pept Sci. 2008;9:1–15. doi: 10.2174/138920308783565741. [DOI] [PubMed] [Google Scholar]
- 18**.Zwart PHAP, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Storoni LC, Terwilliger TC, Adams PD. Automated structure solution with the PHENIX suite. Methods Mol Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 19**.Minor WCM, Otwinowski Z, Chruszcz M. HKL-3000. the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 20*.Kim Y, Quartey P, Volkart L, Hatzos C, Zhou M, Chang C, Cuff M, Nocek B, Osipiuk J, Tan K, Fan Y, Maltseva N, Li H, Wu R, Binkowski A, Zhang Rg, Joachimiak A. Protein Reductive Methylation: Improving Protein Crystallization - a Large-Scale Evaluation. 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21**.Consortium; SGCCSG, Northeast Structural Genomics Consortium GS. Nordlund P, Weigelt J, Hallberg BMBJ, Gileadi O, Knapp S, Oppermann U, Arrowsmith C, Hui R, Ming Jd-PS, Park HW, Savchenko A, Yee A, Edwards A, Vincentelli R, Cambillau CKR, Kim SH, Rao Z, Shi Y, Terwilliger TC, Kim CY, Hung LW, Waldo GSPY, Albeck S, Unger T, Dym O, Prilusky J, Sussman JL, Stevens RC, Lesley SAWI, Joachimiak A, Collart F, Dementieva I, Donnelly MI, Eschenfeldt WHKY, Stols L, Wu R, Zhou M, Burley SK, Emtage JS, Sauder JM, Thompson D, Bain KLJ, Gheyi T, Zhang F, Atwell S, Almo SC, Bonanno JB, Fiser A, Swaminathan SSF, Chance MR, Sali A, Acton TB, Xiao R, Zhao L, Ma LC, et al. Protein production and purification. Nat Methods. 2008;5:369. doi: 10.1038/nmeth.f.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22*.Burley SK, Joachimiak A, Montelione GT, Wilson IA. Contributions to the NIH-NIGMS Protein Structure Initiative from the PSI Production Centers. Structure. 2008;16:5–11. doi: 10.1016/j.str.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lesley SA, Wilson I. Protein production and crystallization at the joint center for structural genomics. J Struct Funct Genomics. 2005;6(2–3):71–79. doi: 10.1007/s10969-005-2897-2. [DOI] [PubMed] [Google Scholar]
- 24*.Klock HE, Lesley SA. The Polymerase Incomplete Primer Extension (PIPE) method applied to high-throughput cloning and site-directed mutagenesis. Methods Mol Biol. 2009:498. doi: 10.1007/978-1-59745-196-3_6. [DOI] [PubMed] [Google Scholar]
- 25.Bonanno JBAS, Bresnick A, Chance MR, Fiser A, Swaminathan S, Jiang J, Studier FW, Shapiro L, Lima CD, Gaasterland TM, Sali A, Bain K, Feil I, Gao X, Lorimer D, Ramos A, Sauder JM, Wasserman SR, Emtage S, D’Amico KL, Burley SK. New York-Structural GenomiX Research Consortium (NYSGXRC): a large scale center for the protein structure initiative. J Struct Funct Genomics. 2005;6 (2–3):225–232. doi: 10.1007/s10969-005-6827-0. [DOI] [PubMed] [Google Scholar]
- 26**.Slabinski LJL, Rychlewski L, Wilson IA, Lesley SA, Godzik A. XtalPred: a web server for prediction of protein crystallizability. Bioinformatics. 2007 doi: 10.1093/bioinformatics/btm477. [DOI] [PubMed] [Google Scholar]
- 27.Rosenbaum G, Alkire R, Evans G, Rotella FJ, Lazarski K, Zhang R, Ginell SL, Duke N, Naday I, Lazarz J, Molitsky MJ, Keefe L, Gonczy J, Rock L, Sanishvili R, Walsh MA, Westbrook E, Joachimiak A. The Structural Biology Center 19ID undulator beamline: facility specifications and protein crystallographic results. J Synch Radiation. 2006;13:30–45. doi: 10.1107/S0909049505036721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28*.Berman HM, Westbrook JD, Gabanyi MJ, Tao W, Shah R, Kouranov A, Schwede T, Arnold K, Kiefer F, Bordoli L, et al. The protein structure initiative structural genomics knowledgebase. Nucleic Acids Res. 2009;37:D365–368. doi: 10.1093/nar/gkn790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown G, Singer A, Lunin VV, Proudfoot M, Skarina T, Flick R, Kochinyan S, Sanishvili R, Joachimiak A, Edwards AM, et al. Structural and biochemical characterization of the type II fructose-1,6-bisphosphatase GlpX from Escherichia coli. J Biol Chem. 2009;284:3784–3792. doi: 10.1074/jbc.M808186200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nocek B, Chang C, Li H, Lezondra L, Holzle D, Collart F, Joachimiak A. Crystal structures of delta1-pyrroline-5-carboxylate reductase from human pathogens Neisseria meningitides and Streptococcus pyogenes. J Mol Biol. 2005;354:91–106. doi: 10.1016/j.jmb.2005.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31**.Pantazatos DKJ, Klock HE, Stevens RC, Wilson IA, Lesley SA, Woods VL., Jr Rapid refinement of crystallographic protein construct definition employing enhanced hydrogen/deuterium exchange MS. Proc Natl Acad Sci U S A. 2004;101(3):751–756. doi: 10.1073/pnas.0307204101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konermann LTX, Pan Y. Protein structure and dynamics studied by mass spectrometry: H/D exchange, hydroxyl radical labeling, and related approaches. J Mass Spectrom. 2008;43:1021–1036. doi: 10.1002/jms.1435. [DOI] [PubMed] [Google Scholar]
- 33.Slabinski L, Jaroszewski L, Rodrigues AP, Rychlewski L, Wilson IA, Lesley SA, Godzik A. The challenge of protein structure determination--lessons from structural genomics. Protein Sci. 2007;16:2472–2482. doi: 10.1110/ps.073037907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34**.Donnelly MIZM, Millard CS, Clancy S, Stols L, Eschenfeldt WH, Collart FR, Joachimiak A. An expression vector tailored for large-scale, high-throughput purification of recombinant proteins. Protein Expr Purif. 2006;47:446–454. doi: 10.1016/j.pep.2005.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stols LGM, Dieckman L, Raffen R, Collart FR, Donnelly MI. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr Purif. 2002;25:8–15. doi: 10.1006/prep.2001.1603. [DOI] [PubMed] [Google Scholar]
- 36**.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 37*.Qing GML, Khorchid A, Swapna GVT, Mal TK, Takayama MM, Xia B, Phadtare S, Ke H, Acton T, Montelione GT, Ikura M, Inouye M. Cold-shock induced high-yield protein production in Escherichia coli. Nature Biotechnology. 2004;22:877–882. doi: 10.1038/nbt984. [DOI] [PubMed] [Google Scholar]
- 38*.Malkowski MG, Quartley E, Friedman AE, Babulski J, Kon Y, Wolfley J, Said M, Luft JR, Phizicky EM, DeTitta GT, et al. Blocking S-adenosylmethionine synthesis in yeast allows selenomethionine incorporation and multiwavelength anomalous dispersion phasing. Proc Natl Acad Sci U S A. 2007;104:6678–6683. doi: 10.1073/pnas.0610337104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yokoyama S. Protein expression systems for structural genomics and proteomics. Curr Opin Chem Biol. 2003;7:39–43. doi: 10.1016/s1367-5931(02)00019-4. [DOI] [PubMed] [Google Scholar]
- 40.Vinarov DALB, Peterson FC, Tyler EM, Volkman BF, Markley JL. Cell-free protein production and labeling protocol for NMR-based structural proteomics. Nat Methods. 2004;1 (2):149–153. doi: 10.1038/nmeth716. [DOI] [PubMed] [Google Scholar]
- 41.Vinarov DANC, Tyler EM, Markley JL, Shahan MN. Wheat germ cell-free expression system for protein production. Curr Protoc Protein Sci. 2006 doi: 10.1002/0471140864.ps0518s44. [DOI] [PubMed] [Google Scholar]
- 42.Wernimont AEA. In situ proteolysis to generate crystals for structure determination: an update. PLoS ONE. 2009;4:e5094. doi: 10.1371/journal.pone.0005094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43**.Dong A, Xu X, Edwards AM, Chang C, Chruszcz M, Cuff M, Cymborowski M, Leo RD, Egorova O, Evdokimova E, et al. In situ proteolysis for protein crystallization and structure determination. Nat Methods. 2007 doi: 10.1038/nmeth1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44*.Cooper DRBT, Grelewska K, Pinkowska M, Sikorska M, Zawadzki MZD. Protein crystallization by surface entropy reduction: optimization of the SER strategy. Acta Crystallogr D Biol Crystallogr. 2007;63(Pt 5):636–645. doi: 10.1107/S0907444907010931. [DOI] [PubMed] [Google Scholar]
- 45*.Goldschmidt LCD, Derewenda ZS, Eisenberg D. Toward rational protein crystallization: A Web server for the design of crystallizable protein variants. Protein Sci. 2007;16(8):1569–1576. doi: 10.1110/ps.072914007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rayment I. Reductive alkylation of lysine residues to alter crystallization properties of proteins. Methods Enzymol. 1997;276:171–179. [PubMed] [Google Scholar]
- 47*.Kim Y, Quartey P, Li H, Volkart L, Hatzos C, Chang C, Nocek B, Cuff M, Osipiuk J, Tan K, et al. Large-scale evaluation of protein reductive methylation for improving protein crystallization. Nat Methods. 2008;5:853–854. doi: 10.1038/nmeth1008-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48*.Sauder MJRM, Bain K, Rooney I, Gheyi T, Atwell S, Thompson DA, Emtage SBS. High throughput protein production and crystallization at NYSGXRC. Methods Mol Biol. 2008;426:561–575. doi: 10.1007/978-1-60327-058-8_37. [DOI] [PubMed] [Google Scholar]
- 49.Kim YDI, Zhou M, Wu R, Lezondra L, Quartey P, Joachimiak G, Korolev O, Li H, Joachimiak A. Automation of protein purification for structural genomics. J Struct Funct Genomics. 2004;5:111–118. doi: 10.1023/B:JSFG.0000029206.07778.fc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stols LZM, Eschenfeldt WH, Millard CS, Abdullah J, Collart FR, Kim Y, Donnelly MI. New vectors for co-expression of proteins: structure of Bacillus subtilis ScoAB obtained by high-throughput protocols. Protein Expr Purif. 2007;53:396–403. doi: 10.1016/j.pep.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 51*.Bochkarev A, Tempel W. High throughput crystallography at SGC Toronto: an overview. Methods Mol Biol. 2008;426:515–521. doi: 10.1007/978-1-60327-058-8_34. [DOI] [PubMed] [Google Scholar]
- 52.Chayen NE, Saridakis E. Protein crystallization for genomics: towards high-throughput optimization techniques. Acta Crystallogr D Biol Crystallogr. 2002;58:921–927. doi: 10.1107/s0907444902005322. [DOI] [PubMed] [Google Scholar]
- 53*.Gerdts CJTV, Yadav MK, Dementieva I, Collart F, Joachimiak A, Stevens RCKP, Kossiakoff A, Ismagilov RF. Time-controlled microfluidic seeding in nL-volume droplets to separate nucleation and growth stages of protein crystallization. Angew Chem Int Ed Engl. 2006;45(48):8156–8160. doi: 10.1002/anie.200602946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54*.Zheng B, Gerdts CJ, Ismagilov RF. Using nanoliter plugs in microfluidics to facilitate and understand protein crystallization. Curr Opin Struct Biol. 2005;15:548–555. doi: 10.1016/j.sbi.2005.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55*.Terwilliger TCAP, Read RJ, McCoy AJ, Moriarty NW, Grosse-Kunstleve RW, Afonine PV, Zwart PH, Hung LW. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr D Biol Crystallogr. 2009;65:582–601. doi: 10.1107/S0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dauter Z. Current state and prospects of macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2006;62:1–11. doi: 10.1107/S0907444905034050. [DOI] [PubMed] [Google Scholar]
- 57.Hendrickson WA, Horton JR, LeMaster DM. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. Embo J. 1990;9:1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walsh MA, Dementieva I, Evans G, Sanishvili R, Joachimiak A. Taking MAD to the extreme: ultrafast protein structure determination. Acta Crystallogr D Biol Crystallogr. 1999;55:1168–1173. doi: 10.1107/s0907444999003698. [DOI] [PubMed] [Google Scholar]
- 59.Williams WA, Zhang RG, Zhou M, Joachimiak G, Gornicki P, Missiakas D, Joachimiak A. The membrane-associated lipoprotein-9 GmpC from Staphylococcus aureus binds the dipeptide GlyMet via side chain interactions. Biochemistry. 2004;43:16193–16202. doi: 10.1021/bi048877o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60*.Fischetti RFXS, Yoder DW, Becker M, Nagarajan V, Sanishvili R, Hilgart MC, Stepanov S, Makarov O, Smith JL. Mini-beam collimator enables microcrystallography experiments on standard beamlines. J Synchrotron Radiat. 2009;16:217–225. doi: 10.1107/S0909049508040612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cowan JA, Nave C. The optimum conditions to collect X-ray data from very small samples. J Synchrotron Radiat. 2008;15:458–462. doi: 10.1107/S0909049508014623. [DOI] [PubMed] [Google Scholar]
- 62.Stern EA, Yacoby Y, Seidler GT, Nagle KP, Prange MP, Sorini AP, Rehr JJ, Joachimiak A. Reducing radiation damage in macromolecular crystals at synchrotron sources. Acta Crystallogr D Biol Crystallogr. 2009;65:366–374. doi: 10.1107/S090744490900540X. [DOI] [PubMed] [Google Scholar]
- 63*.Otwinowski ZBD, Majewski W, Minor W. Multiparametric scaling of diffraction intensities. Acta Crystallogr A. 2003;59:228–234. doi: 10.1107/s0108767303005488. [DOI] [PubMed] [Google Scholar]
- 64.Terwilliger TCG-KR, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, Adams PD. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr. 2007;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65*.Badger J, Hendle J, Burley SK, Kissinger CR. Deposit3D: a tool for automating structure depositions to the Protein Data Bank. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2005;61:818–820. doi: 10.1107/S1744309105026126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Koth CMOS, Larson SM, Edwards AM. Use of limited proteolysis to identify protein domains suitable for structural analysis. Methods Enzymol. 2003;368:77–84. doi: 10.1016/S0076-6879(03)68005-5. [DOI] [PubMed] [Google Scholar]