Conspectus
Electron transfer (ET) reactions provide a nexus among chemistry, biochemistry, and physics. These reactions underpin the “power plants” and “power grids” of bioenergetics, and they challenge us to understand how evolution manipulates structure to control ET kinetics. Ball-and-stick models for the machinery of electron transfer, however, fail to capture the rich electronic and nuclear dynamics of ET molecules: these static representations disguise, for example, the range of thermally accessible molecular conformations. The influence of structural fluctuations on electron-transfer kinetics is amplified by the exponential decay of electron tunneling probabilities with distance, as well as the delicate interference among coupling pathways. Fluctuations in the surrounding medium can also switch transport between coherent and incoherent ET mechanisms—and may gate ET so that its kinetics is limited by conformational interconversion times, rather than by the intrinsic ET time scale. Moreover, preparation of a charge-polarized donor state, or of a donor state with linear or angular momentum, can have profound dynamical and kinetic consequences. In this Account, we establish a vocabulary to describe how the conformational ensemble and the prepared donor state influence ET kinetics in macromolecules. This framework is helping to unravel the richness of functional biological ET pathways, which have evolved in within fluctuating macromolecular structures.
The conceptual framework for describing nonadiabatic ET seems disarmingly simple: compute the ensemble averaged (mean-squared) donor–acceptor (DA) tunneling interaction, <HDA2>, and the Franck–Condon weighted density of states, ρFC, to describe the rate: (2π/ħ) <HDA2> ρFC. Modern descriptions of the thermally averaged electronic coupling and of the Franck–Condon factor establish a useful predictive framework in biology, chemistry, and nanoscience. Describing the influence of geometric and energetic fluctuations on ET allows us to address a rich array of mechanistic and kinetic puzzles. How strongly is a protein’s fold imprinted on the ET kinetics, and might thermal fluctuations “wash out” signatures of structure? What is the influence of thermal fluctuations on ET kinetics beyond averaging of the tunneling barrier structure? Do electronic coupling mechanisms change as donor and acceptor reposition in a protein, and what are the consequences for the ET kinetics? Do fluctuations access minority species that dominate tunneling? Can energy exchanges between the electron and bridge vibrations generate vibronic signatures that label some of the D-to-A pathway traversed by the electron, thus eliminating unmarked pathways that would otherwise contribute to the DA coupling (as in other “which way” or double-slit experiments)? Might medium fluctuations drive tunneling–hopping mechanistic transitions? How does the donor-state preparation—in particular its polarization toward the acceptor and its momentum characteristics (which may introduce complex rather than pure real relationships among donor orbital amplitudes)—influence the electronic dynamics?
In this Account, we describe our recent studies that address puzzling questions of how conformational distributions, excited-state polarization, and electronic dynamical effects influence ET in macromolecules. Indeed, conformational and dynamical effects arise in all transport regimes, including the tunneling, resonant transport, and hopping regimes. Importantly, these effects can induce switching among ET mechanisms.
Keywords: electron transfer, tunneling pathways, vibronic interactions, fluctuating barriers, disorder, inelastic tunneling, bioenergetics
Time-scales in biological ET
The golden rule, Born-Oppenheimer, Franck-Condon (FC) formulation of ET (with rate constant ) is appropriate when the ET mechanism is coherent electron tunneling from D to A, when the FC times (the amount of time D and A electronic states remain resonant) are shorter than ET coupling fluctuation times,1–13 when medium relaxation of the product state is fast compared to the ET rate14 and when conformational exchange among the “dominant” conformations is rapid on the ET timescale (i.e., the process is not gated).15 Although we have found that fast bond angle fluctuations (tens of fsec) and slower torsion angle fluctuations of the tunneling bridge have a significant effect on the ET coupling,13 their contribution to the ET rate for most biological ET reactions is well described by the quantity because such fluctuations are faster than most long-distance ET rate times. Other slow events on the time scale of ET may lead to kinetic gating.16
Are these regimes satisfied in all biological ET systems? In the case of Ru-azurins,13 decay times of the DA coupling autocorrelation function are tens of fsec, an order of magnitude larger than the time scale for nuclear motion out of the crossing region of the DA potential energy surfaces ,17 where λ is the reorganization energy, a few fsec for azurin). Thus, HDA does not change during nuclear wave packet motion through the crossing point of the DA potential energy surfaces, and the ET rate is proportional to ,12,13,18,19 where the averaging is over the ensemble of protein structures.
The shortest decay time of in azurin was found to arise largely from valence angle fluctuations.13 Since these valence angle vibrations occur on similar time scales in proteins, the Condon approximation is likely to be valid more broadly for protein ET. However, the validity of the Condon approximation for DNA ET remains an open question.
The persistence of structural memory in biological electron transfer
The tunneling pathway model of biological ET estimates the DA interaction based on the decay through a sequence of covalent and noncovalent contacts between donor and acceptor. The decay parameters for the contacts along the pathway are based on simple rules derived from the physical principle that bond-mediated interactions are generally larger than through-space interactions.20 Although simple, this model successfully predicts the dependence of rate on medium structure in dozens of biological and semi-biological systems,8,21 and provides a quick survey of electronic communication for newly reported protein X-ray and NMR structures.
Going beyond the pathway description of electron tunneling mediation, in order to incorporate more specific chemical and multi-path effects, especially quantum interferences among paths, necessitates the inclusion of multiple coupling routes and ensemble averaging.22 Inclusion of quantum interference effects in HDA is usually accomplished using effective Hamiltonian and Green’s function methods,23,24 the generalized Mulliken-Hush scheme,25,26 and the tunneling current approach.27.
The mean-squared coupling in kET is the sum of the average coupling squared and the squared coupling variance:
| (1) |
where σ2 = 〈(HDA − 〈HDA〉)2〉. To what extent is the mean-squared coupling dominated by the average coupling compared to fluctuations away from the average? Balabin and Onuchic first posed this question in the context of photosynthetic reactions and examined the relative values of σ vs <HDA>.28 through the “coherence parameter” C = 〈HDA〉2/〈HDA2〉 = [1 + (σ2 /〈HDA〉2 )]−1 (not to be confused with in Figure 1). When 〈HDA〉2 >> σ2, the average medium structure defines the coupling. However, when 〈HDA〉2 << σ2, protein geometries that differ from the average protein structure provide coupling interactions that are much larger than the interaction provided by the average structure.
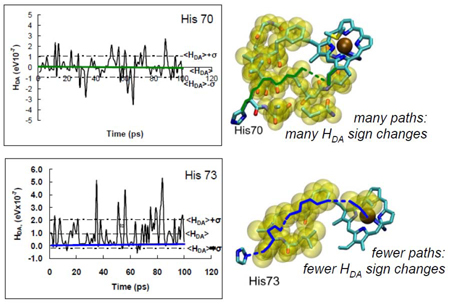
Figure 1.
(left) The normalized electronic coupling autocorrelation function in Ru-His 83 modified azurin decays on the time scale of tens of fsec,13 one order of magnitude longer than the Franck-Condon decay time.17 During each DA curve crossing event, the DA interaction is essentially unchanged. These fluctuations, rapid on the time scale of the ET rate, enter the rate through the mean squared coupling, .
Assessment of mean vs. fluctuation-dominated couplings has provided new insights to the field of biological ET in the last 5 years. For example, fluctuation-dominated ET was suggested as a possible means to control charge flow and to introduce novel temperature dependencies to the kinetics.28 We find that ET reactions to edges of hemes or chlorophylls favor a multi-pathway mechanism (σ2 dominated), while axial coupling to the coordinated metals in these rings favors a dominant-pathways (<HDA>2) mechanism.11 The edge-coupled multi-pathway mechanism helps to rationalize the near exponential decay as a function of distance for photosynthetic reaction center ET,29,30 while pathway-specific couplings dominate for systems with axial pathways, including some semi-biological Ru-protein systems.11 The emergence of average medium and few pathway behavior deduced here remains a fascinating topic for research. In the tunneling-limited Ru-modified protein systems of Gray and Winkler,8 all derivatives with anomalously slow rates for their DA distance are in the larger C few pathway (〈HDA〉2 >> σ2) regime and are axially coupled to the heme donor. “Fast” and “average” rate derivatives (for their distance) fall in the multi-path regime (〈HDA〉2 << σ2) with effective interactions determined by multiple alternative pathway structures, rather than by a few dominant paths.11 The conspectus figure shows examples of a σ2 -dominated regime (top, edge coupled to heme) and a 〈HDA〉2 -dominated regime (bottom, axial ligand coupled to heme).
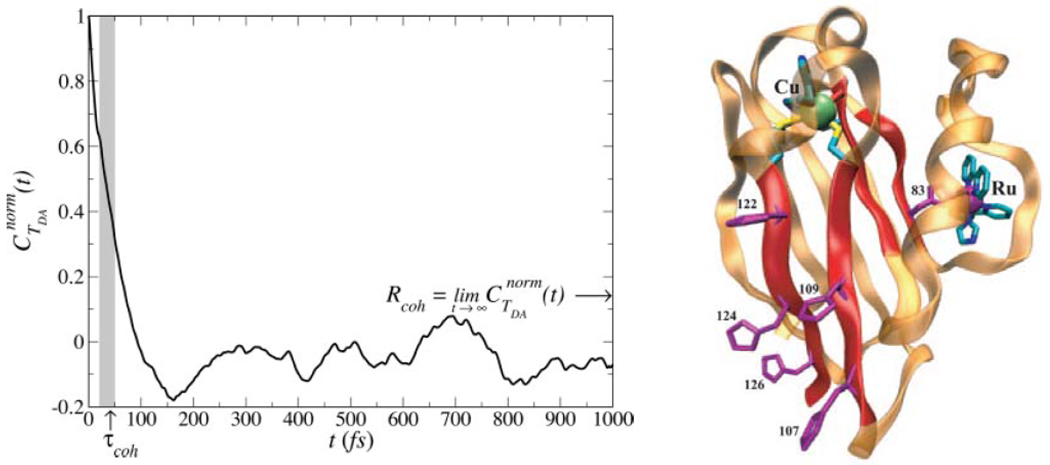
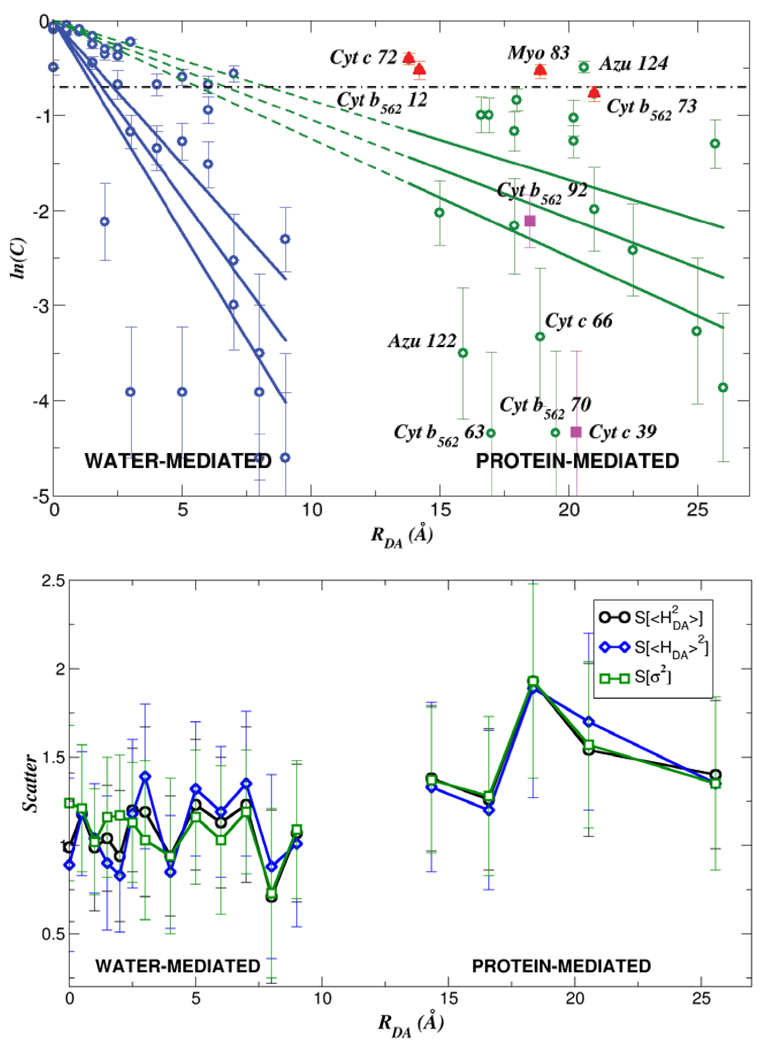
Comparing <HDA>2 to σ2 values in proteins has led us to pose a statistical question regarding coupling: is there a distance that characterizes, on average, a transition between 〈HDA〉2 and σ2 dominated kinetics?31 An answer emerges from coupling analysis for classical molecular dynamics sampled geometries of Ru-modified proteins,8 as well as for water-mediated cytochrome b5 self-exchange.32 We find a transition between the 〈HDA〉2 regime and the σ2 regime for protein ET at distances of ~ 6–7 Å, about the size of an amino acid residue (see Figure 2). This analysis assumes that the coherence parameter C is anchored to unity at donor-acceptor contact (ln C = 0 in Figure 2 at RDA=0). It will be interesting to explore trends in C at distance under 6–7 Å, when tunneling mediation accesses fewer coupling pathways, and also to examine the case of DNA ET. For water-mediated self-exchange, the transition to a fluctuation-dominated coupling regime occurs at distances about the size of a water molecule, ~ 2–3 Å (Figure 2).31
Figure 2.
(top) Dependence of ln(C) on the DA distance RDA where C = 〈HDA〉2 /〈HDA2〉 = [1 + (σ2 /〈HDA〉2)]−1. The dot-dash line denotes the value of C where 〈HDA〉2 = σ2 (bottom) (“Scatter”) vs distance. V of the scale of unity indicates that the specific protein structure largely determines the observable rate.31
Do the medium fluctuations described above wash out the influence of the protein fold on ET kinetics within the database of protein and water-mediated tunneling examined here? If this were the case, in the σ dominated regime, all proteins could be described as creating an effective structureless tunneling barrier. To address this central question, which has deep links to the role of evolution in determining coupling pathway structure,9,33,34 we computed the metric 31 for multiple ET protein sets characterized by the same transfer distance RDA (using the Gray-Winkler Ru-proteins8 as a test set). The variance (var) and the average are taken over all protein ET species at the same transfer distance. We found that V was of order unity for all transfer distances (Figure 2, bottom), indicating that the influence of structural differences between proteins on mean squared ET couplings is large. Even at large RDA, where σ2 dominates, the diversity of values at any given distance is not eliminated by structural fluctuations.31 Thus, the protein structure defines the tunneling rates (for the dataset of Ru-proteins examined here) even when fluctuations dominate the ensemble of coupling values. Recent theoretical studies of Ru-proteins11 and of cytochrome bo321, indicate that the structure of the underlying coupling medium indeed controls the kinetics. Dynamic docking studies of protein-protein ET provide further evidence of the structure sensitivity of ET reactions; these studies find that a very small fraction of the ensemble of docked protein structures is ET active.35
Water-mediated electron tunneling
Water-mediated ET,36 as noted above, is fluctuation dominated at distances beyond 2–3 Å.31 Our studies of electron self-exchange in cytochrome b5 indicate a “coupling plateau” in the distance dependence of water-mediated tunneling.32 In this plateau regime, structured water bridges between proteins provide a population of strong tunneling routes that include a few water molecules between cofactors (Figure 3). In this regime, structured water bridges establish coupling pathways across otherwise unfavorable van der Waals gaps, enhancing the DA coupling and establishing the plateau.
Figure 3.
Mean-squared donor-acceptor interaction as a function of distance for self-exchange in cytochrome b5 with protein and water (P,W) produces a coupling plateau compared to the case where only the protein (P) is included.32
The model calculations described above provide a framework for understanding an otherwise puzzling array of biological ET studies, including tunneling across water interfaces in the hydroxylating domain of peptidylglycine α-amidating monooxygenase.37 Water-mediated tunneling effects have been suggested recently in the Mo-heme ET kinetics of mutant sulfite dehydrogenase38 and in conducting polymers,39 as well as in small C-clamp style molecules that were designed to bind waters.40 Questions of water-mediated tunneling mechanism link to foundational experiments in ET theory (e.g., Fe(II)/Fe(III) self-exchange) that continue to provide test beds for new descriptions of tunneling interactions.41–43
Donor state polarization and ET kinetics
In biological ET, a growing body of data indicates that the polarization of the donor state may tune the rate of photoexcited ET. In addition to inducing donor-bridge mixing by the excitation pulse (in the small tunneling energy gap regime44), excited-state polarization itself may influence ET kinetics. For example, the excited state of the primary donor in the bacterial photosynthetic reaction center is believed to be strongly polarized in the direction of charge separation.45 Other ET reactions in photobiology appear to be accelerated by excited states that are polarized toward the acceptor. Our recent theoretical studies46 of the DNA repair protein photolyase47 have revealed that photoexcited ET between a reduced flavin adenine dinucleotide (FADH−) and the protein bound thymine dimer lesion is strongly accelerated by the FADH− excited-state polarization in the direction of the dimer lesion. In contrast to earlier studies, we found that the π−π* flavin excited state charge denisty is polarized strongly toward the acceptor (the “proximal” direction) and that tunneling is dominantly mediated by a methyl group (Figure 4).46,48 Because of the excited-state’s polarization, the nearby adenine, a natural candidate for superexchange mediation due to its unsaturation,49 appears to play a largely structural role (i.e., a docking site for the thymine dimer), rather than to provide tunneling-mediation pathways.46,48 The excited state D polarization helps to explain the fast (sub-nanosecond) ET from the photolyase excited state to the dimer.
Figure 4.
Electronic energy level diagram for photolyase. The S1 state is predicted to be polarized strongly in the direction of the thymine dimer.46
Studies of Skourtis and Nitzan further highlight the potential role of excited-state preparation on ET kinetics.44 In DNA ET, where the energy gap between D/A and bridge states is small, the initially prepared excited state may contain a considerable admixture of bridge amplitude. Similarly, in molecular conductance experiments, the width of the Fermi-Dirac distribution may populate bridge states, even though the center of the distribution lies in the bridge electronic energy gap gap. ET in nucleic acids is explored further below.
Pathway Phase, Chirality, and Electron Transfer
Donor-acceptor interactions are generally computed in the energy domain as the splitting of two quasi-degenerate states.24 Two interfering coupling pathways with pure real amplitudes of magnitude |A| and |B| produce a squared DA interaction proportional to (|A|±|B|)2, where the + sign enters if the paths interfere constructively and – sign enters if the path interfere destructively. If the first pathway instead has the complex amplitude A eiϕ, where A and ϕ are pure real numbers, the net interaction is proportional to A2+B2+2AB cos(ϕ). The phase, ϕ, changes the D-bridge interaction, but a simple sign reversal of ϕ has no effect on the magnitude of the net DA coupling. In this energy (splitting) framework, the net interaction expected for D and for its complex conjugate D* is thus identical. As we will see, this energy domain analysis does not indicate that the ET dynamics of D and D* may in fact be very different in ways that may determine ET rates and yields. A more complete analysis of the electronic dynamics will be required.
Indeed, Waldeck, Naaman, and coworkers recently showed that experimental ET yields are different for DBA systems with different bridge (B) handedness when the donor state is prepared with circularly polarized light (CPL).50,51 This result is not explained within the two-state energy-domain analysis above: the energy eigenstate spectra for mirror image systems are essentially identical, and reversing the phase relationship of the D-B interactions (equivalent to reversing the relative phase of the two pathways described in the paragraph above) does not change the magnitude of the coupling. Something is missing from the stationary-state (energy domain) perspective on ET.
The time-dependent transition amplitude from an initial complex (momentum carrying) state D or D* to a real final state A in a system with Hamiltonian H is:
| (2) |
where |Ψm〉 (Em) are the eigenstates (energy eigenvalues) of the Hamiltonian. Writing 〈A|Ψm〉〈Ψm|D〉 = |Rm|exp[iΘm] and 〈A|Ψm〉〈Ψm|D*〉 = |Rm|exp[−iΘm], one finds that the probability of being on the acceptor as a function of time is:
| (3) |
Importantly, switching the donor state to D* (accomplished by reversing the polarization of the CPL) introduces phase shifts in the time evolution. Reversing the helicity of a bridge that interacts with a ring-like donor state has an equivalent effect. Even in achiral systems, bridges that interact with the complex phase donor at more than one site will display different time evolutions for the D to A vs the D* to A transitions.52,53
Introducing D and A state lifetimes in eq 3 (ħ/γD, ħ/γA) produces phase dependent ET yields, , and a yield asymmetry:
| (4) |
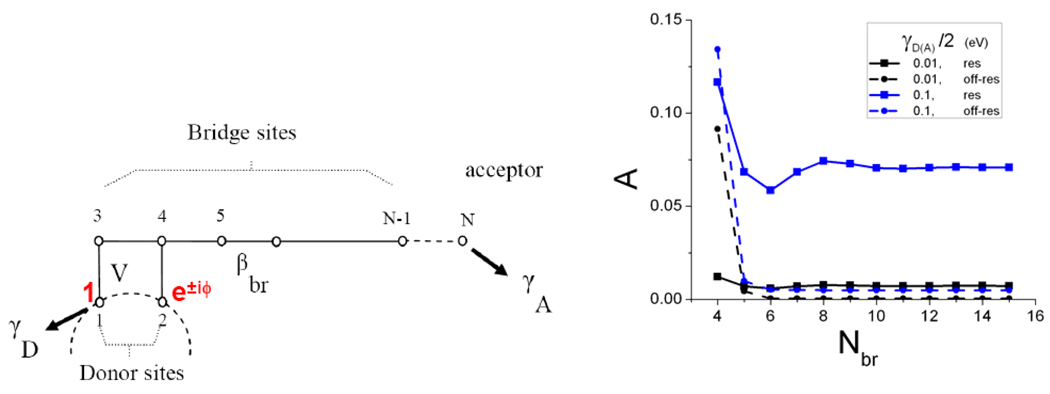
This predicted asymmetry was validated in tight-binding calculations for the model of Figure 5,52 which produces yield asymmetries consistent with the experimental results.50,51 Figure 5 shows that the yield asymmetry grows with γD (zero when γD = 0) because donor relaxation “locks in” the difference in the propagation from the prepared states of opposite phase.
Figure 5.
(left) A model for ET from angular momenta states D or D*. (right) Plot of the yield asymmetry for D (blue) vs. D* (black) as a function of the number of bridging sites.52
It is remarkable that changes in the initial state orbital angular momentum or bridge handedness introduce detectable differences in ET yields. Introducing donor state angular momentum necessitates introducing complex amplitudes, and bridges that interact with the complex donor state at more than one site produce pathway interferences that reflect the phase relationship among donor sites. It will be fascinating to expand this framework for describing “momentum transfer” to the regime of molecular-junctions, as well as to unimolecular ET kinetics.
A molecular double-slit experiment
In the multi-pathway ET systems discussed above, the electron propagates coherently from D to A. That is, HDA is computed by adding the amplitudes arising from all bridging pathways. The coherent donor-acceptor interaction mediated by a bridge can be written as a sum over products of amplitudes propagating among chains of orbitals with interaction Vnm. This sum is known as a Dyson expansion and carries the McConnell model5,24,54 for DA interactions to infinite order. The lowest-order contribution to the coupling along pathway n from D to A is:
| (5) |
and the total coupling (summed coherently over all pathways) is:
| (6) |
where Et is the tunneling energy of the electron and Ei is the energy of orbital i along the pathway, and there is no energy exchange between the electron and bridge vibrations.
Imagine that, when the electron occupies orbital j′ in one of the tunneling pathways contributing to the sum, it interacts with a localized vibrational mode (e.g., a carbonyl vibration) enabling inelastic tunneling. If the electron tunneling event changes the quantum vibrational state of the mode localized on orbital j′ , that vibrational excitation is evidence that the electron visited orbital j′ while tunneling. Thus, only pathways in the summation of eq 6 that include orbital j′ (where energy is exchanged between the electron and nuclear vibrations) enter the DA coupling (eq 7), in contrast to the elastic case where all pathways enter the summation.55–57
| (7) |
Since pathways may interfere constructively or destructively, inelastic events can result in either increasing or decreasing HDA. The inelastic effect described here arises for the same reason that detecting the electron’s path in a double-slit experiment destroys interference between alternative paths.10 In ET molecules, this inelastic effect may be particularly dramatic if the elastic coupling pathways interfere destructively (as in orbital symmetry forbidden ET). As such, turning on an inelastic transport mechanism may switch on an otherwise forbidden kinetic process. This effect is different from other effects of inelastic interactions on the tunneling matrix element and on the nuclear Franck-Condon factor, which have been studied.18,58
The inelastic tunneling pathway effect described here is only predicted when a fully quantum treatment is made of bridge-mediated inelastic tunneling (i.e., when coupling pathways are defined as sequences of vibronic states, such as products of electronic orbitals and bridge vibrational states).55–57
For the model in Figure 6, to lowest order in the electronic coupling interactions tij, the elastic coupling is (ΔE is the D/A to bridge orbital electronic energy gap), and the inelastic coupling is (where γ is the electron-vibrational coupling on the bridge). The inelastic path coupling thus has only one bridge pathway contribution, while the elastic coupling has two parallel path contributions. The inelastic path is expected to be weaker by about one order of magnitude compared to the elastic path in many systems.55 As such, inelastic effects will be most pronounced in systems with symmetry forbidden elastic ET kinetics that will be allowed in the inelastic regime.
Figure 6.
A model DBA system with vibronic coupling to the upper bridge orbital. D, U, L, and A denote donor, bridge and acceptor electronic states, respectively. The spring represents a bridge localized vibration that is perturbed when the electron visits orbital U. 55–57
A detailed quantum analysis of intramolecular tunneling in more realistic model molecular systems confirms the argument that inelastic tunneling via localized vibrational modes will allow the selection of coupling pathways.57 Indeed, we have proposed a scheme for driving local normal modes with IR radiation, thus promoting inelastic tunneling57 and experimental tests of this conjecture are in progress. Pathway manipulation via inelastic ET provides an example where bridge vibrational dynamics enables control of the bridge-mediated donor-acceptor electronic communication and, hence, of ET kinetics. The goal of designing, synthesizing, and experimentally probing unimolecular inelastic tunneling systems (with spatially localized vibrational modes) represents a grand challenge.3,27,59–61
ET in nucleic acids and the effects of thermal fluctuations
Conformational fluctuations play a central role in nucleic acid (NA) ET. The accessibility of many viable transport mechanisms, including superexchange, incoherent multi-step hopping, and polaronic transport, rely upon medium fluctuations (see ref.62 and references therein). The reported distance dependences of ET in NAs, from nearly distance-independent to exponentially decaying, are linked both to the average energetics of the donor, bridge, and acceptor states, and to fluctuations in their energies and couplings.62
In the deep-tunneling regime, fluctuations cannot provide access to hopping or other transport mechanisms, despite fluctuations in base energetics and inter-base couplings.63 In this tunneling regime, the thermally averaged ET rates decay exponentially with distance. Despite a significant spread due of base energies and interactions due to fluctuations, a model based on the average energetics describes the ET very well (Figure 7d).63
Figure 7.
(a) G localized hole states of the terminal Gs in the CATG DNA segment.64 (b) Hole states shared between G and A. (c) Histogram of fluctuating HOMO and HOMO-1 energies. Note the significant overlap of the G and A orbital energies. (d) The conductance (σ) computed for individual snapshots of the single strand PNA segment TTTXTTT (X=A,C,T,G) shows scatter over orders of magnitude because of fluctuation in both base energies and inter-base couplings.63
The influence of fluctuations on ET kinetics and mechanism is very different as the DA-bridge barrier height shrinks, as in the case of G-G hole transport across NAs.64 For a G base bound hole, the energy gap to the nearest bridge hole states (A) is on the scale of tenths of eV for the bases in their average geometries. As a consequence, most ET models assume a tunneling mechanisms for short distance transfer and a hopping mechanism over larger distances (typically, above 3–4 base pairs). Indeed, tunneling-hopping transitions are observed in conjugated polymers65 as a function of length, and the crossover presumably arises when the propensity for carrier injection into bridge states exceeds the tunneling probability. Our recent simulations, however, indicate that the two mechanisms may not be so clearly separated in NA ET.64 ET theory predicts electronic energy fluctuations on the scale of where λ is the reorganization energy. For NAs, λ ~ 1eV so δE ~ 0.1–0.2eV at room temperature, which is on the same scale as the difference in the oxidation potential difference for G and A bases.64 Indeed, a 0.1–0.2 eV thermal spread in frontier orbital energies was computed in our recent analysis of the electronic structure fluctuations for short DNA and PNA (peptide nucleic acid) segments (Figure 7c,d).64 The principal fluctuations leading to the energy spread appear to arise from covalent bond length changes of the individual bases, rather than from fluctuations in NA helicoidal parameters.
Thermal fluctuations thus bring the G and A bases into frequent degeneracy (see Figure 7), despite the fact that their redox potentials differ by ten times thermal energies or more. The resulting hole delocalization between the G and A (Figure 7a,b) indicates a possibility of hopping transport even at short G-to-G distances. In our analysis of the CATG DNA/PNA sequence, we found about 10% of the structural snapshots displayed significant hole delocalization away from the Gs.64 These studies suggest that fluctuations may limit the viability of pure superexchange or pure hopping models for hole transfer in NAs, even at short distances. Previous theoretical studies (see refs.66,67 and references therein) have shown that, in sufficiently low gap ET systems, the tunneling matrix element and the two-state golden rule description of ET are not appropriate for describing the observed kinetics because of degeneracy, or near degeneracy, with bridge states. It will be interesting to reexamine kinetic data associated with DNA and PNA hole transport in light of such mixed mechanisms.
Conclusions and Prospects
In proteins, the motif and ET distance determine whether fluctuations or average structure controls the coupling mechanism. Water structure and fluctuations add additional surprises – organized water between proteins near contact appears to provide enhanced tunneling propagation despite the large bridge orbital energy gap for water. Excited-state preparation also influences ET kinetics and dynamics. Donor states with different angular momenta propagate differently, as a consequence of the complex valued donor state amplitudes, giving rise to new ET effects that are not predicted by analyzing energy splittings. Further, polarized excited donor states were shown to select specific coupling pathways in proteins and to have large effects on ET rates, especially for large donors. This principle is particularly important in the protein photolyase, where polarization of the donor appears to accelerate DNA repair, likely enhancing the protein’s function.
DNA and PNA provide examples where fluctuations of bridge energetics influence ET coupling interactions as well as the transport mechanism. Since energy fluctuations are on the same scale as the differences among the redox potentials of the bases, fluctuations will likely mix superexchange, hopping and polaronic transport regimes.
Finally, a perspective that includes fluctuating coupling pathways may provide schemes to control ET. Indeed, leaving behind vibrational “breadcrumbs” that label tunneling pathways should change the donor-acceptor interaction and the ET kinetics. It seems likely that schemes being developed to control coherence and entanglement in meso and nanoscale quantum structures will find intriguing mappings in the world of molecular ET.
The authors thank the National Institutes of Health, the National Science Foundation, the Department of Energy and the University of Cyprus for support of our research programs. We are grateful to our students and postdocs – Horacio Carias, Liz Hatcher, Tsutomu Kawatsu, Igor Kurnikov, Jianping Lin, and Tatiana Prytkova – and to our collaborators – Catalina Achim, Harry Gray, Brian Hoffman, Fred Lewis, Marcela Madrid, Ron Naaman, Abe Nitzan, José Onuchic, Igor Rubtsov, Jon Sessler, Dave Waldeck, and Jay Winkler – for their enthusiastic interactions on these projects.
Biographies
David Beratan is the R.J. Reynolds Professor of Chemistry and Professor of Biochemistry at Duke.
Spiros S. Skourtis is Associate Professor and Chair in the Department of Physics, University of Cyprus.
Ilya A. Balabin studied at the Moscow State University and at the University of California, San Diego. He is a Postdoctoral Associate at Duke.
Alexander Balaeff studied at Moscow Institute of Physics and Technology and at the University of Illinois at Urbana-Champaign. He is a Postdoctoral Associate at Duke.
Shahar Keinan studied at the Hebrew University of Jerusalem. She is a Postdoctoral Associate at Duke.
Ravindra Venkatramani studied at Pune University, India and the University of Rochester. He is a Postdoctoral Associate at Duke.
Dequan Xiao studied at Sichuan University, the University of Central Florida, and Duke University; he is currently a post-doctoral associate at Yale University.
References Cited
- 1.Bendall DS, editor. Protein Electron Transfer. Oxford: BIOS Scientific Publishers, Inc.; 1996. [Google Scholar]
- 2.Skourtis SS, Lin J, Beratan DN. In: Modern Methods for theoretical Physical Chemistry of Biopolymers. Starikov EB, Lewis JP, Tanaka S, editors. Boston: Elsevier; 2006. [Google Scholar]
- 3.Nitzan A. Electron transmission through molecules and molecular interfaces. Annu. Rev. Phys. Chem. 2001;52:681–750. doi: 10.1146/annurev.physchem.52.1.681. [DOI] [PubMed] [Google Scholar]
- 4.Bixon M, Jortner J, editors. Adv. Chem. Phys. 1999. pp. 106–107. Vol. [Google Scholar]
- 5.Newton MD. In: Electron Transfer in Chemistry. Balzani V, editor. Vol. 1. New York: Wiley-VCH; 2001. pp. 3–63. [Google Scholar]
- 6.Barbara PF, Meyer TJ, Ratner MA. Contemporary issues in electron transfer research. J. Phys. Chem. 1996;100:13148–13168. [Google Scholar]
- 7.Adams DM, Brus L, Chidsey CED, Creager S, Creutz C, Kagan CR, Kamat PV, Lieberman M, Lindsay S, Marcus RA, Metzger RM, Michel-Beyerle ME, Miller JR, Newton MD, Rolison DR, Sankey O, Schanze KS, Yardley J, Zhu XY. Charge transfer on the nanoscale: Current status. J. Phys. Chem. B. 2003;107:6668–6697. [Google Scholar]
- 8.Gray HB, Winkler JR. Electron tunneling through proteins. Q. Rev. of Biophys. 2003;36:341–372. doi: 10.1017/s0033583503003913. [DOI] [PubMed] [Google Scholar]
- 9.Edwards PP, Gray HB, Lodge MTJ, Williams RJP. Electron transfer and electronic conduction through an intervening medium. Angew. Chemie - Int. Ed. 2008;47:6758–6765. doi: 10.1002/anie.200703177. [DOI] [PubMed] [Google Scholar]
- 10.Imry Y. Introduction to Mesoscopic Physics. 2 ed. Oxford: Oxford University Press; 2002. [Google Scholar]
- 11.Prytkova TR, Kurnikov IV, Beratan DN. Coupling coherence distinguishes structure sensitivity in protein electron transfer. Science. 2007;315:622–625. doi: 10.1126/science.1134862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Troisi A, Ratner MA, Zimmt MB. Dynamic nature of the intramolecular electronic coupling mediated by a solvent molecule: A computational study. J. Am. Chem. Soc. 2004;126:2215–2224. doi: 10.1021/ja038905a. [DOI] [PubMed] [Google Scholar]
- 13.Skourtis SS, Balabin IA, Kawatsu T, Beratan DN. Protein dynamics and electron transfer: Electronic decoherence and non-Condon effects. Proc. Nat. Acad. Sci. U.S.A. 2005;102:3552–3557. doi: 10.1073/pnas.0409047102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nitzan A. Chemical Dynamics in Condensed Phases. Oxford: Oxford University Press; 2006. [Google Scholar]
- 15.Hoffman BM, Ratner MA. Gated electron-transfer - when are observed rates controlled by conformational interconversion. J. Am. Chem. Soc. 1987;109:6237–6243. [Google Scholar]
- 16.Davidson VL. Protein control of true, gated, and coupled electron transfer reactions. Accounts of Chem. Res. 2008;41:730–738. doi: 10.1021/ar700252c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lockwood DM, Cheng YK, Rossky PJ. Electronic decoherence for electron transfer in blue copper proteins. Chem. Phys. Lett. 2001;345:159–165. [Google Scholar]
- 18.Daizadeh I, Medvedev ES, Stuchebrukhov AA. Effect of protein dynamics on biological electron transfer. Proc. Nat. Acad. Sci. U.S.A. 1997;94:3703–3708. doi: 10.1073/pnas.94.8.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishioka H, Kimura A, Yamato T, Kawatsu T, Kakitani T. Interference, fluctuation, alternation of electron tunneling in protein media. 2. Non-Condon theory for the energy gap dependence of electron transfer rate. J. Phys. Chem. B. 2005;109:15621–15635. doi: 10.1021/jp051606i. [DOI] [PubMed] [Google Scholar]
- 20.Beratan DN, Betts JN, Onuchic JN. Protein electron-transfer rates set by the bridging secondary and tertiary structure. Science. 1991;252:1285–1288. doi: 10.1126/science.1656523. [DOI] [PubMed] [Google Scholar]
- 21.Beratan DN, Balabin IA. Heme-copper oxidases use tunneling pathways. Proc. Nat. Acad. Sci. U.S.A. 2008;105:403–404. doi: 10.1073/pnas.0711343105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolfgang J, Risser SM, Priyadarshy S, Beratan DN. Secondary structure conformations and long range electronic interactions in oligopeptides. J. Phys. Chem. B. 1997;101:2986–2991. [Google Scholar]
- 23.Skourtis SS, Beratan DN, Onuchic JN. The 2-state reduction for electron and hole transfer in bridge-mediated electron-transfer reactions. Chem. Phys. 1993;176:501–520. [Google Scholar]
- 24.Skourtis SS, Beratan DN. Theories of structure-function relationships for bridge-mediated electron transfer reactions. Adv. Chem. Phys. 1999;106:377–452. [Google Scholar]
- 25.Cave RJ, Newton MD. Generalization of the Mulliken-Hush treatment for the calculation of electron transfer matrix elements. Chem. Phys. Lett. 1996;249:15–19. [Google Scholar]
- 26.Voityuk AA. Assessment of semiempirical methods for the computation of charge transfer in DNA pi-stacks. Chem. Phys. Lett. 2006;427:177–180. [Google Scholar]
- 27.Stuchebrukhov AA. Toward ab initio theory of long-distance electron tunneling in proteins: Tunneling currents approach. Adv. Chem. Phys. 2001;118:1–44. [Google Scholar]
- 28.Balabin IA, Onuchic JN. Dynamically controlled protein tunneling paths in photosynthetic reaction centers. Science. 2000;290:114–117. doi: 10.1126/science.290.5489.114. [DOI] [PubMed] [Google Scholar]
- 29.Page CC, Moser CC, Chen XX, Dutton PL. Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature. 1999;402:47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 30.Jones ML, Kurnikov IV, Beratan DN. The nature of tunneling pathway and average packing density models for protein-mediated electron transfer. J. Phys. Chem A. 2002;106:2002–2006. [Google Scholar]
- 31.Balabin IA, Beratan DN, Skourtis SS. Persistence of structure over fluctuations in biological electron-transfer reactions. Phys. Rev. Lett. 2008;101:158102. doi: 10.1103/PhysRevLett.101.158102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin JP, Balabin IA, Beratan DN. The nature of aqueous tunneling pathways between electron-transfer proteins. Science. 2005;310:1311–1313. doi: 10.1126/science.1118316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Onuchic JN, Kobayashi C, Miyashita O, Jennings P, Baldridge KK. Exploring biomolecular machines: energy landscape control of biological reactions. Philos. Trans. R. Soc. London, Ser. B. 2006:1439–1443. doi: 10.1098/rstb.2006.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Moser CC, Page CC, Dutton PL. Darwin at the molecular scale: selection and variance in electron tunnelling proteins including cytochrome coxidase. Philos. Trans. R. Soc. London, Ser. B. 2006:1295–1305. doi: 10.1098/rstb.2006.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang ZX, Kurnikov IV, Nocek JM, Mauk AG, Beratan DN, Hoffman BM. Dynamic docking and electron-transfer between cytochrome b(5) and a suite of myoglobin surface-charge mutants. Introduction of a functional-docking algorithm for protein-protein complexes. J. Am. Chem. Soc. 2004;126:2785–2798. doi: 10.1021/ja038163l. [DOI] [PubMed] [Google Scholar]
- 36.Balabin I, Skourtis SS, Beratan DN. In: Wiley Encyclopedia of Chemical Biology. Begley TP, editor. 2009. [Google Scholar]
- 37.Francisco WA, Wille G, Smith AJ, Merkler DJ, Klinman JP. Investigation of the pathway for inter-copper electron transfer in peptidyglycine alpha-amidating monooxygenase. J. Am. Chem. Soc. 2004;126:13168–13169. doi: 10.1021/ja046888z. [DOI] [PubMed] [Google Scholar]
- 38.Emesh S, Rapson TD, Rajapakshe A, Kappler U, Bernhardt PV, Tollin G, Enemark JH. Intramolecular electron transfer in sulfite-oxidizing enzymes: Elucidating the role of a conserved active site arginine. Biochemistry. 2009;48:2156–2163. doi: 10.1021/bi801553q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leary E, Hobenreich H, Higgins SJ, van Zalinge H, Haiss W, Nichols RJ, Finch CM, Grace I, Lambert CJ, McGrath R, Smerdon J. Single-molecule solvation-shell sensing. Phys. Rev. Lett. 2009;102:086801. doi: 10.1103/PhysRevLett.102.086801. [DOI] [PubMed] [Google Scholar]
- 40.Chakrabarti S, Parker MFL, Morgan CW, Schafmeister CE, Waldeck DH. Experimental evidence for water mediated electron transfer through bis-amino acid donor-bridge-acceptor oligomers. J. Am. Chem. Soc. 2009;131:2044–2045. doi: 10.1021/ja8079324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller NE, Wander MC, Cave RJ. A theoretical study of the electronic coupling element for electron transfer in water. J. Phys. Chem. A. 1999;103:1084–1093. [Google Scholar]
- 42.Cukier E, Daniels S, Vinson E, Cave RJ. Are hydrogen bonds unique among weak interactions in their ability to mediate electronic coupling? J. Phys. Chem. A. 2002;106:11240–11247. [Google Scholar]
- 43.Migliore A, Sit PHL, Klein ML. Evaluation of electronic coupling in transition-metal systems using DFT: Application to the hexa-aquo ferric-ferrous redox couple. J. Chem. Theory Comput. 2009;5:307–323. doi: 10.1021/ct800340v. [DOI] [PubMed] [Google Scholar]
- 44.Skourtis S, Nitzan A. Effects of initial state preparation on the distance dependence of electron transfer through molecular bridges and wires. J. Chem. Phys. 2003;119:6271–6276. [Google Scholar]
- 45.Blankenship RE. Molecular Mechanisms of Photosynthesis. Oxford: Blackwell Science; 2002. [Google Scholar]
- 46.Prytkova TR, Beratan DN, Skourtis SS. Photoselected electron transfer pathways in DNA photolyase. Proc. Nat. Acad. Sci. U.S.A. 2007;104:802–807. doi: 10.1073/pnas.0605319104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sancar A. Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Revs. 2003;103:2203–2237. doi: 10.1021/cr0204348. [DOI] [PubMed] [Google Scholar]
- 48.Skourtis SS, Prytkova T, Beratan DN. Flavin charge transfer transitions assist DNA photolyase electron transfer. AIP Conf. Proc. 2007;963:674–677. doi: 10.1063/1.2836174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Antony J, Medvedev DM, Stuchebrukhov AA. Theoretical study of electron transfer between the photolyase catalytic cofactor FADH(-) and DNA thymine dimer. J. Am. Chem. Soc. 2000;122:1057–1065. [Google Scholar]
- 50.Ray K, Ananthavel SP, Waldeck DH, Naaman R. Asymmetric scattering of polarized electrons by organized organic films of chiral molecules. Science. 1999;283:814–816. doi: 10.1126/science.283.5403.814. [DOI] [PubMed] [Google Scholar]
- 51.Wei JJ, Schafmeister C, Bird G, Paul A, Naaman R, Waldeck DH. Molecular chirality and charge transfer through self-assembled scaffold monolayers. J. Phys. Chem. B. 2006;110:1301–1308. doi: 10.1021/jp055145c. [DOI] [PubMed] [Google Scholar]
- 52.Skourtis SS, Beratan DN, Naaman R, Nitzan A, Waldeck DH. Chiral control of electron transmission through molecules. Phys. Rev. Lett. 2008;101:238103. doi: 10.1103/PhysRevLett.101.238103. [DOI] [PubMed] [Google Scholar]
- 53.Cave RJ. Inducing chirality with circularly polarized light. Science. 2009;323:1435–1436. doi: 10.1126/science.1169338. [DOI] [PubMed] [Google Scholar]
- 54.Ratner M, Jortner J, editors. Molecular Electronics. Oxford: Blackwell Science; 1997. [Google Scholar]
- 55.Skourtis SS, Waldeck DH, Beratan DN. Inelastic electron tunneling erases coupling-pathway interferences. J. Phys Chem. B. 2004;108:15511–15518. [Google Scholar]
- 56.Skourtis SS, Beratan DN. A molecular double slit paradigm. AIP Conf. Proc. 2007;963:809–812. doi: 10.1063/1.2836174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xiao D, Skourtis SS, Rubtsov IV, Beratan DN. Turning charge transfer on and off in a molecular interferometer with vibronic pathways. Nano Lett. 2009;9:1818–1823. doi: 10.1021/nl8037695. [DOI] [PubMed] [Google Scholar]
- 58.Medvedev ES, Stuchebrukhov AA. Inelastic tunneling in long-distance biological electron transfer reactions. J. Chem. Phys. 1997;107:3821–3831. [Google Scholar]
- 59.Goldsmith RH, Wasielewski MR, Ratner MA. Electron transfer in multiply bridged donor-acceptor molecules: Dephasing and quantum coherence. J. Phys. Chem. B. 2006;110:20258–20262. doi: 10.1021/jp0639187. [DOI] [PubMed] [Google Scholar]
- 60.Maddox JB, Harbola U, Liu N, Silien C, Ho W, Bazan GC, Mukamel S. Simulation of single molecule inelastic electron tunneling signals in paraphenylene-vinylene oligomers and distyrylbenzene[2.2]paracyclophanes. J. Phys. Chem. A. 2006;110:6329–6338. doi: 10.1021/jp061590b. [DOI] [PubMed] [Google Scholar]
- 61.Troisi A, Beebe JM, Picraux LB, van Zee RD, Stewart DR, Ratner MA, Kushmerick JG. Tracing electronic pathways in molecules by using inelastic tunneling spectroscopy. Proc. Nat. Acad. Sci. U.S.A. 2007;104:14255–14259. doi: 10.1073/pnas.0704208104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schuster GB, editor. Topics in Current Chemistry. Vol. 2004. pp. 236–237. and references therein. [Google Scholar]
- 63.Paul A, Bezer S, Venkatramani R, Kocsis L, Wierzbinski E, Balaeff A, Keinan S, Beratan DN, Achim C, Waldeck DH. Role of nucleobase energetics and nucleobase interactions in single stranded peptide nucleic acid charge transfer. J. Am. Chem. Soc. 2009;131:6498–6507. doi: 10.1021/ja9000163. [DOI] [PubMed] [Google Scholar]
- 64.Hatcher E, Balaeff A, Keinan S, Venkatramani R, Beratan DN. PNA versus DNA: Effects of structural fluctuations on electronic structure and hole-transport mechanisms. J. Am. Chem. Soc. 2008;130:11752–11761. doi: 10.1021/ja802541e. [DOI] [PubMed] [Google Scholar]
- 65.Davis WB, Svec WA, Ratner MA, Wasielewski MR. Molecular-wire behaviour in p-phenylenevinylene oligomers. Nature. 1998;396:60–63. [Google Scholar]
- 66.Skourtis SS. Electron transfer through time-dependent bridges: Tunneling by virtual transitions that break the Born-Oppenheimer approximation. Chem. Phys. Lett. 2003;372:224–231. [Google Scholar]
- 67.Teklos A, Skourtis SS. Electron transfer through time dependent bridges: Differences between Franck-Condon and Born-Oppenheimer breakdown. Chem. Phys. 2005;319:52–68. [Google Scholar]