Summary
The CATHGEN study reported associations of chromosome 3q13-21 genes (KALRN, MYLK, CDGAP, and GATA2) with early-onset coronary artery disease (CAD). This study attempted to independently validate those associations. Eleven single nucleotide polymorphisms (SNPs) were examined (rs10934490, rs16834817, rs6810298, rs9289231, rs12637456, rs1444768, rs1444754, rs4234218, rs2335052, rs3803, rs2713604) in patients (N=1,618) from the Intermountain Heart Collaborative Study (IHCS). Given the higher smoking prevalence in CATHGEN than IHCS (41% vs 11% in controls, 74% vs 29% in cases), smoking stratification and genotype-smoking interactions were evaluated. Suggestive association was found for GATA2 (rs2713604, p=0.057, OR=1.2). Among smokers, associations were found in CDGAP (rs10934490, p=0.019, OR=1.6) and KALRN (rs12637456, p=0.011, OR=2.0) and suggestive association in MYLK (rs16834871, p=0.051, OR=1.8, adjusting for gender). No SNP association was found among non-smokers, but smoking/SNP interactions were detected for CDGAP (rs10934491, p=0.017) and KALRN (rs12637456, p=0.010). Similar differences in SNP effects by smoking status were observed on re-analysis of CATHGEN. CAD associations were suggestive for GATA2 and among smokers significant post hoc associations were found in KALRN, MYLK, and CDGAP. Genetic risk conferred by some of these genes may be modified by smoking. Future CAD association studies of these and other genes should evaluate effect modification by smoking.
Keywords: coronary disease, genetic association, replication study, smoking
Introduction
Chromosome 3q13-21 has been linked to coronary artery disease (CAD) in the GENECARD study and the Diabetes Heart Study (Hauser et al. 2004; Bowden et al. 2006). Subsequent fine-mapping and association testing provided evidence of multiple genes within the broad linkage peak that are associated with early-onset CAD in the CATHGEN dataset, including the Kalirin-Rho GTPase pathway (KALRN, MYLK, and CDGAP) and the transcription factor GATA2 (Wang et al. 2007; Connelly et al. 2006).
The Rho GTPase signal-transduction pathway is involved in a variety of cell regulation functions, including cell proliferation, migration, and adhesion as well as the cellular inflammatory responses (Knaus 2000; Cernuda-Morollon & Ridley 2006). Rho GTPases may influence cardiac function (Ren & Fang 2005; Shimokawa & Takeshita 2005) and kalirin has been implicated in the inhibition of inducible nitric oxide synthase and thus may restrict the cardioprotective action of nitric oxide (Ratovitski et al. 1999; Cooke & Losordo 2002). GATA2 is a transcription factor that is expressed in hematopoietic stem cells and the cells composing the aortic vasculature and may be involved in vascular disease susceptibility (Connelly et al. 2006).
While further mechanistic insights will assist in understanding the potential effects of these genes on CAD, this study sought to validate the CATHGEN associations with CAD of KALRN, MYLK, CDGAP, and GATA2 in a large independent but similarly constructed dataset of patients undergoing coronary angiography in Salt Lake City, UT, and who were enrolled in the cardiac catheterization registry of the Intermountain Heart Collaborative Study (IHCS).
Materials and Methods
Population
Patients were selected from the IHCS cardiac catheterization registry (Taylor et al. 1998). This registry includes consenting patients undergoing coronary angiography at tertiary-care hospitals within the Intermountain Healthcare system who were enrolled and donated a DNA sample between April, 1997, and February, 2003. The study was approved by the Intermountain Healthcare Institutional Review Board.
Angiographically-determined CAD was defined by physician report from standard coronary angiography. Early-onset cases, or “young affected” (YA), were defined as patients with a Duke CAD index ≥32 and an age of onset ≤55 years for females or ≤50 years for males. Later-onset cases were labeled “old affected” (OA) and defined as patients with a Duke CAD index ≥74 with an age of onset >55 years for females or >50 years for males. The category of “all affected” (AA) included the joint set of YA and OA cases. All patients with a history of cardiac transplant were excluded. These definitions are consistent with previously published work (Wang et al. 2007; Connelly et al. 2006).
Controls were older angiographic normals with age at coronary angiography ≥61 years, Duke CAD index ≤23, no significant disease (i.e., no stenosis ≥75%) in any vessel, and no other history of cardiovascular disease. Controls were the comparison group for each of the three affected case definitions, as in previous work (Wang et al. 2007; Connelly et al. 2006). Because only 24 patients of African ancestry were available among the >5,000 samples genotyped, study evaluations were restricted to those of Caucasian ancestry.
Genotyping
Eleven previously-studied single nucleotide polymorphisms (SNPs) in CDGAP (rs10934490), MYLK (rs16834817), KALRN (rs6810298, rs9289231, rs12637456, rs1444768, rs1444754, rs4234218), and GATA2 (rs2335052, rs3803, rs2713604) selected as the strongest SNPs from fine-mapping studies (Wang et al. 2007; Connelly et al. 2006) were tested. Genotyping of all SNPs was performed using 5′ exonuclease (Taqman®) chemistry on the ABI Prism® 7000 (Applied Biosystems, Foster City, CA). All assays were validated by direct sequencing using Big Dye v 3.1 terminator chemistry (Applied Biosystems).
Study Covariables
Data reported by physicians and acquired from patients included patient age at catheterization, age of CAD onset, sex, body mass index (BMI), ethnicity, smoking, diabetes, hypertension, and hyperlipidemia. Age at catheterization was defined as the patient's age at the time of consent to participate in the registry. Age of onset was defined as participant age at the time of first CAD diagnosis for affected cases or age at the time of the first recorded cardiac catheterization procedure for controls.
Diabetes, hypertension, and hyperlipidemia were obtained from physician report and based on clinical and laboratory findings and/or current medication. Diabetes was defined as a fasting glucose ≥126 mg/dL; hypertension as systolic blood pressure ≥140 mmHg or diastolic blood pressure ≥90 mmHg; and hyperlipidemia as fasting total cholesterol ≥200 mg/dL or low-density lipoprotein cholesterol (LDL-C) ≥130 mg/dL. Smoking was defined as present for current smokers or those with a >10 pack-year smoking history. Duke CAD index was computed as previously described (Smith et al. 1991).
Statistical Considerations
Single SNP associations with CAD were evaluated using an a priori-defined additive genetic model (all odds ratios [OR] are quoted as per additional variant allele) among only those of Caucasian ancestry. YA was the primary case phenotype in the study, and OA and AA were secondary phenotypes studied for completeness of comparisons. Analysis utilized multivariable logistic regression with basic models controlling for sex and full multivariable models adjusting for sex, BMI, diabetes, hypertension, hyperlipidemia, and smoking. In analyses of AA, fully adjusted models also entered age at catheterization.
The primary difference between the IHCS and CATHGEN samples was the much lower prevalence of smoking in IHCS (11% vs. 41% in controls, 29% vs. 74% in cases). Thus, smoking-stratified analyses to evaluate potential effect modification were performed post hoc and a regression model was fit for each SNP using a SNP-smoking interaction analysis. Due to the small number of smokers in IHCS, smoking-stratified analyses were performed among the CATHGEN sample for validation purposes (Wang et al. 2007; Connelly et al. 2006).
Results
All baseline characteristics in YA, OA, and AA were significantly different from controls (Table 1), except for BMI among OA. Characteristics also differed between YA and OA for smoking (p=0.026), hypertension (p=0.003), BMI (p<0.001), and CAD Index (p<0.001), while ages differed by design.
Table 1.
Baseline patient demographics.
| Characteristic | Controls (n=773) |
Young Affected (n=552) |
Old Affected (n=293) |
All Affected (n=845) |
|---|---|---|---|---|
| Age-at-cath | 70±6 | 49±9* | 72±9† | 57±14* |
| Age-of-onset | 70±7 | 44±10* | 66±9* | 52±14* |
| Sex (male) | 42% | 75%* | 78%* | 76%* |
| BMI | 28.6±5.9 | 30.4±6.6* | 28.7±5.5 | 29.8±6.3* |
| Smoking | 11% | 29%* | 21%* | 26%* |
| Hypertension | 56% | 56% | 67%* | 60% |
| Hyperlipidemia | 37% | 68%* | 66%* | 67%* |
| Diabetes | 13% | 29%* | 28%* | 28%* |
| CAD index | 1.8±5.6 | 48.2±17.3* | 81.1±9.6* | 59.2±21.7* |
p<0.001 vs. control;
p<0.05 vs. control.
Raw minor allele frequencies for each SNP are provided by phenotype in Table 2, and Figure 1 illustrates the linkage disequilibrium pattern between the SNPs. Overall, suggestive association was found for GATA2 in rs2713604 (YA: OR=1.21 [all ORs are per additional variant allele], p=0.057; OA: OR=1.22, p=0.10; AA: OR=1.22, p=0.046), with the univariate YA result being modified by adjustment for BMI, diabetes, and hyperlipidemia which were all less prevalent among variant allele carriers. No other SNP association was significant in the YA (Table 3) or secondary phenotypes (Supplement 1).
Table 2.
Minor allele frequencies in each phenotype group.
| GENE | SNP name | Controls (n=773) | Young Affected (n=552) | Old Affected (n=293) | All Affected (n=845) | SNP Label in Figure 1 |
|---|---|---|---|---|---|---|
| CDGAP | rs10934490 | 0.417 | 0.427 | 0.452 | 0.435 | 1 |
| MYLK | rs16834817 | 0.117 | 0.129 | 0.108 | 0.121 | 2 |
| KALRN | rs6810298 | 0.293 | 0.305 | 0.288 | 0.298 | 3 |
| rs9289231 | 0.090 | 0.083 | 0.068 | 0.080 | 4 | |
| rs12637456 | 0.261 | 0.259 | 0.260 | 0.260 | 5 | |
| rs1444768 | 0.376 | 0.390 | 0.387 | 0.390 | 6 | |
| rs1444754 | 0.393 | 0.389 | 0.413 | 0.399 | 7 | |
| rs4234218 | 0.417 | 0.402 | 0.413 | 0.406 | 8 | |
| GATA2 | rs2335052 | 0.153 | 0.142 | 0.150 | 0.144 | 9 |
| rs3803 | 0.192 | 0.209 | 0.170 | 0.195 | 10 | |
| rs2713604 | 0.312 | 0.328 | 0.327 | 0.328 | 11 |
Figure 1.
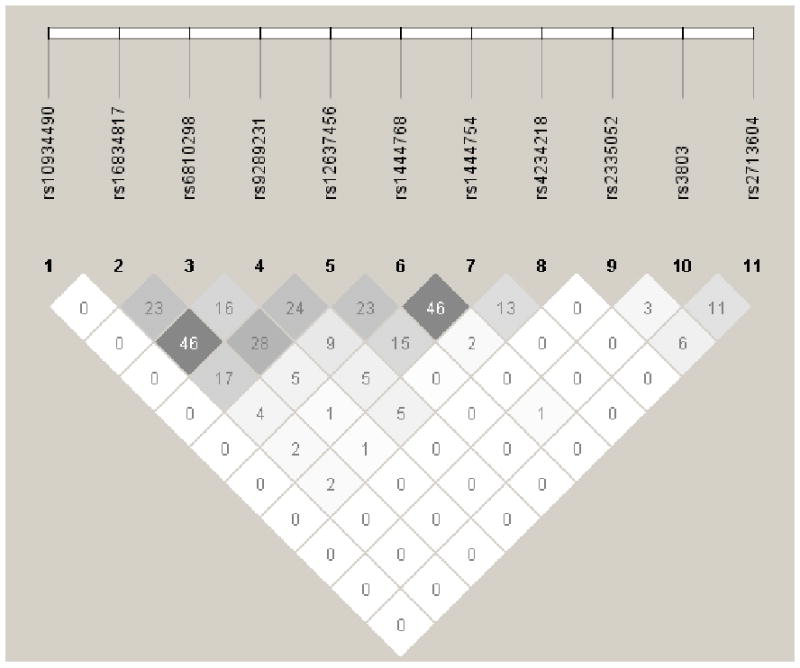
Linkage disequilibrium (r2) between SNPs in MYLK, CDGAP, KALRN, and GATA2 among all study participants.
Table 3.
Association of SNPs with CAD in basic and full models among the Young Affected (n=552 vs. n=773 Controls; minor allele frequencies are provided in Table 2).
| Basic Model* | Full Model† | ||||
|---|---|---|---|---|---|
| GENE | SNP name | OR‡ (95% CI) | p-value | OR‡ (95% CI) | p-value |
| CDGAP | rs10934490 | 1.05 (0.89, 1.24) | 0.59 | 1.06 (0.88, 1.27) | 0.55 |
| MYLK | rs16834817 | 1.18 (0.92, 1.51) | 0.21 | 1.13 (0.86, 1.48) | 0.38 |
| KALRN | rs6810298 | 1.09 (0.91, 1.30) | 0.36 | 1.12 (0.92, 1.36) | 0.25 |
| rs9289231 | 0.97 (0.72, 1.30) | 0.83 | 0.95 (0.69, 1.31) | 0.75 | |
| rs12637456 | 1.03 (0.85, 1.24) | 0.80 | 1.09 (0.89, 1.35) | 0.41 | |
| rs1444768 | 1.04 (0.88, 1.23) | 0.64 | 1.08 (0.90, 1.29) | 0.40 | |
| rs1444754 | 0.93 (0.79, 1.10) | 0.41 | 0.99 (0.82, 1.19) | 0.88 | |
| rs4234218 | 0.87 (0.74, 1.04) | 0.12 | 0.89 (0.74, 1.07) | 0.20 | |
| GATA2 | rs2335052 | 0.89 (0.70, 1.12) | 0.31 | 0.90 (0.70, 1.16) | 0.42 |
| rs3803 | 1.07 (0.88, 1.32) | 0.50 | 1.03 (0.83, 1.29) | 0.78 | |
| rs2713604 | 1.11 (0.93, 1.33) | 0.25 | 1.21 (0.99, 1.46) | 0.057 |
Basic model adjusted for sex only;
Full model adjusted for sex, BMI, diabetes, hypertension, hyperlipidemia, and smoking;
All odds ratios are per additional variant allele under the additive genetic model.
Smokers were younger than non-smokers, more likely to be male and to have hypertension and diabetes, and had a higher average CAD Index (Supplement 2). Although only about 1 in 5 participants in IHCS was a smoker, associations with YA were found (Table 4) in the KALRN-MYLK-CDGAP cluster, for rs10934490 in CDGAP (OR=1.64, p=0.019), and for rs12637456 in KALRN (OR=1.96, p=0.011). Suggestive association for rs16834871 in MYLK (OR=1.65 p=0.10) was also found. Risk levels above OR=1.5 per genotype were observed for rs9289231 in KALRN (OR=1.55, p=0.20). Findings among smokers for OA and AA are provided in Supplement 3.
Table 4.
Association of SNPs with CAD by smoking status among the Young Affected.
| Basic Model* | Full Model† | ||||
|---|---|---|---|---|---|
| GENE | SNP name | OR‡ (95% CI) | p-value | OR‡ (95% CI) | p-value |
| SMOKERS (n=246 [151 YA, 95 Controls]) | |||||
| CDGAP | rs10934490 | 1.56 (1.06, 2.31) | 0.025 | 1.64 (1.09, 2.48) | 0.019 |
| MYLK | rs16834817 | 1.80 (0.99, 3.26) | 0.051 | 1.65 (0.90, 3.03) | 0.10 |
| KALRN | rs6810298 | 1.23 (0.81, 1.88) | 0.34 | 1.19 (0.76, 1.86) | 0.45 |
| rs9289231 | 1.54 (0.80, 2.94) | 0.19 | 1.55 (0.79, 3.02) | 0.20 | |
| rs12637456 | 1.98 (1.21, 3.23) | 0.006 | 1.96 (1.17, 3.29) | 0.011 | |
| rs1444768 | 1.18 (0.80, 1.74) | 0.40 | 1.21 (0.81, 1.82) | 0.36 | |
| rs1444754 | 0.96 (0.66, 1.39) | 0.82 | 0.99 (0.66, 1.48) | 0.96 | |
| rs4234218 | 0.89 (0.62, 1.27) | 0.51 | 0.90 (0.62, 1.32) | 0.60 | |
| GATA2 | rs2335052 | 1.00 (0.59, 1.69) | 0.99 | 1.03 (0.59, 1.80) | 0.92 |
| rs3803 | 1.33 (0.83, 2.13) | 0.23 | 1.34 (0.81, 2.20) | 0.25 | |
| rs2713604 | 1.44 (0.96, 2.16) | 0.08 | 1.39 (0.90, 2.14) | 0.14 | |
| NON-SMOKERS (n=1,079 [393 YA, 686 Controls]) | |||||
| CDGAP | rs10934490 | 0.96 (0.79, 1.16) | 0.65 | 0.95 (0.77, 1.16) | 0.59 |
| MYLK | rs16834817 | 1.04 (0.78, 1.39) | 0.79 | 1.02 (0.75, 1.39) | 0.90 |
| KALRN | rs6810298 | 1.08 (0.88, 1.32) | 0.46 | 1.11 (0.89, 1.38) | 0.36 |
| rs9289231 | 0.81 (0.57, 1.16) | 0.25 | 0.80 (0.54, 1.17) | 0.24 | |
| rs12637456 | 0.92 (0.74, 1.15) | 0.46 | 0.96 (0.76, 1.22) | 0.75 | |
| rs1444768 | 1.03 (0.86, 1.24) | 0.73 | 1.05 (0.86, 1.29) | 0.63 | |
| rs1444754 | 0.94 (0.77, 1.14) | 0.53 | 0.98 (0.79, 1.21) | 0.83 | |
| rs4234218 | 0.89 (0.73, 1.08) | 0.22 | 0.88 (0.72, 1.09) | 0.25 | |
| GATA2 | rs2335052 | 0.85 (0.65, 1.12) | 0.26 | 0.87 (0.65, 1.16) | 0.34 |
| rs3803 | 1.01 (0.80, 1.28) | 0.95 | 0.96 (0.74, 1.23) | 0.74 | |
| rs2713604 | 1.05 (0.86, 1.28) | 0.64 | 1.17 (0.94, 1.46) | 0.15 | |
Basic model adjusted for sex only;
Full model adjusted for sex, BMI, diabetes, hypertension, and hyperlipidemia;
All odds ratios are per additional variant allele under the additive genetic model.
No association was found in any SNPs among non-smokers in the IHCS sample (Table 4, supplement 4). The SNP that showed the most promise was GATA2 rs2713604 (OR=1.17, p=0.15). Interactions between SNPs and smoking status (see Supplement 5) were detected in CDGAP (rs10934491, p-interaction=0.026) and KALRN (rs12637456, p-interaction=0.004), with suggestive interactions for KALRN (rs9289231, p-interaction=0.09) and MYLK (rs16834817, p-interaction=0.08).
Similar differences in risk based on smoking status were found among YA in the CATHGEN sample. Smokers had higher ORs (Table 5) for SNPs in KALRN and MYLK (but not CDGAP) than were found overall or among the non-smokers (Table 5), and all these were associated with CAD at p<0.05 in the CATHGEN smokers. The GATA2 association was significant only in non-smokers (rs2713604, OR=2.10, p=0.004), and only one of the SNPs in KALRN-MYLK-CDGAP had fully-adjusted p<0.05 in CATHGEN non-smokers (rs9289231, OR=3.05, p=0.001). In this smaller CATHGEN sample, no statistically-significant SNP by genotype interaction for any SNP was detected (all p-interaction>0.10).
Table 5.
CATHGEN: Association of SNPs with CAD by smoking among the Young Affected.
| Basic Model* | Full Model† | ||||
|---|---|---|---|---|---|
| GENE | SNP name | OR‡ (95% CI) | p-value | OR‡ (95% CI) | p-value |
| SMOKERS (n=353 [238 YA, 115 Controls]) | |||||
| CDGAP | rs10934490 | 1.23 (0.87, 1.73) | 0.24 | 1.27 (0.87, 1.85) | 0.22 |
| MYLK | rs16834817 | 2.09 (1.16, 3.77) | 0.014 | 2.45 (1.30, 4.60) | 0.006 |
| KALRN | rs6810298 | 1.74 (1.18, 2.54) | 0.005 | 1.71 (1.13, 2.60) | 0.011 |
| rs9289231 | 2.69 (1.35, 5.37) | 0.005 | 3.22 (1.52, 6.82) | 0.002 | |
| rs12637456 | 2.38 (1.59, 3.55) | <0.001 | 2.28 (1.47, 3.52) | <0.001 | |
| rs1444768 | 1.93 (1.34, 2.79) | <0.001 | 1.94 (1.30, 2.89) | 0.001 | |
| rs1444754 | 1.57 (1.12, 2.19) | 0.009 | 1.45 (1.02, 2.08) | 0.041 | |
| rs4234218 | 1.49 (1.06, 2.09) | 0.022 | 1.46 (1.00, 2.13) | 0.0498 | |
| GATA2 | rs2335052 | 0.99 (0.61, 1.60) | 0.95 | 0.87 (0.51, 1.49) | 0.61 |
| rs3803 | 0.87 (0.57, 1.33) | 0.52 | 0.76 (0.47, 1.22) | 0.26 | |
| rs2713604 | 1.27 (0.90, 1.81) | 0.18 | 1.29 (0.88, 1.89) | 0.20 | |
| NON-SMOKERS (n=268 [93 YA, 175 Controls]) | |||||
| CDGAP | rs10934490 | 1.41 (0.94, 2.13) | 0.10 | 1.42 (0.91, 2.22) | 0.13 |
| MYLK | rs16834817 | 1.50 (0.84, 2.70) | 0.17 | 1.75 (0.91, 3.37) | 0.10 |
| KALRN | rs6810298 | 1.00 (0.66, 1.53) | 1.00 | 1.24 (0.76, 2.03) | 0.39 |
| rs9289231 | 2.71 (1.45, 5.06) | 0.002 | 3.05 (1.54, 6.05) | 0.001 | |
| rs12637456 | 1.26 (0.81, 1.96) | 0.30 | 1.46 (0.89, 2.39) | 0.13 | |
| rs1444768 | 1.68 (1.09, 2.58) | 0.018 | 1.54 (0.95, 2.50) | 0.08 | |
| rs1444754 | 1.36 (0.91, 2.03) | 0.13 | 1.31 (0.84, 2.05) | 0.23 | |
| rs4234218 | 1.37 (0.91, 2.05) | 0.13 | 1.33 (0.84, 2.10) | 0.23 | |
| GATA2 | rs2335052 | 1.21 (0.69, 2.12) | 0.50 | 1.11 (0.60, 2.08) | 0.74 |
| rs3803 | 0.80 (0.50, 1.29) | 0.37 | 0.82 (0.48, 1.39) | 0.46 | |
| rs2713604 | 1.69 (1.08, 2.63) | 0.021 | 2.10 (1.28, 3.46) | 0.004 | |
Basic model adjusted for sex only;
Full model adjusted for sex, BMI, diabetes, hypertension, hyperlipidemia, and smoking;
All odds ratios are per additional variant allele under the additive genetic model.
Further sub-analyses by sex strata were undertaken among the IHCS patient sample, but no significant associations were detected. Suggestive associations among females were found for rs16834817 in MYLK (females: OR=1.45, p=0.059; males: OR=1.02, p=0.90; p-interaction=0.17), and rs3803 in GATA2 (females: OR=1.34, p=0.09; males: OR=0.96, p=0.78; p-interaction=0.12) were found. Adjustment in full models reduced the associations among females for rs16834817 (p=0.09) and rs3803 (p=0.14).
Discussion
Summary of findings
Overall, suggestive evidence of association with early-onset CAD was found for just one SNP (non-significant p=0.057 in GATA2) among 11 SNPs evaluated for replication in the IHCS sample. SNP rs2713604 was the GATA2 SNP that previously replicated in the CATHGEN cohort after initial evidence was found in the GENECARD probands (Connelly et al. 2006).
For the other three genes, though, findings for KALRN, MYLK, and CDGAP were unimpressive when the hypothesis was prospectively defined and was tested in a geographically distant population. Association studies of the 8 SNPs in these genes were performed previously among multiple CATHGEN cohorts with evidence among Caucasians of strong association with early-onset CAD (Wang et al. 2007). With the care that was taken in that previous investigation, including the replication in multiple cohorts, it seems unlikely that the failure to replicate associations herein was due to the traditional issues that have plagued replication attempts of initially significant genetic associations (Ioannidis et al. 2001; Hegele 2002).
Effect modification by smoking
Given the high prevalence of smoking exposure in the GENECARD and CATHGEN cohorts, it was hypothesized that the risk associations of genes in the 3q13-21 linkage peak such as KALRN, MYLK, and CDGAP may have been discovered because of interactions with smoking. A polymorphism may have a small effect on CAD risk among individuals free from smoking exposures, but some of these same genetic factors may endow an individual with large elevations in risk in the context of smoking exposure (Talmud 2007). For example, the effect of the ApoE ε2/ε3/ε4 SNP is likely present only among smokers (Talmud et al. 2005, Talmud 2007), and a similar finding was reported for the lipoprotein lipase gene (Talmud et al. 2000). Other environmental risk factors may cause effect modifications on genetic risk variants (Talmud 2007).
In the IHCS sample, the sample size of smokers was low compared to overall, but the risk estimates for multiple SNPs in KALRN, MYLK, and CDGAP were markedly higher among smokers compared to non-smokers. Several of these SNPs had significant statistical interactions with smoking, and re-analysis of the CATHGEN sample suggested a potentially similar effect modification by smoking although tests of interaction were not significant in the sample from which they were originally selected as the best SNPs for study. These hypothesis-generating risk differences based on smoking deserve further investigation (including for interactions with second-hand smoking exposure which was not available for the IHCS or CATHGEN samples).
SNPs in GATA2 did not demonstrate the smoking/SNP interaction. In fact, among the CATHGEN sample, odds ratios for GATA2 SNPs were qualitatively higher among the non-smokers. Since GATA2 SNPs were not in LD with the SNPs in KALRN, MYLK, and CDGAP, it may be that GATA2 influences CAD risk via a different pathway, as previously suggested (Wang et al. 2007; Connelly et al. 2006).
Importance of gene-environment interactions
Genetic contributions to the risk of complex prevalent diseases such as CAD are complex and little attention has been paid historically to gene-environment interactions. Heterogeneity of genetic effect arising from modification by environmental causes is increasingly recognized as a potential source of variability between genetic findings, particularly for a strong environmental risk factor such as smoking (Talmud 2007).
In contrast to the multiplicative smoking interactions described above, other genetic associations may exist primarily among non-smokers. The high risk effect of environmental exposure to smoking may obscure rather than modify the often relatively small genetic contributions to risk from some genes (Sen-Banerjee et al. 2000; Freeman et al. 2003; Horne et al. 2007). Taken together, the differential effects of smoking on genetic risk suggest that genetic associations with CAD and MI should be a priori stratified by smoking status, as previously suggested (Luo et al. 2007). Subsequently, if no effect modification is detected, data from smokers and non-smokers could be pooled to achieve a larger sample size (although this may not be advisable since smoking may increase the background noise in the data and thus hide small genetic effects on risk).
Limitations
This study was an observational cohort study and may suffer from the various limitations inherent in non-randomized medical studies. Population stratification, uncontrolled confounding, and differences in the LD compared to previous studies between tested SNPs and causal variants may obscure genetic associations with CAD. Care was taken to adjust for standard cardiac risk factors, though, and SNP associations were not notably affected by these adjustments. The focus of this study on individuals of Caucasian ancestry limits generalizability to other ethnic populations, thus further studies in other ethnic groups are indicated. Care should be taken in interpretation of smoking sub-analyses since, despite similarity of results among smokers in IHCS and CATHGEN, potential issues of multiple testing and post hoc hypothesis selection may require further confirmation of the potential smoking effect modification.
A strength of the study was that the CAD phenotype was determined from coronary angiography, the gold-standard assessment. Cases had clinically-significant coronary stenoses and the primary endpoint was early-onset disease. Controls, although not a random population sample, were definitively free from coronary lesions and were those who had lived to older age without developing CAD or its complications, as is suggested to be an optimal control sample (Luo et al. 2007) The age-discordant phenotype should improve clarity of genetic contributions to CAD.
Conclusions
This study provided suggestive evidence of association of GATA2 sequence variants with early-onset CAD, while among smokers the association with early-onset CAD of SNPs in KALRN, MYLK, and CDGAP was potentially greater in both the IHCS and CATHGEN cohorts. These data suggest that the genetic risk conferred by KALRN, MYLK, and CDGAP may be modified by cigarette smoking, which should be tested in other populations. Interaction between smoking and genotype could explain all or part of the difference in observed genetic effects and may explain the differential evidence for genetic association in different datasets for these and potentially other genes. Together with prior evidence of effect modification of SNPs among smokers in other genes, this study's findings suggest that genetic epidemiologic studies should be designed to explicitly account for participant smoking status.
Supplementary Material
Acknowledgments
This research was supported by NIH grants P01 HL73042 (Goldschmidt-Clermont), R01 HL073389-01 (Hauser), the Department of Medicine at Duke University, and the Cardiovascular Department at Intermountain Medical Center.
References
- Bowden DW, Rudock M, Ziegler J, Lehtinen AB, Xu J, Wagenknecht LE, Herrington D, Rich SS, Freedman BI, Carr JJ, Langefeld CD. Coincident linkage of type 2 diabetes, metabolic syndrome, and measures of cardiovascular disease in a genome scan of the diabetes heart study. Diabetes. 2006;55:1985–1994. doi: 10.2337/db06-0003. [DOI] [PubMed] [Google Scholar]
- Cernuda-Morollon E, Ridley AJ. Rho GTPases and leukocyte adhesion receptor expression and function in endothelial cells. Circ Res. 2006;98:757–767. doi: 10.1161/01.RES.0000210579.35304.d3. [DOI] [PubMed] [Google Scholar]
- Connelly JJ, Wang T, Cox JE, Haynes C, Wang L, Shah SH, Crosslin DR, Hale AB, Nelson S, Crossman DC, Granger CB, Haines JL, Jones CJ, Vance JM, Goldschmidt-Clermont PJ, Kraus WE, Hauser ER, Gregory SG. GATA2 is associated with familial early-onset coronary artery disease. PLoS Genet. 2006;2:e139. doi: 10.1371/journal.pgen.0020139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke JP, Losordo DW. Nitric oxide and angiogenesis. Circulation. 2002;105:2133–2135. doi: 10.1161/01.cir.0000014928.45119.73. [DOI] [PubMed] [Google Scholar]
- Freeman DJ, Samani NJ, Wilson V, McMahon AD, Braund PS, Cheng S, Caslake MJ, Packard CJ, Gaffney D. A polymorphism of the cholesteryl ester transfer protein gene predicts cardiovascular events in non-smokers in the West of Scotland Coronary Prevention Study. Eur Heart J. 2003;24:1833–1842. doi: 10.1016/j.ehj.2003.07.001. [DOI] [PubMed] [Google Scholar]
- Hauser ER, Crossman DC, Granger CB, Haines JL, Jones CJ, Mooser V, McAdam B, Winkelmann BR, Wiseman AH, Muhlestein JB, Bartel AG, Dennis CA, Dowdy E, Estabrooks S, Eggleston K, Francis S, Roche K, Clevenger PW, Huang L, Pedersen B, Shah S, Schmidt S, Haynes C, West S, Asper D, Booze M, Sharma S, Sundseth S, Middleton L, Roses AD, Hauser MA, Vance JM, Pericak-Vance MA, Kraus WE. A genomewide scan for early-onset coronary artery disease in 438 families: the GENECARD Study. Am J Hum Genet. 2004;75:436–447. doi: 10.1086/423900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegele RA. SNP judgments and freedom of association. Arterioscler Thromb Vasc Biol. 2002;22:1058–1061. doi: 10.1161/01.atv.0000026801.56080.14. [DOI] [PubMed] [Google Scholar]
- Horne BD, Camp NJ, Anderson JL, Mower CP, Kolek MJ, Clarke JL, Carlquist JF. Multiple less common genetic variants explain the association of the cholesteryl ester transfer protein gene with coronary artery disease. J Am Coll Cardiol. 2007;49:2053–2060. doi: 10.1016/j.jacc.2007.02.039. [DOI] [PubMed] [Google Scholar]
- Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29:306–309. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- Knaus UG. Rho GTPase signaling in inflammation and transformation. Immunol Res. 2000;21:103–109. doi: 10.1385/IR:21:2-3:103. [DOI] [PubMed] [Google Scholar]
- Luo AK, Jefferson BK, Garcia MJ, Ginsburg GS, Topol EJ. Challenges in the phenotypic characterisation of patients in genetic studies of coronary artery disease. J Med Genet. 2007;44:161–165. doi: 10.1136/jmg.2006.045732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratovitski EA, Bao C, Quick RA, McMillan A, Kozlovsky C, Lowenstein CJ. An inducible nitric-oxide synthase (NOS)-associated protein inhibits NOS dimerization and activity. J Biol Chem. 1999;274:30250–30257. doi: 10.1074/jbc.274.42.30250. [DOI] [PubMed] [Google Scholar]
- Ren J, Fang CX. Small guanine nucleotide-binding protein Rho and myocardial function. Acta Pharmacol Sin. 2005;26:279–285. doi: 10.1111/j.1745-7254.2005.00059.x. [DOI] [PubMed] [Google Scholar]
- Sen-Banerjee S, Siles X, Campos H. Tobacco smoking modifies association between Gln-Arg192 polymorphism of human paraoxonase gene and risk of myocardial infarction. Arterioscler Thromb Vasc Biol. 2000;20:2120–2126. doi: 10.1161/01.atv.20.9.2120. [DOI] [PubMed] [Google Scholar]
- Shimokawa H, Takeshita A. Rho-kinase is an important therapeutic target in cardiovascular medicine. Arterioscler Thromb Vasc Biol. 2005;25:1767–1775. doi: 10.1161/01.ATV.0000176193.83629.c8. [DOI] [PubMed] [Google Scholar]
- Smith LR, Harrell FE, Jr, Rankin JS, Califf RM, Pryor DB, Muhlbaier LH, Lee KL, Mark DB, Jones RH, Oldham HN. Determinants of early versus late cardiac death in patients undergoing coronary artery bypass graft surgery. Circulation. 1991;84(suppl 5):III245–III253. [PubMed] [Google Scholar]
- Talmud PJ, Bujac SR, Hall S, Miller GJ, Humphries SE. Substitution of asparagines for aspartic acid at residue 9 (D9N) of lipoprotein lipase markedly augments risk of ischaemic heart disease in male smokers. Atherosclerosis. 2000;149:75–81. doi: 10.1016/s0021-9150(99)00309-3. [DOI] [PubMed] [Google Scholar]
- Talmud PJ, Stephens JW, Hawe E, Demissie S, Cupples LA, Hurel SJ, Humphries SE, Ordovas JM. The significant increase in cardiovascular disease risk in APOE ε4 carriers is evident only in men who smoke: potential relationship between reduced antioxidant status and ApoE4. Ann Hum Genet. 2005;69:613–622. doi: 10.1111/j.1529-8817.2005.00205.x. [DOI] [PubMed] [Google Scholar]
- Talmud PJ. Gene-environment interaction and its impact on coronary heart disease risk. Nutr Metab Cardiovasc Dis. 2007;17:148–152. doi: 10.1016/j.numecd.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Taylor GS, Muhlestein JB, Wagner GS, Bair TL, Li P, Anderson JL. Implementation of a computerized cardiovascular information system in a private hospital setting. Am Heart J. 1998;136:792–803. doi: 10.1016/s0002-8703(98)70123-1. [DOI] [PubMed] [Google Scholar]
- Wang L, Hauser ER, Shah SH, Pericak-Vance MA, Haynes C, Crosslin D, Harris M, II, Nelson S, Hale AB, Granger CB, Haines JL, Jones CJH, Crossman D, Seo D, Gregory SG, Kraus WE, Goldschmidt-Clermont PJ, Vance JM. Peakwide mapping on chromosome 3q13 identifies the Kalirin gene as a novel candidate gene for coronary artery disease. Am J Hum Genet. 2007;80:650–663. doi: 10.1086/512981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


