Abstract
Background
Edwardsiella tarda is the etiologic agent of edwardsiellosis, a devastating fish disease prevailing in worldwide aquaculture industries. Here we describe the complete genome of E. tarda, EIB202, a highly virulent and multi-drug resistant isolate in China.
Methodology/Principal Findings
E. tarda EIB202 possesses a single chromosome of 3,760,463 base pairs containing 3,486 predicted protein coding sequences, 8 ribosomal rRNA operons, and 95 tRNA genes, and a 43,703 bp conjugative plasmid harboring multi-drug resistant determinants and encoding type IV A secretion system components. We identified a full spectrum of genetic properties related to its genome plasticity such as repeated sequences, insertion sequences, phage-like proteins, integrases, recombinases and genomic islands. In addition, analysis also indicated that a substantial proportion of the E. tarda genome might be devoted to the growth and survival under diverse conditions including intracellular niches, with a large number of aerobic or anaerobic respiration-associated proteins, signal transduction proteins as well as proteins involved in various stress adaptations. A pool of genes for secretion systems, pili formation, nonfimbrial adhesions, invasions and hemagglutinins, chondroitinases, hemolysins, iron scavenging systems as well as the incomplete flagellar biogenesis might feature its surface structures and pathogenesis in a fish body.
Conclusion/Significance
Genomic analysis of the bacterium offered insights into the phylogeny, metabolism, drug-resistance, stress adaptation, and virulence characteristics of this versatile pathogen, which constitutes an important first step in understanding the pathogenesis of E. tarda to facilitate construction of a practical effective vaccine used for combating fish edwardsiellosis.
Introduction
Edwardsiella tarda, a Gram-negative bacteria belonging to Enterobacteriaceae, is the etiological agent for edwardsiellosis, a devastating fish disease prevailing in worldwide aquaculture industries and accounting for severe economical losses [1], [2]. The organism commonly affects more than 20 species of freshwater and marine fishes including carp, tilapia, eel, catfish, mullet, salmon, trout, turbot and flounder, causing systemic hemorrhagic septicemia and emphysematous putrefactive disease with swelling skin lesions, as well as ulcer and necrosis in internal organs such as liver, kidney, spleen, and musculature [1]. Besides piscine species, E. tarda also inhabits and infects a broad range of cold or warm -blooded hosts such as reptiles, amphibians, birds, mammals and even humans [2], [3], raising a concern about E. tarda being a significant zoonotic pathogen.
Edwardsiellae bacterium resides in subgroup 3 in γ-group of Proteobacteria [2] and contains 3 species, E. tarda, E. hoshinae and E. ictaluri, the notorious pathogen relatively strictly inhabiting and causing enteric septicemia in Channel catfish [4]. Like phylogenetically related Enterobacteriaceae bacteria Salmonella spp. [5], E. tarda possesses the capacity of invading epithelial cells [6], [7] and macrophages [8], and multiplies in the cells, which is implicated to be one of the critical steps in its pathogenesis by subverting the fish immune system and causing systemic hemorrhagic septicemia [9]. For the present, the scant knowledge about the genetic basis for the intracellular lifestyle and molecular pathogenesis of E. tarda infection has largely hindered the development of a practical effective vaccine used for combating fish edwardsiellosis. Moreover, the criticized indiscriminate long-term application of antibiotics is marginally effective [10] and raises the increasing concern of multi-drug resistant E. tarda strains [11], [12], leaving satisfactory control methods of the disease currently unavailable.
To unravel the genetic properties for habitat adaptation, virulence determinants, invasive nature and multi-drug resistance of E. tarda and to facilitate the construction of a practical effective vaccine used for combating fish edwardsiellosis, we utilized the high-throughput pyrosequencing (454 Life Sciences Corporation) together with conventional sequencing method (PCR-based sequencing on ABI3730 automated capillary electrophoresis sequencer, Applied Biosystem Inc.) to quickly determine the complete genome sequence of E. tarda EIB202, a chloramphenicol, tetracycline, rifampicin, and streptomycin-resistant and highly virulent strain isolated from a recent outbreak in farmed turbot in Shandong province of China [12]. E. tarda EIB202 has 50% lethal doses (LD50) of 3.8×103 colony forming units (CFU) g−1 for swordtail fish [12], 5×102 CFU g−1 for zebra fish, and 4.5×102 CFU g−1 for turbot, and displays fast growth rates in a wide range of sodium chloride concentrations (0.5%–5%) as well as temperature shifts (20°C–37°C) (our unpublished data), presenting as a versatile fish pathogen. Analysis of the complete genome sequence of EIB202 revealed a number of gene hallmarks in E. tarda for adaptation to broad host niches and shed lights on the mechanisms underlying the intracellular colonization of the bacterium in host cells.
Results and Discussion
General features of the complete chromosome sequence
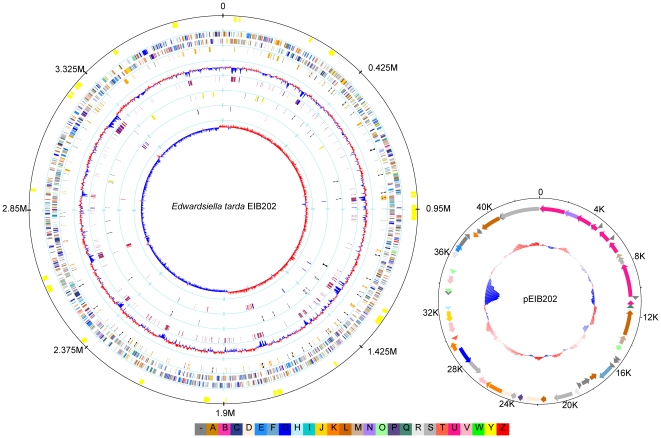
E. tarda EIB202 contains a single circular chromosome of 3,760,463 bp with an average G+C content of 59.7% (Table 1). The chromosome is predicted to distinctly harbor 8 rRNA operons, 95 tRNA genes, and 8 stable noncoding RNAs, relatively higher than that of other sequenced enterobacteria (Table S1) and in consistent with the rapid growth of the bacterium [12]. The eight rRNA operons, among which one operon contains a duplication in 5S rRNA gene, are scattered in the circular genome except for two locating in tandem as previously reported [13] (Figure 1). In addition to 77 pseudogenes (including 32 phage and 31 transposase genes), 3,486 coding sequences (CDSs) with an average length of 906 bp were encoded in the chromosome, representing 83.9% of the genome. Among all the protein-coding genes, 79.2% of the CDSs (n = 2,823) were assigned to a functional category of Cluster of Orthologous Groups (COG). Approximately 28% (980/3563) of the chromosomal genes are hypothetical in nature, accounting for the majority of genes (597/852) that are specific to the E. tarda genome among the enterobacterium genome samples.
Table 1. Overall features of the genome of E. tarda EIB202.
| Chromosome | Count or percent |
| Size | 3,760,463 bp |
| C+G content (%) | 59.7 |
| CDSa | 3,486 |
| Coding percentage | 83.9% |
| Pseudogenes or gene fragmentsb | 77 |
| IS elements | 19 |
| rRNA genes | 7*(16S+23S+5S), 1*(16S+23S+5S+5S) |
| tRNA genes | 95 |
| Other RNA gene | 8 |
| Average CDS length | 906 |
| Plasmid | |
| Size | 43,703 bp |
| CDS | 53 |
| C+G content(%) | 57.3 |
| IS elements | 1 |
Total not including pseudogenes.
Pseudogenes include transposase and phage-related genes.
Figure 1. Circular atlas of E. tarda EIB202 genome and plasmid pEIB202.
Left, chromosome; Right, plasmid. Circles range from 1 (outer circle) to 9 (inner circle) for chromosome and I (outer circle) to III (inner circle) for plasmid, respectively. Circle 1, genomic islands; Circles 2/I and 3/II, predicted coding sequences on the plus and minus strands, respectively; Circle 4, variable number of tandem repeats (VNTRs) (black) and direct repeat sequences (DRs) (orange); Circles 5/III, G+C percentage content: above median GC content (red), less than or equal to the median (blue); Circle 6, potential horizontally transferred genes and EIB202-specific genes with respect to Enterobacteriaceae bacteria; Circle 7, stable RNA molecules: tRNA (black), rRNA (yellow); Circle 8, phage-like genes and transposases; Circle 9, GC skew (G−C)/(G+C): values>0 (red), values<0 (blue). All genes are colored by functional categories according to COG classification: gold for translation, ribosomal structure and biogenesis; orange for RNA processing and modification; light orange for transcription; dark orange for DNA replication, recombination and repair; antique white for cell division and chromosome partitioning; pink for defense mechanisms; tomato for signal transduction mechanisms; peach for cell envelope biogenesis and outer membrane; deep pink for intracellular trafficking, secretion and vesicular transport; pale green for posttranslational modification, protein turnover and chaperones; royal blue energy production and conversion; blue for carbohydrate transport and metabolism; dodger blue for amino acid transport and metabolism; sky blue for nucleotide transport and metabolism; light blue for coenzyme metabolism; cyan for lipid metabolism; medium purple for inorganic ion transport and metabolism; aquamarine for secondary metabolites biosynthesis, transport and catabolism; gray for function unknown.
A conjugative plasmid pEIB202
A circular plasmid (designated as pEIB202) of 43,703 bp was identified from the assembled sequences. The plasmid pEIB202 carries 53 predicted CDSs, among which around 27% encode hypothetical proteins (Figure 1). The open reading frames (ORFs) of putative replication initiator protein (RepA) and plasmid partition proteins (KorA, IncC, KorB, TopA, and ParA) were found on this plasmid, suggesting that this plasmid might belong to IncP plasmid and was capable of replication and stable inheritance in a wide variety of gram-negative bacteria [14].
In the sequence of pEIB202, six genes were probably involved in resistance to antibiotics, including tetA and tetR for tetracycline, strA and strB for streptomycin, sulII for sulfonamide, and catA3 for chloramphenicol resistance, providing genetic properties for previously described multi-drug resistance in EIB202 [12]. A complex transposon ISSf1 containing IS4 family transposase [15] and the catA3 gene was identified. The average G+C content of this region was observed to be comparatively extremely low (37.4%) (Figure 1), and differed by above 3σ (standard deviation) from the average G+C content of the plasmid (57.3%) or of the genome (59.7%) (σ = 0.053 for pEIB202; σ = 0.062 for the genome; window length 1.2 kb), suggesting that the chloramphenicol resistance might be recently acquired by the plasmid. Notably, the plasmid encodes an incomplete set of components involved in the type IV A secretion system (T4ASS) (virB2, -B4, -B5, -B6, -B8, -B9, -B10, -B11, -D2, and -D4). The VirB/VirD4 T4ASS was well documented in various pathogens to be involved in horizontal DNA transfer, and in secretion or injection of protein effectors into the medium milieu or into host cells [16]. In addition, several genes associated to plasmid conjugation (mobC, traC, traD, traL, traN, traX) are present in the pEIB202 sequence, demonstrating the genetic basis for its capability to transfer between bacteria in the laboratory system with a conjugation frequency of 1.6×10−6 (data not shown).
Genomic plasticity and genomic islands
As illustrated by Figure 1, the G+C content of the E. tarda EIB202 genome is highly variable. A large portion (15%) of the genome is composed of mobile genetic elements or related to special genomic islands, displaying a mosaic structure of the genome. EIB202 contains 46 genes which are shown to be phage-like products, integrases or recombinases. In addition, a large quantity (n = 599, a total of 560 kb) of variable number of tandem repeats (VNTRs) or direct repeat sequences were detected in the genome. It also harbors 15 complete and 4 disrupted insertion sequences (IS) including 10 intact IS100, 2 truncated IS100 and a copy of IS1414I that might lost its transposition activity as a consequence of the nonsense mutation in this insertion sequence (data not shown). Given the reported continued transposition activity of IS100 [17], we postulated that the particular IS100 copies were due to duplicated translocation or multiple integration events of this element occurred within this strain. Interestingly, EIB202 and E. ictaluri 93–146 share an insertion sequence ISSaen1, which could also be found in S. enterica serovar Enteritidis [18]. All these genes may represent tremendous potential for generating genetic diversity within protein-coding genes over a very short evolutionary time for its adaptation to various niches.
In the genome sequence of EIB202, a total of 24 genomic islands (GI) were discerned (Table 2) to scatter throughout the chromosome and contain a total of 852 EIB202-specific genes that were not found in the other Enterobacteriaceae bacteria investigated so far. The previously described type III secretion system (TTSS) [19] and type IV secretion system (T6SS) [19], [20] are included in the genomic islands (GI7 and GI17). The GI10 contains a mammalian Toll-like/IL-1 receptor (TIR) domain protein, a novel virulence factor implicated in the intracellular survival and lethality of S. enteric [21], and may also contribute to the intracellular colonization of E. tarda in host cells. In addition to these GIs, the regions, including GI4, GI6, GI9, and GI22 encoding type I secretion system (T1SS), hemagglutinin, O-polysaccharide (OPS) biosynthesis enzymes, and type I restriction-modification system, respectively, maybe consist of the major pathogenicity islands (PAIs) of the bacterium. Some of the GIs are flanked by tRNAs or contain transposases and prophage proteins (Table 2), indicating that these GIs are still involved in the evolution of the bacterium. Among these GIs, GI2 and GI4 are absent in the genome of E. ictaluri 93–146 and might be characteristics of the main differences between the two species. Interestingly, all of the GIs except for GI7 and GI17, which encodes TTSS and T6SS, are absent in the phylogenetically related Salmonella spp., suggesting that E. tarda, as discussed below, the descendent of a lineage that diverged from the ancestral trunk before Salmonella and Escherichia split, might acquire these genome regions from its evolution events or Salmonella and its predecessors might not have acquired these GIs at the first place.
Table 2. Overview of the genomic islands of EIB202.
| No. | CDS | Characteristics or putative functions |
| GI1 | ETAE_0032-ETAE_0037 | Peroxidase, peptidase |
| GI2 | ETAE_0049–ETAE_0058 | Hypothetical proteins |
| GI3 | ETAE_0252–ETAE_0256 | Nitrate/nitrite transporter |
| GI4 | ETAE_0315–ETAE_0326 | T1SS, invasin |
| GI5 | ETAE_0798–ETAE_0805 | Oxidoreductase, integrase |
| GI6 | ETAE_0808–ETAE_0822 | IS, hemagglutinin, haemolysin secretion system |
| GI7 | ETAE_0839–ETAE_0892 | TTSS |
| GI8 | ETAE_1166–ETAE_1177 | IS, iron uptake |
| GI9 | ETAE_1192–ETAE_1214 | OPS , CRISPR |
| GI10 | ETAE_1390–ETAE_1396 | IS, Toll-like/IL-1 receptor |
| GI11 | ETAE_1586–ETAE_1602 | IS, hypothetical proteins |
| GI12 | ETAE_1608–ETAE_1613 | Prophage, O-antigen polymerase protein |
| GI13 | ETAE_1759–ETAE_1762 | IS, acetyltransferase |
| GI14 | ETAE_1811–ETAE_1829 | Choline/carnitine/betaine transporter |
| GI15 | ETAE_2025–ETAE_2037 | Carnitine dehydratase |
| GI16 | ETAE_2243–ETAE_2255 | P-pilus related proteins |
| GI17 | ETAE_2428–ETAE_2443 | T6SS |
| GI18 | ETAE_2465–ETAE_2476 | Prophage Sf6. Flanked by tRNA-Arg |
| GI19 | ETAE_2742–ETAE_2748 | Integrase, bacteriophage proteins |
| GI20 | ETAE_3032–ETAE_3043 | Recombinase, invasin. |
| GI21 | ETAE_3046–ETAE_3052 | Transposase, chorismate mutase. Flanked by IS100 |
| GI22 | ETAE_3069–ETAE_3074 | Type I restriction-modification system |
| GI23 | ETAE_3078–ETAE_3091 | Transposase. Flanked by tRNA-Leu and IS100 |
| GI24 | ETAE_3405–ETAE_3428 | Transposase, phage proteins. Flanked by tRNA selC |
Relationship of E. tarda to other bacterial taxa
E. tarda shows its specific taxonomic position in Enterobacteriaceae as inferred from the sequence similarities of the housekeeping genes (Figure 2). At variance from the previous description that Trabulsiella guamensis and Enterobacter sakazakii were the closest relatives of Edwardsiella based on analysis of the limited 16S rDNA sequences [2], E. tarda presents as the sister clad with the phytopathogenic bacterium Erwinia carotovora atroseptica SCRI1043 (branch length value = 0.173), the endophytic bacterium Serratia proteamaculans 568 (value = 0.174), as well as human pathogen Yersinia pestis (value = 0.182). In addition, E. tarda is the most deeply diverging lineage among some notorious enteric pathogenic bacteria such as Escherichia, Salmonella, Shigella, and Klebsiella, but after the divergence of Vibrio cholera and Pseudomonas aeruginosa. The clustering of E. tarda EIB202 is also supported by the previous described biochemical pathways of aromatic amino acid biosynthesis and their regulation in most of the enteric bacteria [22]. Therefore, Edwardsiella species comprise a lineage that diverged from the ancestral trunk before the divergence of some other extensively researched enteric pathogenic bacteria, such as Salmonella and Escherichia. E. tarda adopts both of the intracellular and extracelluar lifestyles as its relatives such as pathogenic S. typhimurium, Y. pestis as well as symbiont Sodalis glossinidius, further suggesting that they experienced independent and divergent evolution driven by their specific hosts and inhabitant niches.
Figure 2. Phylogenetic relationship of EIB202.
Phylogenies of Enterobacteriaceae species inferred from concatenated alignments of the protein sequences encoded by 44 housekeeping genes as described in the Materials and Methods. Bacillus cereus ATCC 14579 was used as the outgroup. Accession numbers for the selected bacterial genome sequences are as following: Bacillus cereus ATCC 14579, NC_004722; Burkholderia mallei ATCC 23344, NC_006348; Citrobacter koseri ATCC BAA-895, NC_009792; E. ictaluri 93-146, NC_012779; E. sakazakii ATCC BAA-894, NC_009778; Enterobacter sp. 638, NC_009436; E. carotovora atrosepticum SCRI1043, NC_004547; E. tasmaniensis Et1/99, NC_010694; E. coli K-12 substr MG1655, NC_000913; E. coli O157:H7 str. Sakai O157:H7, NC_002695; E. fergusonii ATCC 35469, NC_011740; K. pneumoniae 342, NC_011283; K. pneumoniae subsp. pneumoniae MGH 78578, NC_009648; P. luminescens subsp. laumondii TTO1, NC_005126; Proteus mirabilis HI4320, NC_010554; Pseudomonas aeruginosa PAO1, NC_002516; S. enterica subsp. enterica serovar Typhi str. Ty2, NC_004631; S. typhimurium LT2, NC_003197; S. proteamaculans 568, NC_009832; Shewanella oneidensis MR-1, NC_004347; S. flexneri 2a str. 2457T, NC_004741; S. flexneri 5 str. 8401, NC_008258; Sodalis glossinidius str. morsitans, NC_007712; Vibrio cholera O1 biovar eltor str. N16961, NC_002505; Wigglesworthia brevipalpis endosymbiont of Glossina brevipalpis, NC_004344; Xanthomonas campestris pv. campestris str. ATCC 33913, NC_003902; Y. pestis CO92, NC_003143 ; and Y. pestis KIM, NC_004088.
Comparative genomics analysis with other enterobacteria
Utilizing the COG database [23], about 64.3% of the E. tarda proteins were grouped into three functional groups (Table 3), and only 14.8% were assigned to the “poorly characterized” group. The differences between E. tarda EIB202 and other Enterobacteriaceae bacteria were overviewed in Table 3. Among the whole-genome sequenced enterobacteria, E. tarda EIB202 contains a genome of the minimum size (Table S1), which may correspond to the previous suggestion that E. tarda may not be present as a free-living microorganism in natural waters but multiply intracellularly in protozoan and transmission to fish, reptile and other animals or humans [2]. Despite of the minor variations in all areas, the most obvious differences where E. tarda EIB202 consistently varied from all the other Enterobacteriaceae bacteria were discerned with the counts of E. tarda proteins for translation, ribosomal structure and biogenesis (J), cell envelope biogenesis, outer membrane (M), signal transduction mechanisms (T), nucleotide transport and metabolism (F), and coenzyme metabolism (H) as the highest and that for carbohydrate transport and metabolism (G) as the lowest (Table 3). The significant differences of these COG distributions were also statistically supported by the Chi-square tests using pair-wise comparisons with EIB202 (χ2>3.84, P<0.05) (Table 3). The relatively high proportion of genes in the J and M group in E. tarda EIB202 is consistent with the high growth rate of the bacterium as previously described [12]. Moreover, the abundance of genes in F and H group as well as the relative paucity of genes in G group may reflect that the organism is well adapted to the aquatic ecosystems and intracellular niches, where may exist relatively mean carbohydrates and wealth of nucleic acid molecules. Again, the comparatively high level of genes in signal transduction mechanisms (T) is a well manifestation of its capacities to cope with various growth conditions and to enhance its survival and persistence under a series of stresses (Table 3).
Table 3. Comparison of COG category distributions of EIB202 with Enterobacteriaceae *.
| Functional categories | E. tarda | S. typhimurium | E. carotovora | E. sakazakii | E. coli | K. pneumoniae | P. luminescens | S. proteamaculans | S. flexneri | Y. pestis |
| Information storage and processing | ||||||||||
| Translation, ribosomal structure and biogenesis (J) | 171 (4.80%) | 185 (3.98, 3.90%) | 176 (4.95, 3.80%) | 179 (3.61, 3.91%) | 184 (3.03, 4.01%) | 199 (5.88, 3.75%) | 184 (6.95, 3.65%) | 197 (4.24, 3.89%) | 173 (3.51, 3.94%) | 175 (2.42, 4.08%) |
| Transcription (K) | 222 (6.23%) | 329 (1.66, 6.94%) | 333 (2.95, 7.19%) | 290 (0.02, 6.34%) | 308 (0.75, 6.71%) | 445 (14.22, 8.39%) | 279 (1.83, 5.54%) | 449 (20.25, 8.87%) | 249 (1.11, 5.67%) | 237 (1.79, 5.52%) |
| DNA replication, recombination and repair (L) | 163 (4.58%) | 167 (5.89, 3.52%) | 188 (1.30, 4.06%) | 156 (6.90, 3.41%) | 215 (0.05, 4.68%) | 227 (0.45, 4.28%) | 260 (1.53, 5.16%) | 175 (6.96, 3.46%) | 513 (127.79, 11.69%) | 346 (39.02, 8.06%) |
| Cellular processes | ||||||||||
| Cell division and chromosome partitioning (D) | 34 (0.95%) | 37 (0.72, 0.78%) | 50 (0.31, 1.08%) | 39 (0.13, 0.85%) | 36 (0.68, 0.78%) | 41 (0.84, 0.77%) | 44 (0.15, 0.87%) | 35 (1.83, 0.69%) | 34 (0.75, 0.77%) | 35 (0.43, 0.82%) |
| Defense mechanisms (V) | 35 (0.98%) | 49 (0.05, 1.03%) | 47 (0.02, 1.02%) | 43 (0.01, 0.94%) | 49 (0.14, 1.07%) | 74 (2.98, 1.39%) | 69 (2.62, 1.37%) | 57 (0.41, 1.13%) | 44 (0.01, 1.00%) | 43 (0.01, 1.00%) |
| Posttranslational modification, protein turnover, chaperones (O) | 119 (3.34%) | 161 (0.02, 3.40%) | 140 (0.66, 3.02%) | 142 (0.29, 3.11%) | 138 (0.74, 3.01%) | 147 (2.38, 2.77%) | 118 (7.76, 2.34%) | 150 (0.99, 2.96%) | 131 (0.82, 2.98%) | 134 (0.30, 3.12%) |
| Cell envelope biogenesis, outer membrane (M) | 209 (5.87%) | 259 (0.61, 5.47%) | 240 (1.81, 5.18%) | 221 (4.08, 4.83%) | 239 (1.69, 5.21%) | 241 (7.77, 4.54%) | 203 (15.46, 4.03%) | 246 (4.26, 4.86%) | 213 (4.04, 4.85%) | 205 (4.64, 4.78%) |
| Cell motility and secretion (N) | 78 (2.19%) | 125 (1.71, 2.64%) | 116 (0.87, 2.51%) | 117 (1.01, 2.56%) | 116 (0.98, 2.53%) | 68 (10.86, 1.28%) | 105 (0.11, 2.08%) | 115 (0.06, 2.27%) | 89 (0.25, 2.03%) | 137 (7.34, 3.19%) |
| Inorganic ion transport and metabolism (P) | 167 (4.69%) | 202 (0.86, 4.26%) | 244 (1.43, 5.27%) | 191 (1.13, 4.18%) | 223 (0.13, 4.86%) | 317 (6.83, 5.97%) | 151 (16.76, 3.00%) | 281 (3.15, 5.55%) | 194 (0.33, 4.42%) | 210 (0.18, 4.89%) |
| Intracellular trafficking and secretion (U) | 84 (2.36%) | 142 (3.13, 3.00%) | 73 (6.53, 1.58%) | 109 (0.00, 2.38%) | 135 (2.60, 2.94%) | 116 (0.29, 2.19%) | 128 (0.29, 2.54%) | 140 (1.37, 2.77%) | 97 (0.20, 2.21%) | 157 (11.06, 3.66%) |
| Signal transduction mechanisms (T) | 151 (4.24%) | 183 (0.75, 3.86%) | 121 (16.56, 2.61%) | 191 (0.01, 4.18%) | 184 (0.27, 4.01%) | 200 (1.24, 3.77%) | 137 (14.89, 2.72%) | 191 (1.20, 3.77%) | 151 (3.44, 3.44%) | 130 (8.27, 3.03%) |
| Metabolism | ||||||||||
| Energy production and conversion (C) | 214 (6.01%) | 292 (0.09, 6.16%) | 232 (3.88, 5.01%) | 203 (9.81, 4.44%) | 291 (0.38, 6.34%) | 296 (0.72, 5.58%) | 169 (34.52, 3.35%) | 280 (0.88, 5.53%) | 245 (0.66, 5.58%) | 183 (12.34, 4.26%) |
| Carbohydrate transport and metabolism (G) | 237 (6.65%) | 393 (7.81, 8.29%) | 353 (2.85, 7.63%) | 340 (1.74, 7.43%) | 377 (6.99, 8.21%) | 529 (29.70, 9.97%) | 189 (37.33, 3.75%) | 424 (8.75, 8.37%) | 319 (1.14, 7.27%) | 320 (1.90, 7.46%) |
| Amino acid transport and metabolism (E) | 291 (8.17%) | 356 (1.22, 7.51%) | 380 (0.00, 8.21%) | 326 (2.95, 7.13%) | 367 (0.08, 7.99%) | 468 (1.15, 8.82%) | 312 (12.50, 6.19%) | 482 (4.67, 9.52%) | 306 (4.07, 6.97%) | 303 (3.43, 7.06%) |
| Nucleotide transport and metabolism (F) | 89 (2.50%) | 89 (3.73, 1.88%) | 85 (4.25, 1.84%) | 85 (3.62, 1.86%) | 97 (1.34, 2.11%) | 97 (4.67, 1.83%) | 79 (9.43, 1.57%) | 110 (0.99, 2.17%) | 82 (3.72, 1.87%) | 75 (5.37, 1.75%) |
| Coenzyme metabolism (H) | 151 (4.24%) | 177 (1.36, 3.74%) | 131 (12.03, 2.83%) | 155 (3.76, 3.39%) | 155 (4.14, 3.38%) | 200 (1.24, 3.77%) | 188 (1.42, 3.73%) | 178 (2.98, 3.52%) | 151 (3.44, 3.44%) | 149 (3.12, 3.47%) |
| Lipid metabolism (I) | 75 (2.11%) | 94 (0.15, 1.98%) | 113 (1.01, 2.44%) | 88 (0.25, 1.92%) | 103 (0.18, 2.24%) | 125 (0.61, 2.36%) | 108 (0.01, 2.14%) | 143 (4.39, 2.82%) | 84 (0.37, 1.91%) | 81 (0.48, 1.89%) |
| Secondary metabolites biosynthesis, transport and ccatabolism (Q) | 44 (1.24%) | 67 (0.49, 1.41%) | 64 (0.34, 1.38%) | 64 (0.30, 1.40%) | 66 (0.62, 1.44%) | 107 (7.77, 2.02%) | 116 (13.02, 2.30%) | 121 (14.86, 2.39%) | 54 (0.00, 1.23%) | 62 (0.64, 1.44%) |
| Poorly characterized | ||||||||||
| General function prediction only (R) | 285 (8.00%) | 440 (9.29%) | 370 (7.99%) | 400 (8.75%) | 401 (8.73%) | 501 (9.44%) | 362 (7.19%) | 515 (10.17%) | 362 (8.25%) | 318 (7.41%) |
| Function unknown (S) | 279 (7.83%) | 374 (7.89%) | 329 (7.11%) | 376 (8.22%) | 328 (7.14%) | 390 (7.35%) | 366 (7.26%) | 397 (7.84%) | 314 (7.15%) | 369 (8.60%) |
Comparison of the ORFs in each COG category (other than Chromatin structure and dynamics (B), Extracellular structures (W), Nuclear structure (Y) and Cytoskeleton (Z) due to zero values) of EIB202 and the other Enterobacteriaceae bacteria by using Chi-square test were described in the related text. The Chi-square value (χ2) and the percentage of the ORF counts in each COG category were shown within brackets. When compared with EIB202, statistically significant differences in some Enterobacteriaceae bacteria in each of the COG category were shown with χ2>3.84 (P<0.05).
Predicted metabolic pathways
EIB202 genome encodes the complete sets of enzymes necessary for glycolysis, the tricarboxylic cycle, the pentose phosphate pathway and Entner-Doudoroff pathway (Figure 3). In contrast, the glyoxylate shunt is not complete because isocitrate lyases (icl1 and icl2) and malate synthases are missing. Except for the gene encoding pyruvate carboxylase, the complete set of genes for gluconeogenesis is present in the EIB202 genome (Figure 3). The strain also encodes a putative citrate lyase synthetase complex (ETAE_0223–0228), which may be involved in the lysis of citrate into acetate and oxaloacetate or the reverse reaction. Though genes encoding for oxalate decarboxylase, alanine transaminase and LL-diaminopimelate aminotransferase which are involved in synthesizing L-alanine were not identified in the EIB202 genome, the growth test of the bacterium indicated that it could synthesize L-alanine in an unidentified mechanism (data not shown), suggesting that the bacterium might be highly self-sufficient in amino acid biosynthesis (Figure 3 and Table S2).
Figure 3. Overview of metabolism and transport in EIB202.
Different transport families are distinguished by different colors and shapes. From top left going clockwise: ABC-2 and other transporters (yellow); phosphate and amino acid transporters (green); Siderophore-iron (III) receptors (brick red); TonB-dependent receptors (rosybrown); P-type ATPase (chocolate); mineral and organic ion transporters (violet); ion efflux (green); secretion systems (pink); drug/metabolite efflux (red); nucleotides transporters (orange); the major facilitator superfamily (MFS) (purple); the resistance-nodulation-cell division family (RND) (blue); phosphotransferase system (PTS) (black). Arrows indicate the direction of transport. All the amino acid biosynthesis genes are listed in Table S2.
EIB202 is able to produce adenosine triphosphate (ATP) through a complete respiratory chain as well as an ATP synthetase complex (ETAE_3528–3534). The genome encodes a variety of dehydrogenases (n = 80, Table 4) that enable it to draw on a variety of substrates as electron donors, such as NADH, succinate, formate, isocitrate, proline, acyl-CoA, D-amino acids and so on. The genome also encodes a number of reductases [fumarate reductase (ETAE_0335–0338), nitrate reductase (ETAE_0248–0252), dimethylsulfoxide (DMSO) reductase (ETAE_2192–2195), arsenate reductase (ETAE_1091), anaerobic sulfide reductase (ETAE_1738–1740), thiosulfate reductase (ETAE_1843–1845), anaerobic ribonucleoside triphosphate reductase (ETAE_0422–0423) and tetrathionate reductase (ETAE_1647–1649)], which may contribute to the respiration with alternative electron acceptors to oxygen (fumarate, nitrate, DMSO arsenate, thiofulfate and tetrathionate) under anaerobic conditions, which is in agreement with its facultative anaerobic lifestyle in intracellular niches.
Table 4. Dehydrogenases in EIB202.
| CDS | Gene | Annotation |
| ETAE_0085 | tdh | L-threonine 3-dehydrogenase |
| ETAE_0104 | wecC | UDP-N-acetyl-D-mannosaminuronate dehydrogenase |
| ETAE_0240 | Putative iron-containing alcohol dehydrogenase | |
| ETAE_0386 | mdh | Malate/lactate dehydrogenases |
| ETAE_0565 | thrA | Bifunctional aspartokinase I/homeserine dehydrogenase I |
| ETAE_0571 | gabD | Succinate-semialdehyde dehydrogenase I |
| ETAE_0587 | xdhC | Xanthine dehydrogenase, Fe-S binding subunit |
| ETAE_0588 | xdhB | Xanthine dehydrogenase, FAD-binding subunit |
| ETAE_0589 | xdhA | Xanthine dehydrogenase subunit |
| ETAE_0601 | pdxA | 4-Hydroxythreonine-4-phosphate dehydrogenase |
| ETAE_0620 | leuB | 3-Isopropylmalate dehydrogenase |
| ETAE_0658 | pdhR | Pyruvate dehydrogenase complex repressor |
| ETAE_0659 | aceE | Pyruvate dehydrogenase subunit E1 |
| ETAE_0662 | lpdA | Dihydrolipoamide dehydrogenase |
| ETAE_0770 | Putative alcohol dehydrogenase | |
| ETAE_0899 | gldA | Glycerol dehydrogenase |
| ETAE_0967 | sdr | Short-chain dehydrogenase/reductase |
| ETAE_0968 | gutB | L-iditol 2-dehydrogenase |
| ETAE_1048 | dld | D-lactate dehydrogenase |
| ETAE_1161 | sfcA | Malate dehydrogenase (oxaloacetate-decarboxylating) |
| ETAE_1202 | ugd | UDP-glucose 6-dehydrogenase |
| ETAE_1212 | gnd | 6-Phosphogluconate dehydrogenase |
| ETAE_1248 | pyrD | Dihydroorotate dehydrogenase 2 |
| ETAE_1334 | Iron-containing alcohol dehydrogenase | |
| ETAE_1364 | Pyruvate/2-Oxoglutarate dehydrogenase complex | |
| ETAE_1416 | D-isomer specific 2-hydroxyacid dehydrogenase NAD-binding | |
| ETAE_1449 | zwf | Glucose-6-phosphate 1-dehydrogenase |
| ETAE_1474 | dadA | D-amino-acid dehydrogenase |
| ETAE_1483 | gapA | Glyceraldehyde-3-phosphate dehydrogenase |
| ETAE_1508 | adhE | Aldehyde-alcohol dehydrogenase |
| ETAE_1549 | Short chain dehydrogenase | |
| ETAE_1658 | putA | Proline dehydrogenase |
| ETAE_1724 | Short-chain alcohol dehydrogenase of unknown specificity | |
| ETAE_1753 | Short-chain dehydrogenase/reductase | |
| ETAE_1771 | ldhA | D-lactate dehydrogenase |
| ETAE_1899 | kduD | 2-Deoxy-D-gluconate 3-dehydrogenase |
| ETAE_1922 | 3-Hydroxyisobutyrate dehydrogenase and related beta-hydroxyacid dehydrogenases | |
| ETAE_2050 | Isocitrate dehydrogenase, specific for NADP+ | |
| ETAE_2070 | ndh | NADH dehydrogenase, FAD-containing subunit |
| ETAE_2118 | D-beta-hydroxybutyrate dehydrogenase | |
| ETAE_2276 | hisD | Histidinol dehydrogenase |
| ETAE_2380 | nuoG | NADH dehydrogenase/NADH:ubiquinone oxidoreductase 75 kD subunit (chain G) |
| ETAE_2416 | asd | Aspartate-semialdehyde dehydrogenase |
| ETAE_2417 | pdxB | Erythronate-4-phosphate dehydrogenase |
| ETAE_2583 | sucB | 2-Oxoglutarate dehydrogenase, E2 subunit, dihydrolipoamide succinyltransferase |
| ETAE_2584 | sucA | Component of the 2-oxoglutarate dehydrogenase complex,thiamin-binding |
| ETAE_2585 | sdhB | Succinate dehydrogenase iron-sulfur subunit |
| ETAE_2586 | sdhA | Succinate dehydrogenase catalytic subunit |
| ETAE_2587 | sdhD | Succinate dehydrogenase cytochrome b556 small membrane subunit |
| ETAE_2588 | sdhC | Succinate dehydrogenase cytochrome b556 large membrane subunit |
| ETAE_2663 | caiA | Crotonobetainyl-CoA dehydrogenase |
| ETAE_2695 | folD | Methylenetetrahydrofolate dehydrogenase (NADP(+)) |
| ETAE_2705 | dltE | Short chain dehydrogenase |
| ETAE_2787 | guaB | Inositol-5-monophosphate dehydrogenase |
| ETAE_2836 | tyrA | Bifunctional chorismate mutase/prephenate dehydrogenase |
| ETAE_2856 | sdhA | Succinate dehydrogenase flavoprotein subunit |
| ETAE_2857 | sdhD | Succinate dehydrogenase hydrophobic membrane anchor protein |
| ETAE_2858 | sdhC | Succinate dehydrogenase cytochrome b-556 subunit |
| ETAE_2934 | aroE | Shikimate 5-dehydrogenase |
| ETAE_2939 | gcvP | Glycine dehydrogenase |
| ETAE_2949 | serA | D-3-phosphoglycerate dehydrogenase |
| ETAE_2958 | epd | D-erythrose 4-phosphate dehydrogenase |
| ETAE_2986 | Short chain dehydrogenase | |
| ETAE_3113 | glpC | Sn-glycerol-3-phosphate dehydrogenase subunit C |
| ETAE_3114 | glpB | Anaerobic glycerol-3-phosphate dehydrogenase subunit B |
| ETAE_3115 | glpA | Sn-glycerol-3-phosphate dehydrogenase subunit A |
| ETAE_3141 | Probable zinc-binding dehydrogenase | |
| ETAE_3173 | xdhC | Xanthine dehydrogenase accessory factor, putative subfamily |
| ETAE_3194 | aroE | Shikimate 5-dehydrogenase |
| ETAE_3312 | glpD | Glycerol-3-phosphate dehydrogenase |
| ETAE_3336 | fdhD | Formate dehydrogenase accessory protein |
| ETAE_3337 | Formate dehydrogenase-O, major subunit | |
| ETAE_3338 | Anaerobic dehydrogenases, typically selenocysteine-containing | |
| ETAE_3339 | fdnH | Formate dehydrogenase-N beta subunit |
| ETAE_3340 | fdnI | Formate dehydrogenase-N subunit gamma |
| ETAE_3341 | fdhE | Formate dehydrogenase accessory protein |
| ETAE_3344 | asd | Aspartate-semialdehyde dehydrogenase |
| ETAE_3353 | gdhA | Glutamate dehydrogenase/leucine dehydrogenase |
| ETAE_3429 | thrA | Homoserine dehydrogenase |
| ETAE_3457 | gpsA | Glycerol-3-phosphate dehydrogenase (NAD(P)+) |
Stress adaptation and signal transduction
E. tarda has been implicated to inhabit diverse host niches [2], where it encounters and responds to ecological changes, such as temperature change, osmolarity variation, UV/oxidative stress, pH shift, famine as well as the responsive reactions of hosts, before and during survival, invasion and cause diseases in the hosts. In E. tarda EIB202, an array of sigma factor (σ70), alternative sigma factors or extracytoplasmic function (ECF) sigma factors (σ54, −28, −24, −32, −38, −54) as well as anti-sigma factors were identified (Table 5), illuminating its basis to respond to various environmental or host stimuli and drive the expression of related functional genes for cellular fitness.
Table 5. Sigma factors and anti-simga factors in EIB202.
| Type | CDS | Gene | Annotation |
| Sigma factor | ETAE_0454 | rpoD | DNA-directed RNA polymerase, subunit sigma-70 RpoD |
| ETAE_2728 | rpoE | RNA polymerase factor sigma-24 RpoE | |
| ETAE_2126 | rpoF | Flagellar biosynthesis factor sigma-28 FliA | |
| ETAE_3326 | rpoH | RNA polymerase factor sigma-32 RpoH | |
| ETAE_0497 | rpoN | RNA polymerase factor sigma-54 RpoN | |
| ETAE_2873 | rpoS | RNA polymerase factor sigma-38 RpoS | |
| Anti-sigma factor | ETAE_0508 | Putative anti-sigma B factor antagonist | |
| ETAE_0576 | dnaK | Molecular chaperone | |
| ETAE_1223 | flgM | Anti-sigma 28 factor | |
| ETAE_1867 | pspA | Phage shock protein A, suppresses σ54-dependent transcription | |
| ETAE_2684 | Anti-sigma regulatory factor (Ser/Thr protein kinase) |
The organism is well equipped to cope with the first main obstacle, temperature fluctuations, in aquatic ecosystems. Six homologues of cold shock proteins (CspA, −B, C, D, G, H, I), among which two copies of cspC were included, were discerned to represent one of the largest paralogue gene family in EIB202. Closer investigation indicated that the established cold adaptation-related proteins RpoE (ETAE_2728), RseA (ETAE_2727), Rnr (ETAE_0360), DeaD (ETAE_0411), RbfA (ETAT_0406), NusA (ETAE_0404), and PNP (ETAE_0409) were all encoded in the EIB202 genome, consisting a reservoir to cope with the physically extreme cold in the environment, and may help the organism to persist in a previously described dormant state known as viable but not culturable state (VBNC) [24]. In line with the versatility in coping with the cold scenarios, EIB202 genome also has an arsenal of 34 shock proteins (GroEL, GroES, IbpAB, GrpE, etc.) or chaperons for other environmental or host changes (Table 6). One operon, sspAB, as well as another conserved ORF (ETAE_2419) which was known to play essential roles in acid tolerance response, were found in the chromosome, as was the operon pspFABCD, pspE and pspG genes known as encoding phage shock proteins responding to various membrane stimuli other than phage induction.
Table 6. Shock proteins in EIB202.
| Shock protein | Gene | Function |
| ETAE_0008 | ibpA | Molecular chaperone (small heat shock protein) |
| ETAE_0009 | ibpB | Molecular chaperone (small heat shock protein) |
| ETAE_0086 | Putative ATPase with chaperone activity | |
| ETAE_0235 | pspG | Phage shock protein G |
| ETAE_0297 | torD | Chaperone protein |
| ETAE_0313 | groES | Co-chaperonin GroES (HSP10) |
| ETAE_0314 | groEL | Chaperonin GroEL (HSP60 family) |
| ETAE_0576 | dnaK | Molecular chaperone |
| ETAE_0577 | dnaJ | DnaJ-class molecular chaperone with C-terminal Zn finger domain |
| ETAE_0746 | ompH | Periplasmic chaperone |
| ETAE_0867 | escB | Type III secretion system chaperone protein B |
| ETAE_0871 | escA | Type III secretion low calcium response chaperone |
| ETAE_0945 | yegD | Putative chaperone |
| ETAE_1031 | Fimbrial chaperon protein | |
| ETAE_1420 | torD | Cytoplasmic chaperone TorD family protein |
| ETAE_1657 | htpX | Heat shock protein |
| ETAE_1774 | hslJ | Heat shock protein HslJ |
| ETAE_1864 | pspD | Phage shock protein D |
| ETAE_1865 | pspB | Phage shock protein B |
| ETAE_1867 | pspA | Phage shock protein A (IM30), suppresses sigma54-dependent transcription |
| ETAE_1868 | pspF | Phage shock protein operon transcriptional activator |
| ETAE_2048 | hchA | Molecular chaperone |
| ETAE_2146 | fliJ | Flagellar biosynthesis chaperone |
| ETAE_2218 | fimC | Periplasmic chaperone |
| ETAE_2251 | papD | Chaperone protein |
| ETAE_2362 | Hydrogenase 2-specific chaperone | |
| ETAE_2419 | Acid shock protein precursor | |
| ETAE_2466 | Phage tail assembly chaperone gp38 | |
| ETAE_2735 | grpE | Heat shock protein |
| ETAE_2811 | hscA | Chaperone protein |
| ETAE_2812 | hscB | Co-chaperone Hsc20 |
| ETAE_2829 | clpB | Protein disaggregation chaperone |
| ETAE_3271 | Heat shock protein 15 | |
| ETAE_3272 | Disulfide bond chaperones of the HSP33 family |
Regarding to osmotic stress, EIB202 has developed the ability to tolerate high concentrations of sodium chloride (up to 5%) [12]. The genes responsible for the synthesis and uptake of several compatible solutes (osmolytes), such as ectABC for ectoine biosynthesis, bccT and betABI for the transport of betaine, as well as proVWX for the uptake and transport of proline and glycine betaine, which often reside on the genome of halophilic bacteria, are absent in EIB202. In contrast, the caiTABCDC (ETAE_2658–2664) and caiF (ETAE_2672) genes involved in carnitine/betaine uptake and metabolism are present in the genome. Carnitine is a ubiquitous substance in eukaryotes and, in coupling with its metabolic intermediates crotononbetaine and γ-butyrobetaine, may be served as osmoprotectant and stimulate growth in anaerobic and starvation conditions. Spermidine and putrescine uptake system potABCD (ETAE_1881–1886) and biosynthesis related genes (ETAE_2962–2963) (Figure 3) are also present in the genome which may be involved in acid resistance and biofilm formation and can act as a free radical ion scavenger [25]. The lacking of genes encoding osmolarity responding proteins OmpC and OmpF also supports the idea that EIB202 might utilize unusual mechanisms to cope with the osmosis challenges.
Two-component signal transduction system (TCS), comprising of a sensor histidine kinase (HK) protein and a response regulator (RR) protein, is well documented to regulate various biophysical processes as well as virulence in bacteria [26]. EIB202 harbors dozens of TCSs including 31 HK genes and 33 RR genes (including ETAE_1502 as an orphan RR protein) (Table 7). Most of the HK genes reside adjacent to RR genes on the chromosome and are likely to be functional pairs involved in responses to environmental changes. However, the order of these gene pairs (5′-HK/3′-RR or 5′-RR/3′-HK) and the transcriptional direction relative to the chromosome (direct or complementary) appear to be random. In this respect, four particular pairs (i.e., ArcB/ArcA, CheA/CheB/CheY, BarA/UvrY and YehU/YehT) are exceptional in the sense that each corresponding partner resides at a different location of the chromosome, although each pair is known to function together in a certain signaling pathway. In E. tarda, TCSs may mediate adaptive responses to a broad range of environmental stimuli, including phosphate/Mg2+ limitation (Pho), anaerobic condition (Cit and Arc), heavy metal overload (Cus), osmosis change (EnvZ/OmpR), and motility/chemotaxis (Che), etc. It is probably safe to conclude that most two-component regulation is used for enhancing the versatility of the response of the organism to environmental stimuli by the regulation of normally unexpressed genes, while some TCSs, such as the previously described EsrA/EsrB [19] and PhoP/PhoQ [20], may also contribute to the virulence in EIB202.
Table 7. Two component signal transduction system in EIB202.
| Histidine protein kinase (HK) | Response regulator (RR) | HK gene | RR gene | Putative functions |
| ETAE_0228 | ETAE_0229 | citA | citB | Citrate uptake and metabolism |
| ETAE_0302 | ETAE_0300 | torS | torR | Trimethylamine N-oxide respiration |
| ETAE_0393 | ETAE_0394 | basS | basR | Modification of lipopolysaccharide |
| ETAE_0447 | ETAE_0448 | qseC | qseB | Flagellar biogenesis and motility |
| ETAE_0775 | ETAE_0777 | yfhK | yfhA | Unknown function |
| ETAE_0885 | ETAE_0886 | esrA | esrB | Protein secretion and virulence |
| ETAE_1068 | ETAE_1067 | baeS | baeR | Multidrug efflux |
| ETAE_1081 | ETAE_1080 | phoR | phoB | Phosphate limitation |
| ETAE_1112 | ETAE_1111 | narQ | narP | Nitrogen metabolism |
| ETAE_1319 | ETAE_1318 | pgtB | pgtA | Phosphoglycerate transport |
| ETAE_1329 | ETAE_1330 | uhpB | uhpA | Hexose phosphate transport |
| ETAE_1646 | ETAE_1645 | Unknown function | ||
| ETAE_1662 | ETAE_1663 | cusS | cusR | Heavy metal efflux |
| ETAE_1755 | ETAE_1754 | rstB | rstA | Unknown stress |
| ETAE_1912 | ETAE_1913 | Unknown function | ||
| ETAE_2010 | ETAE_2011 | atoS | atoC | Short chain fatty acid metabolism |
| ETAE_2060 | ETAE_2059 | phoQ | phoP | Mg2+ starvation and virulence |
| ETAE_2333/2335 | ETAE_2334 | yojN/rcsC | rcsB | Unknown function |
| ETAE_2603 | ETAE_2604 | kdpD | kdpE | Potassium transport |
| ETAE_2684 | ETAE_2686 | rsbU | Unknown function | |
| ETAE_3010 | ETAE_3009 | lytS | Autolysis regulation | |
| ETAE_3278 | ETAE_3279 | envZ | ompR | Osmosis regulation |
| ETAE_3354 | ETAE_3355 | Unknown function | ||
| ETAE_3397 | ETAE_3398 | uhpB | uhpA | Hexose phosphate uptake |
| ETAE_3454 | ETAE_3453 | cpxA | cpxR | Cell envelop protein folding and degradation |
| ETAE_3494 | ETAE_3495 | ntrB | ntrC | Nitrogen assimilation |
| ETAE_0529 | arcB | Anaerobic respiration | ||
| ETAE_0561 | arcA | |||
| ETAE_0672 | creB | |||
| ETAE_1340 | cheA | Bacterial chemotaxis | ||
| ETAE_1346 | cheB | |||
| ETAE_1347 | cheY | |||
| ETAE_2717 | barA | Carbon storage regulation | ||
| ETAE_2045 | uvrY | |||
| ETAE_3345 | yehU | Unknown function | ||
| ETAE_2838 | yehT | |||
| ETAE_1502 | Unknown function, orphan RR protein |
Quorum sensing (QS) is the signal transduction system that responds to cell density for inter- and intra- species communication under various conditions or stresses [27]. E. tarda EIB202 carries AI-1/AHL dependent EdwI/EdwR (ETAE_2593/2594) system [28], AI-2/LuxS (ETAE_2854) system [29] and a putative AI-3/epinephrine/norepinephrine system which might be sensed by the QseB/QseC (ETAE_0447/0448) TCS system to activate the expression of flagellar operons and virulence related genes, the way as that in E. coli [30]. Also, EIB202 contains dozens of proteins that may be involved in the c-di-GMP mediated signal transduction system, including the effector protein with a PilZ domain (ETAE_3384), and 10 proteins with c-di-GMP biosynthesis related GGDEF or degradation associated EAL domain (Table 8). It is intriguing to detect another 5 proteins carrying both GGDEF and EAL domain (Table 8). These proteins may be involved in the signal transduction networks controlling the “make and break” of the second messenger c-di-GMP, which in turn binds to an unprecedented range of effector components and controls diverse targets necessary for virulence and different bacterial lifestyles in various niches [31]. In all, the abundant repertoire (5% of all the EIB202 CDS) of signal transduction related genes in terms of their numbers and families (Table 3) fundamentally contribute to the survival of EIB202 in various hosts.
Table 8. Proteins involved in the c-di-GMP mediated signal transduction system in EIB202.
| CDS | EAL* | GGDEF* | PilZ* | Gene | Annotation |
| ETAE_0946 | Y | Y | Hypothetical protein | ||
| ETAE_0950 | Y | Y | Diguanylate cyclase/phosphodiesterase | ||
| ETAE_1159 | Y | Y | Hypothetical protein | ||
| ETAE_1561 | Y | Hypothetical protein | |||
| ETAE_1829 | Y | Hypothetical protein | |||
| ETAE_1905 | Y | Diguanylate cyclase | |||
| ETAE_1913 | Y | Response regulator receiver modulated diguanylate cyclase | |||
| ETAE_2054 | Y | Hypothetical protein | |||
| ETAE_2294 | Y | hypothetical protein | |||
| ETAE_2320 | Y | Hypothetical protein | |||
| ETAE_2699 | Y | ycdT | Diguanylate cyclase | ||
| ETAE_2759 | Y | Hypothetical protein | |||
| ETAE_3138 | Y | Y | csrD | Regulatory protein | |
| ETAE_3375 | Y | Hypothetical protein | |||
| ETAE_3380 | Y | Y | Diguanylate cyclase/phosphodiesterase | ||
| ETAE_3384 | Y | bcsA | Cellulose synthase catalytic subunit |
Y indicates presence of EAL, GGDEF or PilZ domain in the listed proteins.
Surface structures and putative virulence factors
Previous studies have determined that E. tarda infects fish via the following three principle entry sites: skin, gill and intestine [6]. A variety of surface structures mediating motility, adherence and pathogen-host recognition seem to be the most important properties for the initiation of infection process in E. tarda. The gene clusters for P pilus (pap genes), type 1 fimbriae (fim genes) as well as several genes for other nonfimbrial adhesins, invasins and hemagglutinins (Table 9) are present in the EIB202 genome, suggesting its ability to bind to specific receptors distributed in its various hosts and therefore defining the site of entry and colonization. Interestingly, dozens of these surface structure related proteins are encoded in the EIB202 GIs such as GI4 (invasion), GI6 (hemagglutinin), GI9 (OPS biosynthesis cluster), GI12 (OPS biosynthesis protein) and GI16 (P pilus related proteins) (Table 2), further suggesting that its surface structures might be shaped by the evolution events to acquire colonization and fitness when approaching various hosts. These observations also underlie the previous descriptions regarding the various mannose-resistant hemagglutination (MRHA) and mannose-sensitive hemagglutination (MSHA) phenotypes as well as serotypes in E. tarda strains [2], [32], [33].
Table 9. Partial of surface structures and virulence related genes in EIB202.
| CDS | Characteristics |
| ETAE_0315 | Hypothetical protein, putative BAP type adhesins |
| ETAE_0323 | Putative invasin, shdA, non-fimbrial adhesin |
| ETAE_0613 | Putative hemolysin secretion ATP-binding protein |
| ETAE_0817 | Filamentous haemagglutinin family outer membrane protein |
| ETAE_0818 | Putative adhesin/hemagglutinin/hemolysin |
| ETAE_0821 | ShlB/FhaC/HecB family haemolysin secretion/activation protein |
| ETAE_0910 | Hemolysin transporter protein |
| ETAE_0911 | Putative hemolysin precursor |
| ETAE_1008 | Hemolysin expression modulating family protein |
| ETAE_1267 | OmpA, outer membrane protein A |
| ETAE_1528 | OmpW, outer membrane protein W |
| ETAE_2089 | Pic serine protease precursor, FhaB filamentous heamagglutinin |
| ETAE_2842 | Putative adhesin |
| ETAE_2937 | Hemolysin III family |
| ETAE_3034 | Putative invasin |
| ETAE_3045 | Temperature sensitive hemagglutinin |
EIB202 was observed to be of non-motile and deficient in flagellar biosynthesis (Figure 4A). A set of early, middle and late flagellar genes displaying high similarities to S. enterica Serovar Typhimurium were found to be scattered present in the EIB202 genome sequence, encoding components required for flagellar hook basal body and hook-filament junction structures [34]. The main dissimilarities between the two organisms seem to lie in the late stage genes for flagella assembly. In EIB202, though two homologues of S. enterica phase-1 flagellin fliC gene were identified (ETAE_2128 and ETAE_2130) (Figure 4B), genes for S. enterica phase-2 flagellin (fljB), flagellin methylase (fliB), flagellin repressor (fljA), and methyl accepting chemotaxis component (aer) were absent in the EIB202 genome, which might account for the incapacity of flagellar biogenesis and weak motility in EIB202. However, the inability of flagellar biogenesis may enhance its invasion capacity by avoiding the pro-inflammatory responses and escaping the recognition by Toll-like receptor 5 [35] and the attack by caspase-1 and interleukin 1β secreted by host cells which recognize the flagellin of the bacteria mounting the host cells [36].
Figure 4. Flagella and flagellin genes of Edwardsiella strains.
A. E. tarda strains WY37 (isolated from turbot), ATCC15947 (isolated from human feces) and ETV (isolated from human) were overnight cultured on LB liquid medium. Cells were collected by centrifugation at 1,000 rpm for 2 min following removing supernatant and then fixed with 2.5% glutaraldehyde. Scale bars represent 1 µm. B. The aligned putative flagellin sequences from E. tarda EIB202 (ETAE_2128, ETAE_2130), E. tarda PPD130/91 (AAN52540), E. ictaluri str. 93-146 (gi|238920295|, gi|238920297|, gi|238920300|) and S. typhimurium LT2 (NP_461698, NP_460912). Protein sequences are typically highly conserved at their C-terminal and N-terminal ends encoding the flagellar filament backbone while the middle region is generally quite variable, representing the surface-exposed and antigenically variable portion of the filament.
Surface-exposed and secreted proteins are of significance for the niche adaptations and pathogenesis of pathogens. There are various secretion pathways generally including type I to VI secretion systems and other specific protein transduction systems to fulfill the functions of protein secretion in Gram negative bacteria [37]. The Sec-dependent transport system, the components of the main terminal branch of the general secretory pathway (GSP), the signal recognition particle (SRP) and the Sec-independent twin arginine transport (Tat), T1SS, TTSS and T6SS were all identified in the genome of E. tarda EIB202 (Figure 3). Tat mediated protein translocation, which allows for the secretion of folded proteins such as redox proteins and their chaperones, has been demonstrated to mediate various stress responses and virulence in pathogenic bacteria, and is prevalent among halophiles and helps in transport of proteins in their folded state [38], [39]. In EIB202, a total of 33 CDSs harboring the Tat specific N-terminus signal peptide were identified to be the potential substrates of the secretion system (Table S3). These proteins, in combination with their co-translocated substrates, might also contribute to its adaptation to high salt conditions (up to 5%) and other environmental stresses. Unlike Vibrio spp. or other pathogens, EIB202 contains just one copy of TTSS and T6SS, and the components and genetic spatial organizations of the two types of secretion systems in EIB202 were of the same with that described in E. tarda strain PPD130/91 [19], [20]. The various secretion systems in EIB202 may be also involved in the transmembrane transduction of fimbrial or non-fimbrial adhesins/invasions/haemagglutinins to enrich the surface structures of the bacterium.
Survival inside the mucus and fish body seems to be attributed to a vast arsenal of proteins active against the fish tissues, epithelial/endothelial cells, fragile cells and vascular system. In spite of previous description as a mean producer of extracellular proteases [32], [33], EIB202 harbors the genes (n = 22) that encode putative extracellular proteins which may take part in the utilization of extracellular matrix such as nucleotides, and phospholipids for survival and fitness (Table 10). The genes ETAE_2918 and ETAE_2529 encoding putative chondroitinases are present in the genome and may be involved in the formation of the chronic “hole-in-the-head” lesions due to cartilage degradation [40]. With the exception of hlyA encoding β-hemolysin, 6 hemolysins and related function genes are found in the genome, possibly accounting for the usually observed red skin or hemorrhage symptoms in affected fish [2]. The gene encoding collagenase (ETAE_0416) may mediate vascular injury and hemorrhage symptoms during infection. Interestingly, EIB202 genome encodes a protein carrying homology to von Willebrand factor type A domain of humans (ETAE_3520, vwA), which is involved in the normal hemostasis by adhering to the subendothelial matrix following vascular damage [41].
Table 10. Putative extracellular proteins involved in matrix utilization in EIB202.
| CDS | Gene | Function | Location |
| ETAE_0025 | uraA | Putative xanthine/uracil permeases | Cytoplasmic/Membrane |
| ETAE_0130 | pldA | Outer membrane phospholipase A | Outer membrane |
| ETAE_0280 | endA | Deoxyribonuclease I | Periplasmic |
| ETAE_0399 | hflB | ATP-dependent metalloprotease | Cytoplasmic/Membrane |
| ETAE_0511 | degS | Serine endoprotease | Periplasmic |
| ETAE_0512 | degQ | Trypsin-like serine proteases | Periplasmic |
| ETAE_0709 | ptrA | Protease III precursor | Periplasmic |
| ETAE_0744 | Putative membrane-associated zinc metalloprotease | Cytoplasmic/Membrane | |
| ETAE_0950 | Diguanylate cyclase/phosphodiesterase | Cytoplasmic/Membrane | |
| ETAE_1034 | ushA | 5′-Nucleotidase/UDP-sugar diphosphatase | Periplasmic |
| ETAE_1078 | sbcC | Nucleotide exonuclease | Extracellular |
| ETAE_1088 | uraA | Xanthine/uracil permeases | Cytoplasmic/Membrane |
| ETAE_1747 | pntB | Pyridine nucleotide transhydrogenase | Cytoplasmic/Membrane |
| ETAE_2596 | pde | 3′,5′-Cyclic-nucleotide phosphodiesterase | Periplasmic |
| ETAE_2704 | tesA | Acyl-CoA thioesterase I | Periplasmic |
| ETAE_2771 | Serine endoprotease | Periplasmic | |
| ETAE_2918 | Hypothetical chondroitinase | Outer membrane | |
| ETAE_3117 | glpQ | Glycerophosphoryl diester phosphodiesterase | Periplasmic |
| ETAE_3314 | glpG | Predicted intramembrane serine protease | Cytoplasmic/Membrane |
| ETAE_3343 | Putative lysophospholipase | Cytoplasmic/Membrane | |
| ETAE_3490 | rbn | Ribonuclease BN | Cytoplasmic/Membrane |
| ETAE_3545 | Xanthine/uracil permeases family protein | Cytoplasmic/Membrane |
E. tarda has developed abilities to utilize hemin, hemoglobin and hematin as iron source as well as siderophore-mediated iron uptake mechanism [2]. The finding of the clustered genes (ETAE_1793-ETAE_1801) which encode a coproporphyrinogen III oxidase (hemN), heme iron utilization protein, hemin receptor and related ABC transporter proteins underlies its capacities to use hemin related iron sources (Figure 3) [2]. It is interesting to discern a gene cluster (ETAE_1968-ETAE_1974) sharing high similarities to the pvsABCDE-psuA-pvuA operon (Figure 3) which encodes the proteins for the synthesis and utilization of vibrioferrin, an unusual type of siderophore requiring nonribosomal peptide synthetase (NRPS) independent synthetases (NIS) and usually mediating the iron uptake systems in V. parahaemolyticus and V. alginolyticu, normal marine flora as well as opportunistic pathogens for sea animals and humans [42], [43]. In EIB202 genome, genes for ferric uptake regulator, ferric reductase, ferrous iron utilization, ferritin protein and TonB systems are also present (Figure 3), implicating an elaborate iron homeostasis system priming for the inhabitation and invasion of various hosts.
Gene properties for intracellular colonization
It has been demonstrated that EIB202 and other virulent isolates of E. tarda are capable of living and persisting inside the phagocytes before leading a systemic infection [44]. In the facultative intracellular pathogens such as Mycobacterium tuberculosis and Salmonella, fatty acids metabolism and the glyoxylate shunt play important roles in their long-term persistence and infection in hosts [45]. In contrast, as above-mentioned, the genes fadD, fadF, fadE and icl, which are required for fatty acids metabolism and the glyoxylate shunt, are absent in the genome sequence of EIB202, indicating that the bacterium might adopt an unusual intracellular persistent strategy to fulfill colonization and infection in various hosts.
In E. tarda, the ability to produce enzymes including catalase, peroxidase and superoxide dismutase (SOD) to detoxify various reactive oxygen species (ROS) has been implicated to be essential for counteracting phagocyte-mediated killing. The EIB202 genome contains genes putatively for a copper-zinc SOD (ETAE_0247) and an iron-cofactored SOD (ETAE_1676), as well as catalases (ETAE_0889 and ETAE_1368), which were believed to be the genetic marker for the E. tarda virulent strains [46]. Moreover, several genes (n = 9) encoding functions for protecting the cells from ROS damages with peroxidase activities (7) or repairing functions (2) are found, intriguingly including a non-haem peroxidase AhpC (ETAE_0956) for alkyl hydroperoxide reductase and a Dyp-type haem-dependent peroxidase (ETAE_1129) (Table 11). These genes confer the organism broader capacities to cope with the oxidative stresses and may necessarily contribute to the abilities to multiply inside the host cells (e.g. macrophage cells), and further the virulence of the bacterium. Actually, when the alternative sigma factor RpoS is deleted to significantly decrease the expression of SOD and catalase, the bacterium shows deficiency in the internalization and colonization of fish cells [47].
Table 11. ROS related proteins in EIB202.
| CDS | gene | Function |
| ETAE_0034 | Putative iron-dependent peroxidase | |
| ETAE_0099 | trxA | Thioredoxin domain-containing protein |
| ETAE_0247 | sodC | Copper-zinc superoxide dismutase |
| ETAE_0889 | katG | Putative catalase/peroxidase |
| ETAE_0956 | ahpC | Alkyl hydroperoxide reductase, small subunit |
| ETAE_1094 | bcp | Thioredoxin-dependent thiol peroxidase |
| ETAE_1129 | Dyp-type peroxidase family | |
| ETAE_1368 | katE | Putative catalase B |
| ETAE_1484 | msrB | Methionine-R-sulfoxide reductase |
| ETAE_1496 | xthA | Exonuclease III |
| ETAE_1676 | sodB | Superoxide dismutase |
| ETAE_1715 | yhjA | Probable cytochrome C peroxidase |
| ETAE_1859 | tpx | Thiol peroxidase |
In E. tarda, the TTSS and T6SS have been demonstrated to be essential for resistance of phagocytic killing and replicating within the cells, thus important for the full virulence of the organism [19], [48], [49]. The TTSS and T6SS have been suggested to be the genetic hallmarks for the differentiation of virulent and avirulent strains of E. tarda [19], [20], [50]. The preservation of intact genomic islands for TTSS and T6SS in EIB202 genome will definitely potentiate it to live an intracellular life after invading hosts. As the circumstances in Salmonella [51], [52], the DnaK/DnaJ chaperone machinery and the type I restriction-modification system in GI22 in EIB202 may also contribute to its invasion and survival within macrophages and avoid perturbations from host immune cells for a cosy intracellular life. Interestingly, EIB202 genome harbors two separated genes, mgtB (ETAE_3346) and mgtC (ETAE_1776). Their Salmonella homologues are located on the selC locus as a mgtCB operon and are required for its survival within macrophages and growth in low Mg2+ environment [53], while the selC locus on EIB202 is flanked by GI24 containing prophage/transposase/integrase genes. These genetic properties provided strong evidence that EIB202 possesses the capacity of invading macrophages and subverting the fish immune systems, maybe in a manner different from that of extensively studied Salmonella. Further experiments are required to unravel their exact roles in the edwardsiellosis pathogenesis.
Conclusions
E. tarda is well established to be one of the leading fish pathogens haunting the aquaculture industries throughout the world, and its association with high value fish species such as turbot has impelled the attempts for vaccine development against this organism. In this study, we have determined the complete genome sequence of EIB202, a highly virulent and multi-drug resistant isolate. The comprehensive analysis of the genome sequence provides evidences that the bacterium harbors an array of antibiotics-resistance determinants and well prepares for the antibiotics cocktail that might be present in the aquaculture ecosystem, similarly to that described in another E. tarda strain TX01 isolated from moribund turbot (Scophthalmus maximus) in Shandong, China [11]. The self-transmissibility of the plasmid pEIB202 further intensifies the concern that the genome contents of E. tarda are partly shaped by its life in various aquatic ecological niches. The findings of stress responding genes as well as signal transduction systems also confirmed the jack of all trade nature of the bacterium which could survive in a variety of hosts and growth conditions, including intracellular niches. Moreover, analysis of the genome sequence also revealed a virulence arsenal in the bacterium, confirming special pathogenic mechanisms of the organism. The determination of genome sequence of the bacterium will undoubtedly facilitate our understanding of this organism and will set the basis for vaccine development using the “reverse genetics” approach.
Materials and Methods
Bacterial growth and DNA extraction
E. tarda EIB202 (previously referred to as isolate EH202 [12] with a CCTCC No. M208068 and available upon request) was recently isolated from diseased turbot (Scophthalmus maximus) in a mariculture farm in Yantai, Shandong province of China and was routinely cultured on Luria-Bertani (LB) medium at 28°C. Genomic DNA was isolated from 10 ml overnight culture using the TIANamp Bacteria DNA Kit (TIANGEN Biotech, Beijing, China). Genomic DNA was quantified on 0.7% agarose gel stained with ethidium bromide and spectrophotometrically assessed. The stock DNA solution was separated into two aliquots, one for sequencing via pyrosequencing and the other stored at −80°C for further gap closing.
High-density pyrosequencing and sequence assembly of the genome
The complete sequencing work was conducted using Roche GS FLX system [54]. A total of 286,550 reads counting up to 64,706,315 bases (averaged read length as 225 bp), were obtained resulting in a 17-fold coverage of the genome. Assembly was performed using the GS de novo Assembler software (http://www.454.com/) and produced 64 contigs ranging from 500 bp to 337,284 bp (the N50 contig size is 116,367 bp). Relationship of the contigs was determined by multiplex PCR [55]. Gaps were then filled in by sequencing the PCR products using ABI 3730xl capillary sequencers. Phred, Phrap and Consed software packages (http://www.genome.washington.edu) were used for the final assembly and edition, and low quality regions of the genome were resequenced. The assembly of the genome was verified by digestion of the genomic DNA with restriction enzymes and then running the products with pulsed-field gel electrophoresis (PFGE).
Sequence analysis and annotation
Putative CDSs were identified by GeneMark [56] and Glimmer3 [57], and peptides shorter than 30 aa were eliminated. Sequences from the intergenic regions were compared to GenBank's non-redundant (nr) protein database [58] to identify genes missed by the Glimmer or GeneMark prediction and to detect pseudogenes. Insert sequences were first detected using IS Finder database (http://www-is.biotoul.fr/is.html) with default parameters and selected manually. Transfer RNA genes were predicted by tRNAScan-SE [59], while ribosomal DNAs (rDNAs) and other RNA genes were identified by comparing the genome sequence to the rRNA database [60], [61] and by using Infernal program [62]. Functional annotation of CDSs was performed through searching against nr protein database using BLASTP [63]. The protein set was also searched against COG (http://www.ncbi.nlm.nih.gov/COG/; [64]) and the KEGG (Kyoto encyclopedia of genes and genomes; http://www.genome.jp/kegg/) [65] for further function assignment. The criteria used to assign function to a CDS were (1) a minimum cutoff of 40% identity and 60% coverage of the protein length and (2) at least two best hits among the COG, KEGG, or nr protein database. A search for gene families in the genome was performed by BLASTCLUST. Subcellular localization of the proteins was predicted by PSORTb program (v2.0.1) [66]. TatP 1.0 server (v2.0) [67] and TATFIND 1.2 program [39] were used to detect the potential substrates of the Tat secretion system. Pathogenicity islands and anomalous genes were detected by PAI-IDA [68] and SIGI-HMM [69], respectively.
Construction of phylogenetic tree
Phylogenetic position of E. tarda EIB202 within the Enterobacteriaceae was determined based on the protein sequences of 44 housekeeping genes (adk, aroC, dnaA, dnaK, frr, fusA, gapA, gyrA, gryB, infC, nusA, pgk, phoB, phoR, pyrG, recC, rplA, rplB, rplC, rplD, rplE, rplF, rplK, rplL, rplM, rplN, rplP, rplS, rplT, rpmA, rpoA, rpoB, rpoC, rpoE, rpsB, rpsC, rpsE, rpsI, rpsJ, rpsK, rpsM, rpsS, smpB, and tsf) [70]. BLAST algorithm was used when needed and ambiguous regions were trimmed according to an embedded mask. Concatenated protein sequences were aligned by ClustalW [71]. Maximum likelihood tree based on the aligned protein sequences was constructed by using PhyML [72] with 100 bootstrap iterations.
Genome comparison
Orthologs between E. tarda EIB202 and other Enterobacteriaceae bacteria (Escherichia coli K-12 substr MG1655, Erwinia carotovora atrosepticum SCRI1043, Klebsiella pneumoniae subsp. pneumoniae MGH 78578, Salmonella typhimurium LT2, Serratia proteamaculans 568, Shigella flexneri 5 str. 8401, Yersinia pestis CO92, Enterobacter sakazakii ATCC BAA-894 and Photorhabdus luminescens subsp. laumondii TTO1) were detected by all-vs-all reciprocal_BLASTP search against the protein sets of these strains (http://www.ncbi.nlm.nih.gov/RefSeq), respectively. Criteria were as following: (1) E-value = e−20 or less and (2) >40% amino acid sequence identity, then the best hit was selected. Predicted E. tarda EIB202-specific genes were detected by screening EIB202 protein set against orthologs.
Statistical analysis
The  contingency Chi-square tests were performed to detect the significant differences between the counts of ORFs in each COG category for EIB202 and other Enterobacteriaceae bacterium (http://img.jgi.doe.gov). In the significant difference test,
contingency Chi-square tests were performed to detect the significant differences between the counts of ORFs in each COG category for EIB202 and other Enterobacteriaceae bacterium (http://img.jgi.doe.gov). In the significant difference test,
in which a, b were the observed numbers of ORFs in in each COG category for EIB202 and other Enterobacteriaceae bacterium, and c, d were the counts of the rest of all ORFs in each COG category for EIB202 and other Enterobacteriaceae bacterium, respectively. Significant differences were determined at P<0.05 (critical value  ) [73].
) [73].
Data Availability
The nucleotide sequence of the E. tarda EIB202 chromosome and the plasmid pEIB202 were submitted to the GenBank database under accession numbers CP001135 and CP001136, respectively.
Supporting Information
Genomic features of E. tarda EIB202 and other sequenced enterobacteria
(0.04 MB DOC)
Amino acid biosynthesis genes in E. tarda EIB202
(0.17 MB DOC)
The predicted Tat substrates in EIB202
(0.07 MB DOC)
Acknowledgments
We thank Professor Dr. Xiaohua Zhang, Prof. Dr. Li Sun and Dr. Hui Gong for providing E. tarda strains.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by grants from the National High Technology Research and Development Program of China (2008AA092501), Ministry of Agriculture of China (Nos. nyhyzx07-046 and nycytx-50-G08), Shanghai Science and Technology Development Funds (No. 08QA14024) and Shanghai Leading Academic Discipline Project (No. B505) as well as the National Special Fund for State Key Laboratory of Bioreactor Engineering (No. 2060204). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Mohanty BR, Sahoo PK. Edwardsiellosis in fish: a brief review. J Biosci. 2007;32:1331–1344. doi: 10.1007/s12038-007-0143-8. [DOI] [PubMed] [Google Scholar]
- 2.Abbott SL, Janda JM. The genus Edwardsiella. Prokaryotes. 2006;6:72–89. [Google Scholar]
- 3.Bockemuhl J, Pan-Urai R, Burkhardt F. Edwardsiella tarda associated with human disease. Pathol Microbiol (Basel) 1971;37:393–401. [PubMed] [Google Scholar]
- 4.Jarboe HH, Bowser PR, Robinette HR. Pathology associated with a natural Edwardsiella ictaluri infection in channel catfish (Ictalurus punctatus Rafinesque). J Wildl Dis. 1984;20:352–354. doi: 10.7589/0090-3558-20.4.352. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Arias CR. Identification and characterization of an intervening sequence within the 23S ribosomal RNA genes of Edwardsiella ictaluri. Syst Appl Microbiol. 2007;30:93–101. doi: 10.1016/j.syapm.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 6.Ling SHM, Wang XH, Xie L, Lim TM, Leung KY. Use of green fluorescent protein (GFP) to study the invasion pathways of Edwardsiella tarda in in vivo and in vitro fish models. Microbiology. 2000;146:7–19. doi: 10.1099/00221287-146-1-7. [DOI] [PubMed] [Google Scholar]
- 7.Phillips AD, Trabulsi LR, Dougan G, Frankel G. Edwardsiella tarda induces plasma membrane ruffles on infection of HEp-2 cells. FEMS Microbiol Lett. 1998;161:317–323. doi: 10.1111/j.1574-6968.1998.tb12963.x. [DOI] [PubMed] [Google Scholar]
- 8.Srinivasa Rao PS, Yamada Y, Leung KY. A major catalase (KatB) that is required for resistance to H2O2 and phagocyte-mediated killing in Edwardsiella tarda. Microbiology. 2003;149:2635–2644. doi: 10.1099/mic.0.26478-0. [DOI] [PubMed] [Google Scholar]
- 9.Sahoo P, Swain P, Sahoo SK, Mukherjee SC, Sahu AK. Pathology caused by the bacterium Edwarsiella tarda in Anabas testudineus (Bloch). Asian Fish Sci. 2000;13:357–362. [Google Scholar]
- 10.Sommerset I, Krossøy B, Biering E, Frost P. Vaccines for fish in aquaculture. Expert Rev Vaccines. 2005;4:89–101. doi: 10.1586/14760584.4.1.89. [DOI] [PubMed] [Google Scholar]
- 11.Sun K, Wang HL, Zhang M, Xiao ZZ, Sun L. Genetic mechanisms of multi-antimicrobial resistance in a pathogenic Edwardsiella tarda strain. Aquaculture. 2009;289:134–139. [Google Scholar]
- 12.Xiao JF, Wang QY, Liu Q, Wang X, Liu H, et al. Isolation and identification of fish pathogen Edwardsiella tarda from mariculture in China. Aquacult Res. 2009;40:13–17. [Google Scholar]
- 13.Williams ML, Waldbieser GC, Dyer DW, Gillaspy AF, Lawrence ML. Characterization of the rrn operons in the channel catfish pathogen Edwardsiella ictaluri. J Appl Microbiol. 2008;104:1790–1796. doi: 10.1111/j.1365-2672.2007.03704.x. [DOI] [PubMed] [Google Scholar]
- 14.Siddique A, Figurski DH. The active partition gene incC of IncP plasmids is required for stable maintenance in a broad range of hosts. J Bacteriol. 2002;184:1788–1793. doi: 10.1128/JB.184.6.1788-1793.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laine MJ Zhang YP, Metzler MC. IS1237, a repetitive chromosomal element from Clavibacter xyli subsp. cynodontis, is related to insertion sequences from gram-negative and gram-positive bacteria. Plasmid. 1994;32:270–279. doi: 10.1006/plas.1994.1066. [DOI] [PubMed] [Google Scholar]
- 16.Backert S, Meyer TF. Type IV secretion systems and their effectors in bacterial pathogenesis. Curr Opin Microbiol. 2006;9:207–217. doi: 10.1016/j.mib.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 17.Chain PS, Hu P, Malfatti SA, Radnedge L, Larimer F, et al. Complete genome sequence of Yersinia pestis strains Antiqua and Nepal516: evidence of gene reduction in an emerging pathogen. J Bacteriol. 2006;188:4453–4463. doi: 10.1128/JB.00124-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Partridge SR, Hall RM. The IS1111 family members IS4321 and IS5075 have subterminal inverted repeats and target the terminal inverted repeats of Tn21 family transposons. J Bacteriol. 2003;185:6371–6384. doi: 10.1128/JB.185.21.6371-6384.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan YP, Zheng J, Tung SL, Rosenshine I, Leung KY. Role of type III secretion in Edwardsiella tarda virulence. Microbiology. 2005;151:2301–2313. doi: 10.1099/mic.0.28005-0. [DOI] [PubMed] [Google Scholar]
- 20.Rao PS, Yamada Y, Tan YP, Leung KY. Use of proteomics to identify novel virulence determinants that are required for Edwardsiella tarda pathogenesis. Mol Microbiol. 2004;53:573–586. doi: 10.1111/j.1365-2958.2004.04123.x. [DOI] [PubMed] [Google Scholar]
- 21.Newman RM, Salunkhe P, Godzik A, Reed JC. Identification and characterization of a novel bacterial virulence factor that shares homology with mammalian Toll/interleukin-1 receptor family proteins. Infect Immun. 2006;74:594–601. doi: 10.1128/IAI.74.1.594-601.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmad S, Weisburg WG, Jensen RA. Evolution of aromatic amino acid biosynthesis and application to the fine-tuned phylogenetic positioning of enteric bacteria. J Bacteriol. 1990;172:1051–1061. doi: 10.1128/jb.172.2.1051-1061.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du M, Chen J, Zhang X, Li A, Li Y, et al. Retention of virulence in a viable but nonculturable Edwardsiella tarda isolate. Appl Environ Microbiol. 2007;73:1349–1354. doi: 10.1128/AEM.02243-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wortham B, Oliveira M, Patel C. Polyamines in bacteria: pleiotropic effects yet specific mechanisms. Adv Exp Med Biol. 2007;603:106–115. doi: 10.1007/978-0-387-72124-8_9. [DOI] [PubMed] [Google Scholar]
- 26.Hoch JA. Two-component and phosphorelay signal transduction. Curr Opin Microbiol. 2000;3:165–170. doi: 10.1016/s1369-5274(00)00070-9. [DOI] [PubMed] [Google Scholar]
- 27.Waters CM, Bassler BL. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 28.Morohoshi T, Inaba T, Kato N, Kanai K, Ikeda T. Identification of quorum-sensing signal molecules and the LuxRI homologs in fish pathogen Edwardsiella tarda. J Biosci Bioeng. 2004;98:274–281. doi: 10.1016/S1389-1723(04)00281-6. [DOI] [PubMed] [Google Scholar]
- 29.Zhang M, Sun K, Sun L. Regulation of autoinducer 2 production and luxS expression in a pathogenic Edwardsiella tarda strain. Microbiology. 2008;154:2060–2069. doi: 10.1099/mic.0.2008/017343-0. [DOI] [PubMed] [Google Scholar]
- 30.Sperandio V, Torres AG, Kaper JB. Quorum sensing Escherichia coli regulators B and C (QseBC): a novel two-component regulatory system involved in the regulation of flagella and motility by quorum sensing in E. coli. Mol Microbiol. 2002;43:809–821. doi: 10.1046/j.1365-2958.2002.02803.x. [DOI] [PubMed] [Google Scholar]
- 31.Christen B, Christen M, Paul R, Schmid F, Folcher M, et al. Allosteric control of cyclic di-GMP signaling. J Biol Chem. 2006;281:32015–32024. doi: 10.1074/jbc.M603589200. [DOI] [PubMed] [Google Scholar]
- 32.Janda JM, Abbott SL, Kroske-Bystrom S, Cheung WK, Powers C, et al. Pathogenic properties of Edwardsiella species. J Clin Microbiol. 1991;29:1997–2001. doi: 10.1128/jcm.29.9.1997-2001.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong JD, Miller MA, Janda JM. Surface properties and ultrastructure of Edwardsiella species. J Clin Microbiol. 1989;27:1797–1801. doi: 10.1128/jcm.27.8.1797-1801.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chilcott GS, Hughes KT. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica Serovar Typhimurium and Escherichia coli. Microbiol Mol Biol Rev. 2000;64:694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith KD, Andersen-Nissen E, Hayashi F, Strobe K, Bergman MA, et al. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat Immunol. 2003;4:1247–1253. doi: 10.1038/ni1011. [DOI] [PubMed] [Google Scholar]
- 36.Miao EA, Alpuche-Aranda CM, Dors M, Clark AE, Bader MW, et al. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1β via Ipaf. Nat Immunol. 2006;7:569–575. doi: 10.1038/ni1344. [DOI] [PubMed] [Google Scholar]
- 37.Gerlach RG, Hensel M. Protein secretion systems and adhesins: the molecular armory of Gram-negative pathogens. Int J Med Microbiol. 2007;297:401–415. doi: 10.1016/j.ijmm.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 38.Ochsner UA, Snyder A, Vasil AI, Vasil ML. Effects of the twin-arginine translocase on secretion of virulence factors, stress response, and pathogenesis. Proc Natl Acad Sci U S A. 2002;99:8312–8317. doi: 10.1073/pnas.082238299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rose RW, Brüser T, Kissinger JC, Pohlschröder M. Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol Microbiol. 2002;45:943–950. doi: 10.1046/j.1365-2958.2002.03090.x. [DOI] [PubMed] [Google Scholar]
- 40.Thune RL, Fernandez DH, Benoit JL, Kelly-Smith M, Rogge ML, et al. Signature-tagged mutagenesis of Edwardsiella ictaluri identifies virulence-related genes, including a Salmonella pathogenicity island 2 class of type III secretion systems. Appl Environ Microbiol. 2007;73:7934–7946. doi: 10.1128/AEM.01115-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiu C-H, Tang P, Chu C, Hu S, Bao Q, et al. The genome sequence of Salmonella enteric serovar Choleraesuis, a highly invasive and resistant zoonotic pathogen. Nucleic Acids Res. 2005;55:1690–1698. doi: 10.1093/nar/gki297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanabe T, Funahashi T, Nakao H, Miyoshi S-I, Shinoda S, et al. Identification and characterization of genes required for biosynthesis and transport of the siderophore vibrioferrin in Vibrio parahaemolyticus. J Bacteriol. 2003;185:6938–6949. doi: 10.1128/JB.185.23.6938-6949.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang QY, Liu Q, Ma Y, Zhou LY, Zhang YX. Isolation, sequencing and characterization of cluster genes involved in the biosynthesis and utilization of the siderophore of marine fish pathogen Vibrio alginolyticus. Arch Microbiol. 2007;188:433–439. doi: 10.1007/s00203-007-0261-6. [DOI] [PubMed] [Google Scholar]
- 44.Moore RN, Rao PSS, Lim TM, Leung KY. Opsonized virulent Edwardsiella tarda strains are able to adhere to and survive and replicate within fish phagocytes but fail to stimulate reactive oxygen intermediates. Infect Immun. 2001;69:5689–5697. doi: 10.1128/IAI.69.9.5689-5697.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, et al. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J Exp Med. 2003;198:693. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Padros F, Zarza C, Dopazo L, Cuadrado M, Crespo S. Pathology of Edwardsiella tarda infection in turbot, Scophthalmus maximus (L.). J Fish Dis. 2006;29:87–94. doi: 10.1111/j.1365-2761.2006.00685.x. [DOI] [PubMed] [Google Scholar]
- 47.Xiao JF, Wang QY, Liu Q, Xu LL, Wang X, et al. Characterization of Edwardsiella tarda rpoS: effect on serum resistance, chondroitinase activity, biofilm formation, and autoinducer synthetases expression. Appl Microbiol Biotechnol. 2009;83:151–60. doi: 10.1007/s00253-009-1924-9. [DOI] [PubMed] [Google Scholar]
- 48.Okuda J, Arikawa Y, Takeuchi Y, Mahmoud MM, Suzaki E, et al. Intracellular replication of Edwardsiella tarda in murine macrophage is dependent on the type III secretion system and induces an up-regulation of anti-apoptotic NF-κB target genes protecting the macrophage from staurosporine-induced apoptosis. Microbial Pathogenesis. 2006;41:226–240. doi: 10.1016/j.micpath.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 49.Wang X, Wang QY, Xiao JF, Liu Q, Wu HZ, et al. Edwardsiella tarda T6SS component evpP is regulated by esrB and iron, and plays essential roles in the invasion of fish. Fish Shellfish Immunol. 2009;27:469–477. doi: 10.1016/j.fsi.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 50.Sakai T, Matsuyama T, Sano M, Iida T. Identification of novel putative virulence factors, adhesin AIDA and type VI secretion system, in atypical strains of fish pathogenic Edwardsiella tarda by genomic subtractive hybridization. Microbiol Immunol. 2009;53:131–139. doi: 10.1111/j.1348-0421.2009.00108.x. [DOI] [PubMed] [Google Scholar]
- 51.Sakai T, Matsuyama T, Sano MT, Matsui H, Iida T. The DnaK/DnaJ chaperone machinery of Salmonella enterica Serovar Typhimurium is essential for invasion of epithelial cells and survival within macrophages, leading to systemic infection. Infect Immun. 2004;72:1364–1373. doi: 10.1128/IAI.72.3.1364-1373.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Low DA, Weyand NJ, Mahan MJ. Roles of DNA adenine methylation in regulating bacterial gene expression and virulence. Infect Immun. 2001;69:7197–7204. doi: 10.1128/IAI.69.12.7197-7204.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blanc-Potard AB, Groisman EA. The Salmonella selC locus contains a pathogenicity island mediating intramacrophage survival. EMBO J. 1997;16:5376–5385. doi: 10.1093/emboj/16.17.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tettelin H, Radune D, Kasif S, Khouri H, Salzberg SL. Optimized multiplex PCR: efficiently closing a whole-genome shotgun sequencing project. Genomics. 1999;62:500–507. doi: 10.1006/geno.1999.6048. [DOI] [PubMed] [Google Scholar]
- 56.Lukashin AV, Borodovsky M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Delcher AL, Harmon D, Kasif S, White O, Salzberg SL. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 1999;27:4636–4641. doi: 10.1093/nar/27.23.4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protocols. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 59.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wuyts J, Perriere G, van de Peer Y. The European ribosomal RNA database. Nucleic Acids Res. 2004;32:D101–103. doi: 10.1093/nar/gkh065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gardner PP, Daub J, Tate JG, Nawrocki EP, Kolbe DL, et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;37:D136–140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nawrocki EP, Kolbe DL, Eddy SR. Infernal 1.0: inference of RNA alignments. Bioinformatics. 2009;25:1335–1337. doi: 10.1093/bioinformatics/btp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 65.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, et al. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gardy JL, Laird MR, Chen F, Rey S, Walsh CJ, et al. PSORTb v.2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics. 2005;21:617–623. doi: 10.1093/bioinformatics/bti057. [DOI] [PubMed] [Google Scholar]
- 67.Bendtsen J, Nielsen H, Widdick D, Palmer T, Brunak S. Prediction of twin-arginine signal peptides. BMC Bioinformatics. 2005;6:167. doi: 10.1186/1471-2105-6-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tu Q, Ding D. Detecting pathogenicity islands and anomalous gene clusters by iterative discriminant analysis. FEMS Microbiol Lett. 2003;221:269–275. doi: 10.1016/S0378-1097(03)00204-0. [DOI] [PubMed] [Google Scholar]
- 69.Waack S, Keller O, Asper R, Brodag T, Damm C, et al. Score-based prediction of genomic islands in prokaryotic genomes using hidden Markov models. BMC Bioinformatics. 2006;7:142. doi: 10.1186/1471-2105-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu M, Ren Q, Durkin AS, Daugherty SC, Brinkac LM. Life in hot carbon monoxide: the complete genome sequence of Carboxydothermus hydrogenoformans Z-2901. PLoS Genet. 2005;1:e65. doi: 10.1371/journal.pgen.0010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 73.Schneiker S, Perlova O, Kaiser O, Gerth K, Alici A, et al. Complete genome sequence of the Myxobacterium Sorangium cellulosum. Nat Biotechnol. 2007;25:1281–1289. doi: 10.1038/nbt1354. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genomic features of E. tarda EIB202 and other sequenced enterobacteria
(0.04 MB DOC)
Amino acid biosynthesis genes in E. tarda EIB202
(0.17 MB DOC)
The predicted Tat substrates in EIB202
(0.07 MB DOC)
Data Availability Statement
The nucleotide sequence of the E. tarda EIB202 chromosome and the plasmid pEIB202 were submitted to the GenBank database under accession numbers CP001135 and CP001136, respectively.