Synopsis
Objectives
ELB-21 is a pyrrolo[2,1-c][1,4]benzodiazepine dimer with potent anti-staphylococcal activity; it binds covalently to guanine residues on opposing strands of duplex DNA, interfering with regulatory proteins and transcription elongation in a sequence selective manner. Transcriptional and proteomic alterations induced by exposure of Staphylococcus aureus clinical isolate EMRSA-16 to ELB-21 were determined in order to define more precisely the bactericidal mechanism of the drug.
Methods
DNase I footprinting was used to identify high affinity DNA binding sites. Microarrays and gel electrophoresis were used to assess the ELB-21-induced phenotype.
Results
High affinity interstrand binding sites in which guanine residues were separated by four base pairs, and also some intrastrand cross-linking sites of variable length were identified. Exposure of EMRSA-16 to 0.015 mg/L ELB-21 elicited a twofold or greater up-regulation of 168 genes in logarithmic phase and 181 genes in stationary phase; the majority of genes affected were associated with resident prophages φSa2 and φSa3, pathogenicity island SaPI4 and DNA damage repair. ELB-21 induced a marked increase in the number of viable phage particles in culture supernatants. The expression of only a limited number of genes showed more than 50% reduction. Sixteen extracellular and four intracellular proteins were differentially expressed during logarithmic and stationary phases, including RecA, proteins associated with staphylococcal pathogenesis (IsaA, CspA), cell division and wall synthesis.
Conclusions
ELB-21 kills S. aureus by forming multiple interstand and intrastrand DNA cross-links, resulting in induction of the DNA damage response, derepression of resident prophages and modulation of a limited number of genes involved with cell wall synthesis.
Keywords: Staphylococcus aureus, pyrrolobenzodiazepine dimers, DNA cross-linking, DNA damage response, sequence-selective DNA binding
Introduction
Staphylococcus aureus is frequently found as a commensal of the human skin and nares. However, many strains have the capacity to exploit breeches in the integrity of dermal and mucosal barriers to cause a spectrum of potentially life-threatening invasive infections, particularly in the immune deficient host.1 S. aureus is armed with a formidable array of tightly-regulated virulence factors2 and the risk it poses to human health is compounded by an ability to accumulate drug resistance genes in response to antibiotic exposure.3,4 The rapid evolution of resistance, in tandem with economic and regulatory issues, fuels a need for continual development of new treatments for staphylococcal infections, a challenge that is currently not being met in full.5
In this context we are evaluating the antibacterial activity of pyrrolo[2,1-c][1,4]benzodiazepine (PBD) dimers. PBDs are a family of DNA-interactive antibiotics that exert their biological activity through covalent binding to the C2-NH2 of guanine within the minor groove,6 spanning three DNA base pairs with a preference for Pu-G-Pu (Purine-Guanine-Purine; reactive guanine emboldened) and interfering with regulatory proteins and transcription elongation in a sequence-selective manner.7 The potency, binding affinity and sequence specificity of PBDs can be enhanced by linking two PBD units together through their C8/C8′ positions to form dimers that cross-link appropriately separated guanines on opposing DNA strands.8 PBD units tethered through an inert propyldioxy [-O-(CH2)3-O-diether] linker bind predominantly to embedded 5′-Pu-GATC-Py sequences by cross-linking opposite-strand guanines separated by two base pairs, with little or no disruption of DNA secondary structure,8,9 and one such compound10 is currently undergoing Phase 1 clinical evaluation in patients with solid tumours and haematological malignancies. Molecular modelling and electrophoretic assays have indicated that extension of the diether linkage [-O-CH2)5-O-] by two carbons enables the dimer to span an extra base pair and cross-link 5′-Pu-GA(T/A)TC-Py sequences.8 Thus, these molecules have the potential to target either prokaryotic or eukaryotic DNA sequences for therapeutic control of a wide range of conditions, including infectious disease.
We determined the anti-staphylococcal potential of a range of PBD dimers that included symmetric agents spanning two and three base pairs,11 as well as asymmetric molecules spanning up to eleven base pairs,12 and identified the potent symmetric dimer ELB-21 (Figure 1a) as the most cell target-selective compound for evaluation as a potential antibacterial chemotherapeutic agent. ELB-21 was active against a range of Gram-positive bacteria, including methicillin-resistant S. aureus (MRSA) and vancomycin-resistant enterococci (VRE), but ineffective against Gram-negative species, due to the barrier function of the outer membrane.11,13 As will be reported elsewhere, the concentrations required to kill bacteria were lower than those necessary to produce a cytotoxic effect in human tissue culture cell lines. At four times the MIC the compound rapidly killed MRSA and VRE.11 Extensive interstrand cross-linking at multiple sites on the S. aureus EMRSA-16 genome following exposure of bacteria to ELB-21 was demonstrated by probing EcoR1-restricted DNA with mecA and 16S rDNA.13 Four putative oligonucleotide sequences recognised by ELB-21 are found between 182 and 511 times on the EMRSA-16 genome,11 indicating that the compound is likely to affect gene transcription in a selective manner. In order to shed light on the response of S. aureus to ELB-21, we characterised changes in EMRSA-16 global transcription following exposure to subinhibitory concentrations of the PBD dimer and determined the effect of the compound on intracellular and extracellular protein content. The data indicate that ELB-21 invokes a DNA damage response, induces resident S. aureus prophages and inhibits translation of proteins required for cell viability and staphylococcal pathogenesis. In addition, DNase I footprinting methodology identified additional sites of interstrand cross-linking and also provided evidence for covalent intrastrand adducts at specific sites within DNA sequences.
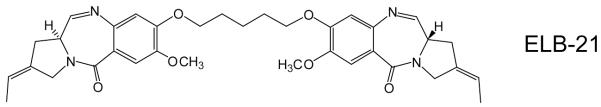
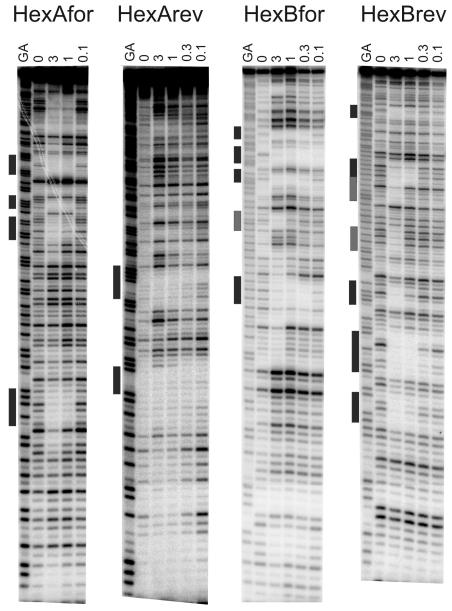
Figure 1.
The pyrrolobenzodiazepine dimer ELB-21: structure (a). DNase I footprinting patterns with the HexA and HexB DNA fragments. The ligand concentration (μM) is shown at the top of the gel lanes. GA indicates a marker lane showing the location of purines; the filled bars show the location of the most pronounced footprints (b). Sequences of HexA and HexB indicating the location of the best binding sites; HexAfor and HexBfor correspond to the labeled upper strands, while HexArev and HexBrev correspond to the lower strands.
Materials and methods
Bacterial strains, culture and reagents
S. aureus strain EMRSA-16 is an epidemic MRSA clinical isolate from the Royal Free Hospital (London, UK) and was provided by Jeremy Hamilton-Miller (University College London). S. aureus 8325-4, a derivative of NCTC 8325 cured of all prophages,14 was used to detect phages and was the kind gift of Friedrich Götz, University of Tübingen, Germany. Bacteria were grown in Mueller-Hinton (MH) broth (Oxoid) or on MH agar plates at 37°C. The PBD dimer ELB-21 was obtained from Spirogen Ltd. and was synthesised as previously described.15 The MIC of ELB-21 against EMRSA-16 was determined by the NCCLS broth microplate assay as previously described.11 In all experiments, ELB-21 was dissolved in DMSO prior to dilution in broth; DMSO was added at the appropriate time point to experimental controls. At the concentrations used, the solvent had no effect on any of the parameters that were measured in this study. Lytic phage particles in bacterial cultures were enumerated using the soft agar overlay method.16
Genome searching
Sequence features of the full genome (2.9 Mb; GC content 32.8%; 2731 genes) of EMRSA-1617 were accessed and visualised with the Artemis genome viewer and annotation tool (http://w.w.w.sanger.ac.uk/Software/Artemis/) developed by the Wellcome Trust Sanger Institute, Hinxton, UK.
DNase I footprinting
Sequence binding preferences of ELB-21 were assessed using the HexA and HexB footprinting substrates (Figure 1b), that between them contain all 64 symmetrical hexanucleotide sequences, as described previously.18 These fragments are each cloned in both orientations as the “forward” and “reverse” sequences so that the regions toward the top of the gel with the forward sequences are located toward the bottom of the reverse sequences. HexAfor and HexArev were obtained by cutting the parent plasmid with HindIII and SacI; HexBfor and HexBrev were obtained by cutting with EcoRI and PstI. These were labeled at the 3′-end of the HindIII and EcoRI sites respectively with [α-32P]dATP using reverse transcriptase. Radiolabeled DNA (1.5 μL; 10 cps/ μL) was mixed with 1.5 μL ELB-21 as described previously.18 After equilibration, the mixture was digested with DNase I and the reaction terminated using formamide – EDTA. The digestion products were resolved on 8% (w/v) denaturing polyacrylamide gels and gels fixed, dried and exposed to a phosphorimager screen (Kodak).
Microarray analysis
The BμG@S SAv1.1.0 microarray used in this study has been described elsewhere19 and contains PCR products representing all predicted open reading frames from the initial seven S. aureus genome sequencing projects. The array design is available in BμG@Sbase (accession number: A-BUGS-17; http://bugs.sgul.ac.uk/A-BUGS-17) and also ArrayExpress (accession number: A-BUGS-17). Flasks containing 50 mL MH broth were inoculated with 500 μL EMRSA-16 overnight culture and incubated at 37 °C with shaking until OD600 reached 0.6 (log phase) or 3.0 (stationary phase). ELB-21 was then added to give a final concentration of 0.015 mg/L (0.5 MIC), cultures incubated for a further 5 min, 15 min, 1 h or 5 h and two volumes of RNAprotect Bacteria reagent (Qiagen) added. Mixtures were incubated for 5 min at RT, centrifuged, RNA isolated from pellets with FastRNA® Pro Blue kit (Qbiogen) and treated with DNase I. RNA aliquots (5 μg) were labeled with Cy5 dye (GE Healthcare) concomitant with reverse transcription into cDNA. EMRSA-16 genomic DNA was used as reference; DNA concentration was measured (OD260) and 1 μg fluorescently labeled with Cy3 dye (GE Healthcare). cDNA was purified using a MinElute kit (Qiagen), probes pooled, hybridised overnight to the S. aureus microarray at 65 °C and subjected to stringent washing.19 Hybridization data was analyzed using an Affymetrix 428 scanner then quantified using Bluefuse for Microarrays 3.5 software (BlueGnome). Data analysis was performed in GeneSpring GX 7.3 (Agilent Technologies) using median-normalized Cy5/Cy3 ratio intensities for three biological replicates. Only genes whose expression ratio showed at least a twofold difference with Benjamini and Hochberg false discovery rate ≤0.05% in the presence of ELB-21 were regarded as being significantly different from the control. Fully annotated microarray data has been deposited in BμG@Sbase (accession number E-BUGS-79 http://bugs.sgul.ac.uk/E-BUGS-79) and also ArrayExpress (accession number E-BUGS-79).
Quantitative real time PCR (qRT-PCR)
RNA samples were treated with the RNase-Free DNase set protocol and RNeasy Minikit protocol (Qiagen). qRT-PCR was carried out using the Brilliant II SYBR® Green qRT-PCR 2-Step kit (Stratagene) according to manufacturer’s instructions. Gene-specific primer pairs (Table 1) were designed for genes of interest to yield amplicons of 100-150 base pairs. The conditions for the PCR reaction were: 95 °C for 15 min, followed by 20-24 cycles of 94 °C for 30 s, 50 °C for 30 s, and 72 °C for 1 min. The comparative threshold method was used to determine relative quantification of mRNA abundance and changes in mRNA expression level were calculated after normalization to the internal control, 16S rRNA. Data shown is the median of three biological replicates; each replicate was performed in duplicate.
Table 1.
Oligonucleotides used for qRT-PCR
| Primers | Sequence |
|---|---|
| ahpC-F | 5′-CAT GGC ATG ACC ATT C-3′ |
| ahpC-R | 5′-GTA CCA CGT TGA GCT A-3′ |
| cspA-F | 5′-GGA TTC GGC TTT ATC G-3′ |
| cspA-R | 5′-CGC CTT CAA CTA CTT C-3′ |
| dnaK-F | 5′-GTA TTC CAG CGG TAC AAG-3′ |
| dnak-R | 5′-CGT CAC CTG TGA TAA CG-3′ |
| fhs-F | 5′-GCT GAT GCA TTC CAT GAG-3′ |
| fhs-R | 5′-CAT AAC CAC CAC CAG TTG-3′ |
| grlB-F | 5′-GTG GTA TGC CAA CAG GTA-3′ |
| grlB-R | 5′-GTG AAG ACC ACC TGA AGT-3′ |
| isaA-F | 5′-CAT GGC TCA ACG TAC TG-3′ |
| isaA-R | 5′-ACC TGA AGC ACC TGA TG-3′ |
| lexA-F | 5′-CAC AGC AGG TGT TCC TA-3′ |
| lexA-R | 5′-TCA TAC TGT CGC CTA CG-3′ |
| mecR1-F | 5′-GCA CCG TTA CTA TCT G-3′ |
| mecR1-R | 5′-CTT GCT CCC GTT CAT T-3′ |
| pgk-F | 5′-GTC CTA GTA CGT GCT GAT-3′ |
| pgk-R | 5′-CCA CCT TGT TCG ATG ATG-3′ |
| recA-F | 5′-CAA CAG TAG CGC TTC AC-3′ |
| recA-R | 5′-CTA CGC CTA ATG CTT GAG-3′ |
| samB-F | 5′-CTG TGG GCA TTG GTT CTA-3′ |
| samB-R | 5′-TCT CAA GGG CTG AAT TGG-3′ |
| sbcC-F | 5′-GGT CTT AGT GGC CTA GAA-3′ |
| sbcC-R | 5′-CTC CTG ATT GCT GCT GTA-3′ |
| sbcD-F | 5′-AGT GGC TCC TTA TTG C-3′ |
| sbcD-R | 5′-AGC GGC TTA AGA GGA A-3′ |
| ssb2-F | 5′-CGT TCA CGA ATG CTC AAG-3′ |
| ssb2-R | 5′-GTA AGC GAC CAT CTA CAC-3′ |
| uvrA-F | 5′-GAG CCA TCA ATT GGA CTG-3′ |
| uvrA-R | 5′-CGC ACG CAT TGT ATA TC-3′ |
| uvrB-F | 5′-TTC GGC GAT GAG ATT G-3′ |
| uvrB-R | 5′-CAC GTG TTA CGA AGT G-3′ |
| 16s rRNA-F | 5′-GAA CCG CAT GGT TCA AAA GT-3′ |
| 16s rRNA -R | 5′-TAT GCA TCG TTG CCT TGG TA -3′ |
Protein extraction
EMRSA-16 was incubated (37 °C) in MH broth with shaking to OD600 0.6 (logarithmic phase) or 3.0 (stationary phase) and ELB-21 added to a final concentration of 0.015 mg/L. Cultures were incubated for a further 1 or 5 h and centrifuged. For preparation of extracellular protein extracts, supernatants were filtered (22 μm) and supernatant proteins concentrated using Vivaspin 20 columns (Sartorius). For preparation of cell protein extracts, bacteria were washed twice with ice-cold TE sample buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0), and suspended in ice-cold base sample buffer (7 M urea, 2 M thiourea, 4% (w/v) CHAPS, Roche protease cocktail tablet). The cells were disrupted by homogenisation with glass beads. Lysates were centrifuged to remove cellular debris and insoluble proteins. Protein concentration was determined by Bradford assay and protein samples prepared for isoelectric focusing (IEF) using a 2D clean-up kit (Bio-Rad).
Protein gel electrophoresis
2D gel electrophoresis was performed using a Protean IEF Cell (BioRad). Proteins were separated using 17 cm immobilized pH (range 4-7) gradient strips (BioRad). Voltage was increased by gradient from 250 to 10 000 V for 2 h with a final phase of 10 000 V for 80 000 Vh. Strips were equilibrated in DTT and iodoacetamide. For identification of proteins by MALDI-TOF-MS, 150 μg of cellular and 250 μg of extracellular proteins were separated using preparative 2D gels. SDS-PAGE was performed with 12% (w/v) polyacrylamide gels and proteins stained with Coomassie Brilliant Blue R-250. Gels were scanned with a Molecular Imager Fx (BioRad), and processed by PDQuest Advanced 2D-image analysis software (BioRad). At least three biological replicates were performed for each experiment and only proteins showing at least a twofold change (p ≤0.05%) following exposure to ELB-21 were considered to be modulated by the agent.
Protein identification
Proteins were identified by MALDI-TOF-MS; Coomassie stained protein spots were cut from gels using an EXQuest spot cutter (BioRad) and transferred into a 96-well microtitre plate. Gel pieces were destained, digested and peptides extracted using a MassPrep robotic liquid handling system (Waters/BioRad Proteome Works). Peptide mixtures were desalted using C18 loaded zip-tips (Millipore) and 2 μL spotted onto sample wells of a stainless steel MALDI target plate that had been previously spotted with 1 μL of matrix solution comprising 1 mg alpha cyano-4-hydroxycinnaminic acid mL (Sigma) in 50% acetonitrile, 50% (v/v) ethanol and an internal standard, adenocorticotrophic hormone (Sigma), at a final concentration of approximately 100 fmol/μL in 0.1% (v/v) formic acid. Samples were analyzed using a Waters MALDI-TOF-MS spectrometer operating at a resolution of >10000 FWHM (Full Width at Half Maximum) in reflectron mode. Spectra (λ337) were acquired at 5 Hz using a nitrogen laser. Ten data collection events were combined to generate each spectrum. Data acquisition was achieved by random sampling from the target well. Peak lists were entered into a MASCOT PMF database search engine; search parameters included a peptide mass accuracy tolerance of 0.2 Da and searches allowed for modifications such as alkylation of cysteine during the tryptic digest procedure and the formation of methionine sulfoxide. A small minority of low-abundance proteins was identified by MALDI-TOF-MS/MS. All experiments were performed in triplicate.
Assays for cell wall integrity
Susceptibility to lysostaphin (Sigma) and induction of autolysis by Triton X-100 (Sigma) were undertaken as described earlier.20 K+ efflux was measured by atomic absorption spectroscopy using a Perkin Elmer ( Waltham, MA) AAnalyst 100 instrument set to Flame Emission Program at a wavelength of 766.5 nm according to Suller and Russell.21
Results
Sequence-selective binding to duplex DNA
Predictions22 that PBD dimers containing a three carbon (n=3) linker would recognize a six base pair sequence embedded in duplex DNA, in which reactive guanines on opposite strands are separated by two base pairs preferentially comprising adenine and thymine, have been confirmed by cross-linking experiments utilizing short (10-13 base pair) oligonucleotides as binding targets.8 These studies also indicated that extension of the linker to n=5 results in optimal cross-linking over a longer sequence, with one further base pair introduced between the two spatially separated and cross-linked guanines. DNase I footprinting confirmed that the PBD dimer SJG-136, containing an n=3 linker, formed high-affinity adducts with 5′-G-GATC-C-3′ sequences.9 In addition, a number of less-favored cross-link sites represented by the 5′-GXXC-3′ motif were also identified and some evidence was found for formation of the mono-adducts, where only one half of the molecule interacted in covalent fashion, with the second PBD residue unreacted. ELB-21 incorporates an extended n=5 diether linkage and is known to bind to 5′-GATTC-3′ and 5′-GAATC-3′ sequences.9,11,13 As ELB-21 is based on a PBD monomeric unit chemically distinct from that in SJG-136 and is significantly more potent than SJG-136, we searched for additional binding sites for ELB-21 using HexA/B footprinting substrates that contain all possible symmetrical hexanucleotide sequences.18
Representative DNase I footprinting gels showing the interaction of ELB-21 with the HexA/B fragments are presented in Figure 1b. Three high affinity hexanucleotide binding sites, 5′-GAATTC-3′, 5′-GTTAAC-3′, 5′-GTATAC-3′, were identified and Artemis searching indicated that these sequences, flanked by a Pu at the 5′ and a Py at the 3′ end, occur 1350, 1574 and 1138 times respectively in the EMRSA-16 genome. Unexpectedly, a number of intrastrand binding sites 5′-GAATTCG-3′ (appearing 254 times), 5′-GACCG-3′ (1080 times), 5′-GTTAACCG-3′ (28 times), 5′-GTACTAG-3′ (148 times), 5′-GCCG-3′ (3968 times), 5′-GCAAG-3′ (3683 times), 5′-GCAATTG-3′ (1056 times), 5′-GATG-3′ (26 995 times) and 5′-GCATATG-3′ (548 times) were identified, indicating a greater number of binding sites than heretofore recognized, due to the capacity of the PBD dimer to span sequences of variable length between reactive guanines.
Effect on S. aureus transcriptome
The capacity of ELB-21 to modulate gene expression in EMRSA-16 was elucidated by comparing the transcriptional profile of logarithmic and stationary phase cultures with those exposed to subinhibitory concentrations of the compound. Logarithmic phase cultures were exposed to 0.015 mg/L ELB-21 (0.5 MIC) for 5, 15 or 60 min and stationary cultures were exposed for 1 or 5 h. ELB-21 induced a marked effect on S. aureus gene expression only after logarithmic and stationary phase cells were exposed to the PBD dimer for 1 h and 5 h respectively (data not shown). The differences between the response time of logarithmic and stationary phase cells is consistent with covalent binding of ELB-21 to the genome; subsequent inhibition of transcription and replication occurs more rapidly in actively dividing cells.
There was a significant twofold or greater change in the expression of 184 genes following exposure of logarithmic phase cells to ELB-21 (0.015 mg/L) for 1 h. Only 16 genes showed more than a 50% reduction in expression, with the majority of genes (168) showing a twofold or greater increase in transcription. Nearly half of the genes affected by ELB-21 were induced prophage genes associated with the resident S. aureus prophages φSa2 and φSa3; all sequenced S. aureus strains, with the exception of COL, carry prophage φSa3, and φSa2 is known to be integrated into EMRSA-16 isolates.23 ELB-21 induced phage release from EMRSA-16. In logarithmic phase cultures containing no drug, 3606 ± 300.3 (mean ± SD) pfu/mL were detected; in the presence of subinhibitory (0.5 MIC) concentrations, a 5.3-fold increase to 18 963 ± 4691.1 pfu/mL was evident 3 h after addition of ELB-21 to mid-logarithmic cultures. An increase of 7.1-fold to 25773 ± 4687.5 pfu/mL was apparent when inhibitory (2 X MIC) concentrations were used.
Also prominent among ELB-21-activated genes were those involved in DNA damage repair (Table 2), including the positive (recA) and negative (lexA) regulators of the SOS response, genes in the LexA regulon involved in DNA metabolism (uvrA, uvrB) and genes encoding a subunit of topoisomerase IV (grlB) and a nonessential DNA polymerase (samB). A number of genes of unknown function were also modulated by subinhibitory concentrations of ELB-21. Three genes encoded by the pathogenicity island SaPI4, tst, hlb and ssp, were also induced by ELB-21; tst encodes the superantigenic toxic shock syndrome toxin-1, the main contributor to staphylococcal toxic shock syndrome. Only a restricted number of genes, 16 in total, were repressed following exposure of logarithmic phase cells to ELB-21 for 1 h. A high proportion of down-regulated genes encoded membrane proteins (e.g., SAR0182, yukA, SAR0870, opuD2) or proteins required for peptidoglycan synthesis, such as femA and murF (Table 3). These changes had little or no impact on cell wall integrity or metabolism: overnight exposure to ELB-21 (0.5 MIC) increased only minimally, from 0.03 mg/L to 0.0078 mg/L, EMRSA-16 susceptibility to the cell wall hydrolase lysostaphin and ELB-21 had no effect on either Triton X-100-induced autolysis up to 3 h after addition of the drug to logarithmic cultures or K+ efflux within 1 h of ELB-21 addition (data not shown).
Table 2.
S. aureus EMRSA-16 DNA damage response genes activated by ELB-21 exposure as determined by microarray
| Gene |
Ratio Log Phase Treated/ Control |
Ratio Stationary Phase Treated/ Control |
Function |
|---|---|---|---|
| recA | 5.24 | 3.41 | Recombination protein, SOS induction |
| lexA | 2.67 | 2.76 | SOS repressor protein |
| uvrA | 5.62 | 2.56 | Nucleotide excision repair |
| uvrB | 5.92 | 4.11 | Nucleotide excision repair |
| sbcD | 4.74 | 4.27 | Homologous recombination repair |
| sbcC | 6.30 | 5.21 | Homologous recombination repair |
| samB | 54.38 | 23.36 | Lesion-replicating DNA polymerase |
| ssb | 26.92 | 7.33 | Single-strand DNA-binding protein |
| ssb2 | 0.85 | 2.64 | Single-strand DNA-binding protein |
| grlB | 2.04 | NDa | Topoisomerase IV, subunit B |
ND= No Data
Table 3.
Genes repressed by logarithmic phase S. aureus EMRSA-16 exposed to ELB-21 as determined by microarray
| Gene | Expression Ratio Treated/ Control |
Function |
|---|---|---|
| SAR0279 | 0.35 | Conserved hypothetical protein |
| asd | 0.43 | Aspartate semialdehyde dehydrogenase |
| SAR0182 | 0.42 | Putative membrane protein |
| yukA | 0.39 | Putative membrane protein |
| sucC | 0.38 | CoA synthetase protein |
| SAR0870 | 0.41 | ABC transporter ATP-binding protein |
| SAR2558 | 0.41 | Conserved hypothetical protein |
| opuD2 | 0.42 | Glycine betaine transporter 2 |
| SAR0280 | 0.40 | Putative membrane protein |
| SAR1343 | 0.44 | Amino acid permease |
| COLB0262 | 0.41 | Predicted CDS |
| SAR2394 | 0.50 | Putative exported protein |
| SAR1423 | 0.46 | Hypothetical protein |
| femA | 0.49 | Peptidogylcan biosynthesis and methicillin resistance |
| murF | 0.49 | Peptidogylcan biosynthesis |
| SAR2428 | 0.49 | Putative membrane protein |
The exposure of stationary phase cells to ELB-21 (0.015 mg/L) for 5 h resulted in altered expression of 183 genes, a similar number to logarithmic phase cells exposed to the agent for 1 h. Only two genes (trxB, glyS) were down-regulated and 181 genes were up-regulated. Consistent with the logarithmic phase data, the majority of genes affected in stationary phase were also phage-related. Overall, these data suggest a correlation between the exposure of S. aureus to ELB-21 and induction of the DNA damage response, with subsequent derepression of resident prophages.
Microarray data indicating changes in expression of nine key genes in logarithmic phase after exposure to ELB-21 for 1 h were validated using qRT-PCR (Table 4). In the large majority of cases, there was good correlation between the two methods.
Table 4.
Comparison of microarray, proteomic and qRT-PCR data after addition of ELB-21 (0.5 MIC) to cultures of EMRSA-16
| Gene | Microarray | Proteomics | qRT-PCRa |
|---|---|---|---|
| recA | 5.24 | 4.39 | 5.27 |
| lexA | 2.67 | 5.99 | |
| uvrA | 5.62 | 3.88 | |
| uvrB | 5.92 | 9.14 | |
| sbcC | 6.30 | 4.42 | |
| sbcD | 4.74 | 23.73 | |
| samB | 54.38 | 404.57 | |
| ssb2 | 0.85 | 0.68 | |
| grlB | 2.04 | 1.52 | |
| ahpC | 0.48 | 0.82 | |
| isaA | 0.23 | 0.86 | |
| cspA | 0.35 | 0.56 | |
| dnaK | 0.43 | 0.68 | |
| fhs | 0.47 | 0.40 | |
| mecR1 | 0.48 | 0.56 | |
| pgk | 3.19 | 0.38 |
mRNA expression levels were normalized to 16s rRNA
Effect on S. aureus protein secretion
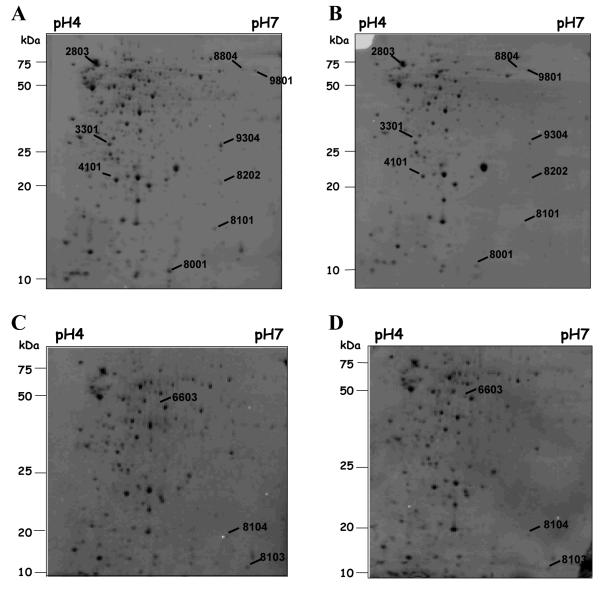
Supernatant proteins from cultures of EMRSA-16 grown in the absence of drug or exposed to ELB-21 at a subinhibitory concentration for 1 h or 5 h were standardised with respect to mass and examined by 2D SDS-PAGE. In total, 12 extracellular proteins were differentially expressed (density increase or decrease at least twofold) by cells exposed to ELB-21, compared to control cells (Figure 2); eight were identified by MALDI-TOF-MS (Table 5). Unidentified spots were low abundance proteins present in insufficient quantities for identification by mass spectrometry. ELB-21 at a concentration of 0.5 MIC enhanced the relative expression of only one protein, phosphoglycerate kinase (Table 5). The remaining seven proteins identified by MS displayed greater than twofold reduction in expression (Table 5). These down-regulated proteins are involved in a variety of key cellular functions, including fatty acid biosynthesis (FabZ), folate biosynthesis (Fhs), β-lactam resistance (MecR1) and cellular stress responses (AhpC, DnaK). Several hypothetical proteins were repressed in the presence of ELB-21, including the protein encoded by locus SAR1457, which is homologous to the Bacillus subtilis protein, DivIVA (Table 5). DivIVA has a critical role in B. subtilis cell division and mutations in DivIVA cause misplacement of the septum during cell division, resulting in irregular shaped bacterial cells,24 and we have previously established11 that ELB-21 alters the morphology of S. aureus cells. These data are consistent with our previous findings that S. aureus cells exposed to ELB-21 show a marked degree of morphological heterogeneity11 and correlate with the observed effect of ELB-21 on the bacterial cell wall and membrane as determined by transcriptional profiling (Table 3).
Figure 2.
Extracellular protein pattern of S. aureus EMRSA-16 grown in MH broth to stationary phase (OD600 ~3.0) and treated or untreated with 0.015 mg/L ELB-21 for 1 or 5 h. Proteins (250 μg) isolated from the supernatant of EMRSA-16 were separated on 2D gels using immobilized pH gradient strips in the range of 4-7. Lettered panels: A and C, controls for 1 and 5 h, respectively; B and D treated with ELB-21 for 1 and 5 h, respectively. The gels were stained with Coomassie Brilliant Blue, scanned and differences in protein abundance detected using PDQuest software. Proteins that differed by at least twofold (p-value >0.5) in amounts between cells grown in the presence and absence of ELB-21 are marked. Each of these spots was excised, treated with trypsin and analyzed by MALDI-TOF-MS. Identified proteins are listed in Table 5.
Table 5.
S. aureus EMRSA-16 extracellular proteins modulated by ELB-21: identification by MALDI-TOF-MS
| Spot No. |
Protein name | Protein function/pathway | Ratio | No. of peptides / Sequence coverage (%) |
Mass/ pI value |
|---|---|---|---|---|---|
| Analysis of extracellular proteins after 1 h exposure | |||||
| 2803 | DnaK | Molecular chaperone/ Stress response | 0.43 | 8/19 | 66.4/4.65 |
| 9801 | Fhs | Formyltetrahydrofolate synthetase/ Folic acid biosynthesis |
0.47 | 9/27 | 60.0/5.69 |
| 9304 | MecR1 | Penicillin-binding protein/ Beta-lactam resistance | 0.48 | 7/27 | 29.2/5.71 |
| 4101 | AhpC | Alkyl hydroperoxide reductase/ Oxidative stress | 0.48 | 8/49 | 20.8/4.88 |
| 3301 | Hypothetical exoprotein |
Function unknown | 0.49 | 5/33 | 24.0/5.09 |
| Analysis of extracellular proteins after 5 h exposure | |||||
| 8104 | Hypothetical protein |
Homologous to the DivIVA protein of Bacillus subtilis |
0.20 | 2/32 | 13.1/5.89 |
| 8103 | FabZ | Beta-hydroxyacyl-acyl carrier protein dehydratase/ Type II fatty-acid biosynthesis |
0.39 | 5/30 | 16.3/5.71 |
| 6603 | PGK | Phosphoglycerate kinase/ Glycolysis, gluconeogenesis |
3.19 | 28/29 | 42.6/5.17 |
Effect on intracellular protein production by S. aureus
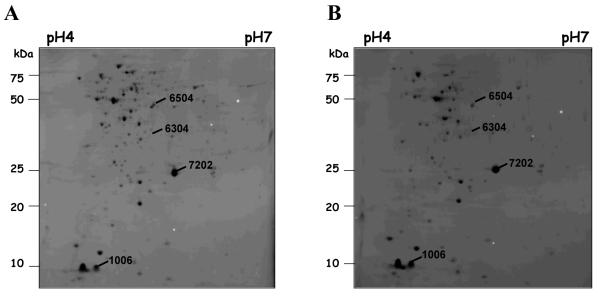
Analysis by 2D SDS-PAGE revealed that only four intracellular proteins were differentially expressed (density ≥2-fold) in log cells exposed to ELB-21 for 1 h, compared to control cells (Figure 3). MALDI-TOF-MS analysis facilitated the identification of three of these proteins (Table 6). Consistent with the microarray data, the RecA protein was up-regulated in ELB-21-exposed cells (Table 5), confirming the induction of the SOS response. The remaining two proteins, IsaA and CspA, were repressed in the ELB-21-treated cells (Table 6) and have been previously identified as immunodominant structures expressed during systemic infection.25 In addition, CspA is required for maximal pigment production in S. aureus; this trait is associated with enhanced staphylococcal pathogenesis and virulence.26 Recently, S. aureus cells lacking the lytic transglycosylase, IsaA have been shown to be mildly attenuated in the mouse septic arthritis model of infection.27 These data suggest that ELB-21 may modulate the pathogenicity of S. aureus at non-lethal concentrations.
Figure 3.
Cellular protein pattern of S. aureus EMRSA-16 grown in MH broth to log phase (OD600 ~0.6) and treated (A) or untreated (B) with 0.015 mg/L ELB-21 for 1 h. Protein extracts (150 μg) were separated on 2D gels using immobilized pH gradient strips in the range of 4-7. The gels were stained with Coomassie Brilliant Blue, scanned and differences in protein abundance detected using PDQuest software. Proteins that differed by at least twofold (p-value >0.5) in amounts between cells grown in the presence and absence of ELB-21 are marked. Each of these spots was excised, trypsinised, and analyzed by MALDI-TOF-MS. Identified proteins are listed in Table 6.
Table 6.
S. aureus EMRSA-16 cellular proteins modulated by ELB-21: identification by MALDI-TOF-MS
| Spot No. |
Protein name |
Protein function/pathway | Ratio | No. of peptides / Sequence coverage (%) |
Mass/ pI value |
|---|---|---|---|---|---|
| 7202 | IsaA | Lytic transglycosylase, major antigen during systemic infection |
0.23 | 4/23 | 24.2/6.11 |
| 1006 | CspA | Cold shock response, major antigen during systemic infection |
0.35 | 2/39 | 7.3/4.52 |
| 6504 | RecA | SOS response/ DNA repair | 4.39 | 13/40 | 37.6/5.13 |
Changes in the expression of genes encoding nine key proteins whose expression was modulated by ELB-21 were evaluated (Table 4). For RecA/recA, there was excellent correlation between qRT-PCR, microarray and proteomic data. The reduction in expression of six proteins (AhpC, IsaA, CspA, DnaK, Fhs, MecR1) appeared to be due to a modest degree of down-regulation of gene expression (Table 4).
Discussion
ELB-21 readily penetrates the staphylococcal cell wall and cytoplasmic membrane; after gaining access to the cytoplasm, the compound is able to efficiently cross-link duplex DNA in a covalent fashion at multiple sites on the chromosome,13 leading to rapid killing at the MIC and above.11 There are several hundred 5′-Pu-GA(T/A)TC-Py primary binding sites for ELB-21 on EMRSA-16 DNA and their distribution on the chromosome indicates that the compound targets sequences within coding regions of both essential and non-essential genes, leading to inhibition of upstream or downstream transcription.13 In addition, binding to sequences located in close proximity to coding regions is likely to affect DNA replication by inhibiting the separation of the two DNA strands and by blocking DNA polymerase activity. In this study we searched for additional ELB-21 binding sites that may contribute to the potent anti-staphylococcal profile of the compound and found that the interaction of the cross-linker with duplex DNA is more extensive than previously envisaged. In addition to three high affinity sites in which guanines on opposing strands are separated by four rather than three base pairs, we identified at least ten sites of intrastrand cross-linking with two to six bases separating same-strand guanines. It is very likely that intrastrand links will contribute further to the interruption of gene transcription and these observations reduce the likelihood that PBD dimers can be chemically engineered to selectively kill bacteria whilst sparing damage to host DNA. Because of the unexpected complexity of the DNA binding profile uncovered in this study, we are now undertaking a detailed kinetic investigation of ELB-21 binding to short (12-13-mer) and long (ca. 200 bp) oligonucleotides using a technique that facilitates structural identification of drug – DNA adducts by HPLC and mass spectrometry.28 Many of the intrastrand sites are common features of the EMRSA-16 genome and the determination of relative ELB-21 DNA binding efficiencies will be of primary importance if we wish to achieve a comprehensive understanding of the relationship between drug – DNA interactions and bactericidal activity.
To investigate the extent of selective inhibition of gene transcription and the degree of modulation of protein expression afforded by exposure to ELB-21, we determined the effect of subinhibitory concentrations using a clinical isolate belonging to the multidrug resistant EMRSA-16 clonal lineage. Exposure to ELB-21 induced 168 ORF transcripts in logarithmic phase cells and 181 in stationary phase cells; the majority could be readily categorized into subsets associated with the SOS response, induction of prophages and selective mobilization of virulence-related factors encoded within pathogenicity island SaPI4. A small number of genes were repressed, some encoding membrane proteins and proteins involved in cell wall synthesis. Induction of SOS-related genes allows the cell to modify transcription in response to environmental stress and is particularly prominent following exposure to agents that cause damage to DNA or interact with proteins involved in DNA replication or in disrupting the integrity of the supercoiled helix during replication and transcription. The SOS response is conserved across a wide of range of bacteria and has been most extensively characterized in Escherichia coli, with more than forty genes under the control of the LexA repressor.29 Response to DNA damage in S. aureus appears to involve a more restricted response. For example, mitomycin C, an antibiotic that causes DNA intrastrand and interstrand cross linking in addition to monofunctional alkyl lesions through binding to the minor groove,30 is a potent inducer of the staphylococcal SOS response: when S. aureus UAMS-1, a methicillin susceptible clinical isolate, was exposed to suprainhibitory concentrations of mitomycin C, a wide range of genes belonging to a variety of functional categories were down-regulated.31 These data are consistent with the general DNA-binding activity of mitomycin C; the more selective binding profile of ELB-21 resulted in a less extensive pattern of inhibition of gene transcription that may reflect fewer binding sites within the EMRSA-16 genome, although it should be noted that in contrast to the study by Anderson et al.31 we employed subinhibitory drug concentrations to ensure a response by viable, metabolically uncompromised bacteria.
A restricted transcriptional response in comparison to mitomycin C was also indicated by the degree of induction of prophage by ELB-21. Lysogenised S. aureus strains are known to release lytic phage with a low frequency and this occurred with the EMRSA-16 isolate used in our study. Addition of ELB-21 to mid-logarithmic phase cultures increased the extent of release of prophages by 5-7-fold. In a study reported by Resch et al.,16 mitomycin C induced a much larger (around 1000-fold) increase in phage release from S. aureus SA113, suggesting a much greater degree of dysregulation of transcriptional control by this agent.
At suprainhibitory concentrations, the topoisomerase IV/gyrase inhibitor ciprofloxacin, known to induce breaks in double-stranded DNA and to promote formation of stalled replication forks, evokes an SOS response that involves the induction of RecA, LexA, and an error-prone DNA polymerase. It has been suggested that this response facilitates bacterial survival and the evolution of drug resistance by alteration of metabolism and the induction of beneficial mutations.32 It has been demonstrated that subinhibitory concentrations of β-lactam antibiotics, agents whose primary mechanism of action does not involve DNA damage, invoke an SOS response similar in nature to that induced by ciprofloxacin and ELB-21.33 In these studies, evocation of the SOS response was accompanied by induction of temperate prophage and we also observed this effect: a large assortment of genes associated with prophages φSa2 and φSa3 were concomitantly induced by ELB-21. Even at the subinhibitory concentrations employed, induction led to the assembly of viable phage particles capable of lysis from within. Phage induction is likely to be SOS dependent, as none was observed in recA-defective lysogenic S. aureus strains.33 The same may hold true for induction of pathogenicity islands, observed in the current and previous studies,32-34 and it has been demonstrated that activation of the expression of temperate phages and pathogenicity islands promotes the spread of staphylococcal virulence genes34 and, potentially, antibiotic resistance genes. Thus, S. aureus may respond to potentially lethal events involving damage to DNA by adopting measures that enhance the dissemination of genes associated with pathogenesis and cell survival through mobilization of elements that are themselves induced by the SOS response. At the drug concentration employed, the expression of only a limited number of genes was down-regulated, some of which were involved in cell wall turnover or associated with membrane-related functions.
There was relatively little modulation of protein expression after exposure to subinhibitory concentrations of ELB-21. Reduced amounts of secreted stress response proteins (DnaK, AhpC), a protein involved in fatty acid biosynthesis (FabZ), a sensor/transducer (MecR1) of penicillin binding protein 2a involved in β-lactam resistance36 and the folate biosynthesis protein formyltetrahydrofolate synthetase (Fhs) were noted and the down-regulation of the encoding genes confirmed by qRT-PCR. Recently, the alkylhydroperoxide reductase protein, AhpC was revealed to have an important role in S. aureus survival and persistence, specifically in nasal colonization.37 Also, antibodies to the heat shock protein, DnaK are present in the sera of patients with S. aureus endocarditis38 and DnaK is known to be induced during early infection of human epithelial cells,39 indicating a putative role for DnaK in S. aureus pathogenesis. Reduced expression of MecR1 raises the possibility that ELB-21 modulates methicillin resistance in MRSA strains and we are currently examining the potential for alteration of β-lactam resistance in EMRSA-16.
Although ELB-21 binds only to a range of specific sequences on duplex DNA, the primary consequence following exposure of EMRSA-16 to this agent appears to be a general DNA damage response that overshadows any selective down-regulation of genes involved in survival or pathogenesis. We continue to explore, in a rational way, the relationships between PBD structure, selectivity of DNA binding and rate and extent of antibacterial activity, to refine these potent molecules and provide a degree of selective toxicity that will facilitate their use as agents for the treatment of life-threatening gram-positive systemic infections.
Aknowledgements
We acknowledge the Bacterial Microarray Group at St George’s for supply of the microarray and the Wellcome Trust for funding the multi-collaborative microbial pathogen microarray facility under its Functional Genomics Resources Initiative. We thank Kevin Bailey of the Queen’s Medical Centre, Nottingham, UK for undertaking protein identification.
Funding This study was supported by Wellcome Trust Project Grant 078669/Z/05/Z.
Footnotes
Transparency declarations None to declare
References
- 1.Gordon RJ, Lowy FD. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis. 2008;46:S350–S359. doi: 10.1086/533591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Molec Microbiol. 2003;48:1429–1449. doi: 10.1046/j.1365-2958.2003.03526.x. [DOI] [PubMed] [Google Scholar]
- 3.Lindsay JA, Holden MTG. Staphylococcus aureus: superbug, super genome? Trends Microbiol. 2004;12:378–385. doi: 10.1016/j.tim.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Grundmann H, Aires-de-Sousa M, Boyce J, et al. Emergence and resurgence of meticillin-resistant Staphylococcus aureus as a public health threat. Lancet. 2006;368:874–885. doi: 10.1016/S0140-6736(06)68853-3. [DOI] [PubMed] [Google Scholar]
- 5.Nathan C, Goldberg FM. The profit problem in antibiotic R&D. Nature Rev Drug Disc. 2005;4:887–891. doi: 10.1038/nrd1878. [DOI] [PubMed] [Google Scholar]
- 6.Thurston DE. Advances in the study of pyrrolo[2,1-c][1,4]-benzodiazepine antitumour antibiotics. In: Waring MJ, Neidle S, editors. Molecular aspects of anticancer drug-DNA interactions. Macmillan; London: 1993. pp. 54–88. [Google Scholar]
- 7.Puvvada MS, Forrow SA, Hartley JA, et al. Inhibition of bacteriophage T7 RNA polymerase in vitro transcription by DNA-binding pyrrolo[2,1-c][1,4]-benzodiazepines. Biochemistry. 1997;36:2478–2484. doi: 10.1021/bi952490r. [DOI] [PubMed] [Google Scholar]
- 8.Smellie M, Bose DS, Thompson AS, et al. Sequence-selective recognition of duplex DNA through covalent interstrand cross-linking: kinetic and molecular modeling studies with pyrrolobenzodiazepine dimers. Biochemistry. 2003;42:8232–8239. doi: 10.1021/bi034313t. [DOI] [PubMed] [Google Scholar]
- 9.Martin C, Ellis T, McGurk CJ, et al. Sequence-selective interaction of the minor-groove interstrand cross-linking agent SJG-136 with naked and cellular DNA: footprinting and enzyme inhibition studies. Biochemistry. 2005;44:4135–4147. doi: 10.1021/bi0479813. [DOI] [PubMed] [Google Scholar]
- 10.Pepper C, Lowe H, Fegan C, et al. Fludarabine-mediated suppression of the excision repair enzyme ERCCI contributes to the cytotoxic synergy with the DNA minor groove crosslinking agent SJG-136 (NSC 694501) in chronic lymphocytic leukaemia cells. Br J Cancer. 2007;97:253–259. doi: 10.1038/sj.bjc.6603853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hadjivassileva T, Thurston DE, Taylor PW. Pyrrolobenzodiazepine dimers: novel sequence-selective, DNA-interactive, cross-linking agents with activity against Gram-positive bacteria. J Antimicrob Chemother. 2005;56:513–518. doi: 10.1093/jac/dki256. [DOI] [PubMed] [Google Scholar]
- 12.Tiberghien AC, Evans DA, Kiakos K, et al. An asymmetric C8/C8′-tripyrrole-linked sequence-selective pyrrolo[2,1-c][1,4]-benzodiazepine (PBD) dimer DNA interstrand cross-linking agent spanning 11 DNA base pairs. Bioorg Med Chem Lett. 2008;18:2073–2077. doi: 10.1016/j.bmcl.2008.01.096. [DOI] [PubMed] [Google Scholar]
- 13.Hadjivassileva T, Stapleton PD, Thurston DE, et al. Interactions of pyrrolobenzodiazepine dimers and duplex DNA from methicillin-resistant Staphylococcus aureus. Int J Antimicrob Agents. 2007;290:672–678. doi: 10.1016/j.ijantimicag.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 14.Novick RP. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology. 1963;33:155–166. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- 15.Gregson SJ, Howard PW, Gullick DR, et al. Linker length modulates DNA cross-linking reactivity and cytotoxic potency of C8/C8′ ether-linked C2-exo-unsaturated pyrrolo[2,1-c][1,4]benzodiazepine (PBD) dimers. J Med Chem. 2004;47:1161–1174. doi: 10.1021/jm030897l. [DOI] [PubMed] [Google Scholar]
- 16.Resch A, Fehrenbacher B, Eisele K, et al. Phage release from biofilm and planktonic Staphylococcus aureus cells. FEMS Microbiol Lett. 2005;252:89–96. doi: 10.1016/j.femsle.2005.08.048. [DOI] [PubMed] [Google Scholar]
- 17.Holden MTG, Feil EJ, Lindsay JA, et al. Complete genomes of two clinical Staphylococcus aureus strains: evidence for the rapid evolution of virulence and drug resistance. Proc Natl Acad Sci USA. 2004;101:9786–9791. doi: 10.1073/pnas.0402521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hampshire AJ, Fox KR. Preferred binding sites for the bifunctional intercalator TANDEM determined using DNA fragments that contain every symmetrical hexanucleotide sequence. Anal Biochem. 2008;374:298–303. doi: 10.1016/j.ab.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 19.Witney AA, Marsden GL, Holden MT, et al. Design, validation, and application of a seven-strain Staphylococcus aureus PCR product microarray for comparative genomics. Appl Environ Microbiol. 2005;71:7504–7514. doi: 10.1128/AEM.71.11.7504-7514.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stapleton PD, Shah S, Ehlert K, et al. The β-lactam-resistance modifier (-)-epicatechin gallate alters the architecture of the cell wall of Staphylococcus aureus. Microbiology. 2007;153:2093–2103. doi: 10.1099/mic.0.2007/007807-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suller MTE, Russell AD. Triclosan and antibiotic resistance in Staphylococcus aureus. J Antimicrob Chemother. 2000;46:11–18. doi: 10.1093/jac/46.1.11. [DOI] [PubMed] [Google Scholar]
- 22.Jenkins TC, Hurley LH, Neidle S, et al. Structure of a covalent DNA-minor groove adduct with a pyrrolobenzodiazepine dimer – evidence for sequence-specific interstrand cross-linking. J Med Chem. 1994;37:4529–4537. doi: 10.1021/jm00052a012. [DOI] [PubMed] [Google Scholar]
- 23.Baba T, Takeuchi F, Kuroda M, et al. Genome and virulence determinants of high virulence community-acquired MRSA. Lancet. 2002;359:1819–1827. doi: 10.1016/s0140-6736(02)08713-5. [DOI] [PubMed] [Google Scholar]
- 24.Cha JH, Stewart GC. The divIVA minicell locus of Bacillus subtilis. J Bacteriol. 1997;179:1671–1683. doi: 10.1128/jb.179.5.1671-1683.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lorenz U, Ohlsen K, Karch H, et al. Human antibody response during sepsis against targets expressed by methicillin resistant Staphylococcus aureus. FEMS Immunol Med Microbiol. 2000;29:145–153. doi: 10.1111/j.1574-695X.2000.tb01517.x. [DOI] [PubMed] [Google Scholar]
- 26.Katzif S, Lee EH, Law AB, et al. CspA regulates pigment production in Staphylococcus aureus through a SigB-dependent mechanism. J Bacteriol. 2005;187:8181–8184. doi: 10.1128/JB.187.23.8181-8184.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stapleton MR, Horsburgh MJ, Hayhurst E, et al. Characterization of IsaA and SceD, two putative lytic transglycosylases of Staphylococcus aureus. J Bacteriol. 2007;189:7316–7325. doi: 10.1128/JB.00734-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Narayanaswarmy M, Griffiths WJ, Howard PW, et al. An assay combining high-performance liquid chromatography and mass spectrometry to measure DNA interstrand cross-linking efficiency in oligonucleotides of varying sequences. Anal Biochem. 2008;374:173–181. doi: 10.1016/j.ab.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 29.Fry RC, Begley TJ, Samson LD. Genome-wide responses to DNA-damaging agents. Ann Rev Microbiol. 2005;59:357–377. doi: 10.1146/annurev.micro.59.031805.133658. [DOI] [PubMed] [Google Scholar]
- 30.Palom Y, Kumar GS, Tang L-Q, et al. Relative toxicities of DNA cross-links and monoadducts: new insights from studies of decarbomoyl mitomycin C and mitomycin C. Chem Res Toxicol. 2002;15:1398–1406. doi: 10.1021/tx020044g. [DOI] [PubMed] [Google Scholar]
- 31.Anderson KL, Roberts C, Disz T, et al. Characterization of the Staphylococcus aureus heat shock, cold shock, stringent, and SOS responses and their effects on log-phase mRNA turnover. J Bacteriol. 2006;188:6739–6756. doi: 10.1128/JB.00609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cirz RT, Jones MB, Gingles NA, et al. Complete and SOS-mediated response of Staphylococcus aureus to the antibiotic ciprofloxacin. J Bacteriol. 2007;189:531–539. doi: 10.1128/JB.01464-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maiques E, Úbeda C, Campoy S, et al. β-lactam antibiotics induce the SOS response and horizontal transfer of virulence factors in Staphylococcus aureus. J Bacteriol. 2006;188:2726–2729. doi: 10.1128/JB.188.7.2726-2729.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Úbeda C, Maiques E, Knecht E, et al. Antibiotic-induced SOS response promotes horizontal dissemination of pathogenicity island-encoded virulence factors in staphylococci. Molec Microbiol. 2005;56:836–844. doi: 10.1111/j.1365-2958.2005.04584.x. [DOI] [PubMed] [Google Scholar]
- 35.Schneider T, Senn MM, Berger-Bächi B, et al. In vitro assembly of a complete, pentaglycine interpeptide bridge containing cell wall precursor (lipid II-Gly5) of Staphylococcus aureus. Molec Microbiol. 2004;53:675–685. doi: 10.1111/j.1365-2958.2004.04149.x. [DOI] [PubMed] [Google Scholar]
- 36.Marrero A, Mallorquí-Fernández G, García-Castellanos TGR, et al. Unbound and acylated structures of the MecR1 extracellular antibiotic-sensor domain provide insights into the signal-transduction system that triggers methicillin-resistance. J Molec Biol. 2006;361:506–521. doi: 10.1016/j.jmb.2006.06.046. [DOI] [PubMed] [Google Scholar]
- 37.Cosgrove K, Coutts G, Jonsson IM, et al. Catalase (KatA) and alkyl hydroperoxide reductase (AhpC) have compensatory roles in peroxide stress resistance and are required for survival, persistence, and nasal colonization in Staphylococcus aureus. J Bacteriol. 2007;189:1025–1035. doi: 10.1128/JB.01524-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qoronfleh MW, Weraarchakul W, Wilkinson BJ. Antibodies to a range of Staphylococcus aureus and Escherichia coli heat shock proteins in sera from patients with S. aureus endocarditis. Infect Immun. 1993;61:1567–1570. doi: 10.1128/iai.61.4.1567-1570.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qoronfleh MW, Bortner CA, Schwartzberg P, et al. Enhanced levels of Staphylococcus aureus stress protein GroEL and DnaK homologs early in infection of human epithelial cells. Infect Immun. 1998;66:3024–3027. doi: 10.1128/iai.66.6.3024-3027.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]