Abstract
Zebra finches are widely used for studying the basic biology of vocal learning. The inability to introduce genetic modifications in these animals has substantially limited studies on the molecular biology of this behavior, however. We used an HIV-based lentivirus to produce germline transgenic zebra finches. The lentivirus encoded the GFP regulated by the human ubiquitin-C promoter [Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D (2002) Science 295:868–872], which is active in a wide variety of cells. The virus was injected into the very early embryo (blastodisc stage) to target the primordial germline cells that later give rise to sperm and eggs. A total of 265 fertile eggs were injected with virus, and 35 hatched (13%); 23 of these potential founders (F0) were bred, and three (13%) produced germline transgenic hatchlings that expressed the GFP protein (F1). Two of these three founders (F0) have produced transgenic young at a rate of 12% and the third at a rate of 6%. Furthermore, two of the F1 generation transgenics have since reproduced, one having five offspring (all GFP positive) and the other four offsping (one GFP positive).
Keywords: lentivirus, song system, zebra finch
Vocal learning is one of the most distinctive characteristics of human behavior. It is not found in other living primates and is rarely encountered in other mammals. It is common in songbirds, however, in which it shares many characteristics with the learning of speech in humans (1, 2). Songbirds are particularly advantageous for research purposes because the neural circuit that mediates the acquisition and production of learned song is anatomically distinct and highly accessible (3–6). Songbirds' research value does not end there; they also provide an excellent model for the study of sensitive periods for learning (7, 8), lateralization of brain function (9, 10), sexual dimorphism (11–16), sensorimotor vocal learning (17–20), the role of sleep in learning (21–24), and the production and replacement of neurons in the adult brain (25, 26).
Various molecular tools for exploring these issues in songbirds are available, including a sequenced cDNA library and microarrays of genes expressed in the zebra finch brain (27–30), the sequence of the zebra finch genome (Songbird Genomics Organization; http://songbirdgenome.org.), and a zebra finch BAC library (31). Other molecular tools not developed specifically for songbirds have been used fruitfully in them, including quantification of immediate early genes to map pathway activation (32–34), laser capture microdissection to compare gene expression between cell types (35), and non–germline-mediated viral manipulation of gene expression to modify behavior (36). However, until now, the ability to manipulate gene expression by using germline transgenic technology has not been available for songbirds. The value of this technology is clearly demonstrated by its use in other model organisms. Here we report the experimental steps that we took to produce transgenic zebra finches.
The zebra finch (Taeniopygia guttata), the most widely studied songbird, is well suited for transgenic development because it breeds year round in captivity and has a relatively short generation time (3 months). Our effort started with attempts to duplicate in zebra finches approaches that have been successful in other species. This involved using a lentiviral vector that was initially used to make transgenic mice (1) and was later shown to be successful in making transgenic chickens (37, 38) and quail (39, 40). Injection of the virus followed a protocol that had produced good results in the quail work. In this protocol, a single injection is made into the subgerminal cavity of embryos from freshly laid eggs (39, 40). At this stage, the quail embryo is a flat disc ≈2.0 mm in diameter comprising thousands of cells. These cells give rise to extra-embryonic and embryonic tissue. The disc also includes the primordial germline cells (PGCs) that later give rise to sperm and oocytes. The PGCs, or their precursors, are the intended targets of the viral infection. The single injections given to quail embryos were efficient; out of the 6 injected embryos that hatched, 5 became germline transgenic founders (39). The success of the technique in quail as well as in chickens (37–39) suggested that it could be readily used to make transgenic zebra finches as well.
The protocol used in quail failed to produce transgenic zebra finches, however. Single injections aimed slightly under the embryonic disc resulted in very sparse, if any, infection of embryonic tissues. The reasons for this were not determined. Instead, we manipulated a number of variables in a search for conditions that might improve infection. Here, we report the conditions that resulted in transgenic founders that produced transgenic offspring.
Results
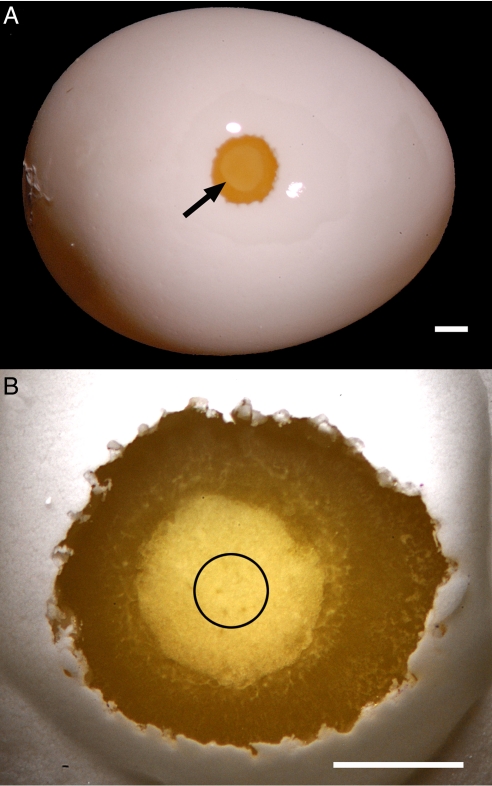
To introduce the transgene, we used a self-inactivating lentiviral vector encoding the GFP regulated by the human ubiquitin-C promoter (1). We made viral injections into embryos (or blastodiscs) of 265 freshly laid zebra finch eggs, of which 35 hatched and 26 were raised to sexual maturity (Table 1); the remaining 11 died soon after hatching. The embryos in the freshly laid eggs were ≈1.0 mm in diameter (Fig. 1) (approximately half the size of the quail embryo) at the time of injection. We successfully produced transgenic individuals under the following conditions:
Instead of making a single injection of virus into the subgerminal cavity below the embryo, as has been described for quail (39), we made multiple injections (10–20) directly into the embryo.
The volume of each injection was ≈15 nL, for a total injected volume of 150–300 nL.
The injections were very shallow and were concentrated in the center of the embryo (Fig. 1B), which is where primordial germ cells are located in chicken (41, 42).
Viral titers ranged from 0.8 to 3.0 × 106 TU/μL, but only titers at the upper end of this range produced transgenics.
Injected eggs were placed in an incubator for 1–3 days and then returned to the nest.
These conditions resulted in numerous embryos that exhibited infection (≈60%) and survived to hatching. In the embryos exhibiting infection, the extent of infection varied considerably, from a few to thousands of cells.
Table 1.
Parameter differences of virally injected eggs
| Viral batch | Group size | Hatched, n (%) | Number of founders (number of offspring) |
|---|---|---|---|
| A | 47 | 10 (21) | 0 |
| B | 77 | 7 (9) | 1 (5) |
| C | 71 | 9 (13) | 2 (4) |
| D | 70 | 9 (13) | 0 |
| Total | 265 | 35 (13) | 3 |
Viral batch A was proximately 1 × 106 TU/μL, batches B and C were 2–3 × 106 TU/μL, and batch D was approximately 0.8 × 106 TU/μL. Group size refers to the number of eggs injected, and founders are those that produced germline transgenic offspring. The number of offspring produced thus far is given in parentheses.
Fig. 1.
(A) View of the zebra finch egg (≈1.5 cm long). A small hole, larger than would normally be made, has been opened to show the embryo of a freshly laid egg (arrow). (Scale bar: 1.0 mm.) (B) A higher-magnification view of an embryo showing the central region in which viral injections were concentrated (black circle). (Scale bar: 1.0 mm.)
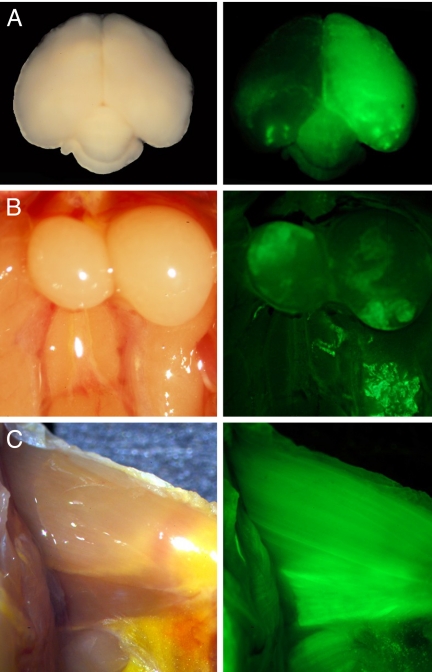
Our initial inspection for GFP expression involved external examination under fluorescence illumination of juveniles at posthatch day 70; the results are presented in Table 2. Our initial inspection did not attempt to quantify the extent of expression, however. The data for these birds suggest that cells giving rise to eye tissue had a higher probability of expressing GFP than cells giving rise to any of the other tissues that we could observe externally. Three of the birds (Blk16, Blk5, and Blk19) were killed and examined in greater detail by opening the body cavity and examining various organs (muscle, gonads, liver, kidney, heart, syrinx, and brain). Blk19, which showed no GFP expression when viewed externally, also lacked GFP expression internally. In Blk16, external examination showed GFP-expressing cells in eye tissue, but internal examination revealed GFP expression in the brain but not in other tissues. Blk5 had a similar external pattern of GFP expression as Blk16, along with expression in the brain, muscles, and testicular tissue (Fig. 2).
Table 2.
Mosaic transgenic birds (age ≈70 days) and tissues that were GFP-positive when visualized under a fluorescence stereomicroscope
| Bird ID | Sex | Legs | Feather base | Breast Muscles | Eyes | Throat | Cloaca |
|---|---|---|---|---|---|---|---|
| Blk14 | F | + | − | − | + | − | − |
| Blk3 | F | (LS) + | + | + | + | − | − |
| Blk11 | M | + | + | − | + | − | − |
| OR682 | F | − | − | − | (RS) + | − | − |
| OR703 | M | − | + | − | + | − | − |
| Blk13 | F | − | − | − | + | − | − |
| Blk15 | F | (LS) + | (LS) + | − | + | − | + |
| *Blk19 | M | − | − | − | − | − | − |
| *Blk5 | M | − | − | − | + | − | − |
| Blk2 | M | + | − | − | + | − | + |
| Blk1 | F | + | − | + | + | − | + |
| Blk12 | F | − | − | + | + | − | − |
| *Blk16 | M | − | − | − | + | − | − |
| OR702 | M | + | + | + | + | − | − |
| OR701 | F | (LS) + | + | − | + | − | − |
| Blk24 | M | + | − | − | + | − | − |
| Blk25 | F | − | − | − | + | − | − |
| Blk26 | F | − | − | − | + | − | − |
| Blk65 | M | − | − | − | + | + | − |
| Blk28 | M | − | (LS) + | − | + | + | − |
| Blk32 | M | + | + | + | + | + | − |
| Blk67 | M | (LS) + | (LS) + | + | + | + | − |
| Blk69 | M | (LS) + | − | − | + | − | − |
| Blk83 | M | + | + | + | + | + | − |
| Blk82 | M | − | − | + | + | − | − |
| Blk47 | F | − | − | − | + | − | − |
“+” indicates the presence of any GFP-positive cells, and “−” indicates that no positive cells were found in that tissue. LS, left side only; RS, right side only. The “Feather base” column refers to the area in and around the base of the primary feathers of the wings, including the papilla. Asterisks identify birds that were not bred. Blk3, OR703, and OR703 were the germline founders.
Fig. 2.
GFP expression in different tissues of Blk5, a mosaic transgenic mouse (FO). Bright field images of tissues are shown in the left column, and the corresponding fluorescent image is presented in the right column. (A) Whole brain as seen from above, looking at the dorsal side of the brain with the caudal end at the bottom of the image. (B) Testes. (C) Muscle tissue of the inner side of the left leg.
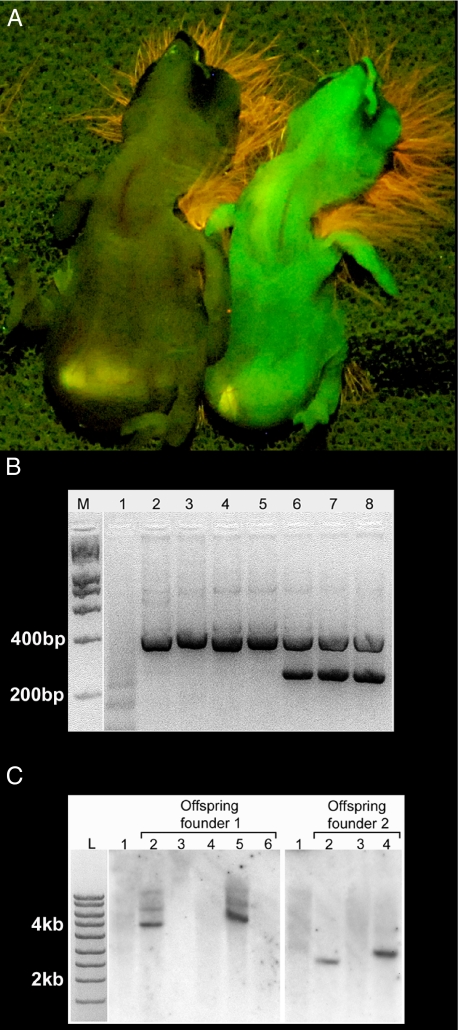
The remaining birds were mated with uninjected partners, and their offspring were screened for expression of the GFP protein. Of 26 potential mosaic founders, 23 were bred, and three (two males and a female) produced respectively, three, five, and one germline transgenic offspring (mean of 12% for two of them and 6% for the third; Fig. 3A). A PCR-based genetic test comparing offspring from one of the founders revealed that all offspring that inherited the transgene from their infected parent also showed expression of the GFP protein under fluorescent illumination (Fig. 3B). Furthermore, Southern blot analysis of two founders and their offspring indicated that a single copy of the transgene was integrated into the genome (Fig. 3C). Although more than one band was observed in the offspring from founder 1 (see lanes 2 and 5), the intensities of the bands differed significantly. Because all bands should be of equal intensity if additional insertions of the transgene occurred, we did not count the upper 2 less-intense bands as additional insertions of the transgene. One possible explanation for the presence of these bands is that they represent incompletely digested genomic fragments. The slight difference in band size between founder 2 offspring (lanes 2 and 4) indicated the possibility of different integration sites of the viral transgene in different infected PGCs, although differences in how the DNA ran in these lanes could be responsible as well. Of the remaining potential founders, 3 have not yet produced eggs, 13 have produced between 10 and 15 eggs each, and 5 have produced 20–30 eggs each, all of which were negative for GFP expression. This shows that 13% of the 23 injected embryos that were bred as adults produced germline transgenic founders (F0). The time from the viral injection to the birth of the first germline transgenic was 5–6 months for two of the transgenic founders and ten months for the third (note: the third bird did not bond and mate until it was months older than the other two founders). The song of the first male germline transgenic (F1) that reached sexual maturity was a good imitation of its tutor (Fig. 4). This male has since bred and produced a single clutch of five babies (F2), all of which were positive for GFP expression by external examination. In addition, a female germline transgenic (F1) has also bred and produced a clutch of four babies (F2), one of which was positive for GFP expression by external examination. Thus, the transgene was passed to a second generation, and the F2 birds continued to express it.
Fig. 3.
(A) Fluorescent illumination of 2 live offspring of a transgenic founder 1 day after hatching. The undersides of the hatchlings are shown; heads are at the top. The soft down on the heads fluoresces orange-red. The hatchling on the left is negative for GFP expression, but its sibling on the right is positive. (B) PCR analysis of genomic DNA from blood. GFP primers (Lower band) test for transgene, and androgen receptor primers (Upper band) test for DNA quality. M,L = Quanti-ladder from Origene. (1) No DNA. (2 and 3) Non–viral-injected adult female and male. (4, 5, 7, and 8) Four siblings from the same clutch (2 negative, 2 positive). All offspring are of the same mosaic founder, which is represented in lane 6. (C) Southern blot analysis of members of 2 clutches of 5 and 3 individuals, each of which was produced by a different F0 individual (represented in lane 1). Note that each clutch has 2 GFP-positive individuals, and that their corresponding lanes show a single dark band, evidence that the transgene is present as a single copy (see Materials and Methods). The GFP-negative individuals lack this band. The absence of a detectable band in the F0 individuals presumably results from the fact that in mosaics, the total number of blood cells that carry the gene is below the detectable limit of Southern blot analysis but not that of PCR analysis.
Fig. 4.
Insertion and expression of the transgene does not inhibit song imitation. Shown are spectrograms of song from the tutor (A) and the germline transgenic male pupil (B) at age 4 months.
Discussion
The lentiviral vector that we used encodes for the GFP protein regulated by the human ubiquitin-C promoter, which is active in a wide range of cells. Mating of the mosaic transgenic animals (F0) resulted in the transgene being inherited and expressed by their offspring (F1) in some cases and, in two cases at least, by the next generation (F2). The 13% efficiency in producing the (F0) founders, although significantly less than that seen in quail (83%) (39) or chickens (50%) (37), was better than that reported in another study on producing transgenic chickens (5%) (38). A mild improvement results if efficiency is calculated based only on those embryos that received injections of virus from batches B and C (13%), from which the three transgenic founders were derived and that had higher viral titers than the other batches. Because the proportion of injected embryos that completed development and hatched was very similar in zebra finches, quail, and chickens, the difference in the ability to produce transgenic zebra finches is likely due to a lower efficiency in the infection of zebra finch PGCs. This interpretation is supported by the need to make multiple injections directly into the zebra finch embryo to get F0 transgenics, whereas a single injection placed below the embryo sufficed in quail and chickens. In addition, the viral titers required to make transgenic quail and chickens (37, 39) were lower than those required to produce transgenic zebra finches. These observations suggest that zebra finch PGCs are not as readily accessible to the virus or as readily infected by the virus, and thus higher viral titers are needed to overcome this. Determining the cause of this relative resistance to infection will make it possible to increase the efficiency with which transgenic zebra finches can be produced. We continue to work on improving the efficiency of our protocol.
Materials and Methods
All animals were cared for in accordance with the standards set by the American Association of Laboratory Animal Care and Rockefeller University's Animal Use and Care Committee.
Lentivirus Production.
The following 3-plasmid system was used to generate lentivirus: transfer vector, pFUGW; packaging vector, Δ8.9; envelope glycoprotein, VSVg (1). Virus production and concentration were performed as described in ref. 43. Titer was determined by making serial dilutions (10-fold steps) of the virus in PBS and infecting HEK 293T cells grown in 96-well plates. After a 72-h incubation, the number of GFP-positive colonies was counted in the well with the most diluted sample of virus that still yielded a minimum of 20 GFP-positive colonies (dilution factor 1 × 105). This number was then used to calculate the titer of virus used for injections.
Injection Apparatus.
Glass pipettes (Drummond Science, catalog no. 5–000-1001) were pulled by using a Narishige model PE-2 pipette puller. Pipettes were polished to a 50- to 60-degree bevel (tip diameter ≈10–20 μm). Approximately 4 μL of viral suspension plus 0.5 μL of phenol red (5% in PBS) were back-loaded into the glass pipette, followed by 1–2 μL of mineral oil, after that a metal plunger supplied with the glass pipettes was inserted. The position of the injecting needle was determined by using a custom-made holder attached to a Narishige UM-3C 3-dimensional manipulator. Movement of the plunger that delivered the injection was controlled by a Narishige MO-10 hydraulic micromanipulator; further details are available on request. This allowed us to position the glass pipette properly and make precise injections of virus into the embryo.
Injection Procedure and Incubation.
An egg candler was used to visualize the embryo through the eggshell. Each egg was placed lengthwise in a silicon mold and oriented so that the embryo was just below the top surface of the egg. A black marker was used to place a dot on the eggshell at this position. With the aid of a stereo zoom microscope (Nikon SMZ800, with an Achromatic 0.5× objective), a 32-gauge needle was used to make many small and shallow holes in the eggshell surface. The holes were made in a circular pattern, creating an approximate 1.0-mm-diameter perforated ring in the shell just above the embryo. Sharp forceps were used to remove the circular piece of shell and expose the embryo. To prevent the embryo from drying, a small amount of albumin from another egg was applied directly to the window opening. [Note: Although not quantified, using standard PBS (pH 7.4) for this part of the protocol seemed to reduce the survival of embryos.] Between 10 and 20 shallow injections of virus (15 nL per injection; 150–300 nL total) were targeted in and around the center of the embryo (Fig. 1B). After the injection, a piece of shell from another egg, slightly larger than the piece removed, was used to patch the hole. Residual albumin around the edges of the patch, when dried, served as a strong adhesive to hold the patch in place. (Note: The egg was allowed to dry at room temperature.) The virally injected eggs were then placed in an incubator for 1–3 days (37–38 °C, 40–50% humidity), after that they were returned to nests to be incubated by birds sitting on eggs of similar age.
GFP Evaluation of Virally Injected Birds.
Examination of GFP expression in different tissues of potential founders (FO) was performed at 70 days. Two different filter sets were used to evaluate GFP expression in tissues, one set to visualize GFP and the other to visualize Texas Red (Chroma Technology 49002 and 49008). False-positives (i.e., non-GFP green spectrum emissions) were readily distinguished from true GFP signals by examining whether the signal also was visible when using the Texas Red filter set. (In most cases, false-positives result from broad-spectrum autofluorescence and thus also appear when viewed with a different spectral filter set, such as that for Texas Red.) The tissues exhibiting the greatest autofluorescence were the intestines, the crop (when full with seed), and the soft down of newly hatched chicks.
Genomic PCR Test for GFP.
Genomic DNA was extracted from blood (≈4 μL), and the presence of the GFP transgene was established using the following GFP-specific primers: forward, GCACGACTTCTTCAAGTCCGCCATGCC; reverse, GCGGATCTTGAAGTTCACCTTGATGCC. The following androgen receptor primers were used as controls: forward, CCTTGTGAGGTGGGAGAGCTTT; reverse, AAGGAGATGCTCAATCCAGGGC. PCR was performed by using an MJ Research PTC200 instrument under the following conditions: 32 cycles of 95 °C for 2 min, followed by 95 °C for 30 s, 67 °C for 1 min, and 68 °C for 1 min. All samples were run on 3–4% Nusieve 3:1 agarose gel.
Southern Blot Analysis of DNA from Blood.
Approximately 75 μL of blood was collected from the wing vein of each bird, and an equal volume of proteinase K buffer [50 mM Tris·HCl (pH 7.6), 0.1 M EDTA (pH 8.0), 0.1 M NaCl, 1% SDS) and 0.5 mg/mL proteinase K was added. The solution was incubated overnight at 50 °C with shaking. A phenol/chloroform extraction was performed, and the genomic DNA was precipitated with ethanol and then resuspended in TE buffer. Then ≈10 μg of DNA was digested with PstI, run on a gel, and transferred to a nylon membrane. The membrane was probed with a 300-bp fragment of the GFP coding sequence amplified using the primers listed above in the presence of radiolabeled p32 dCTP. The membrane was then exposed to radiographic film.
The transgene contains a single PstI restriction site upstream of the GFP coding region. The next downstream PstI site in the zebra finch genome will occur where the transgene integrates itself and will define the size of the band containing the GFP sequence on Southern blot analysis. If more than one transgene is inserted, then the second PstI site likely will be located a different distance from the PstI in the transgene, resulting in a different-sized band on the blot. Thus, the number of bands detected by Southern blot analysis indicates how many copies of the transgene have been integrated in the genome.
Acknowledgments.
We thank the Rockefeller Field Research Center staff for their outstanding care of the birds in this study. This work was funded by McKnight Technological Innovations in Neuroscience Awards, the Whitehall Foundation, and The Rockefeller University.
Footnotes
The authors declare no conflict of interest.
References
- 1.Marler P. Birdsong and speech development: Could there be parallels? Am Sci. 1970;58:669–673. [PubMed] [Google Scholar]
- 2.Doupe AJ, Kuhl PK. Birdsong and human speech: Common themes and mechanisms. Annu Rev Neurosci. 1999;22:567–631. doi: 10.1146/annurev.neuro.22.1.567. [DOI] [PubMed] [Google Scholar]
- 3.Nottebohm F, Stokes TM, Leonard CM. Central control of song in canary, Serinus canarius. J Comp Neurol. 1976;165:457–486. doi: 10.1002/cne.901650405. [DOI] [PubMed] [Google Scholar]
- 4.Bottjer SW, Miesner EA, Arnold AP. Forebrain lesions disrupt development but not maintenance of song in passerine birds. Science. 1984;224:901–903. doi: 10.1126/science.6719123. [DOI] [PubMed] [Google Scholar]
- 5.Bottjer SW, Halsema KA, Brown SA, Miesner EA. Axonal connections of a forebrain nucleus involved with vocal learning in zebra finches. J Comp Neurol. 1989;279:312–326. doi: 10.1002/cne.902790211. [DOI] [PubMed] [Google Scholar]
- 6.Vates GE, Vicario DS, Nottebohm F. Reafferent thalamo-“cortical” loops in the song system of oscine songbirds. J Comp Neurol. 1997;380:275–290. [PubMed] [Google Scholar]
- 7.Eales LA. Song learning in zebra finches: Some effects of song model availability on what is learnt and when. Anim Behav. 1985;33:1293–1300. [Google Scholar]
- 8.Marler P, Tamura M. Culturally transmitted patterns of vocal behavior in sparrows. Science. 1964;146:1483–1486. doi: 10.1126/science.146.3650.1483. [DOI] [PubMed] [Google Scholar]
- 9.Nottebohm F. Neural lateralization of vocal control in a passerine bird, I: Song. J Exp Zool. 1971;177:229–261. doi: 10.1002/jez.1401770210. [DOI] [PubMed] [Google Scholar]
- 10.Nottebohm F. In: Lateralization in the Nervous System. Harnad S, Doty R, Goldstein L, Jaynes J, Krauthamer G, editors. New York: Academic; 1977. pp. 23–44. [Google Scholar]
- 11.Nottebohm F, Arnold AP. Sexual dimorphism in vocal control areas of songbird brain. Science. 1976;194:211–213. doi: 10.1126/science.959852. [DOI] [PubMed] [Google Scholar]
- 12.Gurney ME, Konishi M. Hormone-induced sexual differentiation of brain and behavior in zebra finches. Science. 1980;208:1380–1383. doi: 10.1126/science.208.4450.1380. [DOI] [PubMed] [Google Scholar]
- 13.Ball GF, et al. Seasonal plasticity in the song control system: Multiple brain sites of steroid hormone action and the importance of variation in song behavior. Ann N Y Acad Sci. 2004;1016:586–610. doi: 10.1196/annals.1298.043. [DOI] [PubMed] [Google Scholar]
- 14.Arnold AP, Bottjer SW, Brenowitz EA, Nordeen EJ, Nordeen KW. Sexual dimorphisms in the neural vocal control system in song birds: Ontogeny and phylogeny. Brain Behav Evol. 1986;28:22–31. doi: 10.1159/000118689. [DOI] [PubMed] [Google Scholar]
- 15.Bottjer SW, Johnson F. Circuits, hormones, and learning: Vocal behavior in songbirds. J Neurobiol. 1997;33:602–618. doi: 10.1002/(sici)1097-4695(19971105)33:5<602::aid-neu8>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 16.Wade J, Arnold AP. Sexual differentiation of the zebra finch song system. Ann N Y Acad Sci. 2004;1016:540–559. doi: 10.1196/annals.1298.015. [DOI] [PubMed] [Google Scholar]
- 17.Solis MM, Doupe AJ. Anterior forebrain neurons develop selectivity by an intermediate stage of birdsong learning. J Neurosci. 1997;17:6447–6462. doi: 10.1523/JNEUROSCI.17-16-06447.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konishi M. The role of auditory feedback in the control of vocalization in the white-crowned sparrow. Z Tierpsychol. 1965;22:770–783. [PubMed] [Google Scholar]
- 19.Margoliash D, Konishi M. Auditory representation of autogenous song in the song system of white-crowned sparrows. Proc Natl Acad Sci USA. 1985;82:5997–6000. doi: 10.1073/pnas.82.17.5997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams H, Nottebohm F. Auditory responses in avian vocal motor neurons: A motor theory for song perception in birds. Science. 1985;229:279–282. doi: 10.1126/science.4012321. [DOI] [PubMed] [Google Scholar]
- 21.Dave AS, Margoliash D. Song replay during sleep and computational rules for sensorimotor vocal learning. Science. 2000;290:812–816. doi: 10.1126/science.290.5492.812. [DOI] [PubMed] [Google Scholar]
- 22.Cardin JA, Schmidt MF. Song system auditory responses are stable and highly tuned during sedation, rapidly modulated and unselective during wakefulness, and suppressed by arousal. J Neurophysiol. 2003;90:2884–2899. doi: 10.1152/jn.00391.2003. [DOI] [PubMed] [Google Scholar]
- 23.Deregnaucourt S, Mitra PP, Feher O, Pytte C, Tchernichovski O. How sleep affects the developmental learning of bird song. Nature. 2005;433:710–716. doi: 10.1038/nature03275. [DOI] [PubMed] [Google Scholar]
- 24.Hahnloser RH, Fee MS. Sleep-related spike bursts in HVC are driven by the nucleus interface of the nidopallium. J Neurophysiol. 2007;97:423–435. doi: 10.1152/jn.00547.2006. [DOI] [PubMed] [Google Scholar]
- 25.Goldman SA, Nottebohm F. Neuronal production, migration, and differentiation in a vocal control nucleus of the adult female canary brain. Proc Natl Acad Sci USA. 1983;80:2390–2394. doi: 10.1073/pnas.80.8.2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paton JA, Nottebohm FN. Neurons generated in the adult brain are recruited into functional circuits. Science. 1984;225:1046–1048. doi: 10.1126/science.6474166. [DOI] [PubMed] [Google Scholar]
- 27.Wade J, et al. A cDNA microarray from the telencephalon of juvenile male and female zebra finches. J Neurosci Methods. 2004;138:199–206. doi: 10.1016/j.jneumeth.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 28.Wada K, et al. A molecular neuroethological approach for identifying and characterizing a cascade of behaviorally regulated genes. Proc Natl Acad Sci USA. 2006;103:15212–15217. doi: 10.1073/pnas.0607098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li X, et al. Genomic resources for songbird research and their use in characterizing gene expression during brain development. Proc Natl Acad Sci USA. 2007;104:6834–6839. doi: 10.1073/pnas.0701619104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Replogle K, et al. The Songbird Neurogenomics (SoNG) Initiative: Community-based tools and strategies for study of brain gene function and evolution. BMC Genomics. 2008;9:131. doi: 10.1186/1471-2164-9-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luo M, et al. Utilization of a zebra finch BAC library to determine the structure of an avian androgen receptor genomic region. Genomics. 2006;87:181–190. doi: 10.1016/j.ygeno.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 32.Mello CV, Vicario DS, Clayton DF. Song presentation induces gene expression in the songbird forebrain. Proc Natl Acad Sci USA. 1992;89:6818–6822. doi: 10.1073/pnas.89.15.6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jarvis ED, Nottebohm F. Motor-driven gene expression. Proc Natl Acad Sci USA. 1997;94:4097–4102. doi: 10.1073/pnas.94.8.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimpo RR, Doupe AJ. FOS is induced by singing in distinct neuronal populations in a motor network. Neuron. 1997;18:315–325. doi: 10.1016/s0896-6273(00)80271-8. [DOI] [PubMed] [Google Scholar]
- 35.Lombardino AJ, Li XC, Hertel M, Nottebohm F. Replaceable neurons and neurodegenerative disease share depressed UCHL1 levels. Proc Natl Acad Sci USA. 2005;31:8036–8041. doi: 10.1073/pnas.0503239102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haesler S, et al. Incomplete and inaccurate vocal imitation after knockdown of FoxP2 in songbird basal ganglia nucleus area X. PLoS Biol. 2007;5:e321. doi: 10.1371/journal.pbio.0050321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGrew MJ, et al. Efficient production of germline transgenic chickens using lentiviral vectors. EMBO Rep. 2004;5:728–733. doi: 10.1038/sj.embor.7400171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chapman SC, et al. Ubiquitous GFP expression in transgenic chickens using a lentiviral vector. Development. 2005;132:935–940. doi: 10.1242/dev.01652. [DOI] [PubMed] [Google Scholar]
- 39.Scott BB, Lois C. Generation of tissue-specific transgenic birds with lentiviral vectors. Proc Natl Acad Sci USA. 2005;102:16443–16447. doi: 10.1073/pnas.0508437102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scott BB, Lois C. Generation of transgenic birds with replication-deficient lentiviruses. Nat Protoc. 2006;1:1406–1411. doi: 10.1038/nprot.2006.187. [DOI] [PubMed] [Google Scholar]
- 41.Tsunekawa N, Naito M, Sakai Y, Nishida T, Noce T. Isolation of chicken vasa homolog gene and tracing the origin of primordial germ cells. Development. 2000;127:2741–2750. doi: 10.1242/dev.127.12.2741. [DOI] [PubMed] [Google Scholar]
- 42.Nakamura Y, et al. Migration and proliferation of primordial germ cells in the early chicken embryo. Poultry Sci. 2007;86:2182–2193. doi: 10.1093/ps/86.10.2182. [DOI] [PubMed] [Google Scholar]
- 43.Pease S, Lois C. Mammalian and Avian Transgenesis: New Approaches. New York: Springer; 2006. [Google Scholar]