Abstract
Upon detection of antigen, CD4+ T helper (Th) cells can differentiate into a number of effector types that tailor the immune response to different pathogens. Alternative Th1 and Th2 cell fates are specified by the transcription factors T-bet and GATA-3, respectively. Only a handful of target genes are known for these two factors and because of this, the mechanism through which T-bet and GATA-3 induce differentiation toward alternative cell fates is not fully understood. Here, we provide a genomic map of T-bet and GATA-3 binding in primary human T cells and identify their target genes, most of which are previously unknown. In Th1 cells, T-bet associates with genes of diverse function, including those with roles in transcriptional regulation, chemotaxis and adhesion. GATA-3 occupies genes in both Th1 and Th2 cells and, unexpectedly, shares a large proportion of targets with T-bet. Re-complementation of T-bet alters the expression of these genes in a manner that mirrors their differential expression between Th1 and Th2 lineages. These data show that the choice between Th1 and Th2 lineage commitment is the result of the opposing action of T-bet and GATA-3 at a shared set of target genes and may provide a general paradigm for the interaction of lineage-specifying transcription factors.
Keywords: Genomic map, T helper differentiation, cytokines
The immune response to different pathogens is tailored by the differentiation of CD4+ T helper cell into different effector types (1, 2). Th1 cells are critical for protection against viruses and intracellular pathogens, Th2 cells for the removal of extracellular parasites, and Th17 cells function in the response to extracellular bacteria. T-cell lineage commitment also impacts upon numerous diseases processes, with defects in Th1 and Th17 responses implicated in autoimmunity and Th2 responses in the pathogenesis of allergic disease (3).
Alternative pathways of T-cell differentiation occur through the action of a number of transcription factors. Generation of Th1 cells from naı̄ve precursors requires IL-12 and IFN-γ signaling that leads to activation of STAT4 and STAT1, respectively (1). STAT1 induces expression of T-bet (TBX21) that acts as a key regulator of Th1 cell fate determination (4). T-bet activates Th1 genetic programs whilst simultaneously suppressing Th2 and Th17 programs (5, 6). Loss of T-bet induces default commitment to Th2 and Th17 lineages and T-bet deficient mice have impaired Th1 immunity, are resistant to autoimmune disease and develop spontaneous asthma (7–10).
Generation of Th2 cells requires IL-4, which leads to STAT6 phosphorylation (11) and upregulation of GATA-3, the key regulator of Th2 development (12, 13). Deletion of GATA-3 in peripheral CD4+ T cells prevents differentiation into the Th2 lineage, causing cells to differentiate toward a Th1 phenotype in the absence of polarizing cytokines (14, 15). Conversely, overexpression of GATA-3 in Th1 cells switches their polarity to a Th2 phenotype (13). Interestingly, it has been shown that GATA-3 is also expressed at lower levels in murine Th1 cells and that this expression is necessary for the subsequent induction of a Th2 response via STAT5 (15). We have also reported an interaction between GATA-3 and tyrosine-phosphorylated T-bet in murine thymocytes (16).
Our knowledge of T-bet and GATA-3 function comes mainly from their roles at gene loci that encode IFNG and IL4/IL5/IL13, the signature Th1 and Th2 cytokines, respectively. Th1 differentiation is accompanied by acetylation of histone H3 lysine 9 (H3K9ac) at the IFNG locus and this process is dependent on T-bet (17–23). Conversely, Th2 differentiation is accompanied by hyperacetylation of the IL4/IL5/IL13 locus, dependent on GATA-3 (18, 19, 23–26). Recent murine studies also show that T-bet directly represses the expression of IL4 (23, 27) and that GATA3 directly represses IFNG (22, 28). This suggests that T-bet and GATA-3 may act to promote alternative pathways of T-cell differentiation by acting on the same target genes. However, because only a handful of genes are known to be directly targeted by T-bet and GATA-3 in primary T cells and since Th1 differentiation appears to be only partly dependent on IFN-γ (1), the mechanism by which these two factors direct alternative cell fates remains unclear.
Given the profound influence that T-cell lineage commitment has upon numerous disease processes, it is critical to understand the regulatory mechanisms that contribute to these postdevelopmental cell fate choices in humans. To determine the mechanisms of human T-cell lineage commitment, we have identified the target genes of T-bet and GATA-3 in primary human Th1 and Th2 cells. This work identifies a number of novel pathways with critical relevance to T-cell biology and reveals that master regulator transcription factors can act through a shared set of target genes to control alternative cell fates.
Results
T-bet and GATA-3 target genes in primary human T cells.
To identify genes directly targeted by T-bet and GATA-3 during early human T cell differentiation, we generated Th1 and Th2 cells from primary naïve human T-cells and performed chromatin immunoprecipitation coupled with microarray analysis (ChIP-Chip) (29). We first verified that our Th1 and Th2 cells were appropriately polarized. Human Th1 cells expressed relatively high amounts of T-bet and IFN-γ mRNA and low amounts of IL-4 and GATA-3 and the opposite was the case for Th2 cells (Fig. S1). Only a proportion of human Th1 cells expressed IFN-γ protein but almost all cells expressed T-bet, confirming the cells' Th1 phenotype (Fig. S1) and consistent with previous results (30). Th2 cells did not express T-bet but did express CRTh2 (Fig. S1). GATA-3 could be detected in both Th2 and Th1 cells, consistent with previous studies (Fig. S1) (31–33).
T-bet associates with many genes in Th1 cells.
We performed ChIP for T-bet followed by hybridization to microarrays containing probes for 8kb surrounding the transcription start sites of 18,450 protein-coding genes. Using an error model to analyze data from replicate experiments (29) and using cells from different donors, we detected T-bet at the promoters of 832 protein-coding genes in primary human Th1 cells (Table S1 and Fig. S2). T-bet targets could be confirmed by quantitative PCR (Fig. S2), were not enriched by ChIP with an isotype-matched control antibody in Th1 cells (Fig. 1A and Fig. S2) and, as expected, did not exhibit significant T-bet binding in Th2 cells (Fig. 1A and Fig. S2), providing confirmation that these genes are specific targets of T-bet.
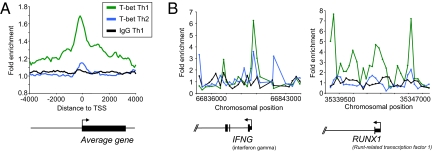
Fig. 1.
T-bet associates with many genes in Th1 cells. (A) Composite T-bet enrichment profile in Th1 cells for genes that show significant binding within 4 kb from their transcription start site (green), compared with the average binding profile for T-bet at the same genes in Th2 cells (blue) and an IgG control antibody ChIP in Th1 cells (black). The plot shows average fold-enrichment (normalized signal from ChIP-enriched DNA divided by the signal from whole cell extract (WCE) DNA. The start and direction of transcription of the average gene is indicated by an arrow. (B) T-bet ChIP signals at IFNG and RUNX1 in Th1 and Th2 cells. The plots show unprocessed enrichment ratios for all probes within a genomic region (ChIP vs. whole genomic DNA) for T-bet in Th1 cells (green), T-bet in Th2 cells (blue), and for an IgG control in Th1 cells (black). Chromosomal positions are from NCBI build 35 of the human genome. Genes are shown to scale below and aligned with the plots by chromosomal position (exons are represented by vertical bars, the start and direction of transcription by an arrow).
The average binding profile shows T-bet binding sites tend to be distributed around the transcription start site (Fig. 1A). The T-bet binding profile peaks just upstream of the transcription start site with a shoulder into the gene. Examining the T-bet binding profile at individual genes, it was apparent that T-bet usually shows two or more distinct binding peaks, with one peak situated at, or upstream of, the transcription start site and the other in the first or second intron (Fig. 1 A and B and Fig. S2).
One T-bet target we identified was IFNG, providing confidence in our dataset (Fig. 1B). T-bet also bound the TNF-α promoter, suggesting a mechanism to ensure co-ordinate regulation of Th1 cytokine production and consistent with regulation of this gene by T-bet in colonic DCs (34). We also detected binding to the promoters of a number of genes with critical functions in T-cell trafficking (CCR5, ITGAL, SELPG, and ICAM1), consistent with our data showing that T-bet controls a specific T-cell migratory program (35). We also detected binding to the promoter of RUNX1 (Fig. 1B). Overexpression of RUNX1 promotes Th1 differentiation by repressing GATA-3, and there is evidence that it may also bind to the IL-4 silencer (23, 36, 37)
To gain a better impression of the cellular functions regulated by T-bet, we used gene ontology (GO) to assign functions to our set of T-bet target genes. This revealed that the set of genes targeted by T-bet were enriched for those involved in metabolism (369 genes, P = 6.4 × 10−8), RNA processing (34 genes, P = 2.4 × 10−5), protein localization (43 genes, P = 6.6 × 10−8), and transcription from RNA polymerase II promoters (37 genes, 9.9 × 10−4). Looking across all GO terms, we identified 100 T-bet target genes with roles in transcriptional regulation, including ATF4, BCL6, CREB1, and IFI16. These functional clusters suggest previously unappreciated roles for T-bet in the biology of Th1 cells. The many transcriptional regulators targeted by T-bet likely form part of the transcriptional regulatory network that operates downstream of T-bet in Th1 cells.
T-bet Activates the Expression of Th1 Genes.
T-bet is known to directly activate expression of IFN-γ to promote Th1 differentiation (4, 17–23). To determine whether the human T-bet gene targets were generally more highly expressed in Th1 cells than Th2 cells, we generated expression data from primary and secondary-stimulated human naı̄ve CD4+ T cells skewed to either Th1 or Th2 lineages (Fig. 2A). We found that although the majority of T-bet target genes showed similar expression levels between Th1 and Th2 cells, a number of genes showed increased gene expression in the former (Fig. 2A and Figs. S2 and S3), including IFNG, NKG7, KSP37, C1QR1, SETBP1, PRF1, CD86, CCL4, CCRL2, and IL18RAP (Fig. S3). Therefore, IFN-γ is but one member of a key set of T-bet target genes that are specifically induced in Th1 cells and likely to play a critical role in Th1 cell biology.
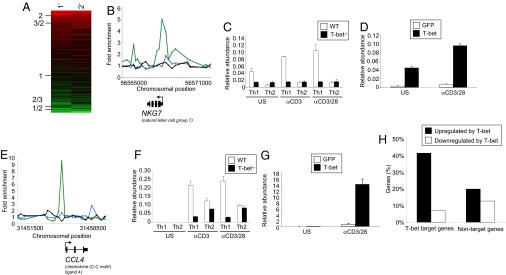
Fig. 2.
T-bet activates the expression of its target genes. (A) Relative levels of expression of T-bet target genes in primary stimulated Th1 cells compared with Th2 cells (column 1) and in secondary stimulated Th1 compared with Th2 cells (column 2). Each row represents a single gene. Genes are ordered by their relative expression level in Th1 cells versus Th2 cells and this average expression ratio is shown on the left hand side. The colors represent relative gene expression levels between Th1 and Th2 cells, with shades of red indicating higher expression in Th1 cells, black equal expression levels and shades of green indicating higher expression in Th2 cells. (B and E) T-bet ChIP signals at NKG7 and CCL4, in human Th1 cells (green) and Th2 cells (blue), and signal from an IgG control IP in Th1 cells (black). Details as for Fig. 1B. (C and F) Real time PCR for NKG7 and CCL4 in WT (open bars) and T-bet−/− (filled bars) murine T cells. Cells were unstimulated (u/s), stimulated with anti-CD3 antibodies (αCD3), or stimulated with anti-CD3 antibodies and anti-CD28 antibodies (αCD3/28). RNA abundance is relative β-actin RNA. (D and G) Real time PCR for NKG7 and CCL4 in CD4+ T cells from T-bet−/−× IFN-γ−/− murine CD4+ T cells transduced with empty vector (open bars) or T-bet (filled bars) expressing retrovirus. (H) Percentage of human Th1 genes whose orthologs are upregulated (at least 1.5-fold, black bars) or downregulated (white bars) in murine T-bet−/−× IFN-γ−/− CD4+ cells upon expression of exogenous T-bet. Th1 genes were defined as those expressed at least 2-fold or higher in Th1 cells than Th2 cells and divided into those bound by T-bet or those not bound by T-bet.
We next sought to determine whether T-bet activated the expression of genes with which it associated and that were overexpressed in Th1 cells (Fig. 2). We used gene targeted mice to test whether the expression of T-bet target genes was altered by the absence and the overexpression of T-bet. Changes in gene expression due to alterations in IFN-γ production were excluded by crossing the T-bet−/− mice to IFN-γ−/− mice. The promoter of the T and NK cell surface cytotoxic molecule, NKG7, is bound by T-bet in human cells (Fig. 2B). This molecule is thought to be important in the regulation of target cells and in the termination of the immune response and it is repressed in murine T-bet−/− T cells (Fig. 2C) and is activated upon T-bet overexpression (Fig. 2D). Chemokines determine site specific migration of immune effector cells, and we found that T-bet bound to and directly activated the expression of CCL3 and CCL4 (RANTES) (Fig. 2 E–G and Fig. S3). These data indicate that T-bet controls a transcriptional program that determines the appropriate migration of lymphocytes to inflammatory sites.
We then extended these studies to test whether T-bet generally acts to induce the expression of genes with which it associates. RNA was isolated from mouse IFN-γ/T-bet double-null CD4+ T cells after transduction with retroviral vectors carrying either a T-bet and a GFP transgene or a GFP transgene only, sorted for GFP expression, quantified using DNA microarrays and gene orthologs mapped between mouse and human using Homologene. We found that T-bet target genes activated in human Th1 cells were often induced upon expression of T-bet in mouse, when compared to Th1 genes that are not associated with T-bet [P < 0.01 (Binomial), Fig. 2H]. Genes directly activated by T-bet in this manner included IFNG, CCL4, NKG7, PRF1, TNF, IL18RAP, IL2RB, SETBP1, and PRDM1. We also identified a smaller number of genes that were bound by and repressed by T-bet, including IL4, PTGER4, FOXO3A, and CD28. Therefore, T-bet directly regulates the expression of a number of genes in Th1 cells in addition to IFNG.
GATA-3 Occupies Immune Response Genes in Th2 Cells.
GATA-3 is the lineage determining transcription factor for murine Th2 cells and seems to perform similar functions in human T cells (38). To identify GATA-3 target genes in Th2 cells, we performed ChIP with a GATA3-specific antibody (D-16) and hybridized the enriched DNA fragments to microarrays. Using our error model, we identified 344 GATA3 target genes in Th2 cells (Fig. 3A, Table S1, and Fig. S4). We also performed replicate experiments using cells from a different donor and with an alternative GATA-3 antibody (HG3–31). This antibody gave a lower enrichment of DNA but otherwise produced similar results (Fig. 3B and Fig. S4).
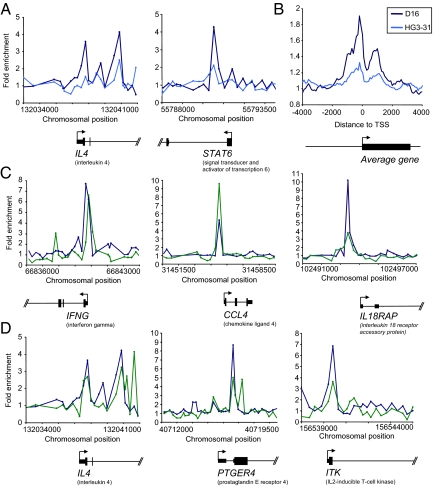
Fig. 3.
GATA-3 gene occupancy in Th2 cells. (A) Examples of GATA-3 ChIP signals in Th2 cells. The plots show unprocessed enrichment ratios for all probes within a genomic region (ChIP vs. whole genomic DNA). D-16 antibody ChIP dark blue, HG3–31 antibody ChIP light blue. (B) Composite GATA-3 enrichment profile in Th2 cells for genes that show significant binding within 4 kb from their transcription start site. D-16 ChIP dark blue, HG3–31 ChIP light blue. (C) Examples of genes with functions in Th1 cells that are bound by both T-bet and GATA-3. The plots show unprocessed enrichment ratios for all probes within a genomic region (ChIP vs. whole genomic DNA) for T-bet in Th1 cells (green) and GATA-3 (D16 ChIP) in Th2 cells (blue). (D) Examples of genes with functions in Th2 cells that are bound by both T-bet and GATA-3. Details as for C.
As we found for T-bet, identification of GATA-3 target genes afforded mechanistic insight into the functional effects of this transcription factor. GO analysis revealed that the targets of GATA-3 were significantly enriched for genes with roles in the immune response (35 genes, P = 1.1 × 10−5) and in signal transduction (79 genes, P = 3 × 10−3). We identified GATA3 binding in the first exon of IL4 (Fig. 3A), a binding site previously identified in murine Th2 cells (26). Importantly, we found that GATA-3 also bound to the transcription factor genes STAT6 (Fig. 3A) and NFATC2IP (NIP45), which we and others have previously shown to be central to the generation of Th2 cells in mice (39). We also found that GATA-3 bound to CCR5 and CCL27, showing that in an analogous manner to T-bet, control of cellular migration is an important function for this transcription factor.
In a pattern similar to T-bet, the average GATA-3 binding profile showed GATA-3 binding peaks occurred just upstream of the transcription start site with a second peak occurring within the gene (Fig. 3B). Examining individual gene plots, some genes showed one peak at the transcription start site (for example ITK), others two peaks (e.g., STAT6) or three peaks (e.g., IL4) (Fig. 3).
GATA-3 and T-bet both target the genes encoding IFN-γ and IL4/IL5/IL13 in mouse cells (22, 23). We therefore compared the sets of T-bet and GATA-3 target genes to investigate whether these factors generally target the same genes in human cells. We identified 76 genes that were associated with both T-bet in Th1 cells and GATA-3 in Th2 cells (Fig. 3 C and D). These genes included those with both Th1 and Th2 functions and expression patterns (Fig. 3 C and D and Fig. S5). For example, GATA-3 and T-bet co-occupied both the Th1-associated genes IFNG, CCL4, IL18RAP, and TNF as well as the Th2-associated genes IL4, PTGER4, ITK, and NFATC2. These results indicate that T-bet and GATA-3 induce alternative T-cell differentiation pathways by acting on many of the same genes.
GATA-3 Occupies Its Target Genes in Th1 Cells.
Assuming that GATA-3 would not associate with genes in Th1 cells (as we saw with the absence of T-bet binding to promoters in Th2 cells), we performed a ChIP-Chip analysis in Th1 cells as a putative negative control. Remarkably, we instead found that GATA-3 also bound to target genes in Th1 cells (Fig. 4, Fig. S6, and Table S1). The set of GATA-3 target genes in Th1 cells encompassed most (78%) of those bound in Th2 cells (Fig. 4A) but with the addition of an extra set of genes that were only bound by GATA3 in Th1 cells (Fig. 4B). Genes bound by GATA-3 in both Th1 and Th2 cells included Th2 genes such as IL4, STAT6, ITK, PTGER4, and CCL27 and Th1 genes such as IFNG, IL18RAP, CCL4, TNF, and NKG7 (Fig. 4 C and D and Fig. S6). Indeed, a total of 92% of Th1 lineage-specific genes occupied by GATA-3 in Th2 cells were also occupied by GATA-3 in Th1 cells. These data show that most genes targeted by GATA-3 in Th2 cells are also occupied by this transcription factor in Th1 cells.
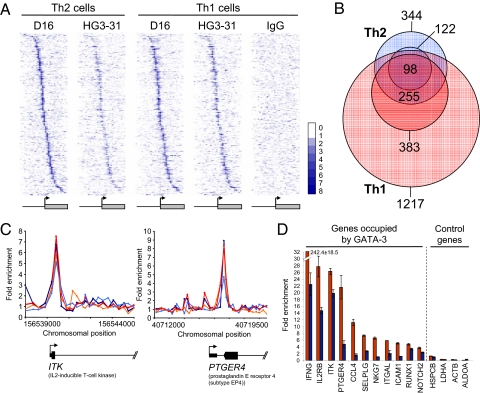
Fig. 4.
GATA-3 occupies its target genes in both Th2 and Th1 cells. (A) Heat maps showing enrichment of GATA-3 at its target genes in Th2 cells and in Th1 cells. Each row represents one gene considered to be occupied by GATA-3 in Th2 cells according to our error model (the same genes are shown in each of the three panels) and each column represents the data from one oligonucleotide probe. Oligos are ordered by their position relative to the transcription start site, as shown by the diagram below. The signal from a control IgG IP in Th1 cells is shown in the last panel. The heat map shows the level of enrichment by color, according to the scale on the right. (B) Venn diagram showing the set of genes bound by GATA-3 in Th2 cells (blue circles) and in Th1 cells (red circles). In both cases, the set of genes identified as targets with the D16 antibody are shown as the lighter-colored circles (1,217 genes in Th1 cells and 344 genes in Th2 cells) and the subsets of genes verified as targets with the HG3–31 antibody as the darker colored circles (383 genes in Th1 cells and 122 genes in Th2 cells) The numbers of genes bound by GATA-3 in both Th1 and Th2 cells are shown in the centre (255 genes identified using D16 alone, with 98 of these also identified with HG3–31). (C) Examples of genes bound by GATA-3 in both Th2 (D16 ChIP data in dark blue, HG3–31 in light blue) and Th1 cells (D16 ChIP data in red, HG3–31 in orange). (D) Fold enrichment of DNA promoter sequences in GATA-3 ChIP material (D16 antibody) from Th1 cells (red) and Th2 cells (blue) relative to whole genomic DNA measured by quantitative PCR. Error bars, standard deviations (n = 3).
T-bet and GATA-3 Share Many Target Genes.
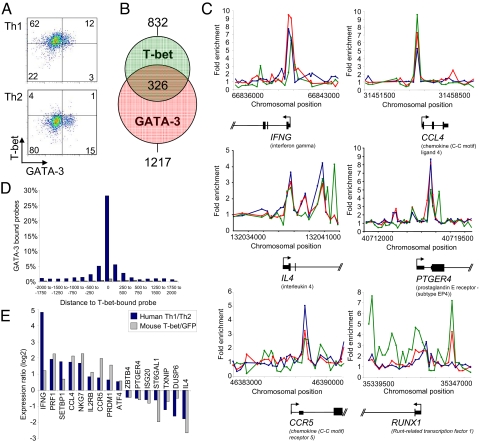
Given that we had unexpectedly found a significant number of genes bound by GATA-3 in Th1 cells and that T-bet and GATA-3 often targeted the same genes, we sought to determine whether GATA-3 and T-bet share target genes when both factors are present within the same cell. We first asked whether T-bet and GATA-3 are co-expressed in Th1 cells (Fig. 5A and Fig. S7). Using intranuclear flow cytometry, we found that T-bet and GATA-3 were co-expressed in Th1 cells and that GATA-3 was generally only expressed in Th1 cells that were positive for T-bet. As expected, in Th2 cells the expression of GATA-3 occurred in the absence of T-bet. We next compared the genes targeted by T-bet in Th1 cells with those targeted by GATA-3 in Th1 cells. We found that T-bet and GATA-3 often bound to the same genes with 39% of T-bet target genes in Th1 cells also being occupied by GATA-3 (P < 10−16, hypergeometric; Fig. 5B and Table S1). Furthermore, when present at the same gene, T-bet and GATA-3 peaks were generally closely spaced, with the majority of these peaks coinciding exactly (within the ≈200bp resolution of the array; Fig. 5 C and D). Genes bound by both T-bet and GATA-3 included those exhibiting differential gene expression between the two cell types (56% of Th1 genes and 37% of Th2 genes bound by T-bet were also bound by GATA-3) (Fig. 5C and Figs. S7 and S8). We also detected GATA-3 binding to these genes in tertiary stimulated Th1 cells and in a Th1 cell clone (40) using quantitative PCR (Fig. S8), indicating that the detection of GATA-3 binding in the primary stimulated Th1 cells was not due to incomplete Th1 polarization or a potentially heterogeneous cell culture. Taken together with our GATA-3 ChIP data from Th2 cells, our Th1 cell data demonstrate that GATA-3 associates with a core set of genes in both Th1 and Th2 cells and that these genes are also targeted in Th1 cells by T-bet.
Fig. 5.
GATA-3 and T-bet co-occupy genes in Th1 cells. (A) GATA-3 (x axis) and T-bet (y axis) expression in primary human Th1 cells (left panel) and Th2 cells (right panel) measured by flow cytometry. (B) Venn diagram showing the overlap between the set of genes bound by T-bet (green circle) and the set of genes bound by GATA-3 (D16 ChIP, red circle) in Th1 cells. The number of genes bound by both T-bet and GATA-3 is shown in the center. (C) Examples of genes bound by both T-bet and GATA-3 in Th1 cells. The plots show unprocessed enrichment ratios for all probes within a genomic region (ChIP vs. whole genomic DNA) for T-bet in Th1 cells (green), GATA-3 (D16 ChIP) in Th2 cells (blue), and GATA-3 in Th1 cells (red). (D) The distribution of distances from oligonucleotide probes reporting GATA-3 binding to oligonucleotide probes reporting T-bet binding (blue). Probes are spaced an average of 250 bp along the genome. The distribution of distances compared with probes reporting binding in an IgG control antibody experiment is shown by comparison (gray). (E) Relative expression of human genes bound by T-bet and GATA-3 between Th1 and Th2 cells (blue bars) and their murine orthologs in T-bet−/−× IFN-γ−/− CD4+ cells in response to exogenous T-bet expressed from a retroviral vector compared to a vector encoding GFP alone (gray bars).
The relatively invariant association of GATA-3 with lineage-specific genes in both Th1 and Th2 cells suggests that their differential expression is not due to differences in GATA-3 binding. This argues that T-bet binding in Th1 cells may be responsible for the differential expression of these genes between the two lineages. To explore this possibility, we used our human and mouse expression data to examine the effects of T-bet upon the expression of genes that were bound by both T-bet and GATA-3 in Th1 cells. Expression of T-bet in T cells from IFNγ−/− × T-bet−/− mice (that express GATA-3, Fig. S8) induced changes in the expression of T-bet/GATA-3 target genes that correlated well with relative expression levels of those genes between human Th1 and Th2 cells (r = 0.49). T-bet and GATA-3 target genes expressed at a higher level in Th1 cells compared with Th2 cells tended to be upregulated by T-bet and genes expressed at a higher level in Th2 cells compared with Th1 cells tended to be downregulated by T-bet (Fig. 5E). These results show that direct binding by T-bet is responsible for the differential expression of shared T-bet/GATA-3 target genes between Th1 and Th2 cells.
Discussion
We have identified the target genes of T-bet and GATA-3 in primary human Th1 and Th2 cells, greatly increasing our understanding of how these two factors govern Th1 and Th2 differentiation. T-bet and GATA-3 occupy many of the same genes, including those that are differentially expressed between Th1 and Th2 cells and play key roles in T-cell biology. This indicates that the choice between Th1 and Th2 lineage commitment is perhaps best viewed as the result of the opposing action of these two transcription factors at a set of shared target genes. These data demonstrate a role for T-bet and GATA-3 in T-cell differentiation that extends beyond the regulation of IFNG and IL4.
Consistent with previous results, we have shown that GATA-3 is present in both human Th2 and Th1 cells (31–33). We have also shown that almost all Th1 cells that express GATA-3 also express T-bet. The co-expression of T-bet and GATA-3 is analogous to recent data showing that FoxP3 and RORγt are co-expressed in murine T cells (41) and that T-bet and FoxP3 function together in a subset of regulatory T cells (42) and this suggests that this co-expression is functionally significant. We find that not only are T-bet and GATA-3 expressed in the same cells, they occupy highly overlapping sets of genes, including those that show differential expression between Th1 and Th2 cells. We have not been able to use ChIP-ReChIP to test whether T-bet and GATA-3 bind to genes simultaneously in Th1 cells due to failure of our positive control ReChIP RNA polymerase II experiments. However, the co-expression of T-bet and GATA-3 in primary human Th1 cells provides each factor with the potential to act upon their shared target genes in these cells. T-bet directly activates IFNG and represses IL4 (Fig. 5E) (23, 27) whereas GATA3 acts in the opposite manner to activate IL4 and represses IFNG (22, 28). Although both factors are coexpressed in human Th1 cells, T-bet activity would appear to be dominant and these cells exhibit an expression pattern that can be recapitulated in murine T-cells by expression of T-bet in the absence of IFNγ. These results indicate that it may be the absence or presence of T-bet that determines T-cell lineage (16, 43) and provides a potential mechanistic rationale that explains why T cells default to the Th2 lineage in the absence of T-bet (3–5). The extension of these findings to vivo polarized human Th1 and Th2 cells is currently under active investigation, but has been limited by the cell numbers required for this type of analysis.
Human T-cell immune responses define pathological outcomes in a wide number of disease states. Identification of novel T-bet and GATA-3 target genes offers the prospect of defining rational therapeutic approaches in these conditions. Indeed, the identification of TNF-α as a direct T-bet target in T cells lead us to the definition of the transcriptional mechanism of a new model of ulcerative colitis (34), providing evidence of the utility of this approach. Other gene targets of T-bet and GATA-3 that we have identified here are also likely to play important roles in infectious disease and autoimmune conditions.
In conclusion, we have shown that Th1 and Th2 lineage-specific genes that play key roles in T-cell biology are targeted by both T-bet and GATA-3. The action of opposing master regulators through a shared set of target genes may prove to be a general mechanism in other cell types and biological systems.
Methods
Cells.
Naïve human CD4+ T cells from the peripheral blood of human donors were sorted for CD4+CD25-CD45RO-HLA-DR- (purity of 98%) and activated with anti-CD3 and anti-CD28 for 48–72 h. Primary Th1 cells were generated over 10–12 days with IL-12 (10 ng/ml) and anti-IL-4 (10 mg/ml) and Th2 cells with IL-4 (10 ng/ml) and anti-IFN-γ (10 mg/ml).
ChIP-Chip.
We followed previously published ChIP-ChIP protocols (29). Binding sites were automatically identified using an algorithm that calculates confidence values for each probe and finds sets of neighboring probes with significant P values (29).
Retroviral Transduction of T-bet.
T-bet and control retroviruses were produced and titred as described (35).
All ChIP-Chip and gene expression microarray data are available at ArrayExpress (accession number E-TABM-759).
Detailed descriptions of methods are available in the SI Text.
Supplementary Material
Acknowledgments.
We thank Francesco Annunziato for the Th1 cell clone, Doris Wilflingseder for culturing the cells, Arshad Kahn for assistance with data analysis, and Esperanza Perucha for Figs. S1k and S7a. This work was supported by the Medical Research Council (R.G.J. and G.M.L.), the National Institute of Health (L.H.G. and R.A.Y.), and an Ellison Medical Foundation grant (to L.H.G.). G.M.L. acknowledges financial support from the National Institute for Health Research Comprehensive Biomedical Research Centre award to Guy's & St Thomas' National Nealth Service Foundation Trust in partnership with King's College London and King's College Hospital National Nealth Service Foundation Trust.
Footnotes
Conflict of interest statement: L.H.G. is a member of the Board of Directors of and holds equity in the Bristol Myers Squibb Corporation.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909357106/DCSupplemental.
References
- 1.Szabo SJ, Sullivan BM, Peng SL, Glimcher LH. Molecular mechanisms regulating Th1 immune responses. Ann Rev Immunol. 2003;21:713–758. doi: 10.1146/annurev.immunol.21.120601.140942. [DOI] [PubMed] [Google Scholar]
- 2.Weaver CT, Harrington LE, Mangan PR, Gavrieli M, Murphy KM. Th17: An effector CD4 T cell lineage with regulatory T cell ties. Immunity. 2006;24:677–688. doi: 10.1016/j.immuni.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Murphy KM, Reiner SL. The lineage decisions of helper T cells. Nat Rev Immunol. 2002;2:933–944. doi: 10.1038/nri954. [DOI] [PubMed] [Google Scholar]
- 4.Szabo SJ, et al. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100:655–669. doi: 10.1016/s0092-8674(00)80702-3. [DOI] [PubMed] [Google Scholar]
- 5.Szabo SJ, et al. Distinct effects of T-bet in TH1 lineage commitment and IFN-gamma production in CD4 and CD8 T cells. Science. 2002;295:338–342. doi: 10.1126/science.1065543. [DOI] [PubMed] [Google Scholar]
- 6.Harrington LE, et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 7.Neurath MF, et al. The transcription factor T-bet regulates mucosal T cell activation in experimental colitis and Crohn's disease. J Exp Med. 2002;195:1129–1143. doi: 10.1084/jem.20011956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bettelli E, et al. Loss of T-bet, but not STAT1, prevents the development of experimental autoimmune encephalomyelitis. J Exp Med. 2004;200:79–87. doi: 10.1084/jem.20031819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finotto S, et al. Development of spontaneous airway changes consistent with human asthma in mice lacking T-bet. Science. 2002;295:336–338. doi: 10.1126/science.1065544. [DOI] [PubMed] [Google Scholar]
- 10.Matsui M, Moriya O, Yoshimoto T, Akatsuka T. T-bet is required for protection against vaccina virus infection. J Virol. 2005;79:12798–12806. doi: 10.1128/JVI.79.20.12798-12806.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaplan MH, Schindler U, Smiley ST, Grusby MJ. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity. 1996;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 12.Zheng W, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for the Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:589–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 13.Zhang DH, Cohn L, Ray P, Bottomly K, Ray A. Transcription factor GATA-3 is differentially expressed in murine Th1 and Th2 cells and controls Th2-specific expression of the intrleukin-5 gene. J Biol Chem. 1997;272:21597–21603. doi: 10.1074/jbc.272.34.21597. [DOI] [PubMed] [Google Scholar]
- 14.Pai SY, Truitt ML, Ho IC. GATA-3 deficiency abrogates the development and maintenance of T helper type 2 cells. Proc Natl Acad Sci USA. 2004;101:1993–1998. doi: 10.1073/pnas.0308697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu J, et al. Conditional deletion of Gata3 shows its essential function in T(H)1-T(H)2 responses. Nat Immunol. 2004;5:1157–1165. doi: 10.1038/ni1128. [DOI] [PubMed] [Google Scholar]
- 16.Hwang ES, Szabo SJ, Schwartzberg PL, Glimcher LH. T helper cell fate specified by kinase-mediated interaction of T-bet with GATA-3. Science. 2005;307:430–433. doi: 10.1126/science.1103336. [DOI] [PubMed] [Google Scholar]
- 17.Mullen AC, et al. Role of T-bet in commitment of TH1 cells before IL-12-dependent selection. Science. 2001;292:1907–1910. doi: 10.1126/science.1059835. [DOI] [PubMed] [Google Scholar]
- 18.Avni O, et al. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol. 2002;3:643–651. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- 19.Fields PE, Kim ST, Flavell RA. Cutting edge: Changes in histone acetylation at the IL-4 and IFN-gamma loci accompany Th1/Th2 differentiation. J Immunol. 2002;169:647–650. doi: 10.4049/jimmunol.169.2.647. [DOI] [PubMed] [Google Scholar]
- 20.Lee DU, Avni O, Chen L, Rao A. A distal enhancer in the interferon–gamma (IFN-gamma) locus revealed by genome sequence comparison. J Biol Chem. 2004;279:4802–4810. doi: 10.1074/jbc.M307904200. [DOI] [PubMed] [Google Scholar]
- 21.Shnyreva M, et al. Evolutionarily conserved sequence elements that positively regulate IFN-gamma expression in T cells. Proc Natl Acad Sci USA. 2004;101:12622–12627. doi: 10.1073/pnas.0400849101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang S, Aune TM. Dynamic changes in histone-methylation ‘marks’ across the locus encoding interferon-gamma during the differentiation of T helper type 2 cells. Nat Immunol. 2007;8:723–731. doi: 10.1038/ni1473. [DOI] [PubMed] [Google Scholar]
- 23.Djuretic IM, et al. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence II4 in T helper type 1 cells. Nat Immunol. 2007;8:145–153. doi: 10.1038/ni1424. [DOI] [PubMed] [Google Scholar]
- 24.Agarwal S, Avni O, Rao A. Cell-type-restricted binding of the transcription factor NFAT to a distal IL-4 enhancer in vivo. Immunity. 2000;12:643–652. doi: 10.1016/s1074-7613(00)80215-0. [DOI] [PubMed] [Google Scholar]
- 25.Yamashita M, et al. Indentification of a conserved GATA3 response element upstream proximal from the interleukin-13 gene locus. J Biol Chem. 2002;277:42399–42408. doi: 10.1074/jbc.M205876200. [DOI] [PubMed] [Google Scholar]
- 26.Tykocinski LO, et al. A critical control element for interleukin-4 memory expression in T helper lymphocytes. J Biol Chem. 2005;280:28177–28185. doi: 10.1074/jbc.M502038200. [DOI] [PubMed] [Google Scholar]
- 27.Koyanagi M, et al. EZH2 and histone 3 trimethyl lysine 27 associated with II4 and II13 gene silencing in Th1 cells. J Biol Chem. 2005;280:31470–31477. doi: 10.1074/jbc.M504766200. [DOI] [PubMed] [Google Scholar]
- 28.Schoenborn JR, et al. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat Immunol. 2007;8:732–742. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee TI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao W, et al. Human T helper (Th) cell lineage commitment is not directly linked to the secretion of IFN-gamma or IL-4: Characterization of Th cells isolated by FACS based on IFN-gamma and IL-4 secretion. Eur J Immunol. 2005;35:2709–2717. doi: 10.1002/eji.200425957. [DOI] [PubMed] [Google Scholar]
- 31.Cousins DJ, Lee TH, Staynov DZ. Cytokine coexpression during human Th1/Th2 cell differentiation: Direct evidence for coordinated expression of Th2 cytokines. J Immunol. 2002;169:2498–2506. doi: 10.4049/jimmunol.169.5.2498. [DOI] [PubMed] [Google Scholar]
- 32.Messi M, et al. Memory and flexibility of cytokine gene expression as separable properties of human T(H)1 and T(H)2 lymphocytes. Nat Immunol. 2003;4:78–86. doi: 10.1038/ni872. [DOI] [PubMed] [Google Scholar]
- 33.De Fanis U, et al. GATA3 up-regulation associated with surface expression of CD294/CRTH2: A unique feature of human Th cells. Blood. 2007;109:4343–4350. doi: 10.1182/blood-2006-05-025940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garrett WS, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lord GM, et al. T-bet is required for optimal proinflammatory CD4+ T-cell trafficking. Blood. 2005;106:3432–3439. doi: 10.1182/blood-2005-04-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Komine O, et al. The Runx1 transcription factor inhibits the differentiation of naı̄ve CD4+ T cells into the Th2 lineage by repressing GATA3 expression. J Exp Med. 2003;198:51–61. doi: 10.1084/jem.20021200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Naoe Y, et al. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbf beta binding to the II4 silencer. J Exp Med. 2007;204:1749–1755. doi: 10.1084/jem.20062456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skapenko A, et al. GATA-3 in human T cell helper type 2 development. J Exp Med. 2004;199:423–428. doi: 10.1084/jem.20031323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hodge MR, Chun HJ, Rengarajan J, Alt A, Lieberson R, Glimcher LH. NF-AT-Driven interleukin-4 transcription potentiated by NIP45. Science. 1996;274:1903–1905. doi: 10.1126/science.274.5294.1903. [DOI] [PubMed] [Google Scholar]
- 40.Annunziato F, et al. Phenotypic and functional features of human Th17 cells. J Exp Med. 2007;204:1849–1861. doi: 10.1084/jem.20070663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou L, et al. TGF-beta-induced Foxp3 inhibits T(H)17 cell differentiation by antagonizing RORgammat function. Nature. 2008;453:236–240. doi: 10.1038/nature06878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koch MA, et al. The transcroption factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Usui T, et al. T-bet regulates Th1 responses through essential effects on GATA-3 function rather than IFNG gene acetylation and transcription. J Exp Med. 2006;203:755–766. doi: 10.1084/jem.20052165. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.