Abstract
The activation of the innate immune responses by DNA exposed within the cytosol has gained much attention and, in this context, several cytosolic DNA sensors have been identified. However, previous studies revealed the operation of redundant and complex mechanisms and it still remains to be clarified how the DNA-mediated evocation of diverse innate immune responses can be achieved. Here we show that two RIG-I-like receptors (RLRs), RIG-I and MDA5, known as cytosolic RNA receptors, nonredundantly function as cytosolic DNA receptors that lead to the selective activation of type I IFN genes. We demonstrate that overexpression of otherwise IFN-inducible RIG-I or MDA5 in IFN signal-deficient cells results in a marked enhancement of type I IFN gene induction upon cytosolic DNA stimulation, while in their absence the induction is impaired. Interestingly, the DNA-mediated induction of other cytokine genes was barely affected by the absence of RLRs. Indeed, unlike the RNA-RLR pathway that activates the transcription factors IRF3 and NF-κB, the DNA-RLR pathway is primarily responsible for the IRF3 activation critical for type I IFN gene transcription, illustrating a deliberate divergence of the DNA signaling pathways. Expectedly, the RLR pathway also contributes to intricate innate immune responses against infection by a DNA virus. Our study may provide insights into the complexity of host defense mechanisms that thwart immune evasion by DNA-containing pathogens.
Keywords: DNA sensor, innate immunity, IRF3, NF-κB
The activation of innate immune responses by nucleic acids is crucial to protective and pathological immunities and is mediated by the transmembrane Toll-like receptors (TLRs) and cytosolic receptors (1–3). DNA is a potent immune activator and its exposure within the cytosol evokes robust type I IFN and other innate immune responses (4, 5). Although molecules such as DNA-dependent activator of IRFs (DAI; also referred to as DLM-1/ZBP1) and absent in melanoma 2 (AIM2) (6, 7) are known to participate in these responses, there is evidence that other cytosolic DNA receptors exist (8, 9). While the role of RIG-I as a cytosolic RNA sensor has been well established (10, 11), its role in the DNA sensing pathway is somewhat controversial. A previous report has indicated that cytosolic DNA-mediated type I IFN responses occur normally in RIG-I-deficient mouse embryonic fibroblasts (MEFs) (4), whereas another report has shown that RIG-I is critical for the responses in a human hepatoma cell line (12). It is uncertain if these discrepancies reflect redundant mechanisms or differences between mice and humans. Moreover, it is unclear if the RIG-I-related RNA sensor melanoma differentiation-associated gene-5 (MDA5) participates in cytosolic DNA sensing.
In the present study, we undertook several approaches to rigorously assess whether two RLR molecules universally function as the DNA sensors across the two species and, if so, which of the signaling pathways is activated by them. We show that both RIG-I and MDA5 nonredundantly participate in both mouse and human cells to the activation of innate immune responses by cytosolic DNA, be it synthetic, pathogen-derived or mammalian DNA. Perhaps surprisingly, while it is well established that the gene induction of both type I IFN and proinflammaroty cytokines is mediated by RLRs upon RNA stimulation, we found that, in at least some cell types, the DNA-RLR pathway is selectively linked to the IRF3-dependent type I IFN gene induction, adducing evidence that the NF-κB-dependent induction of proinflammaroty cytokine genes is mediated, by as yet unknown DNA sensor(s). Our results suggest that the DNA sensing system may consist of more heterogeneous mechanisms than previously anticipated, and that this may have implications in the evolution of the meticulous divergence of the DNA-mediated activation of immune response to effectively cope with DNA-containing pathogens.
Results
Selective Upregulation of Type I IFN Gene Induction by Cytosolic DNAs Upon RIG-I or MDA5 Overexpression.
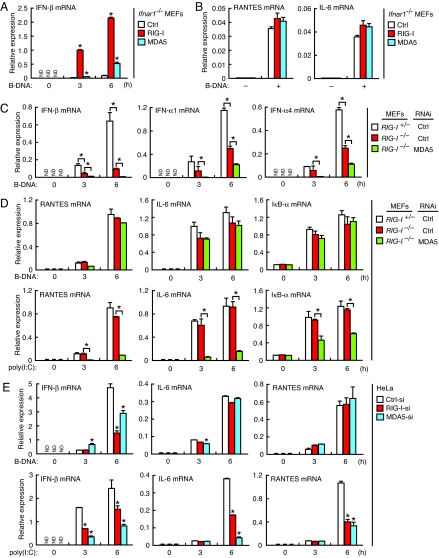
To examine the contribution of RIG-I and MDA5 to the cytosolic DNA-mediated activation of innate immune responses, we expressed mouse RIG-I or MDA5 by retrovirus-mediated gene transfer in MEFs deficient in a component of the type I IFN receptor, IFNAR-1 (Ifnar1−/− MEFs); these cells do not undergo the IFN-dependent induction of endogenous RIG-I and MDA5 (11, 13). As shown in Fig. 1A, the induction of type I IFN mRNA by B-DNA was upregulated by approximately 20-fold in Ifnar1−/− MEFs expressing RIG-I compared with control Ifnar1−/− MEFs. Although less effectively than RIG-I, MDA5 expression also resulted in the upregulation of the mRNA (Fig. 1A). Interestingly, however, the induction of other cytokines, such as IL-6 and RANTES, remained unaffected in these cells, suggesting a selective contribution of RIG-I and MDA5 to the activation of type I IFN responses upon B-DNA stimulation (Fig. 1B). Essentially the same observation was made with other immunogenic DNAs (Fig. S1A).
Fig. 1.
Selective contribution of RIG-I and MDA5 to cytosolic DNA-mediated activation of type I IFN responses. (A and B) RIG-I and MDA5 confer the ability of type I IFN responses against cytosolic DNA. Ifnar1−/− MEFs transduced with empty retrovirus (Ctrl) or retrovirus carrying RIG-I or MDA5 cDNA were lipofected with B-DNA for 6 h, and then mRNA expression levels of the indicated genes analyzed by qRT-PCR. Data are presented as mean ± SD. (n = 3). Asterisk (*), P < 0.01 as compared with Ctrl vector-transduced cells. ND, not detected. (C and D) Analysis of MEFs deficient in RIG-I and MDA5. RIG-I+/− or RIG-I −/− MEFs expressing control (Ctrl-si) or MDA5-targeting siRNA (MDA5-si) were lipofected with B-DNA for the indicated periods, and mRNA expression levels of the indicated genes analyzed by qRT-PCR. Note that we used poly(I:C) with the length ranging from 200 bp to 5 kbp, which would activate both RIG-I and MDA5. Data are mean ± SD. (n = 3). *, P < 0.01. ND, not detected. (E) Analysis of HeLa cells deficient in RIG-I or MDA5. HeLa cells expressing siRNA targeting RIG-I (RIG-I-si), MDA5 (MDA5-si) or control siRNA (Ctrl-si) were lipofected with B-DNA, and then mRNA expression levels of the indicated genes analyzed by qRT-PCR. Data are mean ± SD. (n = 3). *, P < 0.001 as compared with cells expressing Ctrl-si. ND, not detected.
Activation of the Cytosolic DNA-Mediated Innate Immune Responses in the Absence of RIG-I and/or MDA5.
We next examined whether the loss of RIG-I and/or MDA5 in MEFs affects B-DNA-mediated innate immune responses. When MEFs carrying homozygous mutation in RIG-I (RIG-I−/− MEFs) (10) were stimulated by B-DNA, a profound defect was observed in the induction of type I IFN mRNAs as compared with MEFs heterozygous for the RIG-I mutation (RIG-I+/− MEFs) (Fig. 1C). It is worth noting that the same observation was made with other immunogenic DNAs, including viral DNAs and synthetic ISD (IFN stimulatory DNA) that have a sequence different from that of B-DNA (5) (Fig. S1B). Furthermore, the residual mRNA induction was suppressed in RIG-I−/− MEFs expressing a small interfering RNA (siRNA) that specifically targets MDA5 (RIG-I−/−/MDA5-si MEFs) (Fig. 1C). Interestingly, however, the gene induction of proinflammatory cytokines, such as IL-6, RANTES, and IκB-α by B-DNA was diminished only slightly in these cells, while the induction of these genes by poly(I:C) stimulation was profoundly impaired (Fig. 1D). These observations corroborate the RLR overexpression data (Fig. 1B) and further underscore the critical contribution of RLRs, particularly RIG-I, to the cytosolic DNA-mediated activation of type I IFN responses. On the other hand, however, these observations suggest the presence of an additional mechanism(s) for the activation of proinflammatory cytokine genes.
It was shown previously in a human hepatoma cell line that RIG-I is crucial for evoking type I IFN responses following cytosolic DNA stimulation or infection by DNA viruses (12), although neither the induction of proinflammatory cytokines nor the contribution of MDA5 was examined. In view of the above observations, we also examined the contribution of RIG-I and MDA5 in the human HeLa cell line by knocking down one or another of these proteins using siRNAs (RIG-I-si HeLa cells and MDA5-si HeLa cells). As shown in Fig. 1E, the type I IFN mRNA induction by B-DNA stimulation was suppressed strongly in RIG-I-si HeLa cells without affecting the induction of IL-6 and RANTES. The selective suppression of type I IFN mRNA induction was also observed in MDA5-si HeLa cells, albeit more weakly than in RIG-I-si HeLa cells, indicating that the contribution of MDA5 is smaller as compared to RIG-I, a situation also found in mouse cells. These results further demonstrate that RIG-I and MDA5 serve as cytosolic DNA receptors that selectively evoke the type I IFN response pathway in human and mouse cells. The seemingly discrepant conclusion made by a previous study may possibly come from the detection of the low-level of type I IFN mRNA contributed by the B-DNA-MDA5 pathway (see Fig. 1C) by semiquantitative RT-PCR used in the study (4).
Differential Activation of the Transcription Factors by Cytosolic DNA or RNA.
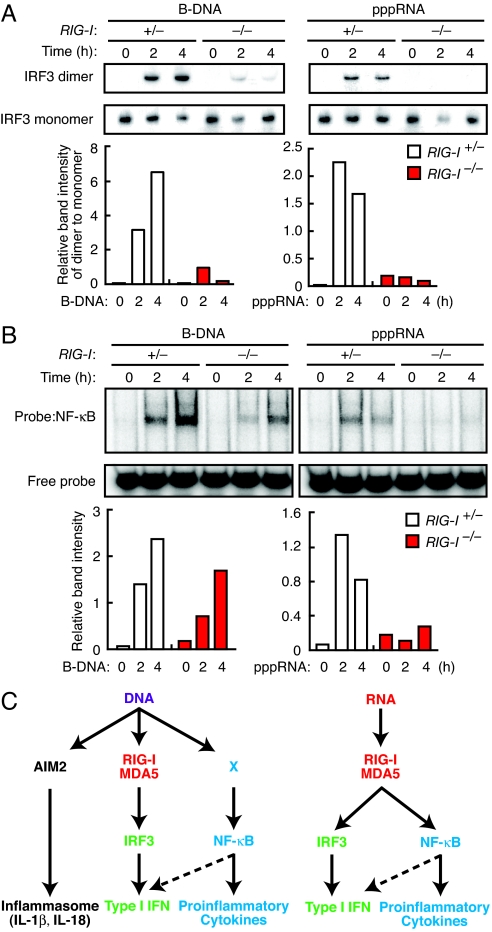
Type I IFN gene induction by B-DNA mainly depends on the activation of IRF3, while proinflammatory gene induction depends on NF-κB (4, 7). Therefore, we examined how the loss of RIG-I affects DNA- or RNA-stimulated activation of these transcription factors. RIG-I+/− and RIG-I−/− MEFs were stimulated either by B-DNA or a 5′-triphosphate-containing single-stranded RNA (pppRNA) that specifically activates RIG-I (14), and the activation of IRF3 and NF-κB was examined by dimerization assay and EMSA, respectively. As shown in Fig. 2A, the IRF3 activation observed in RIG-I+/− MEFs after stimulation by B-DNA or pppRNA was strongly suppressed in RIG-I−/− MEFs. Interestingly, on the other hand, NF-κB activation, which was also severely suppressed in pppRNA-stimulated RIG-I−/− MEFs, was only marginally affected in B-DNA-stimulated RIG-I−/− MEFs (Fig. 2B). We also found that B-DNA-stimulated IRF3 dimerization occurs normally in cells treated by actinomycin D (Fig. S2), further in support of a direct activation of RLRs by DNA, although it is not strictly ruled out that DNA-mediated RLR activation involves RNA possibly transcribed from the delivered DNA. Taken together these findings, we surmise that a RIG-I-independent NF-κB activation pathway, the nature of which still remains to be clarified (therefore denoted as ‘X’ in Fig. 2C), accounts for the induction of proinflammatory cytokine genes in these cells.
Fig. 2.
Differentially diverged signaling pathways activated by cytosolic DNA and RNA. (A and B) Activation status of IRF3 (A) and NF-κB (B). RIG-I+/− or RIG-I−/− MEFs were stimulated by B-DNA or pppRNA. Dimerization of IRF3 was assessed by native PAGE followed by immunoblot analysis. Activation of NF-κB was analyzed by EMSA. The relative band intensities of IRF3 dimer and activated NF-κB were quantified by a densitometer and depicted in graphs at the bottom of each panel. The levels of IRF3 dimer were normalized by those of monomer. (C) A proposed model for the signaling pathways activated by cytosolic DNA and RNA. Innate immune responses against cytosolic DNA and RNA both use RIG-I/MDA5. Upon RNA stimulation, RIG-I/MDA5 activates both IRF3 and NF-κB pathways. In the case of DNA stimulation, three distinct pathways diverge at the receptor level, indicating the existence of an unidentified DNA sensor that activates NF-κB.
Interaction of RIG-I and Immunogenic DNAs.
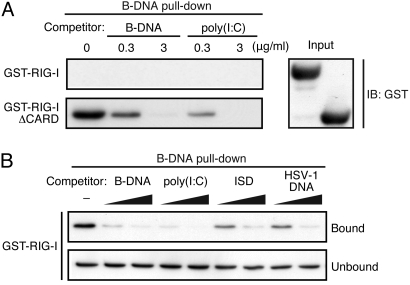
The above observations promoted us to examine whether RIG-I interacts directly with immunogenic DNAs. GST-fused full-length RIG-I (GST-RIG-I) or truncated RIG-I (GST-RIG-IΔCARD) was prepared and subjected to a ‘pull-down’ assay using biotin-conjugated B-DNA. As shown in Fig. 3A, GST-RIG-IΔCARD was pulled down with B-DNA, which was inhibited by unconjugated B-DNA or poly(I:C), indicating that the RIG-I containing the helicase-like domain and C-terminal domain (CTD) binds to both DNA and RNA. The binding of GST-RIG-I with B-DNA was also detected, albeit much more inefficiently (Fig. 3 A and B), indicating that nucleic acids bind to RIG-IΔCARD more easily than full-length RIG-I in vitro. This is expected in view of the fact that RIG-I resides in closed conformation via intramolecular interactions between the CARD domains and the helicase-like and CTD domains (15). We also examined whether other immunogenic DNAs also interact with RIG-I by subjecting them to the above competition assay. As shown in Fig. 3B, the GST-RIG-I interaction with biotin-conjugated B-DNA is inhibited by various DNAs, whose respective immunogenic potential to activate RLRs is shown in Fig. S1B. Thus, these DNAs may also bind RLRs when delivered to cytosol, although their involvement in the activation of RLR by DNA-derived RNA is not ruled out. Whatever the detailed mechanisms, the mode of RIG-I activation by DNA versus RNA may be distinct in several respects. For example, the ensuing responses may be due to the differential usage of downstream adaptor molecules. It has been reported that an RLR-associated adaptor, IFN-β promoter stimulator-1 (IPS-1; also known as MAVS/Cardif/VISA) is partially involved in B-DNA-stimulated evocation of type I IFN response, but not of inflammatory cytokines (16). As a result, IPS-1 may exclusively participate in the B-DNA-RLR-IRF3 limb of the signaling pathway. Other adaptor proteins, such as STING/MITA (17, 18), may differentially contribute to these responses. The exact nature and functional mechanisms of these pathways obviously require further investigation.
Fig. 3.
Interaction of RIG-I with immunogenic DNAs in vitro. (A) In vitro B-DNA pull-down analysis of RIG-I. Recombinant GST-RIG-I and GST-RIG-IΔCARD were incubated with biotin-conjugated B-DNA and streptavidin (SA)-conjugated magnetic beads in the absence or presence of unconjugated B-DNA or poly(I:C). Bound proteins were analyzed by immunoblot analysis using anti-GST antibody (left panel). Input protein levels for this assay are shown in the right panel. (B) Interaction of intact RIG-I with B-DNA and other immunogenic DNAs. The same pull-down assay was performed by incubating GST-RIG-I with biotin-conjugated B-DNA in the presence of increasing amounts of the indicated DNAs. Note that the GST-RIG-I blot is exposed 100-fold longer than that shown in A to visualize the pulled down GST-RIG-I.
Contribution of RLRs in the Activation of Innate Immune Responses by DNA Virus.
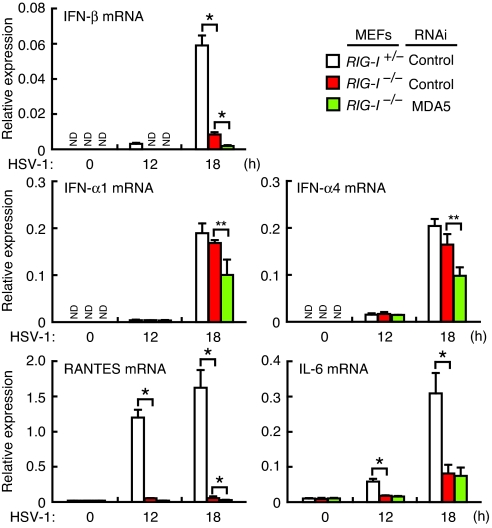
Given that, in both human and mouse cells, the RLR pathway is a principal pathway for the cytosolic DNA-mediated activation of innate immune responses, we next examined to what extent this pathway contributes to innate immune responses to a DNA virus. To this end, we performed HSV-1 infection experiments. Upon infection of RIG-I−/−, RIG-I−/−/MDA5-si, or control RIG-I+/− MEFs, we observed a more complex and subtle requirement for the RLR genes than that for ‘naked’ HSV-1 DNA transfection (Fig. S1B). For one, the induction of IFN-β mRNA is entirely dependent on RIG-I and MDA5, while the induction of IFN-α mRNAs is only partially dependent on these RLRs (Fig. 4): Consequently, virus replication is notably higher, but not dramatically so, in MEFs lacking RIG-I alone or both RLRs (Fig. S3). Moreover, unlike HSV-1 DNA stimulation, the mRNA induction of proinflammatory cytokines by HSV-1 infection is impaired in the absence of RLRs (Fig. 4). Indeed, HSV-1 infection may result in not only the release of DNA into the cytosol, but also the generation of other immunogenic molecules such as viral RNAs, host RNAs cleaved by RNase L and viral proteins (19, 20, 21). It is therefore plausible that these molecules are sensed by RLRs and other sensors in an intricate manner so as to generate the response patterns described above, which are not the same as those found in mere nucleic acid stimulations. Thus, the gene induction for proinflammatory cytokines may be mediated mainly through the activation of RLRs by RNAs generated during virus replication (22), while Ifnb gene may be activated by viral DNA or RNA or by both. As a result, the residual Ifna gene induction in HSV-1-infected RIG-I−/−/MDA5-si MEFs may be mediated by as yet unknown, non-nucleic acid pathway(s); evidence for which may have already been provided (23) and this may involve the activation of IRF7, another transcription factor critical for Ifna gene induction (24). Although TLR9, as well as TLR2 are also involved in the activation of innate immune responses by this virus, the evocation of type I IFN response in MEFs is TLR-independent and requires virus replication (25). In conclusion, although it is clear that the RLR pathway is also critical for evoking innate immune responses to this DNA virus, these results further illustrate the complexity of the innate immune system against infections to DNA-containing pathogens.
Fig. 4.
Requirement of RIG-I and MDA5 in innate immune responses against a DNA virus. RIG-I+/− or RIG-I −/− MEFs expressing control (Ctrl-si) or MDA5-targeting siRNA (MDA5-si) were infected with HSV-1, and mRNA expression levels of the indicated genes analyzed by qRT-PCR. As discussed in more detail in text, the RLR pathway depicted in Fig. 2C operates in an intricate manner during infection by this virus. Error bars indicate ± SD. (n = 3). *, P < 0.01; **, P < 0.05. ND, not detected.
Discussion
Our present study clearly offers genetic evidence for the role of RLRs in the activation of innate immune responses by cytosolic DNA, be it synthetic or pathogen-derived, on the one hand, and further show divergence of the DNA signaling pathways. It is interesting that while the cytosolic activation of innate immune responses by RNA is mediated entirely by RLRs, DNA-mediated activation diverges into multiple pathways. The immunological significance of this finding is an interesting issue. Along this line, it is also worth noting that DNA and not RNA activate the AIM2 pathway that induces formation of inflammasomes (Fig. 2C) (26, 27). One may speculate that unlike the case for RNA viruses, DNA viruses (and other DNA-containing pathogens) with their larger genome sizes have acquired from the host or co-occurring parasites and symbionts additional gene products that interfere with the host's immune responses (28, 29). Thus, the host immune system has had to coevolve multiple activation pathways so as to thwart immune evasion by DNA-containing pathogens. Indeed, numerous sensors other than those for nucleic acids have been identified for these pathogens (1, 21, 28, 30). Therefore, our present findings on the deliberate divergence of the cytosolic DNA-mediated activation pathways, as well as the more versatile responses to DNA virus, may further provide mechanistic insights on the complex nature of the innate immune system activated by nucleic acids and other pathogen-associated molecules. Our study may also have implications for improved manipulation of the immune system by the selective targeting of one or more of the diverged pathways.
After completion of this work, two reports were published that show the activation of RIG-I by B-DNA through an RNA intermediate generated by RNA polymerase III transcription (31, 32). Although not necessarily inconsistent with our findings, a number of aspects of their results differ from ours. First, we and others used different cell types; hence, it is possible that there is a cell type-specific utilization of these two pathways, indicating a more heterogeneous usage of these detection systems than previously thought. For example, pathway X (as depicted in Fig. 2C) functioning in MEFs and HeLa cells may not operate in some cells, such as 293 cells (31, 32). This notion is congruent with our previous reports, which show a major role for DAI in mouse L929 cells, but little or none in MEFs (7, 9). It is also possible that the mode of RLR activation depends on the structure of the immunogenic DNA in which the DNA itself, the DNA-derived RNA, or both may activate RLRs. Finally, it is clear from our results (Fig. 4) and the other reports, that during the course of virus infection, multiple, additional non-nucleic acid sensing system operate in the detection of viral nucleic acids. In summary, our present study, together with other reports, reveal the critical role of RLRs in the cytosolic DNA-mediated activation of innate immune response on the one hand while on the other hand, also opening an avenue of research on the complex nature of the DNA sensing and signaling systems.
Materials and Methods
Cells and Reagents.
RIG-I+/− and RIG-I−/− MEFs and an Ifnar1−/− MEF line (ARF1) and HeLa cells were maintained as previously described (4, 7). B-DNA (7), poly(dG:dC)·(dC:dG), calf thymus DNA, and actinomycin D were purchased from Sigma. Biotin-conjugated B-DNA and other oligo DNAs including ISD were purchased from Hokkaido System Science and Fasmac, respectively. Purified vaccinia virus (MO strain) DNA was kindly provided by A. Kato and M. Kidokoro. HSV-1 DNA was generously provided by Y. Kawaguchi. 5′-triphosphate RNA was gifted by C. Reis e Sousa and J. Rehwinkel. E. coli K12 DNA was purchased from InvivoGen. Poly(I:C) was purchased from GE Healthcare Biosciences. B-DNA, poly(I:C) and other nucleic acid ligands were used at a concentration of 10 μg/mL, unless otherwise mentioned. Transfection of nucleic acid ligands were previously described (7). Antibodies against the following proteins were used: IRF3 (ZM3; Zymed) and GST (B-14; Santa Cruz Biotechnology).
Plasmid Constructions.
Murine RIG-I and MDA5 cDNA were obtained by PCR with reverse transcription (RT-PCR) and then cloned into the SalI and NotI sites of the pMSCVpac-FLAG vector (7). A deletion mutant RIG-IΔCARD (218–925) was isolated by PCR and inserted into the XhoI and NotI sites of the pCAGGS-HA vector (7). To generate GST-tagged RIG-I and RIG-IΔCARD expression vectors, GST cDNA was excised from pGEX4T3-GST and GST, RIG-I and RIG-IΔCARD cDNA were cloned into the EcoRI and NotI sites of the pCXN2 vector (7).
Purification of Recombinant RIG-I and RIG-IΔCARD Proteins.
The fusion proteins of GST-RIG-I and GST-RIG-IΔCARD were purified as described previously (9). The recombinant proteins were isolated to approximately 90% purity, as measured by Coomassie brilliant blue staining.
Quantitative RT-PCR Analysis.
Quantitative RT-PCR (qRT-PCR) analysis was performed as described previously (7). Primer sequences are in the SI Text.
RNA Interference.
Small interfering (si) RNA vectors were constructed by inserting oligonucleotides into EcoRI and HindIII sites of the pSUPER.retro.puro vector (OligoEngine). Retroviruses were prepared by transfection of each pSUPER vector along with pCL-Eco (encoding gag, pol and an ecotropic envelope; Imgenix), or with pMD.OGP (encoding gag and pol) plus pAmpho or pVSV-G (Clontech) into HEK293T cells. Retroviral gene transfer was carried out as described previously (7). Transduced cells were selected by puromycin (2.0 μg/mL; Sigma) for 48 h. The siRNA targeting sequences are in the SI Text.
Pull-Down Assay.
Pull-down assay was performed essentially as described previously (7).
Immunoblot Analysis.
Cell lysis and immunoblot analysis were carried out as described previously (7).
Electrophoretic Mobility Shift Assay (EMSA).
Whole-cell protein extracts (40 μg) were analyzed by EMSA with a 32P-radiolabelled oligonucleotide probe containing a consensus NF-κB binding sequence (7).
Viral Infection.
Cells were infected with 1.0 multiplicity of infection (M.O.I.) of HSV-1 (7). Virus preparation was described previously (7).
Virus Yield Assay.
The yield of HSV-1 was measured by a plaque forming assay that was carried out as described previously (7). All data were reproduced in two additional independent experiments.
Statistical Analysis.
Differences between control and experimental groups were evaluated using the Student's t test.
Supplementary Material
Acknowledgments.
We thank Caetano Reis e Sousa (London Research Institute, London, United Kingdom) and Jan Rehwinkel (London Research Institute, London, United Kingdom) for 5′-triphosphate RNA; Yasushi Kawaguchi (University of Tokyo, Tokyo, Japan) for HSV-1 DNA; Atsushi Katoh (National Institute of Infectious Diseases, Tokyo, Japan) and Minoru Kidokoro (National Institute of Infectious Diseases, Tokyo, Japan) for the vaccinia virus (MO) genome for ligand stimulation; Ruslan Medzhitov, Jeffrey V. Ravetch, Jan Vilcek, Hiroshi Handa, and Yuki Yamaguchi for invaluable advice; and Rie Takeda and Masashi Shishido for technical assistance. This work was supported in part by Grant-in-Aid for Scientific Research on Priority Areas “Integrative Research Toward the Conquest of Cancer,” Grant-in-Aid for Scientific Research (A), and Global Center of Excellence Program “Integrative Life Science Based on the Study of Biosignaling Mechanisms” from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and by the Korea Science and Engineering Foundation Grant. T.B. and Z.W. are research fellows of the Japan Society for the Promotion of Science.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909545106/DCSupplemental.
References
- 1.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. 2007;449:819–826. doi: 10.1038/nature06246. [DOI] [PubMed] [Google Scholar]
- 3.Tamura T, Yanai H, Savitsky D, Taniguchi T. The IRF family transcription factors in immunity and oncogenesis. Annu Rev Immunol. 2008;26:535–584. doi: 10.1146/annurev.immunol.26.021607.090400. [DOI] [PubMed] [Google Scholar]
- 4.Ishii KJ, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- 5.Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 6.Schroder K, Muruve DA, Tschopp J. Innate immunity: Cytoplasmic DNA sensing by the AIM2 inflammasome. Curr Biol. 2009;19:R262–265. doi: 10.1016/j.cub.2009.02.011. [DOI] [PubMed] [Google Scholar]
- 7.Takaoka A, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- 8.Ishii KJ, et al. TANK-binding kinase-1 delineates innate and adaptive immune responses to DNA vaccines. Nature. 2008;451:725–729. doi: 10.1038/nature06537. [DOI] [PubMed] [Google Scholar]
- 9.Wang Z, et al. Regulation of innate immune responses by DAI (DLM-1/ZBP1) and other DNA-sensing molecules. Proc Natl Acad Sci USA. 2008;105:5477–5482. doi: 10.1073/pnas.0801295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 11.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 12.Cheng G, Zhong J, Chung J, Chisari FV. Double-stranded DNA and double-stranded RNA induce a common antiviral signaling pathway in human cells. Proc Natl Acad Sci USA. 2007;104:9035–9040. doi: 10.1073/pnas.0703285104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang DC, et al. Mda-5: An interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc Natl Acad Sci USA. 2002;99:637–642. doi: 10.1073/pnas.022637199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pichlmair A, Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 15.Yoneyama M, Fujita T. Structural mechanism of RNA recognition by the RIG-I-like receptors. Immunity. 2008;29:178–181. doi: 10.1016/j.immuni.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 16.Kumar H, et al. Essential role of IPS-1 in innate immune responses against RNA viruses. J Exp Med. 2006;203:1795–1803. doi: 10.1084/jem.20060792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong B, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29:538–550. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 19.Malathi K, Dong B, Gale M, Jr, Silverman RH. Small self-RNA generated by RNase L amplifies antiviral innate immunity. Nature. 2007;448:816–819. doi: 10.1038/nature06042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rasmussen SB, et al. Herpes simplex virus infection is sensed by both Toll-like receptors and retinoic acid-inducible gene- like receptors, which synergize to induce type I interferon production. J Gen Virol. 2009;90:74–78. doi: 10.1099/vir.0.005389-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kurt-Jones EA, et al. Herpes simplex virus 1 interaction with Toll-like receptor 2 contributes to lethal encephalitis. Proc Natl Acad Sci USA. 2004;101:1315–1320. doi: 10.1073/pnas.0308057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lopez CB, et al. TLR-independent induction of dendritic cell maturation and adaptive immunity by negative-strand RNA viruses. J Immunol. 2004;173:6882–6889. doi: 10.4049/jimmunol.173.11.6882. [DOI] [PubMed] [Google Scholar]
- 23.Paladino P, Cummings DT, Noyce RS, Mossman KL. The IFN-independent response to virus particle entry provides a first line of antiviral defense that is independent of TLRs and retinoic acid-inducible gene I. J Immunol. 2006;177:8008–8016. doi: 10.4049/jimmunol.177.11.8008. [DOI] [PubMed] [Google Scholar]
- 24.Honda K, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 25.Rasmussen SB, et al. Type I interferon production during herpes simplex virus infection is controlled by cell-type-specific viral recognition through Toll-like receptor 9, the mitochondrial antiviral signaling protein pathway, and novel recognition systems. J Virol. 2007;81:13315–13324. doi: 10.1128/JVI.01167-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burckstummer T, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10:266–272. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 27.Hornung V, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iyer LM, Balaji S, Koonin EV, Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117:156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 29.Seet BT, et al. Poxviruses and immune evasion. Annu Rev Immunol. 2003;21:377–423. doi: 10.1146/annurev.immunol.21.120601.141049. [DOI] [PubMed] [Google Scholar]
- 30.Boyle KA, Pietropaolo RL, Compton T. Engagement of the cellular receptor for glycoprotein B of human cytomegalovirus activates the interferon-responsive pathway. Mol Cell Biol. 1999;19:3607–3613. doi: 10.1128/mcb.19.5.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ablasser A, et al. RIG-I-dependent sensing of poly(dA:dT) through the induction of an RNA polymerase III-transcribed RNA intermediate. Nat Immunol. 2009 doi: 10.1038/ni.1779. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiu YH, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138:576–591. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.