Abstract
Glycosylphosphatidylinositol-anchored proteins (GPI-APs) play various roles in cell–cell and cell–environment interactions. GPI is synthesized in the endoplasmic reticulum (ER) from phosphatidylinositol (PI) through step-wise reactions including transfers of monosaccharides and preassembled GPI is transferred en bloc to proteins. Cellular PI contains mostly diacyl glycerol and unsaturated fatty acid in the sn-2 position, whereas mammalian GPI-APs have mainly 1-alkyl-2-acyl PI and almost exclusively stearic acid, a saturated chain, at the sn-2 position. The latter characteristic is the result of fatty acid remodeling occurring in the Golgi, generating GPI-anchors compatible with raft membrane. The former characteristic is the result of diacyl to alkyl-acyl change occurring in the third GPI intermediate, glucosaminyl-inositolacylated-PI (GlcN-acyl-PI). Here we investigated the origin of the sn-1 alkyl-chain in GPI-APs. Using cell lines defective in the peroxisomal alkyl-phospholipid biosynthetic pathway, we demonstrated that generation of alkyl-containing GPI is dependent upon the peroxisomal pathway. We further demonstrated that in cells defective in the peroxisome pathway, the chain composition of the diacyl glycerol moiety in GlcN-acyl-PI is different from those in the first intermediate N-acetylglucosaminyl-PI and cellular PI, indicating that not only diacyl to alkyl-acyl change but also diacyl to diacyl change occurs in GlcN-acyl-PI. We therefore propose a biosynthetic step within GlcN-acyl-PI in which the diacyl glycerol (or diacyl phosphatidic acid) part is replaced by diradyl glycerol (or diradyl phosphatidic acid). These results highlight cooperation of three organelles, the ER, the Golgi, and the peroxisome, in the generation of the lipid portion of GPI-APs.
Keywords: alkyl lipids, glycosylphosphatidylinositol, lipid remodeling
More than 100 human cell surface proteins with various functions are glycosylphosphatidylinositol-anchored proteins (GPI-APs) (1, 2). GPI-APs are typical components of membrane microdomains or lipid rafts (3). GPI-APs exist as nanoclusters or single molecules, and upon clustering induced by ligands, transduce signals through src family tyrosine kinases that are also typical raft components and G proteins (4, 5). GPI-APs include receptors, adhesion molecules, various enzymes, and complement regulatory proteins, and play roles in cell–cell and cell–environment interactions. As demonstrated by knockout mice, complete deficiency of biosynthesis of GPI anchors causes early embryonic lethality, showing the essential roles of GPI-APs in embryogenesis (6). Inherited partial deficiency of GPI biosynthesis causes a disease termed inherited GPI deficiency characterized by portal vein thromboses and seizures (7). Acquired GPI deficiency due to somatic mutations in the hematopoietic stem cells causes paroxysmal nocturnal hemoglobinuria characterized by complement-mediated hemolysis (8).
GPI is synthesized in the endoplasmic reticulum (ER) from phosphatidylinositol (PI) through stepwise reactions including transfers of N-acetylglucosamine (GlcNAc), palmitic, or myristic acid, mannoses, and ethanolaminephosphates, and de-N-acetylation of GlcNAc (1, 9, 10). Preassembled GPI precursor is transferred en bloc as a posttranslational modification to the carboxy-terminus of a protein having a carboxy-terminal GPI-attachment signal peptide (1). GPI-AP is then transported via the secretory pathway to the Golgi where the glycan portion can be further modified by side branch(es) and the PI moiety is subjected to fatty acid remodeling that makes raft-compatible lipid structure (9, 11). Fatty-acid-remodeled GPI-APs are associated with lipid rafts and expressed on the cell surface (9, 12).
In mammalian cells, most free PIs are diacyl glycerol forms, in which the sn-1 chain is usually saturated and the sn-2 chain is unsaturated. Among them, 1-stearoyl-2-arachidonoyl PI is predominant (13). Stearic acid has a saturated C18 chain and arachidonic acid has a polyunsaturated C20 chain. In contrast, mammalian GPI-APs have mainly 1-alkyl-2-acyl PI (alkyl-acyl PI), in which the sn-1 chain is usually saturated C16 or C18 ether and the sn-2 chain is usually stearic acid (13). This unique structure of the PI moiety of GPI-APs is elaborated from cellular PI by means of two distinct remodelings. First is the diacyl to alkyl-acyl change that occurs during biosynthesis of GPI precursors in the ER. The resulting 1-alkyl-2-acyl PI moiety in GPI has an unsaturated (often polyunsaturated) sn-2 chain. The second remodeling (fatty acid remodeling) is an exchange of the unsaturated sn-2 fatty acid, such as arachidonic acid, with stearic acid that occurs in GPI-APs after they are transported into the Golgi. The molecular basis of the second remodeling has been well characterized (12). The unsaturated sn-2 chain is removed to generate lyso-GPI-AP and then stearic acid is transferred back to the sn-2 position, most likely from stearoyl-CoA. The Golgi membrane proteins PGAP3 and PGAP2 are required for the former and latter steps, respectively (12, 14). When PGAP3 is defective, GPI-APs with unremodeled PI having an unsaturated sn-2 chain are expressed on the cell surface. However, those GPI-APs are not recovered in the detergent resistant membrane (DRM) fraction while GPI-APs from wild-type cells are efficiently recovered in the DRM fraction (12). Therefore, fatty acid remodeling is critical for raft association of GPI-APs (12). In contrast, the molecular basis of the first remodeling of the PI moiety, the diacyl to alkyl-acyl change, has not been clarified. We reported previously that the diacyl to alkyl-acyl change occurs in the third biosynthetic intermediate GlcN-acyl-PI (13). However, it is unclear whether the sn-1 acyl chain of GlcN-acyl-PI is exchanged with an alkyl chain or the diacyl glycerol part (or diacyl phosphatidic acid part) is exchanged with 1-alkyl-2-acyl glycerol (or 1-alkyl-2-acyl phosphatidic acid).
In the present study, we asked whether the alkyl-acyl GPI is dependent upon the alkyl phospholipids biosynthetic pathway in the peroxisome. This is the only known pathway that generates sn-1 alkyl glycerol backbone (15). We used two mutant Chinese hamster ovary (CHO) cell lines defective in the peroxisomal pathway (16, 17). These mutant cells generated only diacyl form GPI, indicating that biosynthesis of alkyl-acyl GPI in the ER is dependent upon alkyl-phospholipid biosynthesis in the peroxisome. It is also indicated that the diacyl glycerol or diacyl phosphatidic acid part is exchanged with a diradyl counterpart derived from a putative donor lipid.
Results
Generation of a Protein-Linked Alkyl-Acyl GPI-Anchor Is Dependent Upon the Peroxisomal DHAP-ATase.
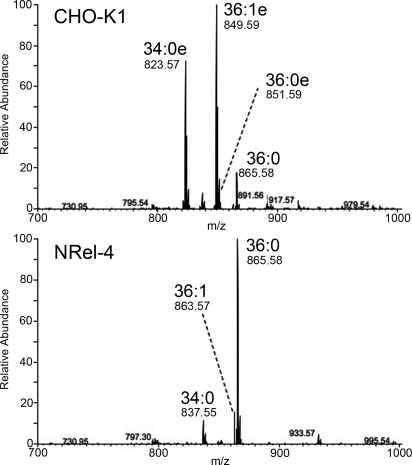
To determine whether the alkyl-acyl form of GPI is generated in cells defective in the peroxisomal alkyl-phospholipid biosynthetic pathway, we used wild-type CHO-K1 cells and mutant NRel-4 cells defective in dihydroxyacetone phosphate acyltransferase (DHAP-ATase), the first enzyme in the pathway (Fig. 1). We generated CHO-K1 and NRel-4 cells stably expressing GPI-anchored CD59 with tandem His- FLAG- GST- and FLAG-tags at the N terminus (HFGF-CD59). HFGF-CD59 was purified with glutathione-Sepharose affinity chromatography and about 1 nM of proteins were obtained. PI moiety was released from HFGF-CD59 by sodium nitrite treatment (18, 19) and analyzed by an LTQ Orbitrap mass spectrometer (Fig. 2). About 90% of the PIs were the alkyl-acyl form in HFGF-CD59 from CHO-K1 cells, whereas almost all of the PIs were the diacyl form in the same protein from NRel-4 cells. The major species were 34:0 alkyl-acyl (34:0e), 36:1 alkyl-acyl (36:1e), and 36:0 alkyl-acyl (36:0e) in CHO-K1 cells, and that in NRel-4 was 36:0 diacyl. Therefore, cells defective in the peroxisomal alkyl-phospholipid biosynthetic pathway did not produce the 1-alkyl-2-acyl form of GPI-anchors.
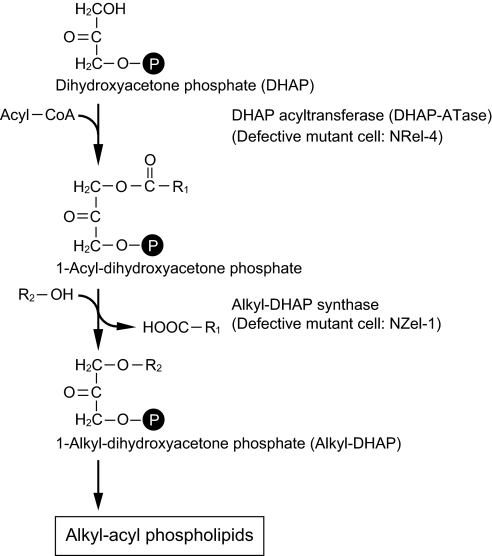
Fig. 1.
Early steps in alkyl-phospholipid biosynthetic pathway in the peroxisome and two defective mutant cells. NRel-4 and NZel-1 derived from CHO cells are defective in dihydroxyacetone-phosphate acyltransferase (DHAP-ATase) and 1-alkyl-dihydroxyacetone-phosphate (alkyl-DHAP) synthase, respectively.
Fig. 2.
GPI-APs from NRel-4 cells have only the diacyl form GPI. Mass analysis of PI moieties in HFGF-tagged CD59 from the plasma membrane of wild-type CHO-K1 (top) and alkyl-phospholipid biosynthesis mutant NRel-4 (bottom) cells. In CD59 from CHO-K1 cells, alkyl-acyl species (34:0e, 36:1e, and 36:0e) were dominant whereas only diacyl species (34:0, 36:1, and 36:0) were found in CD59 from NRel-4 cells.
Peroxisomal DHAP-ATase and Alkyl-DHAP Synthase Are Required for Biosynthesis of Alkyl-Acyl GPI Precursors.
To determine whether alkyl-acyl form GPI precursors are generated in cells defective in the peroxisomal alkyl-phospholipid biosynthetic pathway, we took two approaches to prepare conditions under which GPI-anchor precursors are accumulated in cells so that analysis for alkyl-acyl GPI is possible. In the first approach, we used a chemical compound BE49385A/YW3548, an inhibitor of PIG-N. When PIG-N-mediated transfer of ethanolamine phosphate to the first mannose is inhibited, mannose-containing GPIs are accumulated due to the inefficient transfer of the third mannose (20). In the second approach, we generated DPM2-defective mutants from NRel-4 and CHO-K1 cells. DPM2 is required for synthesis of dolichol-phosphate mannose (21) that donates mannose to GlcN-acyl-PI, hence in DPM2-deficient cells, GlcN-acyl-PI is accumulated, which makes structural analysis feasible (13).
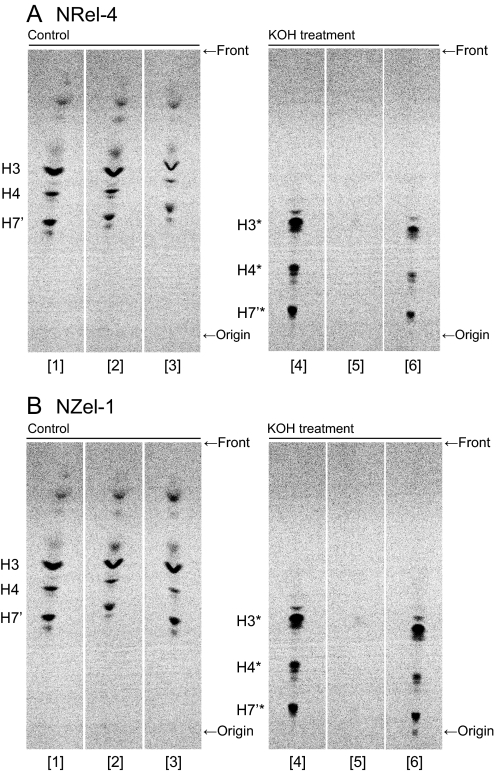
First, we cultured NRel-4, DHAP-ATase-transfected NRel-4, and parental CHO-K1 cells in the presence of radiolabeled mannose and the PIG-N inhibitor (Fig. 3A, lanes 1 to 3). The presence of the alkyl structure in the accumulated GPI precursors (H3 to H7′) was assessed by treatment of 1-butanol-extracted lipids with methanolic KOH for deacylation. In CHO-K1 cells, several mannolipids representing the inositol-deacylated 1-alkyl-2-lyso form of GPI precursors (H3* to H7′*) were observed after KOH treatment, as expected (lane 4), whereas in NRel-4 cells, all spots disappeared (lane 5). When the responsible DHAP-ATase cDNA was transfected into NRel-4 cells, the spots of lyso-forms appeared (lane 6). The quantitative recovery of lipids after KOH-treatment was confirmed by staining the TLC plate with molybdate for phospholipids. Similar profiles of spots were observed for both wild-type and mutant cell samples (Fig. S1). We analyzed NZel-1 and alkyl-DHAP synthase transfected NZel-1 cells in a similar way and obtained similar results (Fig. 3B). These results indicate that cells defective in the first and second enzymes in the peroxisomal alkyl-phospholipid biosynthetic pathway did not produce 1-alkyl-2-acyl forms of mannose-containing late GPI precursors. Therefore, the peroxisomal pathway is required for production of 1-alkyl-2-acyl GPI.
Fig. 3.
Roles of DHAP-ATase (A) and alkyl-DHAP synthase (B) in GPI biosynthesis. (A) Parental CHO-K1 cells (lanes 1 and 4), DHAP-ATase defective NRel-4 cells (lanes 2 and 5), and DHAP-ATase-transfected NRel-4 cells (lanes 3 and 6) were metabolically labeled with D-[2–3H]mannose in the presence of BE49385A, PIG-N-inhibitor, to accumulate late GPI precursors. The extracted lipids were treated with methanol as a control (lanes 1 to 3) or 0.1 N KOH in methanol (lanes 4 to 6) for 1 h and analyzed by TLC with a solvent system of chloroform/methanol/H2O (10:10:3). H3-H7′, mannose-containing GPI precursors; H3*-H7′*, alkali-resistant part of H3-H7′. (B) Parental CHO-K1 cells (lanes 1 and 4), alkyl-DHAP synthase defective NZel-1 cells (lanes 2 and 5), and alkyl-DHAP synthase-transfected NZel-1 cells (lanes 3 and 6) were analyzed in a similar way as panel (A). These are representative data of the similar experiments repeated three times.
Second, to determine the lipid structure of early GPI precursors in CHO-K1 and NRel-4 cells, we established CHO-K1-ΔDPM2 and NRel-4-ΔDPM2 cells from ethylmethane sulfonate (EMS)-treated CHO-K1 and NRel-4, respectively, by sorting and limiting dilution of CD59-negative cells. We know that DPM2-deficient CHO cells are relatively easily generated after EMS-treatment (22). CHO-K1-ΔDPM2 and NRel-4-ΔDPM2 lost the surface expression of CD59 (Fig. S2, closed area). Transfection of DPM2 cDNA restored the CD59 surface expression (open area), showing that the defective surface expression of GPI-AP was due to a defect in DPM2.
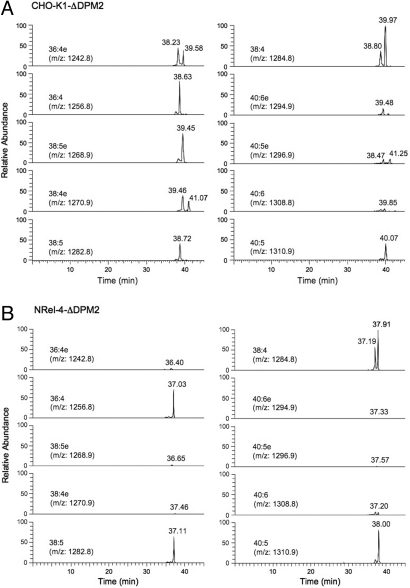
We extracted total lipids from about 108 cells and analyzed them using an LTQ Orbitrap mass spectrometer. GlcN-acyl-PI accumulated in DPM2 defective cells contained either palmitoylated (-Pal) or myristoylated (-Myr) inositol, as reported previously (13). GlcN-acyl-PI as determined by generation of diagnostic fragment ions, [GlcN-Ins-P]−, [GlcN-Ins(-Myr)-P]−, and [GlcN-Ins(-Pal)-P]−, appeared between 37 and 42 min of retention time on liquid chromatography (LC) (Fig. 4). The molecular species were analyzed according to Houjou et al. (13). The parental ions of m/z = 1,242.8 (36:4 alkyl-acyl); 1,256.8 (36:4 diacyl); 1,268.9 (38:5 alkyl-acyl); 1,270.9 (38:4 alkyl-acyl); 1,282.8 (38:5 diacyl); 1,284.8 (38:4 diacyl); 1,294.9 (40:6 alkyl-acyl); 1,296.9 (40:5 alkyl-acyl); 1,308.8 (40:6 diacyl); and 1,310.9 (40:5 diacyl) were assessed in both cells (Fig. 4). As summarized in Table 1, the PI part of GlcN-acyl-PI in CHO-K1-ΔDPM2 cells consisted of 51% alkyl-acyl form and 49% diacyl form. In both alkyl-acyl and diacyl forms, 38:4, 38:5, and 36:4 species were major. In contrast, almost all GlcN-acyl-PI in NRel-4-ΔDPM2 cells were diacyl forms with 38:4 species contributing about 40%, and 40:5, 38:5, and 36:4 species contributing 15–20% each (Fig. 4B and Table 1). These results indicate that DHAP-ATase is required for the production of alkyl-acyl form GlcN-acyl-PI.
Fig. 4.
LC-Orbitrap mass spectrometric analysis of GlcN-acyl-PI accumulated in CHO-K1-ΔDPM2 (A) and NRel-4-ΔDPM2 (B) cells. Species of GlcN-acyl-PI with various fatty chain combinations are indicated as (total number of carbons):(number of unsaturated bonds). Those with e represent alkyl-acyl species and those without e represent diacyl species. Elution time from LC is shown in minutes.
Table 1.
Fatty chain profiles of PI moiety of GlcN-acyl-PI in CHO-K1-ΔDPM2 and NRel-4-ΔDPM2 cells
| PI | CHO-K1-ΔDPM2, percent* | NRel-4-ΔDPM2, percent* |
|---|---|---|
| 36:3† | 1.90 ± 0.10 | 1.39 ± 0.17 |
| 36:4 | 6.11 ± 0.72 | 13.77 ± 2.12 |
| 38:3 | 2.57 ± 0.19 | 3.01 ± 0.67 |
| 38:4 | 15.24 ± 0.96 | 26.79 ± 1.03 |
| 38:5 | 4.93 ± 0.53 | 12.82 ± 1.98 |
| 38:6 | 1.10 ± 0.24 | 2.66 ± 0.38 |
| 40:4 | 1.69 ± 0.31 | 6.20 ± 1.13 |
| 40:5 | 8.17 ± 0.07 | 22.57 ± 1.61 |
| 40:6 | 3.06 ± 0.05 | 6.07 ± 0.21 |
| Subtotal | 44.77 | 95.28 |
| 34:1e‡ | 5.47 ± 0.25 | 0.00 |
| 36:1e | 4.18 ± 0.30 | 0.00 |
| 36:2e | 5.29 ± 0.28 | 0.00 |
| 36:4e | 7.64 ± 0.09 | 0.00 |
| 38:3e | 1.77 ± 0.03 | 0.00 |
| 38:4e | 3.42 ± 0.20 | 0.00 |
| 38:5e | 10.99 ± 0.10 | 0.02 ± 0.01 |
| 38:6e | 3.42 ± 0.13 | 0.00 |
| 40:6e | 4.05 ± 0.10 | 0.00 |
| 40:7e | 3.06 ± 0.92 | 0.00 |
| Subtotal | 49.29 | 0.02 |
| Total | 94.06 | 95.30 |
*Mean ± S.D. of two experiments. Species less than 1% are not listed.
†Number of total carbons: number of unsaturated bonds.
‡e indicates 1-alkyl-2-acyl form.
Different Fatty Chain Compositions of PI Moiety Between GlcNAc-PI/GlcN-PI and GlcN-acyl-PI, Evidence for a Biosynthetic Step.
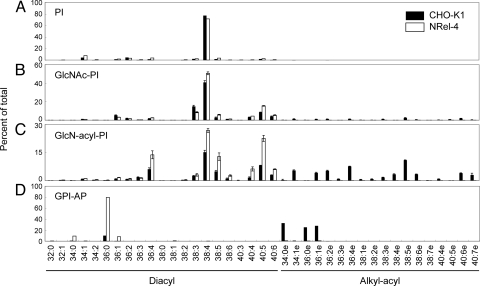
We reported previously that 38:4 diacyl species (1-stearoyl-2-arachidonoyl) is predominant in PI and the first two GPI intermediates, GlcNAc-PI and GlcN-PI, in CHO-K1 cells (Fig. 5 A and B) (13). It is indicated that GPI biosynthesis is initiated using endogenous cellular PI and the PI moiety remains unchanged or only minimally changed through to GlcN-PI. The results shown in Fig. 5C and Table 1 indicate that less than 30% of GlcN-acyl-PI contained 38:4 diacyl PI species in DPM2-defective NRel-4 cells and that the rest of GlcN-acyl-PI had 36:4, 38:5, and 40:5 diacyl PI species, indicating that the diacyl PI moiety in GlcN-acyl-PI is different from those in GlcNAc-PI and GlcN-PI. To confirm this point under the same background of NRel-4 cells, we established a PIG-L-defective NRel-4 cell that accumulates GlcNAc-PI. We know that PIG-L-deficient mutant cell is easily generated from CHO cells. As expected, 38:4 diacyl PI species was predominant in GlcNAc-PI from PIG-L-deficient NRel-4 cells (Fig. 5B). Therefore, the fatty acid compositions in GlcNAc-PI/GlcN-PI are different from that in GlcN-acyl-PI. This is also true in alkyl-phospholipid-sufficient CHO-K1 cells because when only the diacyl PI-containing GlcN-acyl-PI form is counted, 38:4 diacyl PI species accounted for only 34% (Fig. 5C and Table 1), indicating that the diacyl PI moiety also had different chain compositions between GlcNAc-PI/GlcN-PI and GlcN-acyl-PI. These results suggest that the diacyl glycerol or phosphatidic acid portion is changed in GlcN-acyl-PI, that is, there is one more biosynthetic step. This supports the idea that diacyl to alkyl-acyl conversion occurring in GlcN-acyl-PI is the result of exchange of diradyl-glycerol or -phosphatidic acid.
Fig. 5.
Fatty chain profiles of PI and PI moiety of GPI precursors and GPI-APs. (A) Cellular PI; (B) GlcNAc-PI; (C) GlcN-acyl-PI; and (D) HFGF-CD59. Closed bars, CHO-K1 (A and D), PIG-L-defective CHO-K1 (B), and CHO-K1-ΔDPM2 (C); open bars, NRel-4 (A and D), PIG-L-defective NRel-4 (B), and NRel-4-ΔDPM2 (C). Left part, diacyl species; right part, 1-alkyl-2-acyl species. Data in (B) and (C) are mean+/-SD of two experiments.
Fatty Acid Remodeling in the Golgi Occurs in the Absence of the Peroxisomal Alkyl-Phospholipid Biosynthesis.
The PI moiety of GlcN-acyl-PI in NRel-4 cells consisted of 36:4, 38:4, 38:5, 40:5, and 40:6 diacyl species, corresponding to combinations of palmitic, stearic, or oleic acid in the sn-1 position and a polyunsaturated fatty acid in the sn-2 position (Fig. 5C, open bars). The PI moiety of protein-linked GPI in the same cells consisted of 34:0, 36:0, and 36:1 diacyl species, corresponding to combinations of palmitic, stearic, or oleic acid in the sn-1 position and stearic acid in the sn-2 position (Fig. 5D, open bars). These fatty acid profiles represent chain compositions before and after fatty acid remodeling occurring in the Golgi, indicating that fatty acid remodeling takes place even with diacyl GPI-APs.
Discussion
A major finding of this study is that the peroxisomal alkyl-phospholipid biosynthetic pathway is required for biosynthesis of mammalian GPI-APs. Using CHO cell lines defective in DHAP-ATase or alkyl-DHAP synthase, the first two enzymes in the peroxisomal pathway, we demonstrated that both enzymes are required for generation of 1-alkyl-2-acyl-PI-containing GPI precursors and protein-bound GPI anchors. We propose that some 1-alkyl containing lipid generated in the peroxisome is transported to the ER and is used as is or after further modification, as a donor of 1-alkyl-2-acyl glycerol or a phosphatidic acid moiety in GlcN-acyl-PI. These results show that three organelles, the ER, the Golgi, and the peroxisome, are involved in biosynthesis of mammalian GPI-APs.
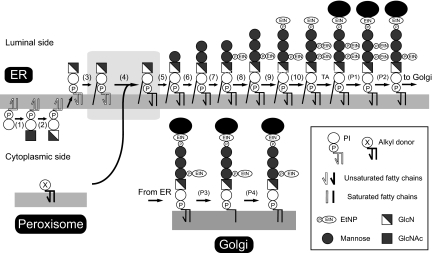
The second finding of this study is that diacyl to alkyl-acyl change in GlcN-acyl-PI is a part of diacyl to diradyl change. Fifty percent or more of GlcN-acyl-PI consists of the 1-alkyl-2-acyl form, while the rest remains as the diacyl form. The chain composition of the diacyl form of GlcN-acyl-PI is different from that of GlcNAc-PI, indicating that even the diacyl form of GlcN-acyl-PI is the product of remodeling. We therefore conclude that there is a biosynthetic step within GlcN-acyl-PI and propose a revised GPI biosynthetic pathway, in which diacyl form GlcN-acyl-PI is the third intermediate and diradyl form GlcN-acyl-PI is the fourth intermediate (Fig. 6).
Fig. 6.
The revised GPI biosynthetic pathway, highlighting cooperation of three organelles. The outlined lipid moieties represent the diacyl form and filled lipid moieties the alkyl-acyl or diacyl form. The GPI biosynthesis is initiated by transfer of N-acetylglucosamine to PI (step 1) followed by de-N-acetylation (step 2) and flipping into the ER lumen, which most likely occurs at this stage. PI and the first two GPI intermediates have an unsaturated acyl chain in the sn-2 position as represented by a kinked line. In step 3, the acyl (mainly palmitoyl or myristoyl) chain is transferred to the inositol ring to generate GlcN-acyl-PI. We propose a reaction step, an exchange of the lipid part (diacylglycerol or phosphatidic acid) (step 4). A major fraction of a putative lipid donor (lipid X) contains 1-alkyl-2-acyl glycerol generated in the peroxisome. GlcN-acyl-PI now bearing diradyl glycerol, entirely different from diacyl glycerol in cellular PI, is further processed to steps 5–10 to generate the complete GPI precursor ready for attachment to proteins by GPI transamidase (TA). The removals of inositol-linked acyl chain (step P1) and ethanolaminephosphate linked to the second mannose (step P2) occur before transportation to the Golgi. In the Golgi, fatty acid remodeling occurs in that an unsaturated sn-2 chain is removed (step P3) and stearic acid is transferred back to the sn-2 position (step P4).
The reaction mechanism of the diacyl GlcN-acyl-PI to diradyl GlcN-acyl-PI change is yet to be determined. It is unlikely that the sn-1-acyl chain is changed to a 1-alkyl chain, a reaction similar to the second step in the peroxisomal alkyl-phospholipid biosynthetic pathway (Fig. 1), because both the first and the second enzymes are required, suggesting that the pathway itself is required for generation of an alkyl donor lipid. Our current model is that the diacyl-glycerol or -phosphatidic acid part is replaced with diradyl-glycerol or -phosphatidic acid. Such a reaction may be mediated by some phospholipase C or D that can mediate an exchange reaction. In case of diacyl to diacyl change in GlcN-acyl-PI, we cannot exclude the possibility that sn-1 and sn-2 fatty acids are individually exchanged.
What could the donor lipid in diacyl to diradyl exchange be? Based on the composition of the product GlcN-acyl-PI (Table 1), the donor lipid would consist of 50–60% alkyl-acyl and 40–50% diacyl forms. The alkyl-acyl form would contain 36:4e, 38:4e, and 38:5e as major species and 40:5e and 40:6e as minor species. The diacyl form would contain 36:4 and 38:4 as major species and 38:5, 40:5, and 40:6 as minor species. We analyzed the chain compositions of phosphatidylcholine (PC) and phosphatidylethanolamine (PE) from ER-enriched membranes of CHO-K1 cells. PC did not seem to be a good candidate because only about 30% is the alkyl-acyl form and its major species are 34:1e, 36:1e, and 36:2e. PE was not excluded as a candidate because about 50% is the alkyl-acyl form and it contains 38:5e as a major species and 40:5e and 40:6e as minor species. Establishment of an in vitro reaction system is required to determine the donor lipid.
About 50–60% of GlcN-acyl-PI contains alkyl-acyl PI, whereas about 90% of GPI-APs have alkyl-acyl PI (Fig. 5 C and D). It appears that enrichment of alkyl-acyl form GPI occurs at a step between the first mannosylation and the GPI attachment to proteins. The H3 GPI species shown in Fig. 3 represents Man-Man-GlcN-acyl-PI. Radioactivity in H3 was not lost from the organic phase after alkali treatment, suggesting that most of the Man-Man-GlcN-acyl-PI is the alkyl-acyl form. It is therefore likely that enrichment of the alkyl-acyl form occurs before the step of the second mannose transfer. One possible mechanism of such enrichment is that alkyl-acyl forms of GlcN-acyl-PI and/or Man-GlcN-acyl-PI are better substrates of GPI-mannosyltransferase I and II than diacyl counterparts. Another is that the alkyl-acyl form is recruited more efficiently to GPI-mannosyltransferase I and/or II. There is a report that Arv1p is required for delivery of GlcN-acyl-PI to GPI-mannosyltransferase I in yeast and that Arv1p has a mammalian homolog (23). It was proposed that the function of yeast Arv1p might be to flip GlcN-acyl-PI from the cytoplasmic side to the luminal side of the ER (23). If this is true, diacyl to diradyl exchange could be coupled with or required for flipping. However, when flipping of GPI does occur (GlcN-PI or GlcN-acyl-PI) is not definitively determined. Our view is that GlcN-PI flips and is converted to GlcN-acyl-PI on the luminal side. If this is true, Arv1p may prefer alkyl-acyl form GlcN-acyl-PI in delivery to GPI-mannosyltransferase I.
The biological significance of 1-alkyl chain in GPI-APs is not understood yet. We addressed a possibility that 1-alkyl chain is important for raft association of GPI-APs. Diacyl GPI-APs in NRel-4 mutant cells, which mainly had two saturated acyl chains (Fig. 5), were recovered in DRM fraction as efficiently as GPI-APs in wild-type CHO cells (Fig. S3). Therefore, 1-alkyl chain does not seem to be essential for raft association. A clear difference between the alkyl and acyl chains is sensitivity to hydrolytic enzymes, i.e., an ether bond linking the alkyl chain is resistant whereas an ester bond linking the acyl chain is sensitive. If some phospholipase B enzyme cleaves GPI-APs, diacyl form GPI-APs would result in hydrophilic proteins bearing only GPI glycan, whereas the alkyl-acyl form would become amphiphilic proteins bearing an alkyl chain. Although catalytic specificities are not phopholipase B-like, several lines of evidence suggest biological significance of cleavage in the GPI portion of GPI-APs. Nortum cleaves GPI-anchor and releases glipicans and modulates effects of Wnt (24). Cleavage of sperm GPI-APs by angiotensin converting enzyme (ACE), particularly testis-specific isoform t-ACE, is important for fertilization (25). For understanding roles of 1-alkyl chain in GPI-APs, identification of the gene involved in diacyl to alkyl-acyl conversion and its disruption are clearly required.
Materials and Methods
Cells and Materials.
The alkyl phospholipid biosynthesis mutant CHO cells, NRel-4, and NZel-1, were gifts from R. A. Zoeller (Boston University, MA) (16, 17). Cells were cultured in Ham's F12 medium with 10% FBS. We generated CHO-K1 and NRel-4 cells stably expressing HFGF-CD59. The HFGF-CD59 expressing cell lines were cultured in a medium containing 6 μg/mL of puromycin for maintenance of the HFGF-CD59 expression plasmid.
Complementation of NRel-4 and NZel-1 Cells with DHAP-ATase and Alkyl-DHAP Synthase cDNA, Respectively.
We purchased cDNA clones, which contain DHAP-ATase in pOTB7 plasmid and alkyl-DHAP synthase in pCMV-SPORT6 plasmid from Open Biosystems and Invitrogen, respectively. The sequences were confirmed by sequencing. A fragment containing DHAP-ATase cDNA obtained by EcoRI/XhoI digestion was cloned into pcDNA3.1/Zeo(+) plasmid (Invitrogen). Plasmids were electroporated into corresponding alkyl biosynthesis mutant cells.
Establishment of PIG-L Mutant and DPM2 Mutant Cell Lines.
Ten million each of CHO-K1-HFGF-CD59 and NRel-4-HFGF-CD59 cells in φ150-mm dishes were mutagenized with 300 μg/mL EMS (Sigma-Aldrich) for 24 h at 37 °C. For isolation of PIG-L mutant cells, the cells were harvested 2 weeks after mutagenesis and stained with 5H8 anti-CD59 antibody. The CD59-negative population was sorted by FACSAria (Becton Dickinson). The single clones were isolated by limiting dilution. The mutation was checked by recovery of CD59 expression on the cell surface after transfection of PIG-L expression plasmid, pMEEB-PIG-L-FLAG.
For DPM2 mutant cells, the mutagenized cells were cultured for 3 days and were then treated with 1 μg/mL swainsonine (Merck/Calbiochem) and 12 μg/mL Con A (Seikagaku. Corp.) for 40 days. PHA-E4 FITC (Seikagaku Corp.) positive and CD59 negative populations were sorted by FACSAria. The single clones were isolated by limiting dilution. The mutation was checked by recovery of CD59 expression on the cell surface after transfection of the DPM2 expression plasmid, pME-py-GST-hDPM2.
Purification of PI from the Cell-Surface GPI-Anchored Proteins.
We used 1 nM (47 μg) of HFGF-CD59 for analysis of the PI moiety. The NRel-4 and CHO-K1 cells (109 each) both expressing HFGF-CD59 were lysed in 100 mL of buffer A [60 mM 1-octyl-β-D-glucoside, 20 mM HEPES-NaOH, pH7.4, 150 mM NaCl and 1× protease inhibitor mixture with EDTA (Hoffmann-La Roche Ltd)] and centrifuged at 18,800 × g for 15 min at 4 °C. The supernatant was subjected to centrifugation at 103,745 × g for 1 h at 4 °C and supernatant was applied to a column with a 500-μL bed volume of glutathione-Sepharose 4B (GE Healthcare) at a flow rate of 1 mL/min at 4 °C. The column was washed with 5 mL of PBS with 1% TritonX-100. The HFGF-CD59 was eluted with 5 mL of 20 mM reduced glutathione, 30 mM HEPES-NaOH, pH 7.4 and 1% Triton X-100 and fractions of 1 mL were collected. The HFGF-CD59 containing fractions as assessed by SDS/PAGE were combined and the protein was precipitated with cold trichloroacetic acid. Pellets were washed twice by cold ethanol, air-dried, and resuspended in 200 μL of 2× sample buffer. Samples were applied to SDS/PAGE, and transferred to polyvinylidene difluoride (PVDF) membrane. The membrane was stained with 0.1% Ponceau S in 5% acetic acid, and the band corresponding to HFGF-CD59 was cut out. The membrane strips were washed with water four times, and incubated with a mixture of 500 μL of 0.3 M NaOAc, pH 4.0, buffer, and 500 μL of freshly dissolved 1 M sodium nitrite for 3 h at 37 °C (18, 19). After washing the membrane strips with 1 mL of water four times, PI was extracted in 400 μL of water-saturated butan-1-ol (BuOH) three times. The extracted PI in BuOH phase was dried under a gentle stream of nitrogen, dissolved in 200 μL of chloroform-methanol (C/M) (4:1), and immediately applied to a column (200-μL bed volume) of Si60 silica (Sigma-Aldrich) packed in a glass Pasteur pipette plugged with glass wool and prewashed three times with 1 ml of C/M (1:4) followed by three times with 1 mL of C/M (4:1). After washing the column with 400 μL of C/M (4:1) five times, PI was eluted with five portions of 200 μL of C/M (1:4), dried under a nitrogen stream, and resolved in 100 μL of C/M (2:3). The atmosphere in the tube was changed to nitrogen and it was stored at −80 °C until analysis by mass spectrometry.
In Vivo Labeling of Cells with 3H-Mannose.
The 5 × 105 cells were cultured overnight in a medium containing 10 μM BE49385A (a gift from Banyu Pharmaceutical) (20, 26). Cells were then cultured in 1 mL of 10 μg/mL tunicamycin (Sigma-Aldrich), 100 μg/mL glucose, and 10% dialyzed FBS in glucose-free RPMI medium 1640 (GIBCO/Invitrogen) for 1 h, then 10 μCi/mL of D-[2-3H]mannose (GE Healthcare Ltd.) was added and cells were cultured for 1 h. The total lipids were extracted from the cell pellets using two 300 μL portions of C/M/H2O (10:10:3), and the extracted total lipids were dried by a speed-vac. The GPI precursors were extracted using two 300 μL portions of water-saturated BuOH, back-washed with 200 μL of BuOH-saturated water, and dried. To determine the alkyl-acyl form of PI, the extracted lipids were treated with 300 μL of 0.1 N KOH in methanol for 1 h at 37 °C and 30 μL of 1 M acetic acid was added for neutralization. In the control experiment, extracted lipids were treated with the same volume of methanol and H2O. After treatment, the lipids were dried and GPI precursors were extracted with two 300 μL portions of water-saturated BuOH and back-washed by 200 μL of BuOH-saturated water. After evaporation, the extracted lipids were used as samples for thin layer chromatography (TLC). TLC was performed in C/M/H2O (10:10:3). Mannolipids were identified by testing their sensitivities to Jack bean α mannosidase and comparing their mobilities on TLC with those of authentic GPI mannolipids accumulated in well characterized mutant cells (20).
Extraction of Total Lipids.
For mass analysis, total lipids were extracted from about 1 × 108 cells by the Bligh and Dyer method (27). The total lipid extracts were dried under a gentle stream of nitrogen and then resolved in 100 μL of C/M (1:1). The atmosphere in the tube was changed to nitrogen and it was stored at −80 °C until analysis.
Mass Spectrometry.
The LC system was coupled online to an LTQ Orbitrap mass spectrometer equipped with an electrospray ionization source (Thermo Fisher Scientific). The samples were dried under a gentle stream of nitrogen and then resolved in methanol/water (95:5) containing 0.2% formic acid and 0.028% ammonia (pH 4.8), mixed, and sonicated well. The samples were analyzed in the negative mode. The analytical methods used for the phospholipids, PI, GlcN-acyl-PI, and GlcNAc-PI have been described previously (13, 28).
Supplementary Material
Acknowledgments.
We thank R. A. Zoeller for NRel-4 and NZel-1 cells; F. Mori, Y. Onoe, and K. Kinoshita for excellent technical assistance; K. Nakamura for help with cell sorting; and Y. Morita and M. Fujita for helpful discussions. This work was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. H.R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904762106/DCSupplemental.
References
- 1.Ikezawa H. Glycosylphosphatidylinositol (GPI)-anchored proteins. Biol Pharm Bull. 2002;25:409–417. doi: 10.1248/bpb.25.409. [DOI] [PubMed] [Google Scholar]
- 2.McConville MJ, Menon AK. Recent developments in the cell biology and biochemistry of glycosylphosphatidylinositol lipids (review) Mol Membr Biol. 2000;17:1–16. doi: 10.1080/096876800294443. [DOI] [PubMed] [Google Scholar]
- 3.Orlean P, Menon AK. Thematic review series: Lipid posttranslational modifications. GPI anchoring of protein in yeast and mammalian cells, or: How we learned to stop worrying and love glycophospholipids. J Lipid Res. 2007;48:993–1011. doi: 10.1194/jlr.R700002-JLR200. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki KG, Fujiwara TK, Sanematsu F, Iino R, Edidin M, Kusumi A. GPI-anchored receptor clusters transiently recruit Lyn and G alpha for temporary cluster immobilization and Lyn activation: Single-molecule tracking study 1. J Cell Biol. 2007;177:717–730. doi: 10.1083/jcb.200609174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goswami D, et al. Nanoclusters of GPI-anchored proteins are formed by cortical actin-driven activity. Cell. 2008;135:1085–1097. doi: 10.1016/j.cell.2008.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nozaki M, Ohishi K, Yamada N, Kinoshita T, Nagy A, Takeda J. Developmental abnormalities of glycosylphosphatidylinositol-anchor-deficient embryos revealed by Cre/loxP system. Lab Invest. 1999;79:293–299. [PubMed] [Google Scholar]
- 7.Almeida A, Layton M, Karadimitris A. Inherited glycosylphosphatidyl inositol deficiency: A treatable CDG. Biochim Biophys Acta. 2009 doi: 10.1016/j.bbadis.2008.12.010. doi: 10.1016/j.bbadis.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 8.Parker C, et al. Diagnosis and management of paroxysmal nocturnal hemoglobinuria. Blood. 2005;106:3699–3709. doi: 10.1182/blood-2005-04-1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kinoshita T, Fujita M, Maeda Y. Biosynthesis, remodelling and functions of mammalian GPI-anchored proteins: recent progress. J Biochem. 2008;144:287–294. doi: 10.1093/jb/mvn090. [DOI] [PubMed] [Google Scholar]
- 10.Pittet M, Conzelmann A. Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim Biophys Acta. 2007;1771:405–420. doi: 10.1016/j.bbalip.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 11.Mayor S, Riezman H. Sorting GPI-anchored proteins. Nat Rev Mol Cell Biol. 2004;5:110–120. doi: 10.1038/nrm1309. [DOI] [PubMed] [Google Scholar]
- 12.Maeda Y, et al. Fatty acid remodeling of GPI-anchored proteins is required for their raft association. Mol Biol Cell. 2007;18:1497–1506. doi: 10.1091/mbc.E06-10-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Houjou T, et al. Changes in molecular species profiles of glycosylphosphatidylinositol anchor precursors in early stages of biosynthesis. J Lipid Res. 2007;48:1599–1606. doi: 10.1194/jlr.M700095-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Tashima Y, Taguchi R, Murata C, Ashida H, Kinoshita T, Maeda Y. PGAP2 is essential for correct processing and stable expression of GPI-anchored proteins. Mol Biol Cell. 2006;17:1410–1420. doi: 10.1091/mbc.E05-11-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horrocks LA, Sharma M. Plasmalogens and O-alkyl glycerophospholipids. New Comp Biochem. 1982;4:51–93. [Google Scholar]
- 16.Nagan N, et al. Isolation of a Chinese hamster fibroblast variant defective in dihydroxyacetonephosphate acyltransferase activity and plasmalogen biosynthesis: Use of a novel two-step selection protocol. Biochem J. 1998;332:273–279. doi: 10.1042/bj3320273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nagan N, et al. A fibroblast cell line defective in alkyl-dihydroxyacetone phosphate synthase: A novel defect in plasmalogen biosynthesis. Proc Natl Acad Sci USA. 1997;94:4475–4480. doi: 10.1073/pnas.94.9.4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Serrano AA, Schenkman S, Yoshida N, Mehlert A, Richardson JM, Ferguson MA. The lipid structure of the glycosylphosphatidylinositol-anchored mucin-like sialic acid acceptors of Trypanosoma cruzi changes during parasite differentiation from epimastigotes to infective metacyclic trypomastigote forms. J Biol Chem. 1995;270:27244–27253. doi: 10.1074/jbc.270.45.27244. [DOI] [PubMed] [Google Scholar]
- 19.Fontaine T, Magnin T, Melhert A, Lamont D, Latge JP, Ferguson MA. Structures of the glycosylphosphatidylinositol membrane anchors from Aspergillus fumigatus membrane proteins. Glycobiology. 2003;13:169–177. doi: 10.1093/glycob/cwg004. [DOI] [PubMed] [Google Scholar]
- 20.Hong Y, et al. Pig-n, a mammalian homologue of yeast Mcd4p, is involved in transferring phosphoethanolamine to the first mannose of the glycosylphosphatidylinositol. J Biol Chem. 1999;274:35099–35106. doi: 10.1074/jbc.274.49.35099. [DOI] [PubMed] [Google Scholar]
- 21.Maeda Y, Tanaka S, Hino J, Kangawa K, Kinoshita T. Human dolichol-phosphate-mannose synthase consists of three subunits, DPM1, DPM2, and DPM3. EMBO J. 2000;19:2475–2482. doi: 10.1093/emboj/19.11.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abrami L, et al. Cross-talk between caveolae and glycosylphosphatidylinositol-rich domains. J Biol Chem. 2001;276:30729–30736. doi: 10.1074/jbc.M102039200. [DOI] [PubMed] [Google Scholar]
- 23.Kajiwara K, et al. Yeast ARV1 is required for efficient delivery of an early GPI intermediate to the first mannosyltransferase during GPI assembly and controls lipid flow from the endoplasmic reticulum. Mol Biol Cell. 2008;19:2069–2082. doi: 10.1091/mbc.E07-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kreuger J, Perez L, Giraldez AJ, Cohen SM. Opposing activities of Dally-like glypican at high and low levels of Wingless morphogen activity. Dev Cell. 2004;7:503–512. doi: 10.1016/j.devcel.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 25.Kondoh G, et al. Angiotensin-converting enzyme is a GPI-anchored protein releasing factor crucial for fertilization. Nat Med. 2005;11:160–166. doi: 10.1038/nm1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sutterlin C, et al. Identification of a species-specific inhibitor of glycosylphosphatidylinositol synthesis. EMBO J. 1997;16:6374–6383. doi: 10.1093/emboj/16.21.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 28.Ogiso H, Suzuki T, Taguchi R. Development of a reverse-phase liquid chromatography electrospray ionization mass spectrometry method for lipidomics, improving detection of phosphatidic acid and phosphatidylserine. Anal Biochem. 2008;375:124–131. doi: 10.1016/j.ab.2007.12.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.