Abstract
The rapid specification and differentiation of the embryonic zebrafish gut is essential to provide contractility for the digestion of food. The role of microRNAs in modulating gut epithelial or smooth muscle differentiation is currently not known. Here we show that the microRNA miR-145 is strongly expressed in zebrafish gut smooth muscle and regulates its development. Modulation of miR-145 levels results in gut smooth muscle and epithelium maturation defects. Loss of miR-145 results in defects of smooth muscle function as measured by decreased nitric oxide production but also leads to increased expression of the embryonic smooth muscle markers sm22α-b, nm-mhc-b, and smoothelin. Defects in gut epithelial maturation are also present as observed by immature morphology and a complete loss of alkaline phosphatase expression. Loss or gain of miR-145 function phenocopies defects observed with altered gata6 expression and accordingly, we show that miR-145 directly represses gata6, and that gata6 is a major miR-145 target in vitro and in vivo. miR-145 therefore plays a critical role in promoting the maturation of both layers of the gut during development through regulation of gata6.
Keywords: GATA6, gut, microRNA, smooth muscle
The Gata family of transcription factors plays key roles in differentiation during embryonic development. In vertebrates, Gata family members fall into two subgroups containing Gata1, 2, and 3, which mainly function in hematopoiesis, and Gata4, 5, and 6, which are involved in heart and endodermal development (1). Gata6 is critical for cardiogenesis in Xenopus, zebrafish and mammals (2–4), blood vessel formation (5), and liver development in mouse (6). In vascular smooth muscle and cardiac muscle cells, Gata6 interacts with the serum response factor and the transcription factors of the NK family to activate expression of smooth muscle cell (SMC)-specific genes (7). Even though there is cross-talk among Gata4/5/6 and functional redundancy between Gata4 and Gata6 in some tissues (8), each Gata protein has unique functions (9). Gata6 is expressed in vascular smooth muscle during development, maintains vascular SMCs in a contractile state by activating SMC-specific genes, and inhibits SMC proliferation (10, 11). Depletion of Gata6 in the Xenopus or zebrafish endoderm disrupts normal intestinal morphogenesis and supports roles for Gata6 in endoderm formation and differentiation (3).
The primitive gut tube in zebrafish is formed from the endoderm between 32 and 40 h post-fertilization (hpf) (12). As the gut matures, visceral smooth muscle cells appear and start to express early smooth muscle markers sm22α, α-smooth muscle actin (αsma), non muscle myosin heavy chain (nm-mhc-b), smoothelin, and tropomyosin (13).
MicroRNAs (miRs) are approximately 22-bp noncoding RNAs that regulate eukaryotic gene expression through complementary sequences in the 3′ untranslated regions (UTRs) of target genes and promote mRNA degradation and/or inhibit protein translation. In animals, some miRs have tissue-specific roles in tuning gene expression levels of their targets (14). More broadly expressed miRs have functions in development, cell survival, and metabolism, and are associated with disease. In light of the role of miRs in modulating developmental processes, we are particularly interested in how miRs regulate smooth muscle formation. Here we show that miR-145 controls visceral SMC differentiation through direct targeting of the gata6 mRNA, and also affects gut epithelial differentiation through an indirect mechanism.
Results
Loss of miR-145 Leads to Defects in Heart and Gut Development.
In early zebrafish development between 16 and 24 hpf, miR-145 is expressed ubiquitously at very low levels (Fig. 1A and Fig. S1A) with increasingly specific expression in the heart, ear, and pharyngeal arches between 24 and 72 hpf. miR-145 is specifically localized in both the gut and swimbladder at 96 hpf (Fig. 1 B). Sectioning of 96 hpf embryos reveals that miR-145 expression is strongest within the gut smooth muscle layer but low levels are also seen in the gut epithelium (Fig. 1C). The strongly localized expression of miR-145 at this time point suggests that it might play a role in regulating gut differentiation.
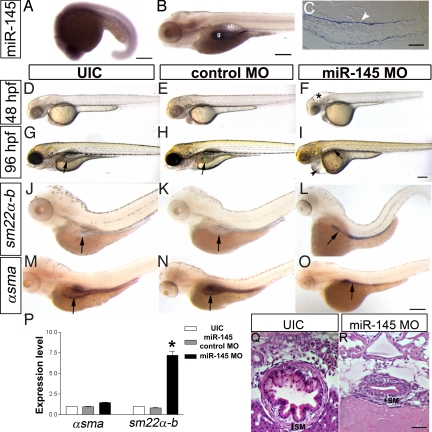
Fig. 1.
miR-145 expression pattern and loss of function phenotype. (A) Lateral view of whole-mount in situ expression of miR-145 at 16–19S shows ubiquitous expression. (B) Later at 96 hpf, miR-145 is expressed in the gut (g) and swimbladder (sb) in wild-type embryos. (C) Longitudinal section of 96 hpf embryo shows miR-145 expressed strongly in the gut smooth muscle cells (white arrowhead) and weakly in gut epithelium. (D–F) Animals injected with miR-145 MO have hydrocephalus in the hind brain ventricle (*) at 48 hpf. (G–I) At 96 hpf, miR-145 morphants have severe pericardial edema (black arrowhead), and an underdeveloped gut and swimbladder (black arrow). In situ hybridization staining of αsma (J–L) and sm22α-b (M–O) in 96 hpf embryos shows an increase of sm22α-b expression but no change of αsma in the miR-145 morphant gut. Gut morphology in the miR-145 morphant is unlooped as compared to the broader tube-like morphology found in UIC and miR-145 control MO embryos. Arrows mark the gut. (P) qPCR of αsma and sm22α-b in miR-145 morphants at 48 hpf. (Q and R) Histology of 96 hpf zebrafish gut. miR-145 morphant shows a thinner layer of SMCs (SM) and lack of villi in the epithelial layer as compared to UIC embryos. (Scale bars, 200 μm except for C, Q, and R, which are 50 μm.)
We analyzed the effects of blocking mature miR-145 function by using a morpholino antisense oligonucleotide (miR-145 MO). To control for the specificity of knockdown, a second morpholino targeting the precursor miR-145 (pre-miR-145 MO) and a 6-bp mismatch to miR-145 MO (control MO) were used. In situ hybridization using an LNA antisense probe against mature miR-145 indicates that 3 ng miR-145 MO or 15 ng pre-miR-145 MO per embryo reduces miR levels to threshold levels of detection (Fig. S1 A–D). In addition, quantitative real-time PCR (qPCR) of the miR itself shows the significant decrease of mature miR-145 after injection of miR-145 MO or pre-miR-145 MO (Fig. S1H). Thus the function of miR-145 is effectively inhibited by both morpholinos.
At 24 hpf, miR-145 morphants are overtly normal, but have a delayed onset of a heart beat. Heart function defects continue, and at 48 hpf, miR-145 morphants display hydrocephalus of the hindbrain ventricle and mild pericardial edema with a stringy, unlooped heart (Fig. 1 D–F). From 96 hpf to 120 hpf, most miR-145 morphants (n > 100) have a markedly underdeveloped gut, severe pericardial edema, and fail to inflate their swimbladder, in contrast to the looped, tube-like gut and inflated swimbladder in wild-type embryos (Fig. 1 G–I). Sections through the anterior intestine at 6 dpf reveal the smooth muscle layer of miR-145 morphants is thinner and the gut epithelial layer lacks finger-like villi (Fig. 1 Q and R). The specificity of the phenotype is demonstrated by similar heart and gut phenotypes after injection of pre-miR-145 MO (Fig. S1L).
To test whether miR-145 regulates the maturation of gut smooth muscle we investigated αsma, sm22α-b, nm-mhc-b, and smoothelin expression (13). In situ hybridization and reverse-transcription (RT-PCR) shows increased sm22α-b in miR-145 morphants as compared to uninjected controls (UIC) or control morphants at 96 hpf (Fig. 1 J–L), whereas αsma expression remains unchanged (Fig. 1 M–O). The staining pattern of these smooth muscle markers in whole-mount embryos highlights the unlooped immature gut morphology of miR-145 morphants. qPCR demonstrates a more than 7-fold increase in sm22α-b expression in miR-145 morphants compared to controls at 48 hpf (Fig. 1P; P < 0.001), whereas expression of αsma remains unchanged. Similar expression changes of sm22α-b, but not αsma, are found in pre-miR-145 MO treated embryos at 48 hpf (Fig. S2B). Up-regulation is not unique to sm22α-b as the early smooth muscle markers nm-mhc-b and smoothelin also show increased expression in miR-145 morphants at 96 hpf (Fig. S2A). Significant up-regulation of nm-mhc-b is also observed by qPCR of both miR-145 and pre-miR-145 morphants at 48 hpf (Fig. S2C).
Increase in miR-145 Expression Leads to Opposite Gene Expression Changes.
To up-regulate miR-145 expression, we injected a double-stranded miR-145 RNA mimic into embryos. In situ hybridization shows that miR-145 mimic-injected embryos have ubiquitously increased expression of miR-145 compared to their wild-type siblings (Fig. S1 E–G). Co-injection of miR-145 MO and the miR-145 mimic led to a normalization of development, indicating that both are acting specifically on miR-145 (Fig. S1 I–K). At 24 hpf, 54.7% of miR-145-mimic injected embryos have mild pericardial edema (n = 315; Fig. 2 A–C), which progresses in severity through to 96 hpf and is accompanied by abnormal gut morphology (Fig. 2 D–F). These phenotypes are specific as 95% of wild-type and 97% of the control-mimic injected embryos show normal development. Consistent with the previous observation of increased expression of sm22α-b in miR-145 morphants, sm22α-b is almost 10-fold down-regulated in miR-145 mimic treated embryos at 48 hpf, with no observed change in αsma expression (Fig. 2G).
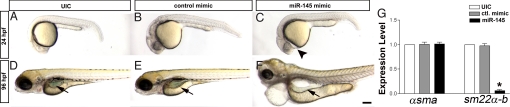
Fig. 2.
miR-145 overexpression affects gut and heart development. (A–C) Overexpression of miR-145 causes mild pericardial edema (arrowhead) in 24 hpf embryos, (D–F) which then develops into severe pericardial edema accompanied by a unlooped gut at 96 hpf. The control mimic RNA injection has no effect. (G) Overexpression of miR-145 leads to strong down-regulation of sm22α-b but does not affect αsma levels in 48 hpf embryos as measured by qPCR. Arrows, gut. (Scale bar, 200 μm.)
miR-145 Modulates gata6 Expression.
We hypothesize that miR-145 likely regulates smooth muscle marker expression indirectly because we were unable to find any miR-145 binding sites in the 3′UTRs of sm22α-b, nm-mhc-b, or smoothelin by using the target prediction software DIANA MicroTest (15). Therefore, we used a bioinformatic approach to predict potential miR-145 targets by using miRBase (16) and identified a putative binding site in the gata6 3′UTR with a perfect match to the miR-145 seed region. Loss of miR-145 leads to an up-regulation of gata6 in the gut by in situ hybridization (Fig. 3 A–C), and a 4-fold increase by qPCR at 48 hpf (Fig. 3J), suggesting regulation of gata6 by miR-145. In addition, gata6 is also up-regulated in pre-miR-145 MO treated embryos at 48 hpf (Fig. S2B). Consistently, expression of gata6 decreases by nearly 10-fold after injection of miR-145 mimic, as determined by qPCR (Fig. 3K) and confirmed by in situ hybridization (Fig. 3 D–F).
Fig. 3.
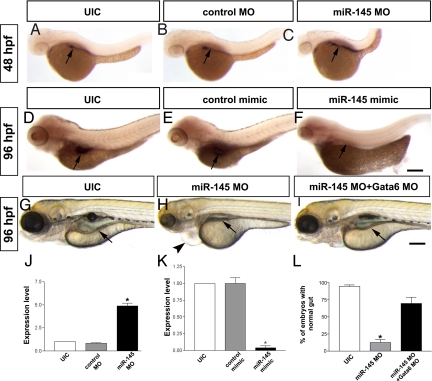
Normalization of gata6 levels rescues loss of miR-145. (A–C) Knockdown of miR-145 results in increased expression of gata6 in zebrafish gut at 48 hpf. (D–F) Overexpression of miR-145 leads to decreased gata6 expression in the gut of 96 hpf embryos. Pericardial edema (arrowhead) is also found in miR-145 morphant. (G–I) gata6 knockdown normalizes the gut morphology (arrow) of miR-145 morphants. (J) qPCR shows strong up-regulation of gata6 in miR-145 morphants of 48 hpf. (K) qPCR indicates significantly decreased expression of gata6 in 48 hpf miR-145 mimic treated embryos. Arrows, gut; bars, mean ± SEM. *, P < 0.01. (L) The number of embryos exhibiting normal gut morphology in the rescue was analyzed by using Student's t test (P < 0.05, n = 286). (Scale bar, 200 μm.)
miR-145 Binds Directly to the gata6 3′UTR.
We next sought to determine whether the regulation of miR-145 on gata6 is direct, in vitro and in vivo. To demonstrate that miR-145 regulates the gata6 3′UTR in vitro, we fused the zebrafish gata6 3′UTR containing the putative miR-145 recognition site behind a luciferase reporter (pLuc-gata6-pA; Fig. 4A). As a negative control, a second construct was generated with luciferase followed by a 3′UTR lacking the miR-145 recognition site (pLuc-pA). miR-145 was expressed from a short hairpin plasmid containing pre-miR-145 (PremiR-145) together with the luciferase reporters. Luciferase expression is strongly repressed when miR-145 is co-expressed with pLuc-gata6-pA, but there is no effect on pLuc-pA (Fig. 4B). As a control, pLuc-gata6-pA transfected in the absence of miR-145 has luciferase expression similar to pLuc-pA. Taken together, these data suggest that miR-145 binds the gata6 3′UTR in vitro.
Fig. 4.
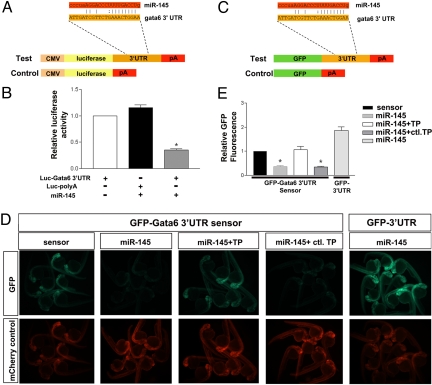
miR-145 directly targets gata6 in vitro and in vivo. (A) Schematic diagram of a luciferase construct with or without the gata6 3′UTR for the in vitro assay. (B) Luciferase reporter activity shows the interaction between miR-145 and gata6 3′UTR. Luciferase activity was normalized to β-galactosidase activity and expressed relative to the gata6 3′UTR conjugated luciferase vector-only transfection. (C) Schematic of EGFP reporters for the in vivo assay which were constructed with or without the gata6 3′UTR. (D) EGFP-gata6-3′UTR sensor assays demonstrating specific regulation of the gata6 3′UTR in vivo by miR-145. EGFP reporter expression (green) and control mCherry expression (red) are shown at 25–28 hpf as modulated by the addition of miR-145 mimic with or without the Gata6-TPmiR145 or control target protector. mCherry mRNA is an injection control. (E) For each experimental group in D, EGFP-gata6-3′UTR (sensor) or EGFP-3′UTR fluorescence is expressed as a percentage of fluorescence observed from the sensor injected alone group (n = 10 embryos per group). Bars, mean ± SEM. *, P < 0.01.
To determine whether miR-145 can regulate gata6 expression in vivo we designed a sensor assay. In the sensor assay, EGFP was fused to the gata6 3′UTR (EGFP: gata6pA), or to a control 3′UTR lacking the gata6 3′UTR element (EGFPpA) (Fig. 4C). To assay miR-145-induced silencing, synthetic mRNAs derived from these reporters were injected into single-cell zebrafish embryos in the presence or absence of a miR-145 mimic, along with mRNA encoding the red fluorescent protein mCherry (which serves as an internal control for injection quantity). When fluorescence levels were examined at 24 hpf, injections of the gata6 sensor (EGFP: gata6pA) mRNA alone resulted in high EGFP expression; however, this fluorescence was dampened over 60% by co-injection of miR-145 mimic (Fig. 4 D and E). This decrease in fluorescence was abolished with co-injection of a target protector morpholino that blocks the putative miR-145 binding site (gata6-TPmiR145) (17, 18). Conversely, a second target protector morpholino (Control TP MO) targeting a different site in the 3′UTR does not prevent miR-145 mimic-induced repression. Lastly, co-injection of miR-145 mimic and EGFPpA mRNA (which does not contain the miR-145 binding site) did not show any dampening of fluorescence (Fig. 4 D and E). Taken together, these results suggest a specific site is present in the 3′UTR of gata6, which binds miR-145 in vivo to control gene expression.
Decrease in gata6 Levels Compensates for Loss of miR-145.
Our data suggests that miR-145 directly binds to the gata6 3′UTR and that loss of miR-145 leads to increased expression of gata6 and subsequent defects in gut development. Thus, we hypothesized that down-regulation of gata6 in miR-145 morphants to restore normal gata6 expression levels would rescue gut defects and allow for normal development. An optimal dose of 0.4 ng Gata6 morpholino (Gata6 MO) was sequentially injected with miR-145 MO into wild-type embryos, resulting in a statistically significant rescue of gut morphological defects (P = 0.033; Fig. 3 G–I and L).
To further validate that miR-145 can regulate gut development by inhibiting gata6 expression in zebrafish, we disrupted the interaction of miR-145 and gata6 3′UTR by injection of the gata6 target protector morpholino into wild-type embryos (18). Twenty-two percent of the Gata6-TPmiR145 embryos displayed cardiac edema at 48 hpf, had a thin, unlooped gut tube and failed to inflate the swim bladder at 96 hpf (Fig. S3 A and B), similar to the phenotype observed in miR-145 morphants. Protection of gata6 mRNA by the Gata6-TPmiR145 increased gata6 and sm22α-b expression at 96 hpf (Fig. S3 C–F). Taken together, these rescue and target protector experiments indicate that gata6 is a predominant target of miR-145.
Loss of miR-145 Affects both Smooth Muscle and Epithelial Maturation.
We find that gata6 is predominantly expressed in gut epithelial cells but also in the SMC layer at 96 hpf (Fig. S4 A and B), whereas miR-145 is predominantly in the smooth muscle layer but also weakly expressed in gut epithelial cells. Thus the miR and its target have partially overlapping expression.
To further investigate the phenotype and determine in which tissue miR-145 and gata6 play critical roles, we examined cell death and proliferation in miR-145 morphant embryos at 96 hpf. TUNEL labeling for apoptotic cells reveals similar sparse TUNEL-positive cells in both gut SMC or epithelial layers in both wild-type embryos and miR-145 morphants at 96 hpf. Similarly anti phospho-histone H3 (pHH3) staining showed a very low proliferation rate in both the SMC and epithelial layer of wild-type, miR-145 MO and control MO-injected guts at 96 hpf (Fig. S4C). Therefore, neither proliferation nor cell death account for maturation defects.
Examination of sections of miR-145 morphants highlights the strikingly abnormal morphology of epithelial cells in the miR-145 morphant gut in comparison to wild-type and control MO embryos (Fig. S4C). In miR-145 morphants there is a lack of lumen formation, epithelial cells are rounded (not columnar), and the SMC layer appears disorganized. To test whether SMCs present in miR-145 morphants had mature function we used 4,5-diaminofluorescein diacetate (DAF-2DA), a fluorescent nitric oxide indicator. After incubation in a solution of DAF-2DA at 120 hpf, strong DAF-2DA fluorescence was found in wild-type embryos but not in miR-145 morphants, indicating a lack of smooth muscle maturation (Fig. S4 D and E). Similarly, alkaline phosphatase activity is an indicator of gut epithelial maturation and normally appears in the zebrafish intestinal lumen by 72 hpf. We show that alkaline phosphatase activity is completely absent in miR-145 morphants at 96 hpf, also indicating a lack of functional maturation (Fig. S4 F and G). Thus both layers of the gut are functionally immature.
A Specific Role for gata6 in Smooth Muscle Development.
gata6 expression begins between 16 and 19 hpf in both endoderm and mesendoderm (8) and continues through 48 hpf. However, the developmental stage at which miR-145 acts on gata6 is not known. We compared gata6 levels by in situ hybridization and qPCR in UIC and miR-145 morphants at both 16–19S and 24 hpf, but found no significant expression level difference of gata6 mRNA at this early stage (Fig. S5 A, B, and E), even though at 48 hpf, there is strong up-regulation of gata6 levels (Fig. S5 C and D). The strong heart developmental phenotype of miR-145 morphants would suggest that miR-145 acts on a second, unknown target gene in the heart before 48 hpf because gata6 levels are unaffected.
Heart function defects in miR-145 morphants are evident on the first day of development and it is possible that gut morphological defects might develop secondarily to these. Thus we undertook to separate the role of gata6 on smooth muscle development and heart development by overexpressing gata6 using the αSMA and CMLC2 promoters in wild-type embryos. Because the αSMA promoter is expressed in both heart and gut smooth muscle, but not gut epithelium (Fig. S6 E and F), phenotypes found in the αSMA -driven rescue, but not the CMLC2-driven rescue represent smooth muscle-specific phenotypes. Overexpression of gata6 using the CMLC2 promoter led to a wild-type level of gut maturation defects (5.4 ± 1.9%, n = 105; Fig. S6 A–D) versus UIC embryos (2.8 ± 1.4%, n = 274; Fig. S6D). However, overexpression of gata6 using the αSMA promoter results in a significant proportion of embryos with an immature, unlooped gut tube (13.2 ± 1.6%, n = 138) at 96 hpf. These data suggest that increased gata6 expression in smooth muscle leads directly to smooth muscle differentiation defects, and indirectly to gut epithelial developmental defects. Heart developmental defects appear to occur independently of gut maturation defects.
Discussion
Our study reveals a key role for miR-145 in regulation of gut formation in vivo through direct regulation and repression of gata6 expression. Gata6 is an important transcription factor regulating the differentiation of heart and intestine. Previous data suggested that miR-145 had strong expression in the maturing zebrafish and Medaka gut (19, 20), in smooth muscle of human and murine blood vessels, and murine colonic muscularis externa and muscularis mucosae (21), all of which is consistent with our finding here that miR-145 is critical for smooth muscle development. In addition, our finding of decreased differentiation of the gut epithelium would be consistent both with a role of mature smooth muscle in promoting epithelial differentiation (22) or, alternatively, of a direct role for miR-145 in the gut epithelium. miR-145 is expressed in the human colonic epithelium and is frequently down-regulated in colon cancer (23).
Identifying targets for a miR is a key step to analyzing the biological function of a microRNA. To date, the only verified target of miR-145 is the insulin receptor substrate-1 (IRS1), which promotes proliferation of cultured colon cancer cells (24), but whether this interaction exists in vivo is unknown. A high degree of sequence complementarity between the 5′ end of the miR, known as the “seed” sequence, and the mRNA is an important determinant of miR targeting, as is the accessibility of the mRNA at the potential binding site (25). In general, perfect or near perfect complementarity between miR and target mRNA will induce the degradation of mRNA; however, imperfect binding can arrest translation of the target gene (26). We have found that the gata6 3′UTR has a perfect binding site for the miR-145 seed region and likely targets gata6 mRNA for degradation, consistent with our findings that expression of gata6 is dramatically increased in miR-145 morphants. Lending further support to the idea that gata6 is a direct target for miR-145 is the observation that miR-145 and gata6 are co-expressed in the gut during development, although miR-145 predominates in the smooth muscle layer and gata6 predominates in the epithelial layer. Importantly, the gut phenotype of miR-145 morphants can be rescued by restoring normal dosage of gata6.
miR targets fall into two classes: phylogenetically conserved and nonconserved (27). Sequence alignment of the gata6 3′UTR shows moderate conservation of the miR-145 target region, as 5/8 residues of the miR-145 seed sequence conserved in both mouse and human. Although the functional significance has not been tested in these species, it is possible that miR-145 function on gata6 expression is phylogenetically conserved.
gata6 expression is first observed in endodermal cells of the gut at 24 hpf and although it is not known when gata6 is first seen in developing visceral SMCs, other smooth muscle markers begin expression at 50 hpf (13). Conversely, miR-145 is first strongly expressed in visceral SMCs at 96 hpf (our above data), consistent with a role of miR-145 in ‘tuning down’ or dampening the expression of preexisting gata6 expression. Thus high levels of miR-145, capable of repressing gata6 expression in the gut are only observed after initial differentiation of gut smooth muscle. Although we do not have molecular tools to measure Gata6 protein levels, we clearly show that gata6 message is decreased after overexpression of miR-145, which would therefore also likely decrease Gata6 protein levels. This is in contrast to early gata6 levels that are unaffected by loss of miR-145, likely because of the weak expression of the miR at this stage, and strong expression of gata6.
Phenotypic defects in the intestine of miR-145 loss-of-function or gain-of-function embryos are found both in the smooth muscle and epithelial layers, and closely resemble defects caused by loss or gain of gata6. For example, miR-145 morphants have increased gata6 expression and a thin, unlooped gut, which is the opposite of the enlarged intestine observed after knockdown of gata6 in Xenopus (3). In contrast, miR-145 mimic injected embryos have decreased gata6 expression and show a similar phenotype to gata6 morphants (9). It is possible that modulation of gata6 expression at early embryonic stages by miR-145 might indirectly influence gut development. However, we found unchanged expression levels of gata6 in miR-145 morphants before 48 hpf as compared to control embryos suggesting that the low ubiquitous expression of miR-145 at early stages is not responsible for later gut defects.
Interestingly, in situ hybridization data shows dramatically elevated gata6 expression in the correct spatiotemporal expression pattern in miR-145 morphants without any evidence of ectopic expression. Thus, our experiments using transgenic overexpression of gata6 under αSMA and CMLC2 promoters accurately phenocopy loss of miR-145. These experiments indicate that gata6 levels in smooth muscle can affect the differentiation of gut as a whole, including that of epithelial cells, suggesting a developmental communication between SMCs and epithelium in promoting their maturation. Further analysis of miR-145 function in SMC will lead to a better understanding of the developmental interrelationships between the two layers and identification of signals passing between them.
During late embryogenesis and postnatal development, SMCs have an extremely high rate of proliferation with subsequent rapid induction of multiple SMC differentiation marker genes (28, 29). In zebrafish, the high proliferation rate of gut epithelial cells and SMC progenitors between 36 hpf and 74 hpf decreases significantly between 74 and 120 hpf, at a time when miR-145 begins to be expressed (12, 30). The miR-145 target gata6 can play both positive and negative roles in differentiation. It is involved in the maintenance of the quiescent phenotype in differentiated vascular SMCs of human (5), but high gata6 levels also inhibit differentiation of the heart (4). In human colon cancer cells, miR-145 inhibits proliferation and is considered anti-oncogenic (31). A fine balance of gata6 levels is therefore required for normal differentiation, and this is controlled in part by miR-145. The differences in miR-145 function in different tissues may be due to the tissue-restricted expression of different cofactors, target genes, or cellular context.
Gata6 regulates a wide variety of smooth muscle-specific genes. Here we show that miR-145 indirectly regulates expression of sm22α-b, nm-mhc-b, and smoothelin, but not αsma, likely through its regulation of gata6 expression. This is consistent with previous in vitro studies where overexpression of gata6 increased expression of sm22α but not αsma, even though Gata6 can activate the αSMA promoter (32). Despite the increased expansion of smooth muscle marker expression in miR-145 morphants we find two lines of evidence for the immaturity of the gut. Staining using the fluorescent nitric oxide indicator DAF-2DA as a marker for functional SMC shows that it is almost absent in the gut of miR-145 morphants. Morphologically, epithelial cells of the miR-145 morphant gut are rounded and do not take on the mature elongated, columnar shape of wild-type epithelial cells at this stage, and luminal alkaline phosphatase staining is almost eliminated in the miR-145 morphant epithelial cells. Thus both gut epithelial and smooth muscle layers are morphologically and functionally abnormal. Normal SMCs are required for the proper development of epithelial cells into a columnar morphology with folds (22, 33). At this point it is unclear whether epithelial defects occur in miR-145 morphants as an indirect result of smooth muscle defects, or whether they are autonomous to the gut epithelium. However we show that expression of Gata6 under the αSMA promoter results in similar gut maturation defects to loss of miR-145 in all tissues, further suggesting that indirect effects from the SMC layer on the epithelial layer can occur.
This report demonstrates that miR-145 targets gata6, resulting in a profound biological effect on development of the zebrafish gut and heart. Given its expression pattern, it is likely that miR-145 plays an autonomous role in the development in these tissues. However because multiple tissues expressing miR-145 interact during development (smooth muscle and gut epithelial cells, or heart and endoderm), it will be interesting to dissect the direct and indirect contributions of miR-145 to the differentiation of cardiac muscle, visceral smooth muscle, and gut epithelium.
Materials and Methods
Zebrafish Lines and Maintenance.
Zebrafish (Danio rerio) were maintained and staged according to standard methods. The wild-type Tübingen Long Fin (TL) strain was used for all experiments.
Morpholino and microRNA Mimic Injections.
Morpholino (MO, Gene Tools LLC) or miR mimic (Dharmacon) sequences and dosages are shown in Table S1, were injected in one- to four-cell stage embryos as described (34). To control for nonspecific neural cell death that can occur from nonspecific activation of p53 with morpholinos, a standard p53 MO was co-injected with all morpholinos (35). As a control, we also confirmed that specific gene expression changes were identical in embryos injected with miR-145 MO alone.
RT-PCR and qPCR Analysis.
Total RNA from zebrafish embryos was isolated by using the RNeasy Mini kit for mRNA and the miRNeasy Mini kit for miR (Qiagen). Three micrograms total of RNA from each sample was reverse transcribed into cDNA for RT-PCR, or mRNA qPCR using iQ SYBR Green Supermix (Bio-Rad), or 10 ng total RNA was transcribed into gene specific cDNA by using miRCURY LNA miRNA PCR system (Exiqon) and assayed in a DNA Engine Opticon 2 system (MJ Research Incorporated). Primer sequences are listed in Table S2. The mRNA qPCR was normalized to elongation factor 1α (EF1α). The relative expression of miR-145 was normalized to miR-26a. The ΔΔCt method was used to calculate the normalized relative expression level of a target gene from triplicate measurements.
Embryo Staining.
Digoxigenin (DIG)-labeled antisense RNA probes for sm22α-b, α-sma, gata6, and nm-mhc-b (13, 36) were used in whole mount in situ hybridization (37). Published protocols (Exiqon, Table S1) for miR in situ hybridization was followed. For immunostaining, embryos were fixed in 4% paraformaldehyde. Rabbit α-pHH3 (Sigma) was used with methanol-acetone permeabilization. Signal was detected with Alexafluor 555 secondary antibodies (Invitrogen), and counterstained with DAPI (Vector Labs). Sections were prepared as described (13). For analysis with DAF-2DA, live embryos were treated in DAF-2DA (Calbiochem) and visualized in vivo as described (38). Embryos were stained for endogenous alkaline phosphatase by incubation in NBT/BCIP.
Luciferase Assay.
Complementary oligonucleotides of the miR-145 mature sequence (IDT Technology) were annealed and inserted into the pcDNA 6.2-GW/EmGFP-miR-145 vector (Invitrogen). The gata6 3′UTR was cloned downstream of the luciferase gene by using HindIII and SacI in the pMIR-REPORT luciferase vector (Ambion Inc.). pMIR-REPORT luciferase-gata6 3′UTR (1.3 μg) and pMIR-REPORT β-gal control plasmid (1.3 μg) were introduced into Cos7 cells (≈5 × 105) with or without the pcDNA 6.2-GW/EmGFP-miR-145 (1.3 μg) using LipofectAmine2000 (Invitrogen). Luciferase activity was measured with the Luciferase assay kit (Promega).
EGFP Sensor Assay and Target Protectors.
For the in vivo sensor test, the gata6 3′UTR was amplified with primers incorporating BamHI sites and inserted into the 3′UTR of the p3E-polyA vector (39). This construct was then recombined into pDestTol2pA2 by Gateway cloning to achieve a SP6 promoter upstream of EGFP: gata6 3′UTR or a SP6 promoter driving EGFP: p3E-polyA 3′UTR. Sensor mRNA and mCherry mRNA were in vitro transcribed from the pCS2 Gateway compatible vector (39) by using the mMessage Machine SP6 kit (Ambion). One-cell zebrafish embryos were injected with 150 pg sensor mRNA and 100 pg mCherry mRNA. When applicable, 30 μM miRNA mimic was added with or without 1 ng Gata6-TPmiR145. miRNA mimic was also used with the control TP morpholino. Live embryos were photographed with an identical exposure time at 24 hpf (n = 10/group). The average pixel intensity for fluorescence was measured as described (17).
Imaging and Statistical Analysis.
Whole-mount embryos were photographed with a Zeiss Axiocam HR or an Optronics Magnafire camera. Images were processed in Adobe Photoshop CS. Data are expressed as mean ± SEM. Three or more treatment groups were compared by one-way ANOVA. Results were considered statistically significant when P < 0.05.
Supplementary Material
Acknowledgments.
We thank Ryan Lamont and Ryan Sobering for helpful comments and Olivera Starovic-Subota for sectioning. L.Z. is funded by the Canadian Institutes of Health Research Strategic Training Program Fellowship in Genetics, Child Development and Health. S.C. is a Senior Scholar of the Alberta Heritage Foundation for Medical Research, and holds a Tier II Canada Research Chair. Operating funding was from the National Science and Engineering Research Council of Canada.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903693106/DCSupplemental.
References
- 1.Patient RK, McGhee JD. The GATA family (vertebrates and invertebrates) Curr Opin Genet Dev. 2002;12:416–422. doi: 10.1016/s0959-437x(02)00319-2. [DOI] [PubMed] [Google Scholar]
- 2.Zhao R, et al. Loss of both GATA4 and GATA6 blocks cardiac myocyte differentiation and results in acardia in mice. Dev Biol. 2008;317:614–619. doi: 10.1016/j.ydbio.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peterkin T, Gibson A, Patient R. GATA-6 maintains BMP-4 and Nkx2 expression during cardiomyocyte precursor maturation. EMBO J. 2003;22:4260–4273. doi: 10.1093/emboj/cdg400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gove C, et al. Over-expression of GATA-6 in Xenopus embryos blocks differentiation of heart precursors. EMBO J. 1997;16:355–368. doi: 10.1093/emboj/16.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wada H, et al. Calcineurin-GATA-6 pathway is involved in smooth muscle-specific transcription. J Cell Biol. 2002;156:983–991. doi: 10.1083/jcb.200106057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morrisey EE, et al. GATA6 regulates HNF4 and is required for differentiation of visceral endoderm in the mouse embryo. Genes Dev. 1998;12:3579–3590. doi: 10.1101/gad.12.22.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nishida W, et al. A triad of serum response factor and the GATA and NK families governs the transcription of smooth and cardiac muscle genes. J Biol Chem. 2002;277:7308–7317. doi: 10.1074/jbc.M111824200. [DOI] [PubMed] [Google Scholar]
- 8.Heicklen-Klein A, McReynolds LJ, Evans T. Using the zebrafish model to study GATA transcription factors. Semin Cell Dev Biol. 2005;16:95–106. doi: 10.1016/j.semcdb.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Holtzinger A, Evans T. Gata4 regulates the formation of multiple organs. Development. 2005;132:4005–4014. doi: 10.1242/dev.01978. [DOI] [PubMed] [Google Scholar]
- 10.Narita N, Heikinheimo M, Bielinska M, White RA, Wilson DB. The gene for transcription factor GATA-6 resides on mouse chromosome 18 and is expressed in myocardium and vascular smooth muscle. Genomics. 1996;36:345–348. doi: 10.1006/geno.1996.0472. [DOI] [PubMed] [Google Scholar]
- 11.Perlman H, Suzuki E, Simonson M, Smith RC, Walsh K. GATA-6 induces p21(Cip1) expression and G1 cell cycle arrest. J Biol Chem. 1998;273:13713–13718. doi: 10.1074/jbc.273.22.13713. [DOI] [PubMed] [Google Scholar]
- 12.Wallace KN, Akhter S, Smith EM, Lorent K, Pack M. Intestinal growth and differentiation in zebrafish. Mech Dev. 2005;122:157–173. doi: 10.1016/j.mod.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 13.Georgijevic S, et al. Spatiotemporal expression of smooth muscle markers in developing zebrafish gut. Dev Dyn. 2007;236:1623–1632. doi: 10.1002/dvdy.21165. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y, Srivastava D. A developmental view of microRNA function. Trends Biochem Sci. 2007;32(4):189–197. doi: 10.1016/j.tibs.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 15.Kiriakidou M, et al. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giraldez AJ, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–79. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 18.Choi WY, Giraldez AJ, Schier AF. Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science. 2007;318:271–274. doi: 10.1126/science.1147535. [DOI] [PubMed] [Google Scholar]
- 19.Wienholds E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- 20.Ason B, et al. Differences in vertebrate microRNA expression. Proc Natl Acad Sci USA. 2006;103:14385–14389. doi: 10.1073/pnas.0603529103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sempere LF, et al. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–11620. doi: 10.1158/0008-5472.CAN-07-5019. [DOI] [PubMed] [Google Scholar]
- 22.Wallace KN, et al. Mutation of smooth muscle myosin causes epithelial invasion and cystic expansion of the zebrafish intestine. Dev Cell. 2005;8:717–726. doi: 10.1016/j.devcel.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 23.Akao Y, Nakagawa Y, Naoe T. MicroRNAs 143 and 145 are possible common onco-microRNAs in human cancers. Oncol Rep. 2006;16:845–850. [PubMed] [Google Scholar]
- 24.Shi B, et al. Micro RNA 145 targets the insulin receptor substrate-1 and inhibits the growth of colon cancer cells. J Biol Chem. 2007;282:32582–32590. doi: 10.1074/jbc.M702806200. [DOI] [PubMed] [Google Scholar]
- 25.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 26.Smalheiser NR, Torvik VI. A population-based statistical approach identifies parameters characteristic of human microRNA-mRNA interactions. BMC Bioinformatics. 2004;5:139. doi: 10.1186/1471-2105-5-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Farh KK, et al. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- 28.Cook CL, Weiser MC, Schwartz PE, Jones CL, Majack RA. Developmentally timed expression of an embryonic growth phenotype in vascular smooth muscle cells. Circ Res. 1994;74:189–196. doi: 10.1161/01.res.74.2.189. [DOI] [PubMed] [Google Scholar]
- 29.Owens GK, Thompson MM. Developmental changes in isoactin expression in rat aortic smooth muscle cells in vivo. Relationship between growth and cytodifferentiation. J Biol Chem. 1986;261:13373–13380. [PubMed] [Google Scholar]
- 30.Wallace KN, Pack M. Unique and conserved aspects of gut development in zebrafish. Dev Biol. 2003;255:12–29. doi: 10.1016/s0012-1606(02)00034-9. [DOI] [PubMed] [Google Scholar]
- 31.Akao Y, Nakagawa Y, Naoe T. MicroRNA-143 and -145 in colon cancer. DNA Cell Biol. 2007;26:311–320. doi: 10.1089/dna.2006.0550. [DOI] [PubMed] [Google Scholar]
- 32.Yin F, Herring BP. GATA-6 can act as a positive or negative regulator of smooth muscle-specific gene expression. J Biol Chem. 2005;280:4745–4752. doi: 10.1074/jbc.M411585200. [DOI] [PubMed] [Google Scholar]
- 33.Kedinger M, et al. Intestinal epithelial-mesenchymal cell interactions. Ann N Y Acad Sci. 1998;859:1–17. doi: 10.1111/j.1749-6632.1998.tb11107.x. [DOI] [PubMed] [Google Scholar]
- 34.Nasevicius A, Ekker SC. Effective targeted gene ‘knockdown’ in zebrafish. Nat Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- 35.Robu ME, et al. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reiter JF, et al. Gata5 is required for the development of the heart and endoderm in zebrafish. Genes Dev. 1999;13:2983–2995. doi: 10.1101/gad.13.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jowett T, Lettice L. Whole-mount in situ hybridizations on zebrafish embryos using a mixture of digoxigenin- and fluorescein-labeled probes. Trends Genet. 1994;10:73–74. doi: 10.1016/0168-9525(94)90220-8. [DOI] [PubMed] [Google Scholar]
- 38.Grimes AC, Stadt HA, Shepherd IT, Kirby ML. Solving an enigma: Arterial pole development in the zebrafish heart. Dev Biol. 2006;290:265–276. doi: 10.1016/j.ydbio.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 39.Kwan KM, et al. The Tol2kit: A multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn. 2007;236:3088–3099. doi: 10.1002/dvdy.21343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.